Abstract
Virus-encoded tumor suppressor p53 transgene expression has been successfully used in vesicular stomatitis virus (VSV) and other oncolytic viruses (OVs) to enhance their anticancer activities. However, p53 is also known to inhibit virus replication via enhanced type I interferon (IFN) antiviral responses. To examine whether p53 transgenes enhance antiviral signaling in human pancreatic ductal adenocarcinoma (PDAC) cells, we engineered novel VSV recombinants encoding human p53 or the previously described chimeric p53-CC, which contains the coiled-coil (CC) domain from breakpoint cluster region (BCR) protein and evades the dominant-negative activities of endogenously expressed mutant p53. Contrary to an expected enhancement of antiviral signaling by p53, our global analysis of gene expression in PDAC cells showed that both p53 and p53-CC dramatically inhibited type I IFN responses. Our data suggest that this occurs through p53-mediated inhibition of the NF-κB pathway. Importantly, VSV-encoded p53 or p53-CC did not inhibit antiviral signaling in non-malignant human pancreatic ductal cells, which retain their resistance to all VSV recombinants. To the best of our knowledge, this is the first report of p53-mediated inhibition of antiviral signaling, and it suggests that OV-encoded p53 can simultaneously produce anticancer activities while assisting, rather than inhibiting, virus replication in cancer cells.
Keywords: vesicular stomatitis virus, VSV, oncolytic, pancreatic cancer, p53, tumor suppressor, antiviral signaling, type I interferon, interferon, gene therapy
INTRODUCTION
Oncolytic virus (OV) therapy is a promising anti-cancer approach that utilizes replication-competent viruses to target and kill cancer cells while leaving normal cells unharmed. Many OVs are being examined in clinical trials in the United States and several are currently in phase III clinical trials, with approval of the first OV in North America expected in the near future (Bauzon and Hermiston, 2014; Russell et al., 2012). This study is focused on vesicular stomatitis virus (VSV, a rhabdovirus), which has been used successfully against many malignancies in preclinical studies [reviewed in (Hastie and Grdzelishvili, 2012; Russell and Peng, 2007; Russell et al., 2012)], and is currently in a phase I clinical trial against hepatocellular carcinoma (trial NCT01628640). In addition to its inherent oncoselectivity, VSV has a robust replication, a ubiquitous cell surface receptor, no risk of host cell transformation, a lack of preexisting immunity in the general population, and an 11-kb genome accommodating to the addition of transgenes to improve its OV efficacy [reviewed in (Hastie and Grdzelishvili, 2012)].
Oncolytic abilities of viruses can be improved by creating recombinant OVs that encode therapeutically beneficial transgenes, including tumor suppressor genes. Restoration of “wild type” (wt) p53 tumor suppressor activity has been exploited extensively in various anticancer approaches and is known to induce cell-cycle arrest, apoptosis, and senescence. Additionally, more recently identified beneficial p53 functions include regulation of metabolism and promotion of enhanced antitumor immunity (Cheok et al., 2011; Lane et al., 2010; Lowe et al., 2013). Not surprisingly, virus-encoded p53 transgene expression has been extensively explored in many replicating OV vectors, including various adenovirus-based OVs (Geoerger et al., 2004; Heideman et al., 2005; Idema et al., 2007; Koo et al., 2012; Mitlianga et al., 2006; Wang et al., 2008; Yamasaki et al., 2012), vaccinia virus (Hardwick et al., 2014), canary pox virus (Odin et al., 2001), and VSV (Heiber and Barber, 2011).
Beyond the observed benefits of p53 due to its anticancer activities, it remains unclear how OV-encoded p53 transgene expression affects viral replication. Several studies showed that in normal cells and some cancer cell lines, p53 inhibited virus replication via enhanced type I Interferon (IFN) antiviral signaling (Dharel et al., 2008; Lazo and Santos, 2011; Munoz-Fontela et al., 2005; Munoz-Fontela et al., 2011; Munoz-Fontela et al., 2008; Shin-Ya et al., 2005; Su et al., 2009; Takaoka et al., 2003; Yonish-Rouach et al., 1991). In agreement with this, many viruses directly target and inhibit p53 activities in infected cells [reviewed in (Sato and Tsurumi, 2013)]. A previous study demonstrated strong antiviral activities of p53 in normal mouse cells against VSV (Takaoka et al., 2003; Yonish-Rouach et al., 1991). The study showed 30-fold higher VSV yield in p53−/− mouse embryonic fibroblasts (MEFs) than in wt MEFs, and that p53 knockout mice succumb to VSV infection with 100-fold increase in VSV found in sera when compared to wt mice (Takaoka et al., 2003; Yonish-Rouach et al., 1991). Such activities of virus-encoded p53 transgenes in cancer cells could dramatically reduce the efficacy of replicating OVs, including VSV.
Here we focus on VSV as a promising OV against pancreatic ductal adenocarcinoma (PDAC) (Moerdyk-Schauwecker et al., 2013; Murphy et al., 2012). PDAC is estimated to be the fourth leading cause of cancer related-deaths in the United States and treatment options to date are largely ineffective (Tarver, 2012). Specific to PDAC, TP53 gene mutations occur in more than 75% of patients and are shown to contribute to proliferation and even drive metastasis through gain of function activities (Iacobuzio-Donahue and Herman, 2014; Weissmueller et al., 2014). Previous studies have shown that exogenous delivery of p53 sensitizes PDAC to chemotherapy (Camp et al., 2013), treatment with adenovirus encoding p53 sensitizes PDAC to radiation (Li et al., 2013), and the use of small molecules like apigenin (King et al., 2012) or mouse double minute 2 homolog (MDM2) inhibitors (Azmi et al., 2010) can restore endogenous p53 function and slow PDAC proliferation. Although VSV encoding p53 has never been studied in PDAC cells, VSV-driven murine p53 gene expression resulted in enhanced antitumor activities in vivo against mammary adenocarcinoma (Heiber and Barber, 2011). The study, however, did not examine the effect of murine p53 transgene expression on antiviral signaling in cancer cells. Also, VSV encoding human p53 has never been studied before, and murine and human p53 may have different activities (Horvath et al., 2007). Here, we sought to examine how virus-encoded human p53 affects antiviral signaling in human PDAC cells. To investigate this issue, we engineered recombinant VSVs to encode human wt p53 or the recently described chimeric p53-CC [tetramerization domain of p53 substituted with the coiled-coil (CC) domain from breakpoint cluster region (Bcr) protein], which evades the dominant-negative activities of endogenously expressed mutant p53. Surprisingly, our data show that both wt p53 and p53-CC downregulate cellular antiviral responses in a variety of PDAC cell lines, and do so through inhibition of the NF-kB pathway.
MATERIALS AND METHODS
Cell lines
The human PDAC cell lines used in this study were: AsPC-1 (ATCC CRL-1682), Capan-2 (ATCC HTB-80), Suit2 (Iwamura et al., 1987) and T3M4 (Okabe et al., 1983). A non-malignant human pancreatic duct epithelial (HPDE) cell line was previously generated by introduction of the E6 and E7 genes of human papillomavirus 16 into normal adult pancreas epithelium. HPDE retains a genotype similar to pancreatic duct epithelium and is non-tumorigenic in nude mice (Furukawa et al., 1996). The baby hamster kidney BHK-21 fibroblasts (ATCC CCL-10) were used to grow viruses. Suit2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Cellgro); AsPC-1, Capan-2, and T3M4 in RPMI 1640 (HyClone); BHK-21 in modified Eagle’s medium (MEM, Cellgro); HPDE in Keratinocyte-SFM (K-SFM, Gibco) without serum. All cell growth media (except for K-SFM) were supplemented with 9% fetal bovine serum (FBS, Gibco), 3.4 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (HyClone). MEM was further supplemented with 0.3% glucose (w/v). Cells were kept in a 5% CO2 atmosphere at 37°C. For all experiments, PDAC cell lines were passaged no more than 10 times. After receipt, the human origin of all PDAC cell lines was confirmed by partial sequencing of KRAS and actin. As expected, all PDAC cell lines (but not HPDE cells) had a mutation in KRAS, as is typical for PDACs (data not shown).
