Abstract
The human kallikrein-related peptidases (KLK) are serine proteases whose concentrations are often abnormal in common human malignancies and contribute to neoplastic progression through multifaceted roles. However, little attention has been paid to their synthesis and involvement in the development and dissemination of lung cancer, the leading cause of cancer mortality worldwide. We have analysed the production of KLK6 in normal lung and tumour tissues from patients with non-small cell lung cancer (NSCLC). KLK6 immunoreactivity was restricted to epithelial cells of the normal bronchi, but most of the cancer samples were moderately or highly immunoreactive, regardless of the histological subtype. In contrast, little or no KLK6 was detected in NSCLC cells. We have developed NSCLC lines expressing wild-type KLK6 in order to investigate the role of KLK6 in lung cancer biology, and analysed its impact on proliferation. Ectopic KLK6 dramatically enhanced NSCLC cell growth and KLK6-producing NSCLC cells had accelerated cell cycles, between the G1 and S phases. This was accompanied by a marked increase in cyclin E and decrease in p21. KLK6 production was also associated with enhanced synthesis of c-Myc, which is known to promote cell-cycle progression. Finally, examination of specimens from patients with NSCLC revealed that KLK6 mRNA is overexpressed in tumour tissue, and high KLK6 concentrations were associated with lower survival rates. We conclude that a high concentration of KLK6 is an indicator of tumour proliferation and an independent predictive factor in NSCLC.
Keywords: NSCLC, kallikrein-related peptidase, KLK6, overexpression, cell proliferation, cell cycle, c-Myc, cyclin, survival
Introduction
Human kallikrein-related peptidase 6 (KLK6) belongs to a family of secreted serine proteases encoded by 15 homologous genes (KLK1–KLK15) that are important factors in cancer proteolysis. KLK production is abnormal, due to disturbed gene transcription or protein synthesis, or both, in many common human malignancies, highlighting the clinical relevance of KLKs as markers for tumour diagnosis and follow-up [1].
KLK6 is present in a myriad of normal tissues including the brain, breast, salivary gland, colon, pancreas, skin, lung and biological fluids [3–6]. Several studies have found that KLK6 synthesis is abnormal in endocrine-related tumours such as ovarian and uterine carcinomas, and non-cancer diseases, suggesting its utility as a marker [1, 7–10]. The concentration of KLK6 is also increased in human skin diseases and this concentration is correlated with tumour progression [11]. The clinical use of KLK6 for diagnosing gastric and colon cancer has been discussed recently [11, 12]. While the biological role of KLK6 in these tissues has not yet been identified, several studies have shown how KLK6 contributes to various phases of cancer biology. KLK6 might promote tumour invasion by breaking down extracellular matrix components and stimulating cell proliferation [11, 13, 14]. However, the molecular mechanism by which it stimulates proliferation remains unclear. Despite these findings, little work has been done on KLK6 synthesis and function in lung cancer, the most common and the largest single cause of death from cancer worldwide [6].
We have analysed the concentration and distribution of KLK6 in normal lung tissue and in 46 tissue samples from patients with a primary non-small cell lung cancer (NSCLC). KLK6 was abundant in both adenocarcinoma and squamous cell carcinoma NSCLC subtypes, but there was little or no KLK6 in a panel of NSCLC lines. We therefore investigated the role of KLK6 in lung cancer biology by transfecting NSCLC cells with the native form of KLK6. Ectopic KLK6 production increased NSCLC cell proliferation, due to the acceleration of cell-cycle progression through the G1-S transition via the induction of cyclin E and repression of p21. KLK6 synthesis was also associated with increased c-Myc production, a key player in cell-cycle progression. Lastly, a quantitative study of matched tumour and non-tumour specimens from NSCLC patients revealed an overabundance of KLK6 transcripts that was correlated with an unfavourable patient outcome. Our findings indicate that KLK6 is involved in lung cancer proliferation and is an indicator of a poor prognosis.
Material and methods
Antibodies
Polyclonal antibodies against KLK6, cyclin E and p21 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) Monoclonal antibody against KLK6 was purchased from Serotec Immunological Excellence (Düsseldorf, Germany). Other antibodies were from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Tissue samples and microarray
Matched tumour and non-tumour tissue samples from 46 patients with primary NSCLC (histological diagnosis and clinical history reported in [15, 16]) were used in this study. All investigations were carried out according to Helsinki principles and French bioethical regulations. Non-necrotic tumour samples, identified by a pathologist from formalin fixed and paraffin embedded tumour blocks, were used to prepare tissue microarrays. Frozen tumour samples from the same cohort of patients and matched tumour and non-tumour tissues from 11 additional patients were used to analyse transcript production.
