Abstract
The hemolysin-determining plasmid pAD1 is a member of a widely disseminated family of highly conjugative elements commonly present in clinical isolates of Enterococcus faecalis. The determinants repA, repB, and repC, as well as adjacent iteron sequences, are believed to play important roles in pAD1 replication and maintenance. The repA gene encodes an initiator protein, whereas repB and repC encode proteins related to stability and copy number. The present study focuses specifically on repA and identifies a replication origin (oriV) within a central region of the repA determinant. A small segment of repA carrying oriV was able to support replication in cis of a plasmid vector otherwise unable to replicate, if an intact RepA was supplied in trans. We demonstrate that under conditions in which RepA is expressed from an artificial promoter, a segment of DNA carrying only repA is sufficient for stable replication in E. faecalis. We also show that RepA binds specifically to oriV DNA at several sites containing inverted repeat sequences (i.e., IR-1) and nonspecifically to single-stranded DNA, and related genetic analyses confirm that these sequences play an important role in replication. Finally, we reveal a relationship between the internal structure of RepA and its ability to recognize oriV. An in-frame deletion within repA resulting in loss of 105 nucleotides, including at least part of oriV, did not eliminate the ability of the altered RepA protein to initiate replication using an intact origin provided in trans. The relationship of RepA to other known initiator proteins is also discussed.
pAD1 is a 60-kb, conjugative plasmid originally identified in Enterococcus faecalis DS16 (12, 23, 49). It encodes a cytolysin (hemolysin/bacteriocin) that contributes to virulence in animal models (8, 37, 40) and is one of numerous plasmids in E. faecalis that facilitate a response to peptide sex pheromones secreted by plasmid-free (recipient) bacteria. pAD1 responds to the pheromone cAD1 and represents a widely disseminated family of cytolysin plasmids commonly associated with clinical infections in humans (38) and which, for the most part, are members of the same incompatibility group (13, 34). (For recent reviews, see references 9 and 10.)
Nucleotide sequence data relating to plasmids from different incompatibility groups (e.g., pAD1, pAM373, pCF10, and pPD1) and responding to four different pheromones have shown that the regions associated with replication and maintenance are organized similarly; in all cases, this region is located adjacent to that involved in regulation of the pheromone response (9, 10). In the case of pAD1 the key determinants associated with plasmid maintenance are repA, repB, and repC (Fig. 1A). On the basis of sequence homology, repA is believed to encode the initiator of vegetative replication, whereas repB and repC most likely represent a partition system (50, 54). When a segment carrying these three determinants was cloned on an E. coli plasmid vector, it enabled the chimera to replicate in E. faecalis (52). Transposon insertion mutations within repA were unable to replicate in E. faecalis, whereas insertions within repB and repC affected stability and copy number. Two series of octanucleotide iterons are located between the divergently oriented repA and repB and, like iteron sequences associated with other plasmid replicons, are believed to play a role in replication and/or maintenance (17). The iterons are a series of 12 and 13 repeats separated by 78 nucleotides (52).
FIG. 1.
(A) pAD1 genetic map (not to scale) showing the replication and maintenance region, along with the adjacent pheromone response regulation region. Putative promoters are indicated by a “P” above the map, and the adjacent arrow indicates the direction of transcription. Transcriptional terminators are represented by t1/t2. Iterons and repA internal direct repeats are represented by “thick” black arrows. Thick gray arrows (above and below) represent the positions and orientations of the traB, repA, and repB primers. Different DNA fragments specifically analyzed are indicated by various lines named accordingly. (B) Nucleotide sequence of the RepA coding sequence showing the internal array of repeats. The numbers above are shown to indicate the nucleotide or amino acid position inside repA. MfeI, RsaI, and DraI restriction sites are indicated. Thin arrows correspond to the RepA internal direct repeats. Thick arrows represent the RepA internal invert repeats. The name of each repeat is also indicated as described previously (3). The asterisk indicates the location of the generated frameshift mutation in repA. Vertical arrows indicate the ends of the repA in-frame deletion obtained. The small gray arrows represent the specific primers used for the construction of clones.
Among the pheromone-responding plasmids, the corresponding repA and repB gene products exhibit significant similarity, and RepA resembles a family of initiator proteins encoded by several low-copy plasmids from Staphylococcus, Lactobacillus, Lactococcus, and Bacillus species (5, 19, 28, 41, 48). For some related plasmids, the replication origin (oriV) has been located within the coding sequences of the initiator proteins (19, 28, 48); however, in the case of the related lactococcal plasmid pCI2000 the oriV appears to be located outside of the initiator determinant (41). Interestingly, the pCI2000 replication region is organized similarly to that of pAD1, with an active partition system transcribed divergently from the initiator determinant. The 70-kb Bacillus natto plasmid pLS32, which has an oriV within the initiator determinant (repN), utilizes a theta replication mechanism, and is the best characterized of the nonenterococcal members of this group (48).
Genes encoding the RepA family of proteins noted here bear interesting, centrally located, directly repeated nucleotide sequences. For example, in the case of repA of pAD1 a 33-bp sequence (DR1) is repeated twice (only one mismatch) and separated by about 75 bp (see Fig. 1B). Similar repeats within the repN of pLS32, but involving different nucleotides, have been suggested to contain the oriV site of that plasmid (48).
pAD1 has two transfer (conjugation) origins, oriT1 and oriT2, that are located about 180° apart on the circular map (3, 11, 23). The oriT2 site is located adjacent to a relaxase (TraX) determinant and is believed to be the preferred site for conjugative transfer (22), whereas oriT1 operates several orders of magnitude less efficiently and is located within repA (3). In addition to the location of oriT1 with respect to repA, another interesting feature that seems to “associate” genes for vegetative replication and transfer functions is a phase variation phenomenon involving transfer functions. This involves the reversible switching on and off of conjugation genes by way of changes in the number of iterons (generally an increase in four iterons [32 nucleotides]) between repA and repB (32, 46). The mechanism by which this phase variation affects conjugation functions remains unknown.