Generation of novel recombinant VSVs
A plasmid containing cDNA copy of recombinant VSV-XN2-ΔM51 genome (VSV Indiana serotype) (Lawson et al., 1995; Wollmann et al., 2010) was kindly provided by Jack Rose (Yale University). A pUC57 plasmid encoding near-infrared fluorescent protein eqFP650 was designed based on the published eqFP650 sequence (accession HQ148301) (Shcherbo et al., 2010) and was purchased from Genscript. The pUC57-eqFP650 plasmid contains a T7 promoter, a XhoI site, and a Kozak consensus sequence upstream of the eqFP650 start site (TAATACGACTCACTATAGGGAGACTCGAGCCACCATG). Downstream of the eqFP650 coding sequence containing a BspEI site there are two stop sites followed by a NheI site (CAGCTCCGGATAATAGCTAGC). Plasmids “GFP-p53” (Cat. no. 12091) (Boyd et al., 2000) and “HA-tagged BCR” (Cat. no. 38189) were purchased from Addgene. Plasmids were amplified in JM109 E. coli. Selection for colonies containing VSV-XN2-ΔM51 or HA-tagged BCR plasmids was done with 50 µg/ml of ampicillin, and GFP-p53 plasmid with 50 µg/ml of kanamycin. To generate VSV-ΔM51-eqFP650, pUC57-eqFP650 was digested using XhoI and NheI restriction enzymes, and the fragment was inserted into a XhoI-NheI digested VSV-XN2-ΔM51 plasmid. To generate VSV plasmids with eqFP650-p53, p53 sequence was first inserted into pUC57-eqFP650 plasmids, and then eqFP650-p53 was cloned into VSV-XN2-ΔM51 plasmid. To generate the p53 gene insert, the p53 sequence from the GFP-p53 plasmid was PCR amplified using primers VG283 and VG284 (Supplemental Table 1). To generate the TP53ΔODRD insert, the TP53ΔODRD sequence from the GFP-p53 plasmid was PCR amplified using primers VG283 and VG285. To generate the CC insert, the CC sequence from the HA-tagged BCR plasmid was PCR amplified using primers VG286 and VG292. To generate TP53-CC, the TP53ΔODRD sequence from the GFP-p53 plasmid was PCR amplified using primers VG283 and VG293 and the CC sequence from the HA-tagged BCR plasmid was PCR amplified using primers VG288 and VG292. The TP53ΔODRD and CC PCR products were used for overlapping PCR along with primers VG283 and VG292. All forward and reverse primers, except those used for overlap PCR, contained a BspEI site and a NheI site, respectively. All PCR products were digested sequentially with NheI and BspEI restriction enzymes and ligated into the NheI-BspEI digested pUC57-eqFP650 vector fragment to generate pUC57-eqFP650-TP53wt, pUC57-eqFP650-TP53ΔODRD, pUC57-eqFP650-CC, and pUC57-eqFP650-TP53-CC. To create the pUC57-eqFP650-TP53-CC/fs, the pUC57-eqFP650-TP53-CC plasmid was digested with BspEI, filled in with T4 DNA polymerase (NEB, MO203S), and re-ligated. All pUC57 plasmids containing the eqFP650-fusion genes were double digested with XhoI and NheI restriction enzymes and the inserts were ligated into the XhoI-NheI digested VSV-XN2-ΔM51 plasmid. All plasmids were sequenced to confirm the ΔM51 deletion and all gene insertions (primers listed in Supplemental Table 1). Recombinant VSVs were rescued as previously described (Grdzelishvili et al., 2005), which is a modification of the original protocol in (Lawson et al., 1995). VSV stocks were prepared using BHK-21 cells infected at a multiplicity of infection (MOI) 0.005 and incubated at 37°C in MEM-based medium containing 5% FBS. Virus-containing medium was collected at 24 hours post infection (h p.i.) and centrifuged at 4,000 × g for 10 min at 4°C to remove large cellular debris. Virus was purified by the method of Kalvodova et al. with slight modifications described previously (Hastie et al., 2013; Kalvodova et al., 2009). Cell specific viral titers were obtained using plaque assay. Following virus rescue, amplification, and purification, RNA was isolated and sequenced confirming the ΔM51 deletion and correct transgene insertions.
Confocal imaging for virus-expressed transgene localization
Suit2 cells were seeded in borosilicate glass chamber slides (Labtek, Cat. No. 155411) to be approximately 80% confluent in 24 h. Cells were mock treated or infected at MOI 5 in growth medium without FBS. Virus-containing medium was aspirated 1 h p.i. and replaced with growth medium containing 5% FBS. At 8 h p.i., growth media was aspirated and cells were stained with 1 µM Hoechst dye (nuclear stain) and HCS CellMask (plasma membrane and cytoplasmic stain) (Life Technologies, H32714) for 30 minutes. Growth media was aspirated, cells were washed twice with PBS, then media with 5% FBS was added and cells were used for confocal imaging (Olympus FluoView 1000) using filters for DAPI/Hoechst (blue), FITC (green), and Alexa Fluor 568 (red).
RNA microarray analysis
Cells (in triplicate) were seeded in 6-well plates so that they reached approximately 80% confluence at 24 h. Cells were mock treated or infected with VSVs at MOI 5. Cellular RNA was extracted with TRIzol (Life Technologies) per the manufacturer protocol with slight modification. In brief, following the first phase separation, the aqueous layer was transferred to a new tube. Then, 500 µl of TRIzol and 100 µl of chloroform were added and phase separation was repeated. Isolated RNA was run on a Bioanalyzer 2100 (Agilent) to check for purity. RNA integrity number (RIN) values were ≥ 7. Samples were reverse transcribed, amplified and labeled using 3’ IVT Express Kit (Affymetrix). The resultant labeled complementary RNA (cRNA) was purified and fragmented as per vendor’s instructions. The cRNA samples together with probe array controls were hybridized onto Affymetrix Human Genome U133+ Plus PM array strips, which cover more than 47,000 transcripts and variants selected from GenBank, dbEST, and RefSeq. Hybridization controls were spiked into the cRNA samples to monitor and troubleshoot the hybridization process. Probes for housekeeping genes were used to assess sample integrity. Hybridization, washing, staining and scanning were performed using Affymetrix GeneChip system instruments. Affymetrix GeneAtlas instrument control software version 1.0.5.267 was used to analyze microarray image data and to compute intensity values. Affymetrix. CEL files containing raw, probe-level signal intensities were analyzed using Partek Genomics Suite version 6.6.12.0713 (Partek). Robust multichip averaging (RMA) was used for background correction, quantile normalization and probe set summarization with median polish (195). Statistical difference was calculated by two-way ANOVA analysis with a false discovery rate (FDR) of 0.05. Pathway analysis was performed using differentially expressed data 2-fold or higher with Ingenuity Pathway Analysis software (Ingenuity Systems).
Cell viability assay
Suit2 cells (in triplicate) were seeded in 96-well plates so that they reached approximately 80% confluence at 24 h. Cells were mock treated or infected with VSVs at MOI 0.002 or 0.00002. Cell viability was analyzed at 1, 24, 48, or 96 h p.i. by a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) cell viability assay (Cat. No. M2128, Sigma-Aldrich).
Western blotting
Cells were seeded in 6-well plates so that they reached approximately 80% confluence at 24 h, then mock treated or infected with VSV at MOI 1. At 8, 12, or 18 h p.i., cells were lysed with cellular lysis buffer (0.0625 M Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.02% (w/v) Bromophenol blue). 20 µl of protein lysate was separated by electrophoresis on 10% SDS PAGE gels and electroblotted to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked using 5% non-fat powdered milk in TBS-T [0.5M NaCl, 20 mM Tris (pH7.5), 0.1% Tween20]. The following antibodies from Cell Signaling Technology were used: p53 (1:2,000; #2524), Phospho-p53 (1:500; #2521), IRF3 (1:1000, #4302), Phospho-IRF3 (1:1000, #4947), Stat1 (1:1000, #42H3), and Phospho-Stat1 (1:500, #9171). In addition, the following primary antibodies were used in TBS-T with 5% BSA and 1% of 2% sodium azide: 1:5,000 rabbit polyclonal anti-VSV antibodies (Grdzelishvili et al., 2005). The following horseradish peroxidase (HRP)-conjugated secondary antibodies in TBS-T with 5% milk antibodies were used: 1:4,000 goat anti-mouse and 1:2,000 goat anti-rabbit (catalog no. 115-035-003 and 111-035-003, respectively; Jackson-ImmunoResearch). The Amersham ECL Western Blotting Detection kit (RPN2106; GE Healthcare) or SuperSignal West Pico (34080; Thermo Scientific) were used for detection.