Immunohistochemistry
Immunohistochemical analysis of KLK6 used a monoclonal antibody and the DakoCytomation EnVision system suitable for rabbit or mouse primary antibodies, according to the manufacturer’s instructions. Staining was revealed with 3,3-diaminobenzidine (Dako Cytomation, Trappes, France). Staining intensity and the percentage of stained cells were graded semi-quantitatively by a pathologist using standard procedures.
Cell lines and culture conditions
The following human NSCLC cell lines were obtained from the American Type Culture Collection (ATCC, LGC Promochem, Nancy, France): bronchioloalveolar carcinoma cell line A549, squamous cell carcinoma cells H520, Calu-1, adenocarcinoma cell lines H23, H1838 and H522, and large cell carcinoma line H460. All the cells were grown in the recommended culture medium. The A549 Flp-In cell line was derived from A549 cells genetically modified in our laboratory. A549 Flp-In cells contain a unique recombinase-mediated DNA integration site (FLP recombination target [FRT]) at a transcriptionally active genomic locus and are resistant to zeocin (data not shown). A3-KLK6 and A5-KLK6 are stable clones derived from A549 Flp-In, which express the native form of KLK6 and are resistant to hygromycin. A549 Flp-In parental or KLK6-expressing cells were grown at 37°C in an atmosphere of 5% CO2 in RPMI-1640 Glutamax I, except Calu-1 that was cultured in McCoy’s 5a medium, containing 10% foetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and if necessary supplemented with either 100 μg/ml zeocin or 100 μg/ml hygromycin.
Plasmids and stable transfections
The complete coding sequence of KLK6 (144–1002 bp from NM_002774) with Stop codon was amplified by PCR and integrated into the T/A cloning site of pcDNA5/FRT/V5-His TOPO (Invitrogen Corp., Cergy Pontoise, France), which contains an FRT site. The sequences of the specific primers are available on request. The expression vector and the plasmid encoding the yeast Flp recombinase were co-transfected into A549 Flp-In using Lipofectamine 2000 (Invitrogen Corp.) according to the manufacturer’s instructions. Stable clones containing the gene of interest inserted into the genome at the FRT site were selected and amplified. KLK6 secretion into the supernatants of clones was analysed by Western blotting.
Cell proliferation assays
Cells were seeded (2.5 × 103 cells/well) in 96-well plates and allowed to grow for 24–96 hrs. Cells were harvested at appropriate times and counted using a Malassez hematimeter.
Cell cycle synchronization
Cells were seeded (4 × 105 cells/well) in six-well plated and grown for 24 hrs. They were then synchronized by starving them of serum in RPMI-1640 for 24 hrs to arrest the cell cycle in G0–G1 [17]. The medium was then changed to RPMI-1640 containing 5% foetal calf serum (FCS) and 3 mM hydroxyurea for another 24 hrs so that the cells accumulated at the end of G1. These cells were extensively washed and placed in fresh medium containing 5% FCS to continue dividing. Cells were harvested at various times after being released, fixed in cold methanol and stored at –20°C. Cell samples taken at the indicated times were also processed for cyclin and other cell cycle regulator analysis by Western blotting.
Cell cycle analysis
The methanol was removed from stored cells and they were treated with the Coulter DNA-Prep reagents kit (Beckman Coulter, Roissy CDG, France). Cell cycle distribution was analysed with the Multicycle-AV software (Phoenix Flow Systems) [17].
Protein extraction and Western blotting
Cells were seeded in six-well plates (5.105 cells/well) or in 75-cm2 flasks and grown in appropriate maintenance cell culture medium for 24 hrs.
They were then harvested and proteins were extracted. The proteins were loaded on gels for SDS-PAGE and then electro-transferred to PVDF membranes [18]. Immunodetection was performed according to the antibody manufacturer’s instructions.
RNA extraction, RT-PCR and quantitative real-time RT-PCR
Total RNA was extracted from NSCLC cells using the RNeasy Minikit from Qiagen, Inc. (Les Ulis, France) according to the manufacturer’s instructions. Total RNA (2 μg) was reverse transcribed using 200 units of SuperScript III RNase H Reverse Transcriptase (Invitrogen Corp.) following the manufacturer’s instructions. One-tenth of the cDNA was amplified by PCR using FastStart Taq DNA Polymerase (Roche Diagnostics, Meylan, France) according to the manufacturer’s instructions. Reactions were performed under the following conditions for both KLK6 and β-actin: an initial denaturation step at 95°C for 3 min., followed by 35 cycles of denaturation at 95°C for 30 sec., annealing at 55.8°C for 30 sec. and extension at 72°C for 30 sec. The following primers were used: KLK6 For 5′-AAG ACA GCA GAT GGT GAT-3′; KLK6 Rev 5′-CTG GCT TCT CCT TTG AT-3′ located on different exons; β-actin For 5′-GGC GGC ACC ACC ATG TAC CCT-3′ and β-actin Rev 5′-AGG GGC CGG ACT CGT CAT ACT-3′.