The close physical association of sequences involved in both vegetative replication and regulation of conjugative transfer prompted us to further characterize the pAD1 region associated with plasmid replication. Here we present data showing that RepA is the only pAD1-encoded protein required to initiate replication and that oriV is located within a small segment of the repA determinant. RepA is shown to bind to small inverted repeat structures (i.e., IR-1) within oriV, and these structures are shown genetically to play an important role in replication. In addition, we identify an intriguing relationship between the internal structure of RepA protein and the recognition of oriV.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The Escherichia coli K-12 and E. faecalis strains, plasmids, and synthetic oligonucleotides used in this study are listed in Table 1. E. faecalis strains were grown in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, Mich.) at 37°C. E. coli strains were grown in Luria-Bertani (LB) broth (47). Plating was on THB or LB agar. The following antibiotics were used at the indicated concentrations with E. faecalis: erythromycin at 20 μg/ml, chloramphenicol at 20 μg/ml, rifampin at 25 μg/ml, and fusidic acid at 25 μg/ml. With E. coli, the concentrations were ampicillin 100 μg/ml, kanamycin at 50 μg/ml, chloramphenicol at 25 μg/ml, erythromycin at 200 μg/ml, and nalidixic acid at 20 μg/ml. All antibiotics were obtained from Sigma Chemical Co. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside) were from Invitrogen and were used at a concentrations of 40 μg/ml and 1 mM, respectively. Synthetic cAD1 peptide was prepared at the University of Michigan peptide synthesis core facility.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence | Source or reference | Plasmid(s) generateda |
|---|---|---|---|
| Strains | |||
| E. faecalis | |||
| JH2-2 | rif fus | 39 | |
| FA2-2 | rif fus | 25 | |
| OG1X | str gel | 36 | |
| FA3333 | FA2-2 defective in cAD1 | 2 | |
| E. coli | |||
| DH5α | endA1 recA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(argF-lacZYA)U169 φ80lacZΔM15 | BRL | |
| BL21(DE3)pLysS | F−ompT rB− mB− DE3 | Invitrogen | |
| Plasmids | |||
| pAM714 | pAD1::Tn917; erm; Agg Tra | 35 | |
| pAM330 | pAD1::pAD2; erm km str | 12 | |
| pAM330Δ9 | pAM330 with deletion repAΔ9 | This study | |
| pET30a | Expression vector | Novagen | |
| pASK60 | Expression vector | Biometra | |
| pSU18 | E. coli cloning vector, cm, p15A | 4 | |
| pAM88 | Suicide vector; cm, p15A, cm+ | 22 | |
| pDAK246ΔE | pAD1 minireplicon; erm par | 53 | |
| pAM3314 | 504-bp internal repA cloned in pAM401 | 3 | |
| pAM3316 | 209-bp internal repA cloned in pAM401 | 3 | |
| pAM3318 | 158-bp internal repA cloned in pAM401 | 3 | |
| pAM88A* | repA (frameshift) cloned in pAM88 | This study | |
| pAM88It | It region cloned in pAM88 | This study | |
| pAM88-3314 | 504-bp internal repA cloned in pAM88 | This study | |
| pAM88-3316 | 209-bp internal repA cloned in pAM88 | This study | |
| pAM88-3318 | 158-bp internal repA cloned in pAM88 | This study | |
| pAM88oriV | MfeI-RsaI 170-bp repA cloned in pAM88 | This study | |
| pSU18b | bac promoter cloned in pSU18 | This study | |
| pSU18b* | bac* promoter cloned in pSU18 | This study | |
| pAM434b* | bac* promoter cloned in pAM434 | This study | |
| pAM434brepA | bacrepA cloned in pAM434 | This study | |
| pAM434brepAΔ9 | bacrepAΔ9 cloned in pAM434 | This study | |
| pAM88ABC | pAD1 rep region cloned in pAM88 | This study | |
| pAM88AΔ9BC | pAD1 rep(Δ9) region cloned in pAM88 | This study | |
| pET30aRepA | repA gene cloned in pET30a | This study | |
| pET30aRepA5′ | 5′ repA gene cloned in pET30a | This study | |
| pET30aRepA3′ | 3′ repA gene cloned in pET30a | This study | |
| pASK60RepA | repA gene cloned in pASK60 | This study | |
| pASK60RepB | repB gene cloned in pASK60 | This study | |
| pAM2603 | 7.5-kb pAD1 replication proficient region | 1 | |
| pBluescript | E. coli cloning vector; ap | Stratagene | |
| pTAd | E. coli cloning vector; ap, km | Clontech | |
| pTAdA | repA cloned in pTAd | This study | |
| pTAdA* | repA (frameshift) cloned in pTAd | This study | |
| pTAdAΔ9 | repAΔ9 cloned in pTAd | This study | |
| pTAdIt | It region in pTAd | This study | |
| pTAdIt5′ | 5′ Iteron repeats (see Fig. 1A) in pTAd | This study | |
| pTAdIt3′ | 3′ Iteron repeats (see Fig. 1A) in pTAd | This study | |
| pBlueScriptoriV | oriV region (see Fig. 1) in pBlueScript | This study | |
| pTAdIR1 | IR1 repeats (see Fig. 5A) in pTAd | This study | |
| pTAdIR1* | IR1* repeats (see Fig. 5A) in pTAd | This study | |
| pTAdItC | ItC repeats (see Fig. 1A) in pTAd | This study | |
| pAM88A*-IR3 + 4 | pAM88A* with mutated IR-1 repeats 3 and 4 | This study | |
| pAM88A*-IR1 + 2 | pAM88A* with mutated IR-1 repeats 1 and 2 | This study | |
| pAM88A*-IR1 + 2 + 3 + 4 | pAM88A* with mutated IR-1 repeats 1, 2, 3, and 4 | This study | |
| pAM88A*-5×IR | pAM88A* with all five IR-1 repeats mutated | This study | |
| Oligonucleotides | |||
| RepA | TTCATTGTAAATACGGTT | pAM88It | |
| RepB | CTTCCCAACGCCGCC | pAM88It, pTAdIt3′ | |
| It1 | TTAAGAATACCAAAACATTATT | pTAdIt5′ | |
| It2 | CCTTTCTACAAAAGGATT | pTAdIt5′ | |
| It3 | CCTTTTGTAGAAAGGTT | pTAdIt3′ | |
| 401A | GAGCAAGAGATTACGCGCAG | pAM88/3314, 6, 8 | |
| 401B | TGCCGGCCACGATGCGTCC | pAM88/3314, 6, 8 | |
| ETrepA.1 | CCCAGATCTGAACAACTTCAAATTTTTTAATGT | pAM88A*, pET30aRepA, and pET30aRepA5′ | |
| ETrepA.2 | CCCAAGCTTATTGATTTTCAACCCAGTT | pAM88A*, pET30aRepA, and pET30aRepA3′ | |
| ETrepA.3 | CCCAAGCTTAACCAAGGGATTCAGCGTT | pET30aRepA5′ | |
| ETrepA.4 | CCCAGATCTAACAAACGCTGAATCCC | pET30aRepA3′ | |
| ASK60repB.1 | TTTAACGGCCGGCATGGTTAAAAAAATTGTATT | pASK60repB | |
| ASK60repB.2 | ATTTTTCGCGACTCATTAGCAGTCGTCCCTTC | pASK60repB | |
| ASK60repA.1 | TTTAACGGCCGGCATGAACAACTTCAAATTTTTTAATGT | pAM434brepA, Δ9, pASK60repA | |
| ASK60repA.2 | ATTTTTCGCGATTGATTTTCAACCCAGTT | pAM434brepA, Δ9, pASK60repA | |
| Bac1 | AGAGCGTCGACTGATTGAA | pAM434b* | |
| Bac2 | GGGGTACCGTCGATCTTATCGCGATT | pAM434b* | |
| ItCs | TTTTTTACTATCTTACTATTTTACTAC | pTAdItC | |
| ItCas | ATTTTTTGTAGTAAAATAGTAAGATAG | pTAdItC | |
| IR1s | AATTGAATCAAGAGGGTATGAAAATCATACCCTGCCAAAAC | pTAdIR1 | |
| IR1as | AATTGTTTTGGCAGGGTATGATTTTCATACCCTCTTGATTC | pTAdIR1 | |
| IR1*s | AATTGAATCAAGAGCCTTTCAAAATGAAAGGCTGCCAAAAC | pTAdIR1* | |
| IR1*as | AATTGTTTTGGCAGCCTTTCATTTTGAAAGGCTCTTGATTC | pTAdIR1* | |
| TraB | CAAGATAATACGTTTTATTAGACAC | ||
| M1s | CAAAACGTTGATAAATCAGAGCCTTTGAAAATCAGT | pAM88A*-5×IR | |
| M1as | ACTGATTTTCAAAGGCTCTGATTTATCAACGTTTTG | pAM88A*-5×IR | |
| M2 | GAGCCTTTCAAAATGAAAGGCTGCCAAAACGTTGATAAATCAG | pAM88A*-IR3 + 4 | |
| M3 | GCAGCCTTTCATTTTGAAAGGCTCTTGATTACCAAGGGATTC | pAM88A*-IR3 + 4 | |
| M4 | GAGCCTTTCAAAATGAAAGGCTCCCCCCAAAGAAAAACAAA | pAM88A*-IR1 + 2, pAM88A*-IR1 + 2 + 3 + 4 | |
| M5 | GGAGCCTTTCATTTTGAAAGGCTCTGATTTATCAACGTTTTC | pAM88A*-IR1 + 2, pAM88A*-IR1 + 2 + 3 + 4 |
For clarity, only relevant plasmids generated by using the specific oligonucleotides are indicated.
Standard molecular techniques.
Recombinant plasmids were generated in E. coli DH5α. Introduction of plasmid DNA into bacterial cells was done by transformation as previously described (16, 31). Electrotransformation of E. faecalis was done as described by Flannagan and Clewell (20). Plasmid DNA was purified from E. coli by using established techniques described elsewhere (47). Isolation of plasmid DNA from E. faecalis was also as previously described (51). When necessary, DNA fragments were purified with silica gel as described by Boyle and Lew (6). Recombinant DNA methodology, as well as analyses of plasmid DNA by using restriction enzymes, agarose gel electrophoresis, and Southern hybridization, involved procedures described by Sambrook et al. (47). Restriction enzymes were purchased from Invitrogen, and reactions were carried out under the conditions recommended. PCR was performed with a Perkin-Elmer Cetus apparatus under conditions recommended by the manufacturer. Specific primers were purchased from Invitrogen, and Taq DNA polymerase was from Roche. PCR-generated fragments were purified by using QIAquick-spin columns (Qiagen). Ligations made use of T4 DNA ligase from New England Biolabs. Nucleotide sequence analyses were carried at the University of Michigan sequencing core facility or using the “fmol DNA Cycle Sequencing System” as specified by the manufacturer (Promega).
Plasmid constructions.
The vector pSU18bac represents a pSU18 (4) derivative in which the bacteriocin (bac) promoter (27) has been cloned as an EcoRI fragment. pSU18bac* contains a point mutation in the promoter −10 box, which results in the sequence CATAAT. From here the SalI/KpnI fragment that contains the bac promoter was subcloned into pAM434 (21), yielding pAM434b*.