RNA RT-PCR analysis
Cells (in duplicate) were seeded in 12-well plates so that they reached approximately 80% confluence at 24 h. Cells were mock treated or infected with VSV at MOI 5 based on titration on Suit2 cells. Total RNA was extracted 8 h p.i. with TRIzol as above. 50 µg of total RNA per reverse transcription (RT) reaction using SMART-Scribe reverse transcriptase (Clontech) was used for the cDNA synthesis as per manufacturer’s protocol. PCR was carried out on cDNA using the following conditions: denaturation at 94°C for 45 seconds (s) annealing at 57°C for 45s, extension at 72°C for 45s for either 25, 30, or 35 cycles and a finishing step at 72°C for 8 min. All primers for PCR are shown in the Supplementary Table 1 and were designed to not amplify genomic DNA. PCR products were electrophoresed on a 2% agarose gel with ethidium bromide and photographed using a GelDoc-It imager (UVP Imaging). Primers for RT-PCR are listed in Supplemental Table 1.
Treatment with inhibitors
Suit2 cell were seeded in 12-well plates so that they reached approximately 80% confluence at 24 h. Cells were mock treated or infected with VSV at MOI 10. After 1 h, virus-containing media was removed and cells were either mock treated (0.3 % DMSO) or treated with 8 µM IKK-2 Inhibitor VIII [purchased from EMD Millipore (cat. No. 401487)] or 2.5 µM ruxolitinib [purchased from Selleck Chemicals (cat. No. S1378)] in growth media containing 5% FBS. Inhibitor treatment also contained 0.3% DMSO. After 8 h treatment, total RNA was extracted with TRIzol as above. cDNA was synthesized and PCR was performed as above.
Cell line genomic mutation profiling
Cellular genomic DNA was isolated from PDAC cell lines using the Life Technologies Purelink Genomic DNA isolation kit. To analyze cellular DNA for frequent cancer mutations, the Ion AmpliSeq™ Cancer Hotspot Panel v2 Kit (Life Technologies) containing 207 primer pairs was used to perform multiplex PCR for the preparation of amplicon libraries from genomic hot spot regions that are frequently mutated in human genes associated with cancer, including approximately 2800 Catalogue of Somatic Mutations in Cancer (COSMIC) mutations of 50 oncogenes and tumor suppressor genes. Sequencing libraries were prepared using an Ion AmpliSeq Library Kit (Life Technologies) per manufacturer’s instructions. Briefly, amplicons were ligated to Ion-compatible adapters, followed by nick repair to complete the linkage between adapters and DNA inserts. The libraries were clonally amplified by emulsion PCR on Ion Sphere Particles (ISPs) using the Ion OneTouch 200 Template Kit (Life Technologies) as directed. Following amplification, the template-positive ISPs were enriched to maximize the number of sequencing reads produced using the Ion PGM Sequencing 200 Kit (Life Technologies) on an Ion PGM Sequencer (Life Technologies) and Ion 314 Chips (Life Technologies). Raw data was transferred to the Ion PGM Torrent Server for base calling, preprocessing 3’ trimming, quality control and assessment, and mapping. Variant calling and annotation was performed using Ion Reporter Software (Life Technologies) and Ingenuity Variant Analysis Knowledge Module (Ingenuity Systems) for Ion Reporter. To confirm the identified p53 mutations, cellular genomic DNA corresponding to TP53 exons 5 through 9 (where a majority of TP53 mutations occur) was amplified by PCR with primers VG 268 and VG 269 (NM_000546.5, Supplemental Table 1) prior to sequencing.
RESULTS
Generation of novel recombinant VSVs encoding p53 and p53-CC
To investigate how p53 affects antiviral signaling in PDAC cells, we engineered recombinant VSVs encoding human p53 (Fig. 1). In addition to VSV encoding wt p53 (p53wt), we designed VSV encoding the recently described chimeric p53-CC, which evades the dominant-negative activities of endogenously expressed mutant p53 (Okal et al., 2013). Transcriptionally active p53 binds to diverse DNA targets as a tetramer. As the C-terminus of p53wt, the CC domain of the Bcr protein allows for formation of antiparallel tetramers. However, unlike the C-terminus of p53wt, the CC domain is no longer a binding site for endogenous mutant p53 (Okal et al., 2013). As a result, the chimeric p53-CC protein was shown to evade endogenous dominant-negative p53 and restore p53wt activity better than p53wt in vitro in human ductal breast epithelial, human breast adenocarcinoma, human epithelial cervical adenocarcinoma, and human non-small cell lung carcinoma cell lines (Okal et al., 2013). To allow for visualization of p53 transgene expression in infected cells and discrimination between virus-encoded and endogenous p53 gene expression, we fused the N terminus of p53 (p53wt or p53-CC) to the C terminus of a near-infrared fluorescent protein, eqFP650 (herein called RFP) (Shcherbo et al., 2007) (Figure 1). The functionality of p53 fused to a fluorescent protein has been demonstrated previously for GFP (green fluorescent protein) (Okal et al., 2013).
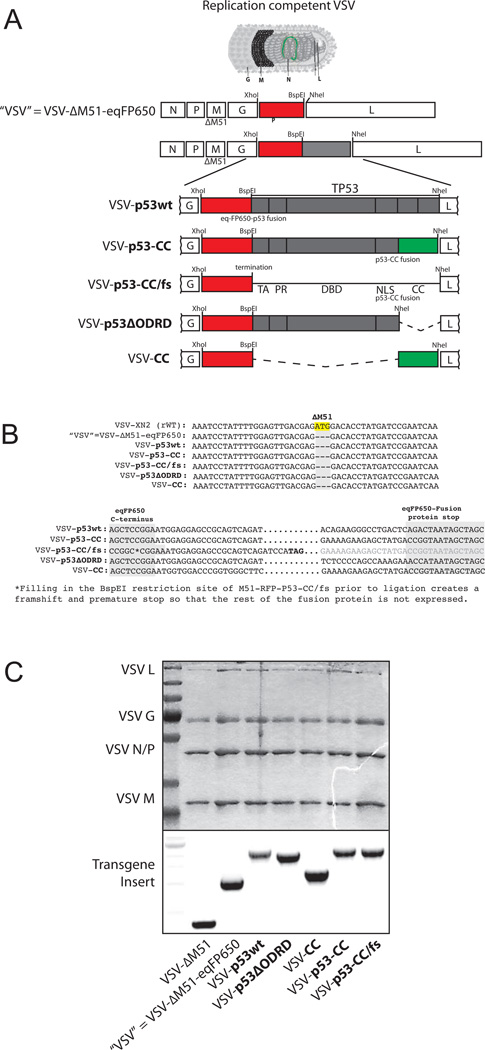
Figure 1. Engineering recombinant VSV, sequencing rescued viruses, and virion protein profile.
(A) Recombinant viruses were engineered using the VSV-ΔM51 backbone, with each transgene inserted between the VSV G and L protein sequence using XhoI and NheI restriction sites. (B) Viral RNA genomes were purified and cDNA was made and sequenced to confirm the ΔM51 deletion and correct transgene insertion. (C) Upper panel: Recombinant viruses were amplified, ultra-purified, and total virion protein was analyzed using SDS-PAGE and Coomassie stained to compare protein profiles. Lower panel: RNA was extracted from purified virions and reverse transcribed into cDNA. Viral cDNA was PCR amplified using primers flanking XhoI and NheI restriction sites to confirm transgene inserts.
We designed all constructs in context of VSV-ΔM51, which has a deletion of the methionine at amino acid position 51 (M51) of the VSV M protein (Fig. 1A). There were three important reasons to use the VSV-ΔM51 backbone. First, wt VSV cannot be utilized as an OV due to its undesirable natural neurotoxicity through an effective evasion of type I IFN mediated antiviral responses in normal cells, including CNS (Jenks et al., 2010; Johnson et al., 2007). VSV M51 mutants lack neurotoxicity but retain good oncolytic abilities (Ahmed et al., 2003; Brown et al., 2009; Ebert et al., 2005; Stojdl et al., 2003; Trottier et al., 2007; von Kobbe et al., 2000; Wollmann et al., 2010). In addition to safety, another advantage of VSV-ΔM51 for p53 transgene expression is that, unlike wt M protein, the ΔM51 M does not prevent cellular gene expression in infected cells (Ahmed et al., 2003; Kopecky et al., 2001; Stojdl et al., 2003). Therefore, VSV-ΔM51-encoded functional p53 would be able to drive beneficial expression of p53-regulated anticancer genes. Finally, this system also allows us to examine the effect of p53 expression on cellular antiviral responses that would be otherwise silenced in the presence of VSV wt M protein.