Total RNA from tissues was extracted and reverse transcribed, and KLK6 transcripts quantified by real-time PCR as previously described [15, 16]. The primer sets were designed to target two exons (KLK6 For 5′-tga tgg tgg tgc tga gt-3′; KLK6 Rev 5′-aca gtg gat gga taa gga c-3′). The cycling conditions were 95°C for 5 min., followed by 50 cycles of 95°C for 10 sec., 65°C for 15 sec. and 72°C for 20 sec. Fluorescent product was measured by a single acquisition mode for 15 sec. after each cycle. Gene expression was normalized to the amount of 18S rRNA.
Statistical analysis
The differences in the KLK6 in non-cancerous and tumour tissues were analysed with the non-parametric Wilcoxon test for matched samples. The results of proliferation and synchronization assays are expressed as medians and clones and parental cells are compared using the Kruskal-Wallis non-parametric test. The survival curves for high-KLK6 and low-KLK6 cells were compared using a log-rank test. Cox proportional hazards models [19] were also used to assess the difference in the overall survival probability between groups with high KLK6 and low KLK6 after adjustment for clinical–pathological variables. These variables included gender, age, histological type, tumour size, nodal tumour status, stage and smoking status (packs/year). We used the Wald test to determine the statistical significance of the survival difference. A P-value below or equal to 0.05 was considered statistically significant.
Results
KLK6 is present in both normal lungs and NSCLC tissues but not in NSCLC cell lines
We determined the KLK6 in normal lung tissues and in 46 tumour specimens from patients with primary NSCLC. There was moderate KLK6 immunoreactivity in the epithelium of the bronchial tree of normal lungs with the signal concentrated at the apical pole of cells, but types I and II pneumocytes were negative (Fig. 1A). There was diffuse cytoplasmic staining for KLK6 in most of the tumour cells of NSCLC. Semi-quantitative assessment of immunoperoxidase labelled KLK6 in a tissue micro-array containing 46 tumour samples from NSCLC patients showed two populations: 25% of cases were low KLK6 and 75% of the cases were high KLK6. Statistical analysis of KLK6 staining scores and clinical–pathological parameters revealed that KLK6 synthesis was not correlated with the histological type, the tumour size, the nodal/tumour status or tumour stage.
Figure 1.
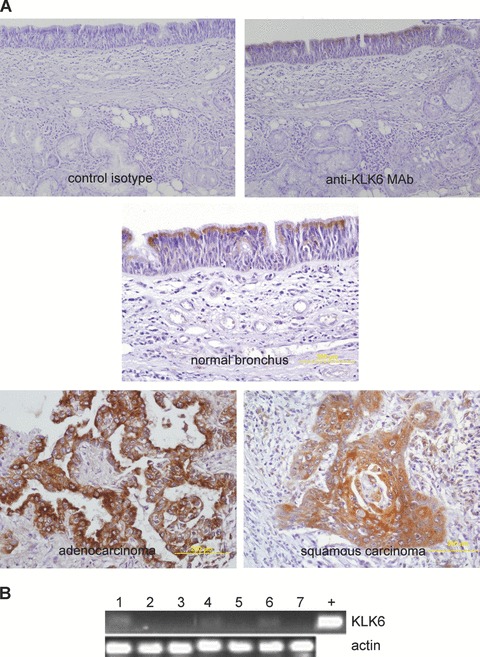
KLK6 in tissues from patients with NSCLC but not in NSCLC-derived cell lines. (A) Immunohistochemistry of KLK6 in normal and malignant lung from 46 patients with primary NSCLC using a monoclonal antibody (original magnification: ×200; haematoxylin counterstain). KLK6 immunostaining (dark) in the apical region of normal bronchial epithelial cells (thin arrow) and in the cytoplasm of most cancer cells (large arrow). (B) KLK6 production in a panel of NSCLC lines by RT-PCR. Total RNA from NSCLC cell lines was extracted and reverse transcribed. KLK6 and β-actin (internal control) mRNAs were amplified from cDNA by PCR and the amplification products were separated on a 1% agarose gel containing ethidium bromide. (1) A549; (2) Calu-1; (3) H23; (4) H460; (5) H520; (6) H522; (7) H1838; +: plasmid containing KLK6 cDNA.
However, analysis of the KLK6 mRNA in a panel of NSCLC cell lines by RT-PCR revealed few or no KLK6 transcripts (Fig. 1B). Similarly, KLK6 protein was not detected in either the cell lysate or the culture supernatants of those lines that were slightly positive by RT-PCR (data not shown). Thus, in contrast to tissue specimens, KLK6 was not produced in the cell lines derived from NSCLC.