Different segments of pAD1 included in the replication-maintenance region were amplified by PCR from template pAM714 (35) or pAM3314, pAM3316, and pAM3318 (3) by using the oligonucleotides indicated in Table 1 and cloned into pTAd via TA cloning. The corresponding clone containing repA was partially MfeI digested and filled with Klenow to obtain a frameshift mutation. From here, XbaI/HindIII fragments were subcloned into the E. faecalis suicide plasmid pAM88 (22), generating the plasmids pAM88A*, pAM88It, pAM88-3314, pAM88-3316, pAM88-3318, and pAM88oriV. The clones containing repA or repAΔ9 coding sequences in pAM434 were cloned in several steps. The fragments repA or repAΔ9 contained in pTAd were obtained by digestion with the restriction enzymes EagI/NruI, purified, and cloned into the BsaI and Eco47III sites of pSU18b*. The SalI/KpnI fragments, which contained genes behind the bac promoter, were subcloned into pAM434, generating the plasmids pAM434brepA and pAM434brepAΔ9. The repA derivatives with point mutations were picked up as “unexpected” variants noticed upon sequencing PCR products, as was the deletion relating to pAM330Δ9and pAM434brepAΔ9.
Fragments of DNA containing sequences from the repA and repB genes were amplified by PCR from pAM714 with the primer pairs ETrepA.1 and ETrepA.2, ETrepA.1 and ETrepA.3, ETrepA.4 and ETrepA.2, ASK60repA.1 and ASK60repA.2, or ASK60repB.1 and ASK60repB.2, respectively (Table 1); digested with BglII and HindIII (repA) or EagI and NruI (repA and repB); and cloned into the same sites of pET30a or pASK60, as indicated, to construct the plasmids pET30aRepA, pET30aRepA5′, pET30RepA3′, pASK60RepA, and pASK60RepB, respectively. In the expression vector pET30, RepA is under the control of the T7 promoter. In pASK60, RepA and RepB are under the control of the lac promoter.
Fragments of DNA containing the iteron sequences were amplified by PCR with the primers It1 and It2 or It3 and RepB with the plasmid pAM714 as a template and cloned into pTAd plasmid vector to construct the plasmids pTAdIt5′ and pTAdIt3′. The complete It region was amplified by PCR with the primers RepA and RepB by using the plasmid pAM714 as a template and then cloned into pTAd plasmid vector to construct the plasmid pTAdIt. The putative oriV site was obtained on a digestion product of MfeI and RsaI and cloned into pBluescript to produce the plasmid pBlueScriptoriV. Plasmids pTAdIR1, pTAdIR1*, and pTAdItC were obtained by annealing the corresponding sense and antisense oligonucleotides (Table 1) and direct ligation into the EcoRI site of the pTAd cloning vector. All of the constructions were confirmed by DNA sequencing.
Stability and incompatibility assays.
Stability assays were performed as previously described by Wirth et al. (55). Incompatibility experiments were performed from single colonies of OG1X strains containing either plasmids pDAK246ΔE and pAM3314 or plasmids pDAK246ΔE and pAM401. Loss of pDAK246ΔE was monitored in the presence of selection for the other plasmid as previously described (52).
Protein purification.
The His-tagged fusion proteins (RepA, RepA5′, and RepA3′) were purified from recombinant E. coli BL21(DE3) induced with 1 mM isopropyl-β-d-thiogalactoside by using an Ni-agarose column as described in the manufacturer's instructions (Qiagen GmbH). The Strep-tagged fusion proteins (RepA and RepB) were purified from recombinant E. coli JM83 induced with 1 mM isopropyl-β-d-thiogalactoside by using a streptavidin-immobilized column as described in the manufacturer's instructions (Boehringer Mannheim). All protein preparations used in DNA binding studies were at least 90% pure based on polyacrylamide gel electrophoresis estimates.
Protein analysis.
Proteins were boiled in sample buffer containing sodium dodecyl sulfate and β-mercaptoethanol and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% gel according to the method of Laemmli (42). Gels were stained with Coomassie brilliant blue R-250. Western blotting was performed as described elsewhere (47), and the His-tagged fusion protein (RepA) was detected by using polyclonal anti-His antibody (Santa Cruz Biotechnology), whereas the Strep-tagged fusion proteins (RepA and RepB) were detected by using the polyclonal anti-Strep antibody (Pierce) and the ECL Western blotting analysis system (Amersham Pharmacia Biotech).
DPAC assays.
DNA-protein tag affinity chromatography was carried out under the conditions described by Fujimoto and Clewell (26). Restriction enzyme-digested DNA plasmid pAM2603 was extracted with phenol-chloroform and precipitated with ethanol. Then, 4 μl corresponding to ca. 2 μg of cleaved DNA in 10 mM Tris (pH 8.0) was used.
Preparation of DNA substrates.
Double-stranded DNA (dsDNA) containing iteron repeat fragments for binding assays were generated by PCR by using the plasmids pTAdIt5′ and pTAdIt3′ as templates. The primers used are indicated in Table 1. dsDNA containing oriV fragment was obtained by digestion from pBlueScriptoriV plasmid. The fragments were labeled with [α-32P]dATP (Amersham) included in the PCR or by filling by Klenow in the oriV-containing fragment. PCR products or digestion bands were separated in an agarose gel, and excised bands were eluted with a QiaQuick gel extraction kit (Qiagen). The DNA samples were loaded on an agarose gel for quantification. S1 digestion and boiling and/or denaturation assays were performed in order to demonstrate the nature of the DNA bands tested (24).
Gel mobility shift assays.
Labeled DNA fragments (1 pmol) were incubated with either RepA (or its putative N- and C-terminal domains), RepB, or control (vector derived) protein fractions (0.1 and 0.5 μg) for 15 min at 30°C in a 20-μl volume containing 50 mM Tris (pH 7.5), 100 mM NaCl, 0.2 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 1.5 μg of poly(dI-dC) DNA, and 0.7 μg of bovine serum albumin. After this incubation period, the binding reaction mixtures were placed on ice, loaded onto a 5% prerun polyacrylamide gel, and electrophoresed at room temperature in 0.5× Tris-borate-EDTA buffer. After electrophoresis, the gel was dried on Whatman 3MM paper and exposed to X-ray film at −70°C with an intensifying screen.
Construction of clones representing the mutated IR-1 sites.
The 525-bp PCR fragment containing the pAD1 oriV site that included modifications in IR3 and IR4 was generated with primers M2 and ETrepA.2 and pAM88A* as a template DNA. The 528-bp PCR fragment also containing the mutated repeats IR3 and IR4 was generated by using M3 and ETrepA.1 as primers and pAM88A* as a template DNA. Both fragments were purified by using QIAquick spin columns (Qiagen), diluted 1:1,000, mixed, and used as a template for a new PCR with ETrepA.1 and ETrepA.2 primers. The resulting 1-kb band contained repA* with the repeats IR3 and IR4 mutated (IR3+4). The 588-bp PCR fragment containing the pAD1 oriV site, including the mutated repeats IR1 and IR2, was generated by using primers M4 and ETrepA.2 and pAM88A* as template DNA. The 465-bp PCR fragment also containing the mutated repeats IR1 and IR2 was generated by using M5 and ETrepA.1 as primers and pAM88A* as template DNA. Again, both fragments were purified (as described above), diluted 1:1,000, mixed, and used as a template for a new PCR by using ETrepA.1 and ETrepA.2 as primers. The resulting 1-kb band contained repA* with the repeats IR1 and IR2 mutated (IR1+2). The 588-bp PCR fragment containing the pAD1 oriV site that included the mutated repeats IR1 and IR2 was generated by using primers M4 and ETrepA.2 and band IR3+4 as template DNA. The 465-bp PCR fragment containing the mutated repeats IR1 and IR2 was generated by using M5 and EtrepA.1 as primers and band IR1+2 as template DNA. Both fragments were purified and diluted (as described above), mixed, and used as template for a new PCR with ETrepA.1 and ETrepA.2 primers. The resulting 1-kb band contained repA* with the repeats IR1, IR2, IR3, and IR4 mutated (IR1+2+3+4). The 501-bp PCR fragment containing the pAD1 oriV site that included the mutated repeat IR5 was generated by using primers M1s and ETrepA.2 and band IR1+2+3+4 as template DNA. The 563-bp PCR fragment containing the mutated repeats IR1, IR2, IR3, IR4, and IR5 was generated by using M1as and ETrepA.1 as primers and band IR1+2+3+4 as template DNA. Both fragments were purified, diluted, mixed (as described above), and used as a template for a new PCR with ETrepA.1 and ETrepA.2 as primers. The resulting 1-kb band contained repA* with all IR-1 repeats mutated (5xIR). The resulting DNA products were purified and ligated to pTAd, and the 1.1-kb XbaI-HindIII fragments were cloned into pAM88 obtaining pAM88A*-IR3+4, pAM88A*-IR1+2, pAM88A*-IR1+2+3+4, and pAM88A*-5xIR, respectively. All clones were confirmed by DNA sequencing.
RESULTS
The oriV of pAD1 is within the repA coding sequence.