We designed and successfully rescued the VSVs, all carrying the ΔM51 deletion and transgene sequences (Fig. 1A). The newly created VSV-ΔM51-eqFP650 virus (herein called “VSV”) is a “parental” genome for all other VSVs expressing the designed transgenes: 1) VSV-p53wt, 2) VSV-p53-CC, 3) VSV-p53-CC/fs where a frameshift (fs) prevents translation of the p53-CC polypeptide but allows for monitoring of any VSV attenuation caused by the increased VSV genome length, 4) VSV-p53ΔODRD that encodes p53, but without the C terminus of wt p53, and 5) VSV-CC that express the CC domain of Bcr alone (no p53 sequences). Viruses were rescued, amplified and ultra-purified. Genomic RNA was isolated from purified virions and analyzed by RT-PCR. Partial sequencing of cDNA confirmed the presence of ΔM51 deletion as well as correct transgene insertion and sequence (Figs. 1B and 1C). Protein composition of the purified virions was examined using SDS-PAGE, and all viruses, as expected, showed a similar banding pattern indicating that transgene insertion did not produce any unexpected alterations in viral genes (Fig. 1C). Importantly, no genomic instability (e.g., loss of any transgene sequences) was seen throughout our studies. All experiments were conducted using ultrapure virion preparations.
Expression, localization and activity of VSV-encoded p53 variants
Most of the experiments were conducted using the human PDAC cell line Suit2 (Iwamura et al., 1987). Suit2 cells, like PDAC cells in 75% of patients, carry a TP53 mutation (Iacobuzio-Donahue and Herman, 2014; Weissmueller et al., 2014). Specifically, Suit2 cells were reported to carry a common dominant-negative R273H mutation in TP53 (Iwamura et al., 1987), which we confirmed by sequencing genomic DNA (Table 3). The presence of this dominant-negative mutation in TP53 allows for comparison of VSV-p53wt to VSV-p53-CC. Second, based on our previous studies using a panel of human PDAC cell lines, the Suit2 cell line represents the majority of the PDAC cell lines that retain inducible type I IFN antiviral responses (Moerdyk-Schauwecker et al., 2013). As such, the Suit2 cell line can be used to examine the effect of p53 on antiviral responses.
Table 3.
TP53 status of PDAC and non-malignant HPDE cell lines.
| CELL LINE | Mutant KRAS? |
Mutant TP53? |
TP53 Sequencing | p53 Protein Detection* |
p53 Dominant Negative** |
p53 Transactivation Class** |
|
|---|---|---|---|---|---|---|---|
| AsPC-1 | x | x | - | p.C135fs | - | - | - |
| T3M4 | x | x | p. P72R | p.Y220C | ++ | Moderate | Non-functional |
| Capan-2 | x | x | - | p.R273H | +++ | Yes | Non-functional |
| Suit2 | x | x | p. P72R | p.R273H | ++ | Yes | Non-functional |
| HPDE | - | - | p. P72R | WT | NA | - | - |
Genomic analysis of cell line p53 status by analyzing cellular genomic DNA by two methods: Ion Ampliseq™ Cancer Hotspot Panel for KRAS and TP53 mutations and PCR with primers VG268 and VG269, which correspond to exons 5 through 9 (where a majority of TP53 mutations occur).
Western blot was conducted to determine cell line p53 protein expression levels (data not shown).
Dominant negative and transactivation status data is from IARC p53 Database (http://p53.iarc.fr/)
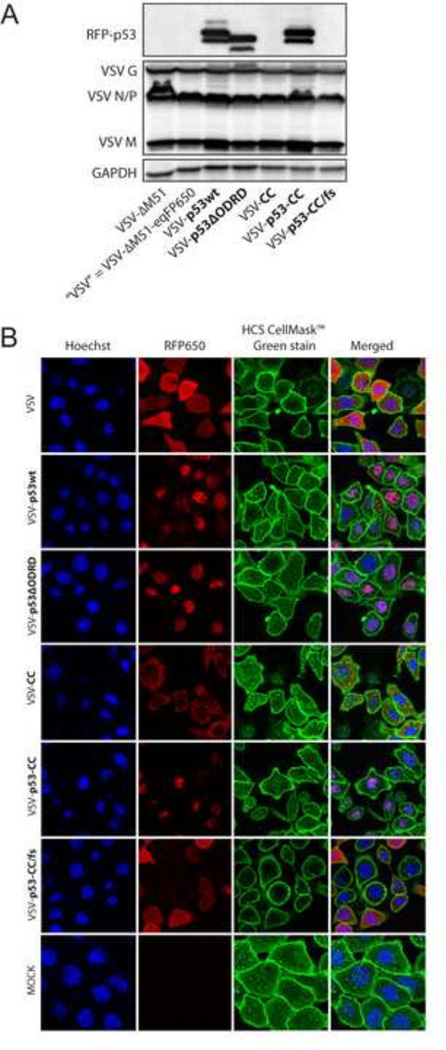
To examine if p53 transgenes are expressed, Suit2 cells were infected at MOI 5, total cell lysates were collected 8 h p.i. and analyzed by Western blot. As shown in Figure 2A, bands for the RFP-p53 fusion proteins were detected in cells infected with the VSV-p53wt, VSV-p53ΔODRD, and VSV-p53-CC viruses. As expected, p53wt and p53-CC fused to RFP appear the same size on the blot (approximately 73 kDa), while p53 with a truncated C terminus encoded by VSV-p53ΔODRD, showed slight downshift on the blot. This data confirms virus-encoded p53 transgene expression.
Figure 2. Recombinant virus transgene expression and localization.
(A) Suit2 cells were infected at MOI 5 and lysed 8 h p.i. Expression of viral proteins and RFP-p53 fusion proteins were analyzed by Western blot. (B) Suit2 cells were infected at MOI 5 for 8 h, and fluorescent microscopy was used to detect RFP-p53 (red) localization in cells stained with Hoechst nuclear stain (blue) and HCS CellMask plasma membrane stain (green).
Nuclear localization is required for p53 function as a transcription factor. Full-length p53wt contains three nuclear localization signals (NLS) in the C terminus: amino acid (aa) residues 305–322, 370–376, and 380–386. Although p53-CC was generated by truncating p53wt (removal of aa residues 323 to 393), it still contains one NLS (aa residues 305–322) and should localize in the nucleus as well. Also, a previous study showed nuclear localization of GFP-p53-CC protein expressed from a plasmid (Okal et al., 2013). To examine nuclear localization of VSV-encoded p53 fused to RFP (eqFP650), Suit2 cells were infected at MOI 5 and fluorescent confocal microscopy was conducted 8 h p.i. (Fig. 2B). Similar RFP-p53 nuclear localization was seen in cells infected with VSV-p53wt, VSV-p53ΔODRD, and VSV-p53-CC. As cell rounding occurs following infection, the cytoplasmic area appears smaller and difficult to detect, compared to mock-infected cells (Fig. 2B), However, the co-localization of RFP (red) and Hoechst (blue) can be clearly seen for these 3 viruses, but not for VSV, VSV-CC and VSV-p53-CC/fs (Fig. 2B). Interestingly, VSV-CC (no p53 sequences) showed a distinct punctate localization for RFP-CC in the cytoplasm, that may be of interest for future studies regarding the function of the CC domain of the BCR gene. In the two viruses encoding transgenes without any NLS, VSV and VSV-p53-CC/fs, RFP localization remained cytoplasmic, as expected. This data confirmed nuclear localization of viral-encoded p53.