Ectopic KLK6 increases the proliferation of NSCLC cells
As there was little or no KLK6 in the NSCLC cells we tested, we produced NSCLC lines synthesizing pre-proKLK6 in order to investigate further the role of KLK6 in lung cancer biology. The cDNA encoding pre-proKLK6 was inserted into the A549 Flp-In by homologue recombination to give stable isogenic clones [20]. Two clones that produced KLK6 and the control parental A549 Flp-In line were selected for further study (Fig. 2A). Because previous reports suggest that KLK6 production influences cell proliferation [11, 12], we assessed the growth of both parental and KLK6-producing A549 cells grown at low density for 96 hrs. The two clones producing KLK6 grew significantly faster than the A549 Flp-In cells after 72 hrs in culture (Fig. 2B). The growth of a non-pulmonary line (HEK-293 Flp-In) genetically modified to produce KLK6 was similarly enhanced, but that of the same cells producing another KLK was not (data not shown).
Figure 2.
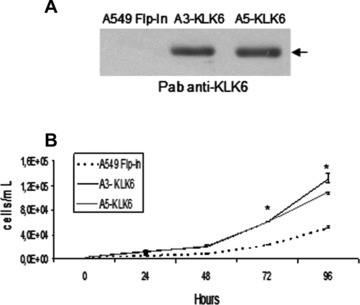
Ectopic KLK6 in NSCLC promotes proliferation. (A) Secretion of KLK6 by A549 Flp-In and stable isogenic clones (A3- and A5-KLK6) assessed by Western blotting using a polyclonal anti-KLK6 antibody. Equal volumes of cell culture supernatant were loaded onto 12% SDS-PAGE gels. (B) Control A549 Flp-In (dashed line) and A3-KLK6 (black full line) and A5-KLK6 (grey full line) (2.5 × 103 cells of each) were seeded and cells were counted at the indicated times. Points, median value of triplicate determinations from one representative experiment (of three). Bars, lower and upper quartiles. P-values were determined using a Kruskal and Wallis test. *P < 0.05.
Ectopic KLK6 accelerates cell cycle progression in NSCLC lines
We synchronized cells at the end of G1 phase and then released them in the cell cycle to analyse their progression. Starvation and hydroxyurea treatment resulted in the arrest of nearly 85% of the cells in G1 phase (Fig. 3A). The cell cycle distribution of cells producing KLK6 5 and 8 hrs after release from hydroxyurea was quite different from that of the control line. Significantly more KLK6-producing A549 cells accumulated in the S and G2 phases (Fig. 3B), suggesting that the KLK6-producing NSCLC cells passed more rapid through the S and next G2 phases. Analysis of the cyclins regulating the G1 to S transition by Western blotting revealed that there was considerably more cyclin E in KLK6-producing cells than in the parental line 5 hrs after release of the cell cycle, while the amounts of cyclins D1 and D3 remained the same in all the cell lines (Fig. 3C). There was also a dramatic decrease in the amount of p21, an inhibitor of cyclin-dependent kinases that limit cell cycle progression, in the KLK6-producing cells both 5 and 8 hrs after release from hydroxyurea.
Figure 3.
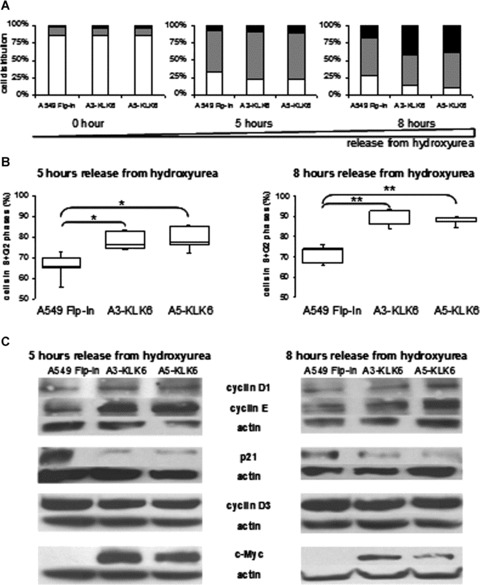
Effect of KLK6 on NSCLC cell growth and regulation of cell cycle progression. (A) Distribution of control A549 Flp-In and A3-KLK6 and A5-KLK6 cells within the G1 (white), S (grey) and G2 (black) phases of the cell cycle 0, 5 or 8 hrs after release from hydroxyurea. Results are expressed as the median values of five independent experiments. (B) Box plot of control A549 Flp-In and A3-KLK6 and A5-KLK6 cells passed from G1 into G2+S phases 5 and 8 hrs after release from hydroxyurea. Results are representative of five independent experiments. P-values were determined using a Kruskal and Wallis test. *P < 0.05, **P < 0.025. (C) Assay of cell-cycle proteins and c-Myc by Western blotting of whole-cell extracts (50 μg) from the control A549 Flp-In, A3-KLK6 and A5-KLK6 lines 5 and 8 hrs after release from hydroxyurea. Actin protein was used to check for equal loading. Results are from one of three experiments.