Although it was previously reported that the pAD1 replicon was located on an ∼3-kb segment of pAD1 carrying repA, repB, and repC and a series of iterons (52), the precise location of oriV was not determined. Although the presence of an array of iterons suggested involvement in replication initiation, recent reports of oriV sequences being located within determinants of repA-like sequences raised the possibility that the origin might be located within repA. Identification of an oriV sequence is generally based on its ability to facilitate replication when present on a plasmid that could otherwise not replicate, if we assume that appropriate replication factors (e.g., initiator protein) are provided in trans. To locate the oriV of pAD1, we cloned specific segments of DNA (either an internal region of repA or the iteron region located between the repA and repB determinants) into a plasmid (pAM88) that is incapable of autonomous replication in E. faecalis.
pAM88 is an E. coli vector carrying a cat determinant able to express in E. faecalis (22) and therefore useful for testing replication. A 1.01-kb segment of pAD1 DNA carrying repA with a frameshift mutation and devoid of a ribosome-binding site was cloned in pAM88, and the resulting chimera, pAM88A* (Table 1 and Fig. 1), was introduced by electroporation into an E. faecalis JH2-2 host harboring a pAD1::pAD2 cointegrate derivative pAM330 (12). (The frameshift mutation in the cloned segment was to assure that there was no expression of RepA from the resulting chimera.) We utilized the cointegrate because it should not be totally dependent on the pAD1 replication origin; that is, it should be able to make use of the pAD2 origin for replication. Selection with chloramphenicol resulted in transformants at a frequency of 3.1 × 10−7 transformants per number of cells per μg of DNA (Table 2). In contrast, a pAM88 chimera designated pAM88It carrying the iteron sequences located between repA and repB (0.5 kb, Table 1 and Fig. 1), as well as the empty vector (pAM88), could not be maintained in (i.e., transform) JH2-2/pAM330 cells (Table 2). Homologous recombination was not involved in the process, as judged by an inability to detect PCR products with primers designed to amplify either of the joint regions that would be expected (Fig. 2A). In addition, restriction bands appropriate for separate (not recombined) plasmids were observed by agarose gel analysis (data not shown). The data imply that a replication origin (oriV) is located within repA and that, since RepA could not be produced by pAM88A* (i.e., because of the frameshift mutation), it responds to RepA supplied in trans from pAM330. Furthermore, the iterons are not part of the pAD1 oriV, since they are not essential for replication.
TABLE 2.
Transformation frequencies of several plasmids in different E. faecalis strains
| Plasmid | E. faecalis strain | Transformation frequencya |
|---|---|---|
| pAM88 | JH2-2/pAM330 | <10−9 |
| pAM88It | JH2-2/pAM330 | <10−9 |
| pAM88A* | JH2-2/pAM330 | 3.1 × 10−7 |
| pAM88A* | JH2-2/pAM330Δ9b | 1.8 × 10−5 |
| pAM88A* | JH2-2/pAM714 | <10−9 |
| pAM88A* | JH2-2 | <10−9 |
| pAM401 | JH2-2/pAM330 | 1.4 × 10−6 |
| pAM401 | JH2-2/pAM330Δ9b | 1.9 × 10−6 |
| pAM401 | JH2-2/pAM714 | 8.2 × 10−6 |
| pAM401 | JH2-2 | 1.0 × 10−6 |
| pAM88 | JH2-2/pAM330 | <10−9 |
| pAM88-3314 | JH2-2/pAM330 | 2.1 × 10−7 |
| pAM88-3316 | JH2-2/pAM330 | <10−9 |
| pAM88-3318 | JH2-2/pAM330 | <10−9 |
| pAM88oriV | JH2-2/pAM330 | 9.9 × 10−8 |
| pAM88oriV | JH2-2 | <10−9 |
| pAM434brepA | JH2-2 | 5.1 × 10−5 |
| pAM434brepAΔ9 | JH2-2 | <10−9 |
| pAM88ABC | JH2-2 | 3.0 × 10−5 |
| pAM88AΔ9BC | JH2-2 | 6.1 × 10−7 |
| pAM401 | JH2-2 | 1.8 × 10−6 |
That is, the number of transformants per total number of competent cells per microgram of DNA.
Derived from JH2-2/pAM330Δ9/pAM88A* by spontaneous curing of pAM88A* during growth in the absence of selection for pAM88A*-encoded chloramphenicol resistance.
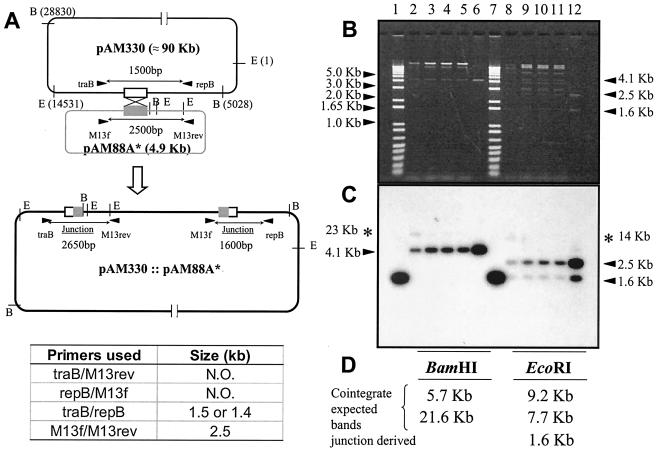
FIG. 2.
Analysis of the replication ability of an E. faecalis suicide vector carrying appropriate fragments of pAD1 DNA. (A) Schematic representation of the process envisioned for cointegrate formation by homologous recombination between the two plasmids pAM330 and pAM88A* carrying the pAD1 repA coding sequence. The related primers used to generate PCR products demonstrating no recovery of the cointegrate plasmid from the chloramphenicol-resistant E. faecalis JH2-2/pAM330 transformants are indicated. The positions of the relevant BamHI and EcoRI sites are also shown. The sizes of the PCR fragments obtained are listed below. N.O., no PCR fragment observed in the assay. (B) Agarose gel electrophoresis representing the BamHI (lanes 2 to 6) and EcoRI (lanes 8 to 12) restriction fragments of the plasmid DNA content of the chloramphenicol-resistant E. faecalis JH2-2/pAM330 transformants containing pAM88oriV plasmid (lanes 2 to 5 and lanes 8 to 11) or the E. coli DH5α/pAM88oriV cells used as a positive control (lanes 6 and 12). Plasmid DNA was obtained in both cases from alkaline lysis preparations as described in Materials and Methods. The molecular mass ladder 1-Kb-Plus (Invitrogen) is shown in lanes 1 and 7, and selected bands or sizes are noted on the left and right. (C) Southern blot analysis of the DNA restriction profiles shown in panel B. The EcoRI restriction fragments of pAM88oriV plasmid DNA were labeled and used as a probe. Black arrowheads indicate the BamHI or EcoRI restriction fragments corresponding to pAM88oriV, and band sizes are noted on the left (BamHI) and right (EcoRI). The asterisks indicate the corresponding pAM330 fragments carrying oriV sequences. (D) The sizes of the restriction fragments that would have been obtained from cointegration events are indicated.
The minimal size of oriV.
To estimate the size of the minimal cis-acting, replication-enabling region within repA, we constructed and examined (as described above) several clones containing different internal fragments of repA designated in Fig. 1A as segments 3324, 3316, and 3318. The smallest replicating fragment was that of pAM88-3314, which corresponded to a 504-bp segment (Table 2). As seen in Fig. 1B, this fragment is the only one containing both DR-1 repeats inside repA, suggesting an involvement for these repeats in replication.
Of likely relevance to the DR-1 sequences is an observation made relating to the earlier-noted experiments involving E. faecalis JH2-2/pAM330 cells transformed with pAM88A*. In the course of examining these transformants we noticed that selected colonies fell into two categories. Approximately 10% of the colonies were “relatively large” while 90% were “small.” Interestingly, when we generated PCR products by using primers flanking repA (primer sequences not located on pAM88A*; see Fig. 2A), we observed that the small colonies gave rise to a product of the expected size (1.5 kb), whereas the large colonies gave rise to a smaller product (1.4 kb). Sequence analysis of the 1.5-kb PCR fragment showed a wild-type repA sequence; however, the 1.4-kb fragment contained a 105-bp in-frame deletion within repA. The deletion removed the region between the two large direct repeats (DR-1), as indicated by the vertical arrows in Fig. 1B. The data indicate that in the large colonies pAM330 had undergone a recombination between the two 33-bp direct repeats (DR-1), leading to a deletion that resulted in the loss of 35 amino acids from an internal portion of RepA. The deletion (pAM330Δ9) did not affect the ability to facilitate replication of pAM88A*. Indeed, pAM88A* replication efficiency may be enhanced under the circumstances since the cells appeared to grow better (i.e., larger colonies) with the deletion. Furthermore, pAM88A* could transform JH2-2/pAM330Δ9 cells at a frequency almost 2 orders of magnitude higher than JH2-2/pAM330 cells (Table 2). A widely used shuttle plasmid pAM401 (55) was able to transform both strains equally well, suggesting that the deletion in pAM330Δ9 decreased incompatibility with pAM88A*, although at this point an elevated expression of the altered RepA also cannot be ruled out.