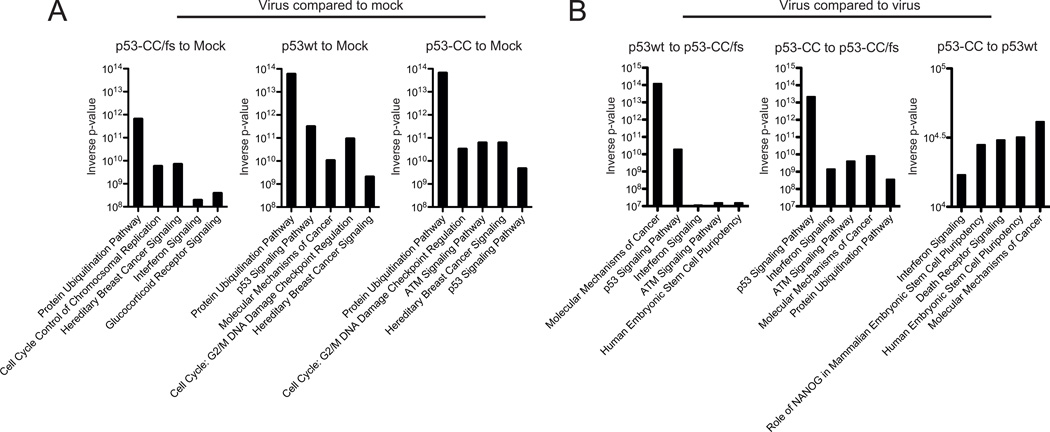
Next, we wanted to examine if VSV-encoded p53wt and p53-CC proteins were active as transcription factors in infected Suit2 cells. Cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs (no p53 production) at MOI 5 and total RNA was collected 8 h p.i. and used for microarray analysis. Microarray results were run though the Ingenuity pathway analysis (IPA) program, which predicts which cellular signaling pathways are most affected based on mRNA transcript levels following treatment. In Fig. 3A (“virus compared to mock”), we compared p53-CC/fs-infected (no p53 transgene is expressed), p53wt-infected, and p53-CC-infected cells to mock-infected cells to determine pathways affected by VSV infection, while in Fig. 3B (“virus compared to virus”) we compared VSV recombinants expressing p53 transgenes to p53-CC/fs (no p53 transgene is expressed) to identify cellular pathways affected by the p53 transgene expression during infection. Both analyses are shown to discriminate between gene expression changes that can be attributed to VSV infection and those attributed specifically to the p53 transgene expression (the Y-axis in Fig. 3 shows the statistical significance for the identification of the affected pathway). Figure 3 shows that the IPA predicts that the p53 signaling pathway is in the top five pathways affected for Suit2 cells infected with VSV-p53wt and VSV-p53-CC, but not for VSV-p53-CC/fs (compared to mock-treated cells) (Fig. 3A). Further, when comparing VSV-p53wt and VSV-p53-CC to VSV-p53-CC/fs, the p53 signaling pathway is also among the top five pathways affected (Fig. 3B). Interestingly, when comparing VSV-p53wt to VSV-p53-CC, the p53 signaling pathway is not highlighted as significantly different, suggesting that the p53wt and p53-CC proteins are both active and behave similarly, at least in Suit2 cells under our experimental conditions (Fig. 3B). Further, we examined changes in expression of many p53 target genes associated with canonical p53 activity. Using a standard cut-off of at least a 2-fold change for mRNA levels, we identified many canonical p53 target genes that were up or downregulated by the p53 expressing viruses, for example: apoptosis [BCL2-Associated X Protein (BAX) and B-Cell CLL/Lymphoma 2 (BCL2)], cell proliferation [Mouse Double Minute 2 (MDM2)], cell cycle arrest [Cyclin-Dependent Kinase Inhibitor 1A (CDKN1A/P21)], angiogenesis inhibition [Thrombospondin 1 (THBS1)), and DNA repair (Topoisomerase (DNA) II Binding Protein 1 (TOPB1)] (see Supplemental Table 2 for a complete list of affected genes).
Figure 3. Top 5 canonical pathways affected in infected Suit-2 cells.
Cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 5. Total RNA was extracted 8 h p.i. Microarray data that indicated more than a 2-fold change in cellular mRNA transcript levels in VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs infected cells (compared to mock or to VSV-p53-CC/fs) was analyzed with Ingenuity pathway analysis software and the top 5 canonical pathways are shown for (A) virus- to mock-treated comparison and (B) virus to virus comparison.
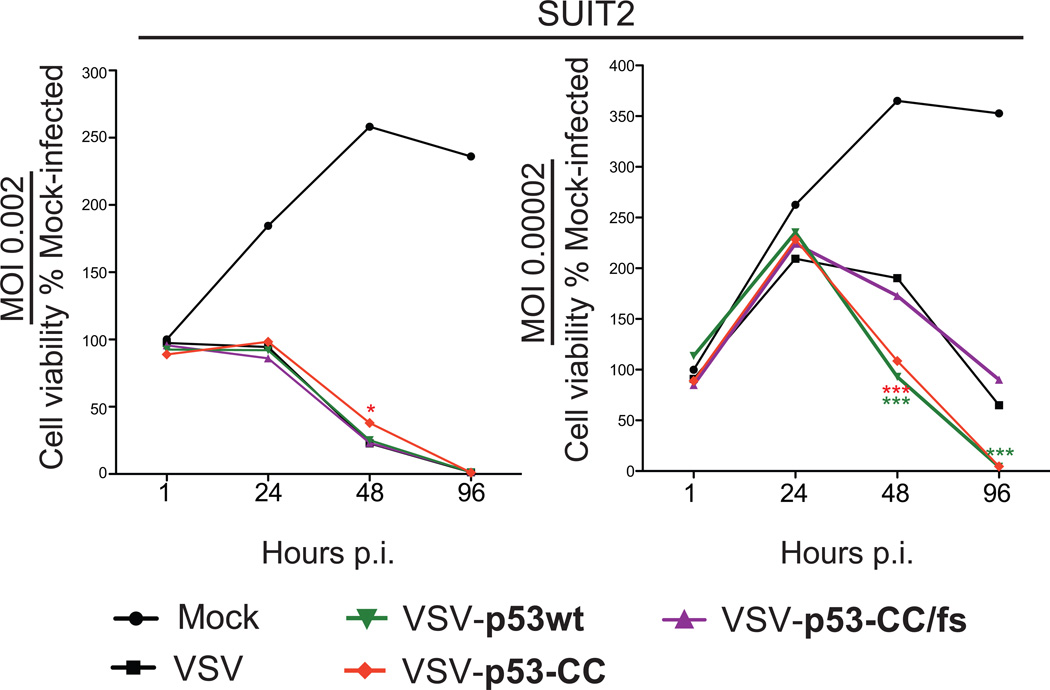
If virus-encoded p53wt and/or p53-CC are active, we expected them to enhance VSV cytotoxicity in cancer cells. Suit2 cells were mock treated or infected MOI 0.00002, 0.002, 0.1 and 1 with VSV, VSV-p53wt, VSV-p53-CC, or VSVp53-CC/fs and cell viability was examined at 1, 24, 48, and 96 h p.i. using an MTT cell viability assay. As shown in Figure 4 for MOI 0.002 and 0.00002, although no difference in cell killing between all viruses was seen at the higher MOI (a similar result was for MOI 0.1 and 1, data not shown), at the lower MOI we observed significantly more oncolysis by the VSVs expressing p53wt or p53-CC transgene, compared to VSV or VSV-p53-CC/fs (at 48 and 96 h p.i.). The inability of virus-encoded p53wt and p53-CC to enhance oncotoxicity of VSV at higher MOIs may be due to strong cytotoxic effect of VSV itself masking the effect of the transgene.
Figure 4. Suit2 cell viability following infection with VSV recombinants.
Suit2 cells were mock treated or infected with VSV, VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs viruses at MOI 0.002 or 0.00002. Cell viability was analyzed 1, 24, 48, and 96 h p.i. by MTT cell viability assay and is expressed as a percentage of mock-treated cells. All MTT assays were done in triplicate, and the data represent the means ±SEM.
Together, our results show that both VSV-p53wt and VSV-p53-CC are able to successfully direct expression of the designed p53 transgene, and that the gene products localize in the nucleus and are active as transcription factors, up or downregulating known p53 targets. Moreover, the virus-encoded p53wt and p53-CC enhance oncotoxicity of VSV, at least under some of our experimental conditions.