KLK6-dependent enhancement of cell cycle progression might be mediated by c-Myc
The dramatic changes in the replication rate and cell cycle progression associated with ectopic KLK6 production indicated that the cell proliferation regulators in KLK6-producing A549 cells were severely disrupted. Therefore, we analysed the expression of c-Myc, a proto-oncogene that is often constitutively overexpressed in lung cancers and promotes cell cycle progression by inducing the genes encoding cyclins D2, -E and -A and repressing cell growth arrest genes like p21[21–23]. KLK6-producing cells had markedly more c-Myc protein than control cells, both when they were grown in complete medium (data not shown) and when released from hydroxyurea in the cell cycle (Fig. 3C).
High KLK6 expression by NSCLC is associated with poor patient survival
Because our findings suggest that KLK6 overproduction plays a dramatic role in lung cancer development by promoting NSCLC cell proliferation, we compared KLK6 gene expression in samples of tumour and non-tumour tissues from 57 patients with primary NSCLC. There was significantly more KLK6 mRNA (median value 23-fold more) in cancer tissue than in normal lung tissue (Fig. 4A). We analysed the relationship between the amount of KLK6 transcripts and clinical–pathological parameters by univariate analysis. A high KLK6 expression was not correlated with any clinicopathological lung tumour variable (data not shown).
Figure 4.
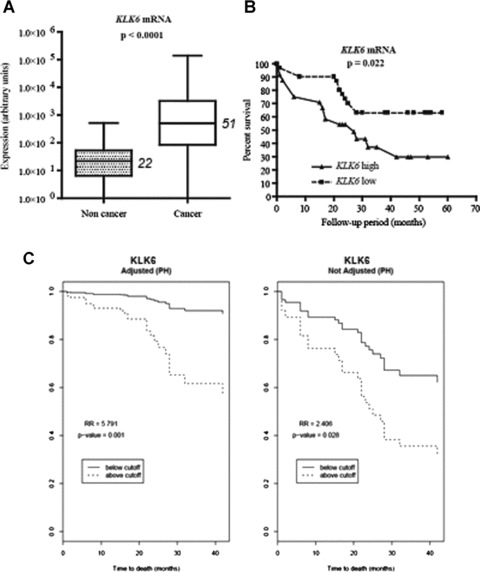
KLK6 overexpression in tumour tissues from patients with NSCLC, and patient survival. (A) Box plot of KLK6 transcript concentration by quantitative real-time RT-PCR in 57 tumours and matched non tumour samples from patients with primary NSCLC undergoing an initial surgical resection of the lung tumour (aged 44–83 years, median: 65). Gene expression was normalized to the amount of 18S rRNA and is reported in arbitrary units. Median values are indicated in italics. The difference between the concentration of KLK6 transcripts in non-tumour and tumour tissues was determined by a Wilcoxon test for matched pairs. (B) Kaplan–Meier curves for overall survival of patients with high and low concentrations of KLK6 mRNA in lung tumour tissues. The optimal cut-off value for survival analysis was identified by the chi-squared test, based on the ability of KLK6 to predict the overall survival of the population studied. Using this cut-off, KLK6 mRNA concentration was categorized as high or low. A cut-off of 600 (in arbitrary units) for KLK6 mRNA was equal to the 56th percentile. (C) Survival curves obtained from Cox regression models after adjusting for gender, age, histological type, tumour size, nodal tumour status, stage and smoking status (packs/ year)
We then studied the association between the KLK6 status and overall patient survival using both Kaplan-Meier survival curves and the Cox proportional hazards regression model. There was a significant correlation between NSCLC cells with a high concentration of KLK6 transcripts and poor patient survival (Fig. 4B and C). The hazard ratio measuring the effect of KLK6 on the overall survival estimated from the Cox model was 5.79, which is highly significant (P= 0.001; 95% CI, 2.147–15.622). The overall survival at 40 months of patients whose NSCLC cells were low-KLK6 was about 95%, while it was about 60% in the patients with a high-KLK6 NSCLC. Because the Cox regression model was adjusted for the clinical–pathological variables, the effect of KLK6 was significantly associated with the overall survival independently of the other risk factors.