To determine whether the origin within repA on pAM88A* contributes to incompatibility against pAD1, the ability of a cloned origin fragment to displace a resident pAD1 replicon was tested. For this purpose we used pDAK246ΔE (53) as the resident plasmid and pAM3314 (3) as the pAD1 oriV-containing plasmid. pDAK246ΔE is a pAD1 minireplicon (Table 1) encoding erythromycin resistance and deleted for the par-encoded postsegregational killing system (located downstream of repC). pAM3314 is a pAM401 clone containing an internal portion (the 3314 segment; Fig. 1A) of repA including the entire putative oriV. (The E. coli-E. faecalis shuttle vector pAM401 replicates independently of pAD1.) Under selection for the pAM3314-encoded chloramphenicol resistance, pDAK246ΔE was consistently lost from ∼80% of cells within 40 generations. In the presence of the empty vector (pAM401), no loss of pDAK246ΔE was observed (the two plasmids are compatible); this is consistent with the view that oriV acts as an incompatibility determinant.
A reasonable interpretation of the above data is that oriV is located, at least in part, in the region between the 33-bp direct repeats (DR-1) in repA and that deletion of this segment eliminated, or greatly reduced, competition between the two plasmids. To further explore this notion, we cloned into pAM88 a 173-bp restriction fragment (MfeI/RsaI) from within repA that contained both repeats (Fig. 1) and introduced it into JH2-2/pAM330; replication was assayed as described above. The chimera (pAM88oriV) was able to transform (selection on chloramphenicol) at a frequency of ∼10−7 (Table 2). (pAM88oriV DNA was not maintained in JH2-2 cells that did not harbor pAM330.) Plasmid isolation from several independently obtained transformants, restriction analyses, and Southern hybridizations (Fig. 2B and C) confirmed the presence of the intact plasmid (pAM88oriV) in JH2-2/pAM330. The data indicate that the cloned 173-bp DNA fragment present in pAM88oriV approximates the minimal segment required in cis to support pAD1 replication.
RepA is the only pAD1-encoded protein necessary for plasmid replication.
We have shown above that pAM330 supplies a “replication factor” able to facilitate in trans the establishment of a chimera carrying the oriV sequence (i.e., the RsaI/MfeI segment). To test the hypothesis that this factor was RepA, we attempted to clone an intact repA gene under a gram-positive bacteriocin promoter (27) in the E. coli vector pAM434b to determine whether autonomous replication occurred in E. faecalis (see Materials and Methods and Table 1). We were not able to clone the repA fragment without generating various mutations in repA; conceivably, expression of repA under these conditions was detrimental to E. coli. However, when we used a vector in which the promoter was altered via mutation in the −10 box (TATAAT changed to CATAAT), we recovered a clone with an intact repA. Assuming that the promoter still functions but is probably less active, we attempted to introduce this derivative, designated pAM434brepA, into E. faecalis JH2-2 cells. Erythromycin-resistant transformants were generated at a frequency of 5.1 × 10−5 (Table 2) and, as shown in Fig. 3, they contained the plasmid. A similarly generated chimera containing the repAΔ9 coding sequence (repA gene derived from pAM330Δ9) was constructed with the same altered −10 box, but this chimera, called pAM434brepAΔ9, was not able to transform E. faecalis cells (Table 2). The data indicate that repA alone comprises the pAD1 minimal replicon and the repA product recognizes an oriV located within its own reading frame. The inability of the repAΔ9 DNA to facilitate replication, despite the apparent ability of RepAΔ9 to recognize an intact oriV (described in the previous section) is again consistent with oriV being located between the two DR-1 repeats in repA. For comparison, transformation values associated with the entire repABC region of pAD1 are also included in Table 2. pAM88ABC (carries the wild-type repABC region) transforms JH2-2 cells, as well as pAM434brepA; however, if a similarly generated clone contains the deletion (pAM88AΔ9BC), the transformation frequency is 2 orders of magnitude lower. This is consistent with the iteron repeats not being a component of the replication origin, since they are present in both plasmids. However, the low level of transformants that does appear with pAM88AΔ9BC may in some way relate to marginal replication enabled by stability functions provided by RepB, RepC, and perhaps the iterons.
FIG. 3.
Analysis of the pAD1 minimal replicon. (A) Schematic diagram showing the construction of pAM434brepA and unique restriction sites are indicated (21); (B) agarose gel electrophoresis representing the BamHI/NsiI (lanes 2 and 3) restriction fragments (black arrowheads) of the DNA plasmid content (pAM434brepA) of two independently obtained erythromycin-resistant E. faecalis JH2-2 transformants. The molecular mass ladder 1-Kb-Plus (Invitrogen) is shown in lane 1. Representative sizes are indicated on the left and right. *, Per Flannagan and Clewell (21).
We were able to detect pAM434brepA DNA from enterococcal transformants as a supercoiled structure and could not detect ssDNA by Southern blot hybridization (not shown), a finding consistent with replication occurring via a theta mechanism (i.e., not a rolling-circle mechanism), as otherwise suggested based on the similarity of repA with replication gene determinants on certain other theta-replicating plasmids (48). pAM434brepA appears at a relatively high copy number (at least 20 to 30 copies per chromosome) based on the amount of DNA detectable in gels compared to the high-copy-number pAM401 (observed in extracts prepared in parallel from similar numbers of cells [results not shown]). This is not surprising since the regulatory machinery normally involved in initiation of the low-copy pAD1 is not present, and RepA is being expressed from an artificial promoter.
The relative stability of pAM434brepA, its small size (∼4.65 kb), and its multiple cloning site (see Fig. 3) make it a good candidate for an E. coli-Enterococcus shuttle plasmid. It appeared at least as stable, if not more stable, than the pAM401 (∼10.4-kb) shuttle plasmid. After unselected growth in THB for 30 generations, the percentage of pAM434brepA-containing cells was 40% compared to 15% for pAM401-containing cells.
RepA binds to its own coding sequence.
If RepA is the replication initiator, it should bind to the oriV within repA. To investigate such behavior, we purified RepA by using an E. coli system that expressed His tag or Strep tag fusions to RepA as described in Materials and Methods. In initial experiments using a previously reported DNA-protein tag affinity chromatography technique (26), we observed that purified Strep-tagged RepA bound specifically to a 0.9-kb DraI restriction fragment generated by cleavage of pAM2603 (which contains 7.9 kb of pAD1 that includes repA cloned in pBluescript [1; data not shown]). As anticipated, this DraI fragment included the region within repA believed to contain oriV (Fig. 1).
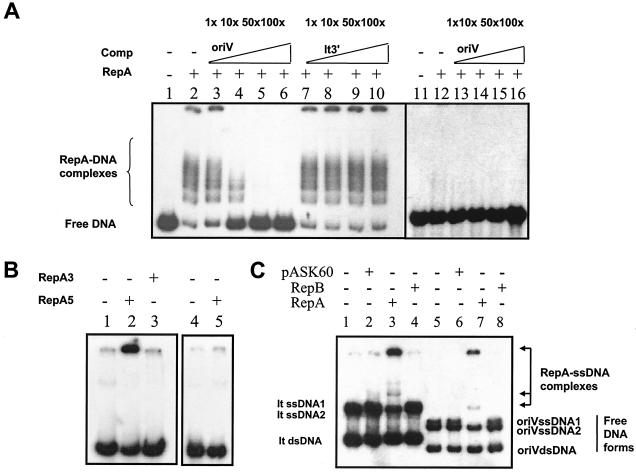
Since the ∼170-bp MfeI/RsaI DNA fragment within repA (Fig. 1A and 2B) was sufficient in cis to allow replication in vivo, we used this segment (designated oriV) in mobility shift assays to examine RepA binding. Comparisons were also made by using segments of DNA containing the set of 12 iterons located adjacent to repB, designated It3′ (see Fig. 1A). As shown in Fig. 4A, purified His-tagged RepA was able to form complexes with the oriV-containing DNA fragment (lane 2) but did not show similar complexes in the case of the iteron It3′ DNA (lanes 12 to 16) or unrelated DNA fragments, such as a 200-bp fragment containing the polylinker of pBlueScript (not shown). A protein preparation generated in the same way as for His-tagged RepA but with E. coli BL21 cells containing an empty expression vector (pET30a) showed no binding activity (not shown), implying that the RepA protein specifically binds to oriV DNA. Confirmation was obtained with competition experiments. As shown in Fig. 4A, the unlabeled oriV DNA fragment greatly reduced RepA interaction with the labeled oriV DNA (lanes 3 to 6), whereas cold It3′ DNA did not compete (lanes 7 to 10), indicating sequence specificity in the binding of RepA. Experiments shown in Fig. 4B suggested this specificity was associated with the N-terminal domain of RepA (RepA5′; see Fig. 1A). Purified His-tagged RepA5′ was able to retard the mobility of the oriV-containing DNA fragment (lane 2) but did not retard an iteron-containing fragment (It5′; lane 5). In contrast to the case observed with the intact (“wild-type”) His-tagged RepA, the DNA-protein complexes remained in the well. Although the nature of these aggregates is currently unknown, the fact that a similar binding did not occur with the iteron-containing fragment (lane 5) or a purified C-terminal (His-tagged RepA3′; lane 3) preparation suggests that this interaction is specific.