p53wt and p53-CC suppress the type I IFN response in Suit2 cells
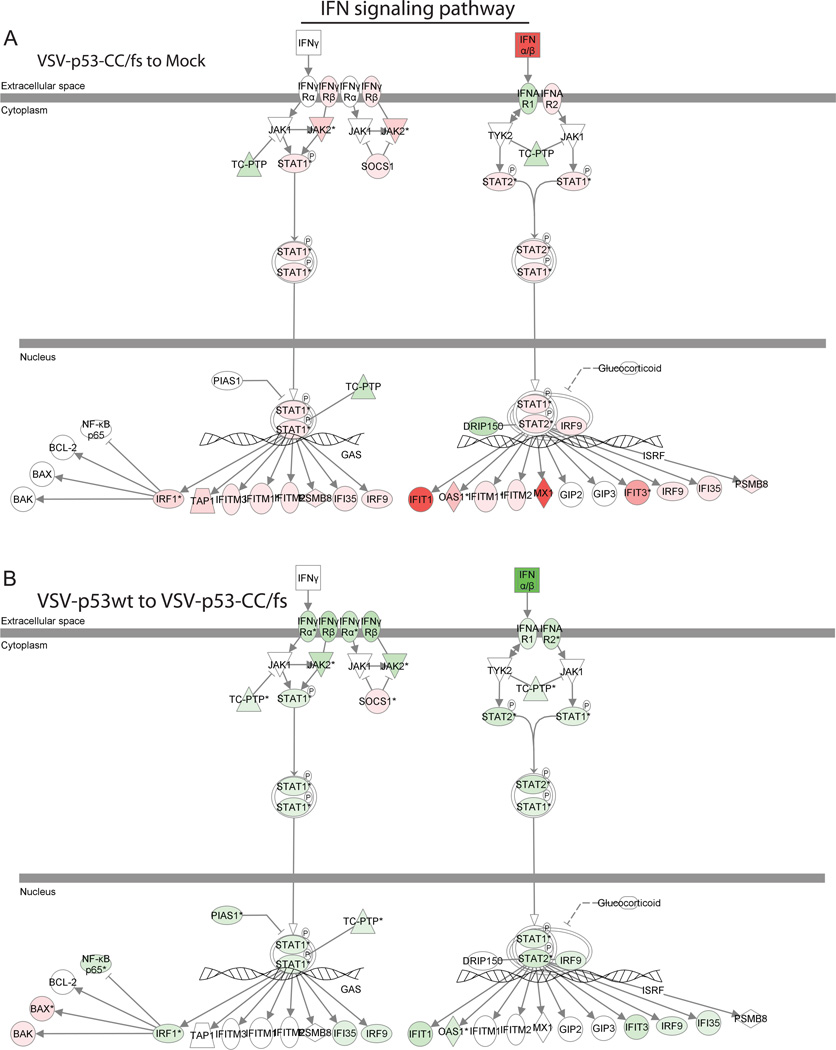
Using the transcriptome profile of Suit2 cells following mock treatment or infection with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs, we analyzed expression levels for genes associated with type I and type II IFN signaling pathways using the Ingenuity pathway analysis. Figure 5 provides a visual interpretation of at least 2-fold up (red) or downregulation (green) of genes that were identified (Fig. 5A shows the data for cells infected with VSV-p53-CC/fs compared to mock-infected cells, while Fig. 5B compares VSV-p53wt- to VSV-p53-CC/fs-infected cells). Table 1 is a quantitative list of the same data. In general, our data show that all the tested VSV induce significant antiviral responses. However, contrary to an expected enhancement of antiviral signaling by p53, when we compare Suit2 cells infected with VSV-p53wt or VSV-p53-CC vs. cells infected with p53-CC/fs (no p53 transgene is expressed), the type I IFN signaling response was dramatically attenuated (Table 1). Specifically, while there was a 291.10-fold increase in IFN-β transcripts by VSV-p53-CC/fs (the recombinant VSV that does not express p53) compared to mock-treated cells, only 25.24-fold and 4.56-fold increases in IFN-β transcript levels were observed for VSV-p53wt or VSV-p53-CC, respectively. Importantly, the transcriptome also showed that p53wt and p53-CC reduced virus-mediated expression of several key cellular antiviral proteins, including Myxovirus resistance 1 (Mx1), 2'-5'-oligoadenylate synthetase-like (OASL), Janus kinase 2 (JAK2), signal transducer and activator of transcription 1 (STAT1), and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) (Fig. 5 and Table 1). Also, infection with VSV-p53wt and VSV-p53-CC caused upregulation of the suppressor of cytokine signaling (SOCS1) that is known to inhibit STAT1 antiviral signaling activity (Song and Shuai, 1998).
Figure 5. Microarray results applied to the Ingenuity IFN signaling pathways.
The Ingenuity pathway analysis identified IFN pathway related transcripts that were differentially expressed in Suit2 cells. The cellular genes are shown with more than a 2-fold change in VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs infected cells (compared to mock or to VSV-p53-CC/fs). (A) Cells infected with VSV-p53-CC/fs are compared to mock-infected cells. (B) Cells infected with VSV-p53wt are compared to VSV-p53-CC/fs-infected cells. Red indicated upregulated transcripts while green represents downregulated transcripts.
Table 1.
Effect of virus infection on interferon signaling pathways
| Fold Induction (virus compared to mock) |
Fold Induction (virus compared to virus) |
||||||
|---|---|---|---|---|---|---|---|
| Symbol | Entrez Gene # |
VSV-p53- CC/fs |
VSV- p53wt |
VSV-p53- CC |
VSV- p53wt to VSV- p53-CC/fs |
VSV-p53- CC to VSV-p53- CC/fs |
VSV-p53-CC to VSV- p53wt |
|
Interferon signaling |
|||||||
| IFNB1 | 3456 | 291.10 | 25.24 | 4.56 | −11.53 | −63.80 | −5.53 |
| IFN Gamma (IL28A) | 282616 | 476.72 | 173.02 | 62.15 | −2.76 | −7.67 | −2.78 |
| IFNAR1 | 3454 | −2.07 | −3.36 | −3.87 | −2.18 | −2.51 | - |
| IFNAR2 | 3455 | 2.74 | −2.96 | −3.21 | −5.12 | −3.22 | - |
| IFNGR1 | 3459 | −5.90 | −7.13 | −10.35 | −3.98 | −5.54 | - |
| IFNGR2 | 3460 | 2.16 | −2.24 | −2.14 | −4.84 | −4.61 | - |
| JAK2 | 3717 | 22.74 | 5.30 | 3.26 | −5.55 | −6.98 | - |
| STAT1 | 6772 | 23.33 | 7.92 | 4.35 | −2.18 | −7.30 | - |
| STAT2 | 6773 | 8.08 | 2.59 | - | −3.11 | −3.79 | - |
| IRF9 | 10379 | 6.08 | 2.84 | 2.05 | −2.14 | −2.96 | - |
|
Inhibitor of Jak/Stat signaling |
|||||||
| SOCS1 | 8651 | 2.24 | 10.34 | 13.74 | 9.32 | 12.38 | |
| PTPN2 (TCPTP) | 5771 | −2.03 | −4.35 | −4.61 | −2.14 | −2.27 | |
| PIAS1 | 8554 | - | −3.46 | −3.38 | −2.78 | −2.79 | |
|
Interferon β/γ Stimulated Genes |
|||||||
| MX1 | 4599 | 71.99 | 37.77 | 27.59 | - | −2.61 | - |
| OASL | 8638 | 170.50 | 59.90 | 29.63 | −2.85 | −5.73 | −2.02 |
| IRF1 | 3659 | 21.52 | 7.83 | 4.21 | −3.39 | −7.51 | −2.21 |
| IFI35 | 3430 | 8.03 | 3.74 | 2.43 | −2.15 | −3.30 | - |
| IFIT1 | 3434 | 71.80 | 19.34 | 9.30 | −3.71 | −7.72 | −2.08 |
| IFIT3 | 3437 | 74.99 | 24.06 | 26.15 | −3.12 | −2.87 | - |
| PSMB8 | 5696 | 3.40 | 2.56 | - | - | -2.49 | - |
|
Downstream of IRF1 |
|||||||
| BAX | 581 | - | 2.46 | - | 4.11 | 3.24 | - |
| BAK1 | 578 | - | 2.64 | - | 2.50 | - | −2.94 |
| BCL2 | 596 | - | - | 2.61 | - | 3.30 | 2.05 |
| RELA | 5970 | - | −2.17 | −2.66 | −2.91 | −2.62 | |
Ingenuity pathway analysis of Interferon Signaling in Suit2 cells. Cellular transcripts were reported if they were more than 2-fold up or downregulated 8 h p.i. Cellular transcript profiles after infection of each virus, VSV-p53-CC/fs, VSV-p53wt, or VSV-p53-CC, are compared to mock as well to each other.