Discussion
The human KLKs have emerged as multifaceted players in cancer proteolysis over the past decade. Numerous studies have shown that KLK synthesis and proteolytic activity is altered in common types of human malignancies [1]. Although most of the studies focused on hormone-dependent tumours, there is recent evidence that KLK production is abnormal in other types of human malignancies [1, 11, 12]. We first demonstrated that several KLKs are produced in the lung and that the synthesis of some of them is deregulated in NSCLC [15, 16, 24]. However, little is known on the involvement of the KLK-related peptidases in this disease. Sher et al. reported that KLK8 suppresses tumour growth and invasion in vitro and in vivo, and confers a favourable clinical outcome for patients with lung cancer [25]. Herein, we examined the KLK6 status and implication in NSCLC. Our immunohistochemical study on a sizeable cohort of patients with lung cancers shows that KLK6 is often present in NSCLC, and that its concentration may be moderate or high. In contrast to Singh et al.[6], we detected KLK6 in both adenocarcinoma and squamous cell carcinomas, the major histological subtypes of NSCLC. This discrepancy might be attributed to the different anti-KLK6 antibodies used for IHC, or a rare phenotype encountered in the single adenocarcinoma sample examined by these authors.
Strikingly, we did not detect KLK6 in a panel of NSCLC cell lines, suggesting that there is little or no KLK6 in the lines derived from NSCLC tumours. Deregulation of KLK6 in NSCLC cell lines might be attributed to multiple mechanisms including genetic alterations (polymorphisms), hypermethylation in permanent cancer cell lines compared to their primary counterpart due to the culture conditions, or lack of regulatory factors in the growing medium as previously described [1, 26, 27]. Recently, hypermethylation of the KLK6 promoter leading to KLK6 gene silencing has been described in metastatic breast cancer cell lines [28]. Moreover, accumulating evidence highlighted the importance of the interactions between cancer cells and the dynamic microenvironment in which they live (containing soluble and non soluble components of the extracellular matrix and ‘activated’ non-neoplastic cells) in the regulation of gene expression in cancer cells [26, 29]. Further studies are now required to elucidate exactly how KLK6 gene expression is controlled in NSCLC tissues and cell lines.
KLKs have recently been implicated in various phases of cancer. They may act alone or as part of a proteolytic cascade in which KLKs collaborate with other KLKs and cancer-associated proteases, to stimulate or inhibit tumour development and dissemination [1, 7, 14, 25, 30]. Recent studies on the overexpression of the mouse orthologue KLK6 gene in keratinocytes, and the silencing of KLK6 gene expression in a gastric cancer line, indicate that KLK6 is involved in cell proliferation. In the light of these findings and our results showing the abundance of KLK6 in NSCLC, we postulate that KLK6 contributes to lung cancer development by promoting cell proliferation [11, 12]. This agrees well with our finding that ectopic KLK6 dramatically accelerates NSCLC cell growth. The results from our group and others indicate that both the human and mouse kallikrein 6 orthologues promote cell growth, and this KLK6-mediated proliferation is cell-type independent.
Because the molecular mechanisms underlying the KLK6-mediated enhancement of cell growth is unknown, we investigated the impact of KLK6 synthesis on the progression of NSCLC cell cycle. We found that KLK6-producing cells passed through the S and the next G2 phases faster than the controls, and that they produced more cyclin E, that regulates the G1–S phase transition of the cell-cycle. And the production of ectopic KLK6 by NSCLC was accompanied by a significant decrease in p21, an inhibitor of cyclin-dependent kinases. These kinases are key regulatory enzymes connected to cyclins. Thus, the promotion of cell growth by KLK6 is due to an acceleration of the G1–S transition following the induction of cyclin E and repression of p21. We also found that the synthesis of KLK6 is associated with an increase in c-Myc synthesis by both synchronized cells and cells cultured in standard conditions (data not shown). The c-Myc gene belongs to the MYC family of oncogenes that are often overexpressed in lung cancers [21]. One of the key functions of c-Myc is to regulate cell-cycle progression by activating cyclin D and E complexes and repressing p21, which governs cell cycle/growth arrest [21–23, 31]. Based on our results, we speculated that KLK6-mediated c-Myc enhancement leads to acceleration of cell cycle progression in lung tumour cells. Further experiments are required to validate this hypothesis, and determine the molecular mechanism associated with higher c-Myc expression.