FIG. 4.
Gel mobility shift assays showing in vitro RepA-DNA-binding properties. PCR or end-labeled dsDNA or ssDNA fragments containing the oriV or iteron repeats were incubated with 0.5 μg of purified RepA, purified RepB, or empty vector (pASK60)-derived control protein extracts in the absence or presence of increasing concentrations of unlabeled competitor DNA fragments. (A) Mobility shift assays showing RepA-specific binding to dsDNA. The substrate DNA used was as follows: lanes 1 to 10, dsDNA corresponding to the repA MfeI/RsaI internal sequence (oriV); lanes 11 to 16, dsDNA corresponding to the iteron repeats upstream of the repB coding sequence (It3′). Purified protein added in each lane is indicated at the top of the figure. The addition of a 1-, 10-, 50-, or 100-fold excess of unlabeled dsDNA fragments (competitor DNA) is also indicated at the top of the figure. (B) Mobility shift assays showing RepA N-terminal domain specific binding to dsDNA. Lanes 1 to 3, dsDNA corresponding to oriV (MfeI/RsaI repA internal fragment); lanes 4 and 5, dsDNA corresponding to the iteron repeats upstream of the repA coding sequence (It5′ PCR product). Purified protein domains (N and C terminal) added in each lane are indicated at the top of the figure. (C) Mobility shift assays showing RepA nonspecific binding to ssDNA. Lanes 1 to 4 represent It3′ PCR product containing both dsDNA and ssDNA forms. In contrast to panels A and B, the DNA used in panel C had not been exposed to S1 nuclease. Lanes 5 to 8 represent oriV PCR product containing both dsDNA and ssDNA forms. Purified protein fractions in each lane are also indicated at the top of the figure. Free DNA forms and RepA-DNA complexes are indicated.
Although RepA was observed to bind to oriV dsDNA, it also bound to ssDNA, but without any sequence specificity. Figure 4C shows RepA binding to both single-stranded oriV DNA (lane 7) and single-stranded It3′ DNA (lane 3). In contrast to the case shown in Fig. 4A and B, the DNA preparations of Fig. 4C were not previously treated with S1 nuclease and thus had a significant amount of ssDNA in the PCR preparations. We also observed that ssDNA of the vector pBluescript and even eukaryotic (human) DNA bound equally well to RepA (not shown), confirming the absence of sequence specificity. The fact that no binding to ssDNA was observed for other protein preparations, including the pAD1-encoded RepB (lanes 4 and 8) or a similar protein preparation derived from E. coli cells containing the empty vector pASK60 (lanes 2 and 6), indicates RepA presence in the protein-DNA complexes. (In addition to not binding to ssDNA, RepB was also not observed to bind to double-stranded DNA (dsDNA) corresponding to It3′ or oriV [not shown].) RepA was able to bind to ssDNA containing oriV sequence with relatively higher affinity compared to the binding to dsDNA, because when both DNA forms were present, RepA showed a preference for the ssDNA (lane 7). (Interestingly, RepA appears to have some preference for one of the two single strands; see lanes 3 and 7.)
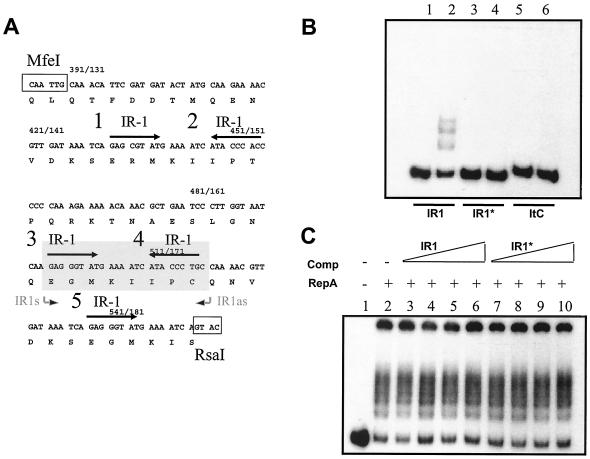
IR-1 sequences in oriV are critical for replication.
The mobility shift assays suggested the formation of at least five RepA/oriV complexes. Inspection of the oriV region reveals the octanucleotide sequence AGGGTATG, noted as IR-1 in Fig. 5A, present as five copies (two with one mismatch). Four of these represent two paired inverted repeat sequences. To determine whether this sequence was involved in RepA binding, two synthetic DNA fragments were prepared as described in Materials and Methods. The first one contains the sequence AATCAAGAGGGTATGAAAATCATACCCTGCCAAAA and corresponds to the region that includes the two central IR-1 repeats (representing sequences 3 and 4 in Fig. 5A); it was cloned in pTAd yielding pTAdIR1. A 155-bp XbaI/HindIII fragment containing the sequence of note was then used in mobility shift experiments. A second chimera, pTAdIR1*, containing the same sequence but with four altered positions in each one of the IR-1 repeats (IR1*),AATCAAGAGCCTTTCAAAATGAAAGGCTGCCAAAA,was also generated in the same way and was designed to conserve both the dyad structure and the G+C percentage. As shown in Fig. 5B, RepA bound to the segment carrying the wild-type (IR1) sequence but did not bind to the DNA carrying the mutated (IR1*) sequence. An additional control DNA representing three iteron sequences cloned in the same way, giving rise to pTAdItC, also did not bind RepA. The data suggest that the IR1 sequence is part of the oriV site to which RepA binds. However, we observed that an excess of unlabeled IR1 DNA did not compete with RepA binding to the larger oriV fragment containing all five IR1 sequences nor did IR1* (Fig. 5C). This suggests that cooperativity or conformation dictated by the presence of more than two IR-1 sequences may play an important role in RepA binding to oriV.
FIG. 5.
Gel mobility shift assays showing the importance of the IR-1 repeats in the putative RepA recognition site. (A) Nucleotide sequence of the repA MfeI/RsaI segment containing the oriV site. IR-1 repeats are indicated by the arrows above the sequence and are numbered 1 through 5 (correlating with specific mutations generated). (B) Gel mobility shift assays with end-labeled dsDNA fragments containing the IR-1 (wild type) (lanes 1 and 2), IR-1* (four point mutations) (lanes 3 and 4), or iteron (ItC) (lanes 5 and 6) repeats. Lanes 1, 3, and 5 represent control dsDNA fragments. Lanes 2, 4, and 6 represent dsDNA plus purified RepA protein. (C) Competition of RepA binding to the MfeI/RsaI oriV dsDNA fragment with increasing concentrations (same as Fig. 4) of unlabeled IR-1 (lanes 3 to 6) or IR-1* (lanes 7 to 10) dsDNA fragments. Lane 1, control oriV DNA; lane 2, RepA control binding reaction to oriV dsDNA; lanes 3 to 10, competition reactions as indicated at the top of the figure.
To further examine the role of IR-1 sequences, we introduced the above modifications (mutations) of IR-1 into plasmid pAM88A* (see Materials and Methods) and generated derivatives with alterations in the first pair (sequences 1 and 2 in Fig. 5A) of IR-1 sequences (pAM88*-IR1+2), the second pair (pAM88A*-IR3+4), the first and second pair (pAM88A*-IR1+2+3+4), and finally with modifications in all five IR-1 sequences (pAM88A*-5xIR). These derivatives were introduced into E. faecalis JH2-2/pAM330Δ9 by electroporation with pAM88A* and pAM401 serving as positive controls. (In contrast to the strain harboring pAM330, this strain was shown above to be much more efficiently transformable by pAM88A*.) The transformation frequencies are shown in Table 3. PCR experiments were done to confirm the presence of autonomously replicating plasmids as per the experiments shown in Fig. 2. Only pAM88A*-IR1+2 and pAM88A*-IR3+4 could be observed as independent plasmids; whereas, transformants deriving from pAM88A*-IR1+2+3+4 and pAM88*-5xIR were the results of cointegration of pAM330Δ9 and the pAM88A* derivative. The much higher transformation frequency exhibited by pAM88A*-IR3+4 (similar to that of the wild-type pAM88A*) compared to pAM88A*-IR1+2 (reduced by 2 orders of magnitude) suggests a more important cis-acting role in replication for the repeats related to the latter derivative. However, substitution of both pairs of IR-1 sequences resulted in complete loss of ability to replicate autonomously. The data are consistent with the in vitro binding studies and show a dependence on IR-1 sequences in cis for replication.