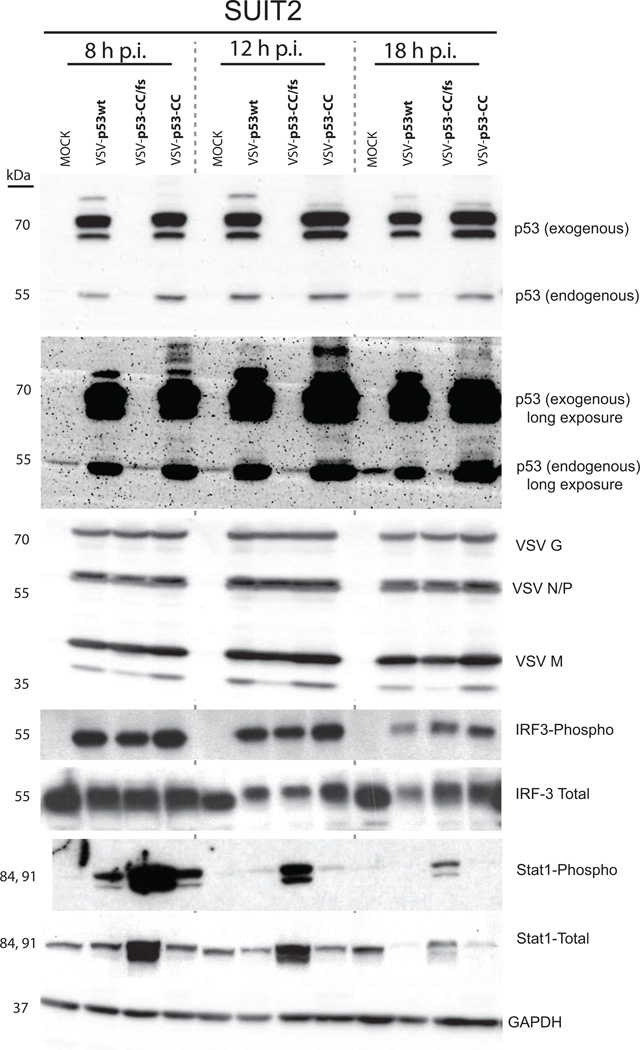
To confirm this surprising effect of p53 inhibition of type I IFN signaling that was seen with the microarray (Table 1), Suit2 cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 5 and total lysates were collected at 8, 12, and 18 h p.i. Western blot confirmed the expression of the p53wt and p53-CC as well as viral proteins at all time points (Fig. 6). Interestingly, there did not appear to be any differences in viral protein accumulation between viruses. Also, the phosphorylation of interferon regulatory factor 3 (IRF3) did not appear different between viruses, suggesting that cellular detection of viruses was similar regardless of expression of p53 transgenes. However, in agreement with the microarray data, in cells infected with VSV-p53wt or VSV-p53-CC, there was significantly reduced phosphorylation of STAT1, which is dependent on IFN production in infected cells. This result confirms that the virus-encoded p53 inhibits, rather than enhances, antiviral responses in VSV-infected Suit2 cells.
Figure 6. Western blot analysis following VSV infection in Suit2 cells.
Cells were infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 5. Cell lysates were collected at the indicated time points and analyzed by Western blot.
p53wt and p53-CC suppress type I IFN responses through inhibition of the NF-κB pathway
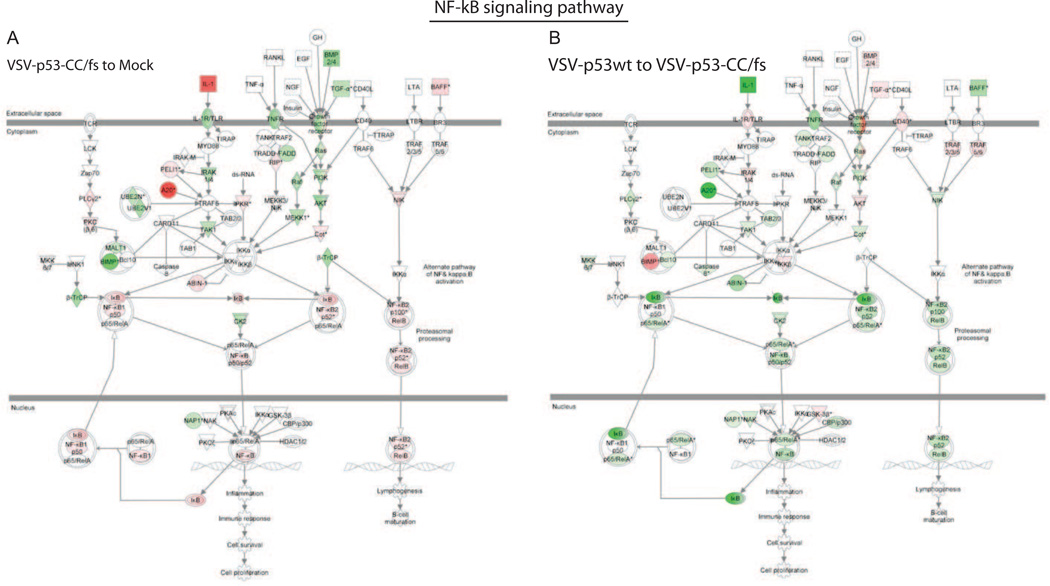
To further investigate the downregulation of IFN signaling by VSV-encoded p53wt and p53-CC, we used the Suit2 cell line transcriptome data to analyze potential effects of p53wt or p53-CC on NF-κB signaling pathways. NF-κB is known to upregulate early antiviral responses including type I IFN expression (Wang et al., 2010) and can be directly or indirectly inhibited by p53 (Ak and Levine, 2010; Culmsee et al., 2003; Heyne et al., 2013; Kawauchi et al., 2008; Murphy et al., 2011; Webster and Perkins, 1999). The Ingenuity pathway analysis detected downregulation of key NF-κB signaling pathway proteins (Fig. 7, Table 2). Importantly, the NF-κB subunit RelA, known to be critical for induction of IFN-β at early time points of infection, was downregulated by both VSV-p53 and VSV-p53-CC (Table 2). Additionally, we found downregulation of TBK1, a cellular protein that links the NF-κB and IRF3 pathways to IFN production (Fitzgerald et al., 2003; Hiscott et al., 2003). Also, there was a 51.99-fold increase in IL-1, an NF-κB induced cytokine, when cells were infected with VSV-p53-CC/fs, but no change after VSV-p53wt or VSV-p53-CC infection (Table 2). This data suggests that the NF-κB pathway is strongly downregulated by expression of the p53 transgenes.
Figure 7. Microarray results applied to the Ingenuity NF-κB signaling pathway.
The Ingenuity pathway analysis identified NF-κB pathway related transcripts that were differentially expressed in Suit2 cells. The cellular genes are shown with more than a 2-fold change in VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs infected cells (compared to mock or to VSV-p53-CC/fs). (A) VSV-p53-CC/fs to mock. (B) VSV-p53wt to VSV-p53-CC/fs. Red indicated upregulated transcripts while green represents downregulated transcripts.
Table 2.