Lung cancer is the leading cause of cancer mortality worldwide, with over one million deaths each year [32]. Most patients are diagnosed with advanced NSCLC, and the overall 5-year survival remains dismal (15% all stages included), despite recent advances in multimodal therapy. Although a minority of patients present with stage I disease and undergo surgery alone, 35–50% relapse within 5 years. Therefore, early diagnosis and identification of specific high-risk patients are crucial for improving the management of patients with NSCLC. Because the synthesis of KLKs is often abnormal in common types of human malignancies, numerous studies have focused on the clinical value of KLKs for tumour diagnosis and follow-up [1]. Several KLKs are approved (like KLK3 or PSA) or emerging markers of endocrine tumours [1]. Recently, a multiparametric panel of serological KLKs has been proposed for lung cancer diagnosis [33]. The present report indicates that the KLK6 gene is markedly overexpressed in lung cancer tissue, and that high KLK6 mRNA expression is significantly correlated with a poor patient outcome, according to both univariate and multivariate analysis. Moreover, the fact that the effect of KLK6 remains significant after adjustment for other clinical–pathological variables indicates that it is an independent prognostic factor. Elevated KLK6 expression, both at the mRNA and protein levels, has been previously described as independent adverse prognostic marker in a subgroup of ovarian carcinomas and in colorectal and gastric cancers [8, 12].
In conclusion, we show that KLK6 is a proliferative factor in NSCLC that might act by inducing c-Myc synthesis, which in turn modulates specific components of the cell cycle machinery to promote cell cycle progression. Our present results, together with those from previous reports showing the influence of KLK6 on cancer cell migration and invasion [11, 13], suggest that KLK6 plays a central role in NSCLC development and progression. Finally, high concentrations of KLK6 might be an unfavourable prognosis factor because a high KLK6 concentration is associated with a poor clinical outcome of patients with NSCLC.
Acknowledgments
We thank Audrey Lorthiois for technical assistance, Dr. Sandra Régina for clinical information support. The English text was edited by Dr. Owen Parkes. We acknowledge financial supports from INSERM; Ligue Contre le Cancer Région Centre; IFR 135 ‘Imagerie Fonctionnelle’; Association pour la Recherche sur le Cancer, grant n° 7935 and Région Centre, grant KalliCap. C.P. benefited a fellowship from the Région Centre and INSERM.
References
- 1.Borgoño CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–90. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson S, Luo LY, Yousef GM, et al. Purification of human kallikrein 6 from biological fluids and identification of its complex with alpha(1)-antichymotrypsin. Clin Chem. 2003;49:746–51. doi: 10.1373/49.5.746. [DOI] [PubMed] [Google Scholar]
- 3.Petraki CD, Karavana VN, Skoufogiannis PT, et al. The spectrum of human kallikrein 6 (zyme/protease M/neurosin) expression in human tissues as assessed by immunohistochemistry. J Histochem Cytochem. 2001;49:1431–41. doi: 10.1177/002215540104901111. [DOI] [PubMed] [Google Scholar]
- 4.Petraki CD, Papanastasiou PA, Karavana VN, et al. Cellular distribution of human tissue kallikreins: immunohistochemical localization. Biol Chem. 2006;387:653–63. doi: 10.1515/BC.2006.084. [DOI] [PubMed] [Google Scholar]
- 5.Shaw JL, Diamandis EP. Distribution of 15 human kallikreins in tissues and biological fluids. Clin Chem. 2007;53:1423–32. doi: 10.1373/clinchem.2007.088104. [DOI] [PubMed] [Google Scholar]
- 6.Singh J, Naran A, Misso NL, et al. Expression of kallikrein-related peptidases (KRP/hK5, 7, 6, 8) in subtypes of human lung carcinoma. Int Immunopharmacol. 2008;8:300–6. doi: 10.1016/j.intimp.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Paliouras M, Diamandis EP. The kallikrein world: an update on the human tissue kallikreins. Biol Chem. 2006;387:643–52. doi: 10.1515/BC.2006.083. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman BR, Katsaros D, Scorilas A, et al. Immunofluorometric quantitation and histochemical localisation of kallikrein 6 protein in ovarian cancer tissue: a new independent unfavourable prognostic biomarker. Br J Cancer. 2002;87:763–71. doi: 10.1038/sj.bjc.6600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santin AD, Diamandis EP, Bellone S, et al. Human kallikrein 6: a new potential serum biomarker for uterine serous papillary cancer. Clin Cancer Res. 2005;11:3320–5. doi: 10.1158/1078-0432.CCR-04-2528. [DOI] [PubMed] [Google Scholar]