TABLE 3.
Transformation frequencies of E. faecalis JH2-2/pAM330Δ9 by IR-1 mutant plasmids
| Plasmid | Transformation frequencya |
|---|---|
| pAM88A* | 1.8 × 10−5 |
| pAM88A*-IR3+4 | 5.2 × 10−5 |
| pAM88A*-IR1+2 | 1.5 × 10−7 |
| pAM88A*-IR1+2+3+4 | 3.5 × 10−8 |
| pAM88A*-5×IR | 2.0 × 10−8 |
| pAM401 | 3.0 × 10−6 |
That is, the number of transformants per total number of competent cells per microgram of DNA.
DISCUSSION
The data presented here demonstrate that the RepA protein of pAD1 is necessary and sufficient for initiation of plasmid DNA replication at an oriV located within its own coding sequence. The oriV region was narrowed down to ca. 170 bp, based on its ability to support replication when RepA was supplied in trans. This segment contains both direct (DR-1 and DR-2) and inverted (IR-1) repeat sequences, and a pair of IR-1 repeats was shown to be essential in cis for oriV function. A segment of DNA containing an intact repA only, and expressing a functional RepA under an artificial promoter, enabled autonomous replication in E. faecalis. The plasmid construct (pAM434brepA) was quite stable; and because of its small size and the presence of a useful polylinker site, it may prove to be a useful E. coli-E. faecalis shuttle plasmid.
Of additional significance was our observation that a spontaneous recombinational event between the two 33-bp DR-1 repeats within repA resulted in an in-frame deletion giving rise to a protein (RepAΔ9) that, despite the absence of 35 centrally located amino acids, remained able to facilitate (in trans) replication of DNA (plasmid suicide vector) containing an intact oriV. The sequence deleted in repAΔ9 contained a significant portion of the oriV sequence; thus, it was not surprising to find that, in contrast to the above-noted pAM434brepA that was able to replicate stably in E. faecalis, a variant containing the repAΔ9 sequence (pAM434brepAΔ9) was not able to generate transformants. When the same deletion (repAΔ9) was present, together with repB, repC, and the iterons (i.e., in the case of pAM88AΔ9BC), however, some degree of replication occurred, presumably owing to stabilizing effects conferred by RepB, RepC, and possibly the iterons, on a remaining portion of oriV. (An analysis of the RepB and RepC roles will be reported elsewhere.)
Our earlier analyses of the pAD1 replicon suggested that RepA was likely to bind to the extensive iterons upstream of the repA coding sequence (32, 52). However, the data of the current study show that RepA targets DNA outside of the iterons. Our in vitro studies showed that RepA (but not a protein [RepB] suspected of functioning in stable inheritance [52]) bound at multiple sites within oriV (inside the repA coding sequence) and indirectly suggested recognition of the five IR-1 sequences. Indeed, an artificially constructed fragment containing one pair of IR-1 sequences bound to RepA, whereas a similar segment containing alterations in the IR-1 did not bind. Subsequent genetic analyses with plasmid constructs with altered IR-1 pairs supported the notion that these sequences play an important cis-acting role in plasmid replication. The likelihood that some degree of cooperativity is involved in RepA binding was apparent from the inability of a DNA fragment containing two copies (inverted) of IR-1 to compete with the ∼170-bp oriV fragment which contained five copies of the repeat. The data would also be consistent with a preferential role for one of the IR-1 pairs, as was indeed suggested from the genetic analyses. Another explanation, also consistent with the in vivo data, could relate to recognition of a specific DNA structure in addition to the IR-1 sequences. We note that numerous attempts at DNase I footprinting experiments under different conditions (unpublished) were not able to identify a region occupied by RepA. Certain DNA-binding proteins that bind within the minor groove of B-type DNA are known to yield poor footprinting data (18); indeed, several replication initiator proteins have been shown to contact DNA via the minor groove (29, 57). Whether RepA is such a protein is not currently known. Our in vitro DNA retardation data showing binding to repeat sequences within repA is to our knowledge the first such evidence for this family of initiator proteins.
The initiation of replication has been widely studied in gram-negative bacteria, in which plasmids utilizing a theta mechanism frequently carry a series of iterons to which the initiator binds (14, 17, 33). A well-known exception is the R1 plasmid, in which the initiator binds to inverted repeat sequences (30). This is also the case for coliphage lambda (15). Interestingly, in the case of lambda phage, as well as several bacteriophages from gram-positive bacteria, oriV sequences are located within the coding sequence of their respective initiator proteins (44, 45, 57). These initiation proteins, however, are not related to those addressed in the present study, despite the fact that certain RepA homologues are associated with bacteriophages. In addition, certain linear plasmids from Streptomyces species have been shown to contain origins within a rep determinant (7).
Initiator proteins of a wide variety of plasmids create a localized melting in an AT-rich region close to their DNA-binding site in the origin (14, 17). It is noteworthy that in the case of pAD1 one of the IR-1 sequences within oriV believed to bind RepA actually overlaps a highly GC-rich sequence (CCCACCCCCC) that only appears once in the entire plasmid and resembles a transfer origin “nick site” of IncF-like plasmids (gram negative) (3). Interestingly, this site is immediately adjacent to a highly AT-rich sequence (AAAGAAAAACAAA), but whether or not this junction of high and low GC content plays a key role in initiation remains to be determined. In addition to binding specifically and facilitating melting within the replication origin, the ability of RepA to strongly bind nonspecifically to ssDNA suggests a possible role in stabilizing a “melted” conformation important in assembly of the replisome. Such a process has been proposed with respect to the E. faecalis plasmid pAMβ1 initiator protein RepE, which has also been shown to bind to ssDNA (43).
RepA of pAD1 is member of a recently described family of replication proteins initially found encoded by the B. natto plasmid pLS32 (48). Generally associated with a theta-type replication mechanism, these “Rep proteins” are encoded by plasmids in gram-positive genera, including Enterococcus, Lactococcus, Lactobacillus, Bacillus, and Staphylococcus and are also associated with the genomes of a number of bacteriophages from Streptococcus spp. (5, 19, 28, 41, 48). Only one sequence in the family relates to a gram-negative bacterium (Fusobacterium nucleatum [EAA24086]). Another member of this family, interferon response binding factor 1 (IREBF-1), appears to have a nonbacterial origin (56); however, Berg et al. (5) have suggested that IREBF-1 may actually represent a contaminant of bacterial origin that was present in the mouse cDNA library.
RepA (pAD1) consists of 336 amino acids, and most members of the related family are similar in size. There are no recognizable motifs such as those representative of ATPase, helicase, or specific HTH-binding domains. There is strong conservation of a number of residues in the N-terminal region, and five amino acids—Y41, D58, L90, L95, and Y116—are absolutely conserved in all of 38 RepA homologues compared (see Fig. 6). These conserved amino acids may relate to a key function and/or represent parts of an active center. (It is worth noting that the spontaneous mutations generated when we attempted to clone repA under the wild-type bacteriocin promoter involved this 5′ region [data not shown].) It has been suggested (56) that this region contains the DNA-binding domain, and binding experiments carried out with the N-terminal domain of RepA were in agreement with this notion. The central region of the RepA homologues shows a conserved presence and organization of repeats, whereas the corresponding amino acid sequences are highly variable (Fig. 6). This is true even among the E. faecalis pheromone-responding plasmids. In the C-terminal part of the RepA family proteins, at least three subgroups are distinguishable, with a number of amino acids uniquely conserved in each group (Fig. 6). These subgroups (Enterococcus, Staphylococcus, and Lactococcus or Lactobacillus) conceivably reflect the different hosts in which the plasmids were originally isolated and may relate to specific functions shared by closely related bacterial species. Although direct evidence for this is not yet available, comparison of the amino acid sequences of different initiator proteins is suggestive of such a possibility. For example, the corresponding Rep proteins from the E. faecalis plasmids pAD1 (A47092) and pCF10 (A53309) exhibit 43% identity in the N-terminal region and 80% in the C-terminal region; whereas, in contrast, a “RepA” protein from E. faecium (ZP_00037682) exhibited 72% identity to that of pAD1 in the N terminus but only 30% identity in the C terminus. Importantly, the greater identity observed in the N-terminal domains of the latter two proteins also correlated with a high conservation in DNA sequence of the direct repeats in the central (oriV) region, which are 92% identical (only 17 differences over 155 bp, and 16 of the 17 differences are outside the DR-1 and IR-1 repeats), pointing again to a DNA-binding function for the N-terminal domain. One might expect that highly conserved DNA-binding domains would share an affinity for DNA sites that are highly homologous; indeed, one can easily envision these two features evolving together. It has been suggested previously (28, 48) for several plasmids (e.g., pLS32 and pSX267), albeit without direct evidence, that the centrally located repeats corresponds to part of the replication origin. Our in vitro and in vivo data, however, represent strong evidence that this is indeed the case for pAD1 and probably the related E. faecium system noted above.