- 10.Zarghooni M, Soosaipillai A, Grass L, et al. Decreased concentration of human kallikrein 6 in brain extracts of Alzheimer’s disease patients. Clin Biochem. 2002;35:225–31. doi: 10.1016/s0009-9120(02)00292-8. [DOI] [PubMed] [Google Scholar]
- 11.Klucky B, Mueller R, Vogt I, et al. Kallikrein 6 induces E-cadherin shedding and promotes cell proliferation, migration, and invasion. Cancer Res. 2007;67:8198–206. doi: 10.1158/0008-5472.CAN-07-0607. [DOI] [PubMed] [Google Scholar]
- 12.Nagahara H, Mimori K, Utsunomiya T, et al. Clinicopathologic and biological significance of kallikrein 6 overexpression in human gastric cancer. Clin Cancer Res. 2005;11:6800–6. doi: 10.1158/1078-0432.CCR-05-0943. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh MC, Grass L, Soosaipillai A, et al. Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumour Biol. 2004;25:193–9. doi: 10.1159/000081102. [DOI] [PubMed] [Google Scholar]
- 14.Klokk TI, Kilander A, Xi Z, et al. Kallikrein 4 is a proliferative factor that is overexpressed in prostate cancer. Cancer Res. 2007;67:5221–30. doi: 10.1158/0008-5472.CAN-06-4728. [DOI] [PubMed] [Google Scholar]
- 15.Planque C, de Monte M, Guyetant S, et al. KLK5 and KLK7, two members of the human tissue kallikrein family, are differentially expressed in lung cancer. Biochem Biophys Res Commun. 2005;329:1260–6. doi: 10.1016/j.bbrc.2005.02.100. [DOI] [PubMed] [Google Scholar]
- 16.Planque C, Aïnciburu M, Heuzé-Vourc’h N, et al. Expression of the human kallikrein genes 10 (KLK10) and 11 (KLK11) in cancerous and non-cancerous lung tissues. Biol Chem. 2006;387:783–8. doi: 10.1515/BC.2006.098. [DOI] [PubMed] [Google Scholar]
- 17.Barascu A, Besson P, Le Floch O, et al. CDK1-cyclin B1 mediates the inhibition of proliferation induced by omega-3 fatty acids in MDA-MB-231 breast cancer cells. Int J Biochem Cell Biol. 2006;38:196–208. doi: 10.1016/j.biocel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Heuzé-Vourc’h N, Liu M, Dalwadi H, et al. IL-20, an anti-angiogenic cytokine that inhibits COX-2 expression. Biochem Biophys Res Commun. 2005;333:470–5. doi: 10.1016/j.bbrc.2005.05.122. [DOI] [PubMed] [Google Scholar]
- 19.Cox DR. Regression models with life tables. J R Stat Soc. 1972;34:187–220. [Google Scholar]
- 20.Heuzé-Vourc’h N, Aïnciburu M, Planque C, et al. Recombinant kallikrein expression: site-specific integration for hK6 production in human cells. Biol Chem. 2006;387:687–95. doi: 10.1515/BC.2006.087. [DOI] [PubMed] [Google Scholar]
- 21.Zajac-Kaye M. Myc oncogene: a key component in cell cycle regulation and its implication for lung cancer. Lung Cancer. 2001;34:S43–6. doi: 10.1016/s0169-5002(01)00343-9. [DOI] [PubMed] [Google Scholar]
- 22.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 23.Gartel AL, Shchors K. Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Exp Cell Res. 2003;283:17–21. doi: 10.1016/s0014-4827(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 24.Planque C, Bléchet C, Ayadi-Kaddour A, et al. Quantitative RT-PCR analysis and immunohistochemical localization of the kallikrein-related peptidases 13 and 14 in lung. Biol Chem. 2008;389:781–6. doi: 10.1515/BC.2008.089. [DOI] [PubMed] [Google Scholar]
- 25.Sher YP, Chou CC, Chou RH, et al. Human kallikrein 8 protease confers a favorable clinical outcome in non-small cell lung cancer by suppressing tumor cell invasiveness. Cancer Res. 2006;66:11763–70. doi: 10.1158/0008-5472.CAN-06-3165. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Goyal J, Dhar S, et al. CpG methylation as a basis for breast tumor-specific loss of NES1/kallikrein 10 expression. Cancer Res. 2001;61:8014–21. [PubMed] [Google Scholar]
- 27.Smiraglia DJ, Rush LJ, Frühwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–9. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 28.Pampalakis G, Sotiropoulou G. Multiple mechanisms underlie the aberrant expression of the human kallikrein 6 gene in breast cancer. Biol Chem. 2006;387:773–82. doi: 10.1515/BC.2006.097. [DOI] [PubMed] [Google Scholar]
- 29.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 30.Pampalakis G, Sotiropoulou G. Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim Biophys Acta. 2007;1776:22–31. doi: 10.1016/j.bbcan.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Blagosklonny MV, Pardee AB. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–10. [PubMed] [Google Scholar]
- 32.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 33.Planque C, Li L, Zheng Y, et al. A multiparametric serum kallikrein panel for diagnosis of non-small cell lung carcinoma. Clin Cancer Res. 2008;14:1355–62. doi: 10.1158/1078-0432.CCR-07-4117. [DOI] [PubMed] [Google Scholar]