FIG. 6.
Map of RepA indicating strongly conserved amino acids among the related family of replication proteins. Identical and similar amino acids conserved in the N-terminal region of all proteins (38 proteins in the database compared) are noted in capital letters. (Similar residues are according to conservative substitutions [V/I/L, T/S, D/E, N/Q, Y/F, and R/K].) Positions of putative motifs reflecting conserved residues are noted by the hashed boxes below the N terminus. Amino acid residues that are strongly conserved among RepA homologues of Enterococcus, Staphylococcus, and Lactococcus spp. are indicated by lowercase letters above the C-terminal region. The “u” denotes hydrophobic amino acids in the noted position. The repeats present in the central region are noted in gray. The white box under the N terminus indicates the fragment shown to have specific DNA-binding properties.
Finally, related to the physical association of sequences involved in replication (oriV) and conjugation (oriT1), it is noteworthy that the DNA fragment marked as 3316 in Fig. 1A provides an origin of transfer (3) when cloned in pAM401 (pAM3316), whereas it did not provide a functional replication origin when cloned in pAM88 (pAM88-3316). Although it is conceivable that there still may be some overlap between the two sites, and therefore possibly some common machinery utilized in the initiation of replication at oriV and conjugation involving oriT1, related processes or their regulation remain the subject of future studies.
Acknowledgments
This study was supported by National Institutes of Health grants GM33956 to D.B.C. and GM55544 to K.E.W. and grant FIS 02/3029 from the Spanish Fondo de Investigación Sanitaria to M.V.F. M.V.F. was also supported in part by grant from NATO.
We thank all members of our laboratories for helpful discussions.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid pAD1 in Enterococcus faecalis. Plasmid 31:215-221. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, F. Y., and D. B. Clewell. 1997. The origin of transfer (oriT) of the enterococcal, pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid 37:87-94. [DOI] [PubMed] [Google Scholar]
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, J. S., and A. M. Lew. 1995. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 11:8. [DOI] [PubMed] [Google Scholar]
- 7.Chang, P. C., E. S. Kim, and S. N. Cohen. 1996. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol. Microbiol. 22:789-800. [DOI] [PubMed] [Google Scholar]
- 8.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clewell, D. B. 1999. Sex pheromone systems in enterococci, p. 47-65. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 10.Clewell, D. B., and G. M. Dunny. 2002. Conjugation and genetic exchange in enterococci, p. 265-300. In S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 11.Clewell, D. B., M. V. Francia, S. E. Flannagan, and F. Y. An. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:265-300. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colmar, I., and T. Horaud. 1987. Enterococcus faecalis hemolysin-bacteriocin plasmids belong to the same incompatibility group. Appl. Environ. Microbiol. 53:567-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodson, M., J. Roberts, R. McMacken, and H. Echols. 1985. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc. Natl. Acad. Sci. USA 82:4678-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa, M., S. Cohen, M. Couturier, G. del Solar, R. Diaz-Orejas, R. Giraldo, L. Jánniere, C. Miller, M. Osborn, and C. M. Thomas. 2000. Plasmid replication and copy number control, p. 1-47. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, London, England.
- 18.Feng, L., B. Wang, B. Driscoll, and A. Jong. 2000. Identification and characterization of Saccharomyces cerevisiae Cdc6 DNA-binding properties. Mol. Biol. Cell 11:1673-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firth, N., S. Apisiridej, T. Berg, B. A. O'Rourke, S. Curnock, K. G. Dyke, and R. A. Skurray. 2000. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 182:2170-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannagan, S. E., and D. B. Clewell. 1991. Conjugative transfer of Tn916 in Enterococcus faecalis: transactivation of homologous transposons. J. Bacteriol. 173:7136-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803-817. [DOI] [PubMed] [Google Scholar]
- 22.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 23.Francia, M. V., W. Haas, R. Wirth, E. Samberger, A. Muscholl-Silberhorn, M. S. Gilmore, Y. Ike, K. E. Weaver, F. Y. An, and D. B. Clewell. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46:117-127. [DOI] [PubMed] [Google Scholar]
- 24.Francia, M. V., J. C. Zabala, F. de la Cruz, and J. M. Garcia Lobo. 1999. The IntI1 integron integrase preferentially binds single-stranded DNA of the attC site. J. Bacteriol. 181:6844-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke, A. E., and D. B. Clewell. 1981. Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harbor Symp. Quant. Biol. 45(Pt. 1):77-80. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto, S., and D. B. Clewell. 1998. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc. Natl. Acad. Sci. USA 95:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujimoto, S., and Y. Ike. 2001. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 67:1262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gering, M., F. Gotz, and R. Bruckner. 1996. Sequence and analysis of the replication region of the Staphylococcus xylosus plasmid pSX267. Gene 182:117-122. [DOI] [PubMed] [Google Scholar]
- 29.Giraldo, R., J. M. Andreu, and R. Diaz-Orejas. 1998. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 17:4511-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giraldo, R., and R. Diaz. 1992. Differential binding of wild-type and a mutant RepA protein to oriR sequence suggests a model for the initiation of plasmid R1 replication. J. Mol. Biol. 228:787-802. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 32.Heath, D. G., F. Y. An, K. E. Weaver, and D. B. Clewell. 1995. Phase variation of Enterococcus faecalis pAD1 conjugation functions relates to changes in iteron sequence region. J. Bacteriol. 177:5453-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helinski, D. R., A. E. Toukdarian, and R. P. Novick. 1996. Replication control and other stable maintenance mechanisms of plasmids, p. 2295-2324. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 34.Ike, Y., and D. B. Clewell. 1992. Evidence that the hemolysin/bacteriocin phenotype of Enterococcus faecalis subsp. zymogenes can be determined by plasmids in different incompatibility groups as well as by the chromosome. J. Bacteriol. 174:8172-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ike, Y., and D. B. Clewell. 1984. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J. Bacteriol. 158:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ike, Y., H. Hashimoto, and D. B. Clewell. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 45:528-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ike, Y., H. Hashimoto, and D. B. Clewell. 1987. High incidence of hemolysin production by Enterococcus (Streptococcus) faecalis strains associated with human parenteral infections. J. Clin. Microbiol. 25:1524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jett, B. D., H. G. Jensen, R. E. Nordquist, and M. S. Gilmore. 1992. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 60:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 43.Le Chatelier, E., L. Janniere, S. D. Ehrlich, and D. Canceill. 2001. The RepE initiator is a double-stranded and single-stranded DNA-binding protein that forms an atypical open complex at the onset of replication of plasmid pAMβ1 from gram-positive bacteria. J. Biol. Chem. 276:10234-10246. [DOI] [PubMed] [Google Scholar]
- 44.Missich, R., F. Weise, S. Chai, R. Lurz, X. Pedre, and J. C. Alonso. 1997. The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol. 270:50-64. [DOI] [PubMed] [Google Scholar]
- 45.Ostergaard, S., L. Brondsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pontius, L. T., and D. B. Clewell. 1991. A phase variation event that activates conjugation functions encoded by the Enterococcus faecalis plasmid pAD1. Plasmid 26:172-185. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Tanaka, T., and M. Ogura. 1998. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 422:243-246. [DOI] [PubMed] [Google Scholar]
- 49.Tomich, P. K., F. Y. An, S. P. Damle, and D. B. Clewell. 1979. Plasmid-related transmissibility and multiple drug resistance in Streptococcus faecalis subsp. zymogenes strain DS16. Antimicrob. Agents Chemother. 15:828-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver, K. E. 2000. Enterococcal genetics, p. 259-271. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 51.Weaver, K. E., and D. B. Clewell. 1988. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J. Bacteriol. 170:4343-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weaver, K. E., D. B. Clewell, and F. An. 1993. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J. Bacteriol. 175:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53-63. [DOI] [PubMed] [Google Scholar]
- 54.Weaver, K. E., L. B. Rice, and G. Churchward. 2002. Plasmids and transposons, p. 219-263. In S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 55.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan, C., and I. Tamm. 1991. Molecular cloning and characterization of interferon alpha/beta response element binding factors of the murine (2′-5′)oligoadenylate synthetase ME-12 gene. Proc. Natl. Acad. Sci. USA 88:144-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuñiga, M., B. Franke-Fayard, G. Venema, J. Kok, and A. Nauta. 2002. Characterization of the putative replisome organizer of the lactococcal bacteriophage r1t. J. Virol. 76:10234-10244. [DOI] [PMC free article] [PubMed] [Google Scholar]