Effect of virus infection on NF-kB signaling pathway
| Fold Induction (virus compared to mock) |
Fold Induction (virus compared to virus) |
||||||
|---|---|---|---|---|---|---|---|
| Symbol | Entrez Gene # |
VSV-p53- CC/fs |
VSV- p53wt |
VSV-p53- CC |
VSV-p53wt to VSV-p53- CC/fs |
VSV-p53- CC to VSV-p53- CC/fs |
VSV- p53-CC to VSV- p53wt |
| Extracellular space | |||||||
| IL1A | 3552 | 51.99 | - | - | −42.09 | −45.11 | - |
| TNFSF13B (BAFF) | 10673 | 19.03 | 2.10 | - | −5.78 | −9.56 | - |
| TGFA | 7039 | −2.39 | - | - | 3.06 | 2.12 | - |
| BMP4 | 652 | −3.75 | −3.00 | −2.43 | - | - | - |
| NFKB Activation / Repression | |||||||
| Cytoplasm | |||||||
| IL1R1 | 3554 | - | 3.48 | - | 3.52 | - | −3.45 |
| PEL1 | 57162 | 4.52 | 2.77 | - | −3.81 | −3.97 | - |
| IRAK1 | 3654 | −2.42 | - | - | 2.24 | - | - |
| TNFAIP3 (A20) | 7128 | 86.24 | 4.87 | 4.18 | −14.14 | −16.47 | - |
| MAP3K7 (TAK1) | 6885 | −2.38 | −2.25 | −4.36 | −2.35 | −2.14 | - |
| EIF2AK2 (PKR) | 5610 | 2.05 | 2.68 | 2.19 | - | −2.04 | - |
| TAB2 | 23118 | - | −3.67 | −3.77 | −2.09 | −2.15 | - |
| CD40 | 958 | - | 4.09 | 3.04 | - | - | - |
| RAS | 3845 | 2.41 | −5.01 | −4.17 | −4.09 | −2.29 | - |
| ARAF | 369 | −2.28 | −3.17 | −3.17 | - | - | - |
| PIK3CA (PI3K) | 5290 | - | - | −2.17 | - | −2.48 | - |
| TNFRSF1A | 7132 | −3.45 | −2.40 | −4.01 | - | - | - |
| TANK | 10010 | - | −2.58 | −7.23 | −2.38 | −4.10 | −2.80 |
| FADD | 8772 | −2.47 | −6.01 | −6.39 | −2.44 | −2.59 | - |
| MAP3K8 (COT) | 1326 | 8.48 | 3.86 | 2.24 | −2.56 | −4.83 | - |
| AKT2 | 208 | −2.05 | −2.29 | −2.16 | - | - | - |
| PLCG1 | 5335 | - | 2.01 | - | 3.00 | - | - |
| TRAF1 | 7185 | 2.76 | - | - | - | −2.12 | - |
| TNIP1 (ABIN1) | 10318 | 10.76 | - | - | −5.83 | −11.58 | - |
| CARD10 (BIMP1) | 29775 | −5.19 | 2.22 | - | 11.54 | 7.04 | - |
| MAP2K6 (MKK6) | 5608 | - | −2.20 | −2.02 | −2.02 | - | - |
| Nuclear | |||||||
| AZI2 (NAP1) | 64343 | 2.24 | −2.46 | −3.35 | −2.40 | −2.12 | - |
| (TBK1) NAK | 29110 | - | −2.07 | −2.82 | −2.92 | −3.98 | - |
| CREBBP | 1387 | - | - | −2.57 | −2.48 | −3.41 | - |
| EP300 (p300) | 2033 | - | −3.63 | −4.43 | −2.94 | −3.72 | - |
| GSK3B | 2932 | - | 2.34 | - | 2.33 | - | - |
| CSNK2A1 | 1457 | −2.65 | −3.34 | −3.09 | −2.91 | −2.38 | - |
| Cytoplasm/Nuclear | |||||||
| RelA (p65) | 5970 | - | −2.17 | −2.66 | −2.91 | −2.62 | - |
| RelB | 5971 | 6.39 | 2.04 | - | −3.13 | −5.10 | - |
| NFKB1 (p50) | 4790 | 3.69 | 3.97 | 2.03 | - | - | - |
| NFKB2 (p52) | 4791 | 2.07 | - | - | −4.24 | −6.21 | - |
| NFKB1A (IkB) | 4792 | 23.41 | - | - | −12.47 | −20.68 | - |
| IKBKB (IKKB) | 3551 | - | 2.20 | 2.20 | 2.20 | 2.32 | - |
| IKKb (IKBKB) | 3551 | - | 2.20 | 2.20 | 2.20 | 2.32 | - |
Ingenuity pathway analysis of NF-kB Signaling in Suit2 cells. Cellular transcripts were reported if they were more than 2-fold up or downregulated 8 h p.i. Cellular transcript profiles after infection of each virus, VSV-p53-CC/fs, VSV-p53wt, or VSV-p53-CC, are compared to mock as well compared to each other.
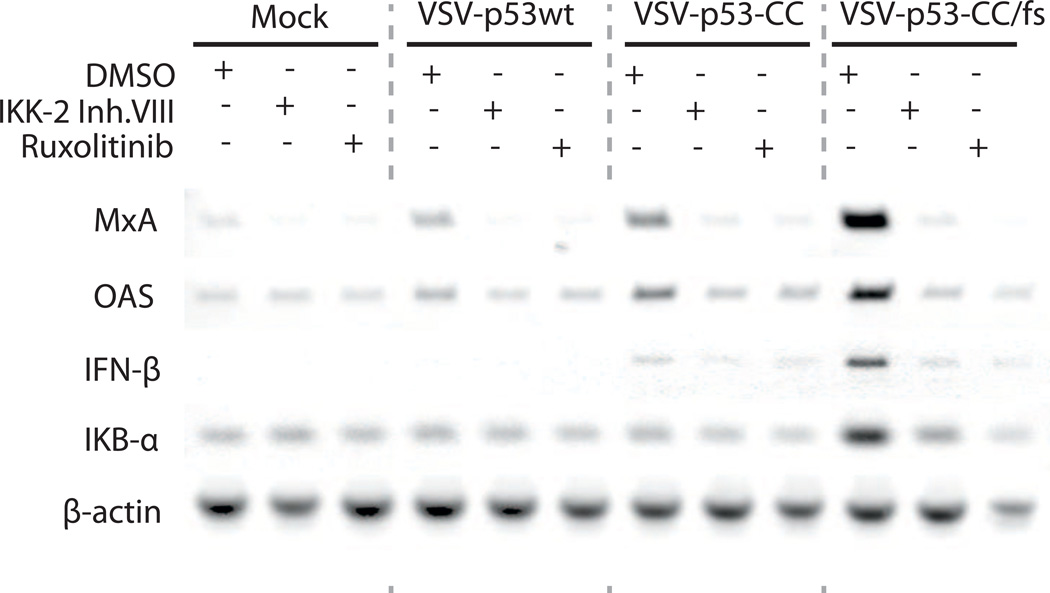
To test the hypothesis that VSV-encoded p53wt and p53-CC inhibit type I IFN responses via inhibition of NF-κB signaling, we conducted an experiment combining VSV infection with treatment of cells either with IKK-2 inhibitor VIII (to specifically prevent NF-κB activation) or JAK1/JAK2 inhibitor Ruxolitinib (to directly inhibit type I IFN-mediated signaling). Thus, if virus-encoded p53 inhibits type I IFN signaling in Suit2 cells via NF-κB inhibition, we would expect to see that i) IKK-2 inhibitor VIII would have a similar effect on antiviral signaling as Ruxolitinib, and that ii) the combination of IKK-2 inhibitor VIII with VSV-p53-CC/fs would produce a similar inhibiting effect on antiviral signaling as VSV-p53wt or VSV-p53-CC alone. Cells were first mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 10, then mock treated (DMSO) or treated with each inhibitor for 8 h. RT-PCR results shows that, compared to mock-treated cells, there was upregulation of MxA, OAS, IFN-β, and IkB-α transcripts by VSV-p53-CC/fs (Fig. 8). And, consistent with the transcriptome data, there was downregulation of all these four cellular genes by VSV-p53wt and VSV-p53-CC (in comparison to VSV-p53-CC/fs). In agreement with our hypothesis, treatment of cells with IKK-2 inhibitor VIII mimicked inhibition of IFN pathway signaling similarly to Ruxolitinib for all 3 viruses. Importantly, administration of the IKK-2 inhibitor VIII inhibited upregulation of antiviral gene transcripts seen by VSV-p53-CC/fs infection, bringing down the expression levels to those induced by VSV-p53wt or VSV-p53-CC alone (Fig. 8). This data suggests that type I IFN signaling in Suit2 is under direct control of NF-κB, and that VSV-encoded p53wt or p53-CC downregulate type I IFN responses via inhibition of NF-κB pathway.
Figure 8. Effect of a NF-κB signaling inhibitor on infected Suit2 cells.
Cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 10, followed by mock treatment (DMSO) or treatment with 8 µM IKK-2 Inhibitor VIII or 2.5 µM Ruxolitinib for 8 h. Total RNA was extracted, reverse transcribed and analyzed by PCR for the indicated genes.
p53-mediated suppression of type I IFN response in other PDAC cells lines
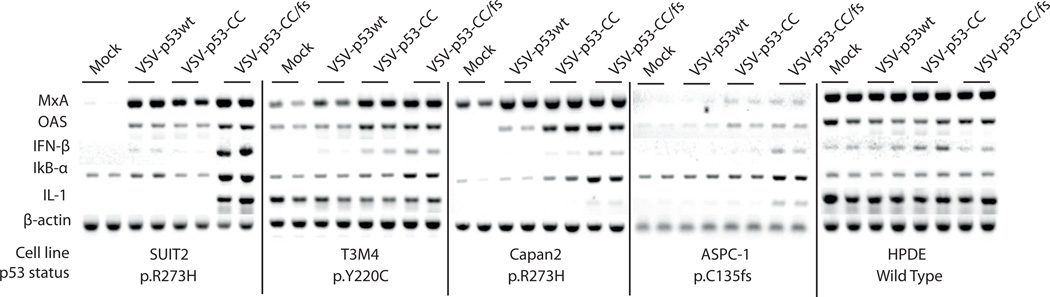
Next, we wanted to see if the inhibition of antiviral signaling by VSV-encoded p53 is observable in other PDAC cell lines, and if the cellular endogenous p53 status plays a role in the observed inhibition. We chose three PDAC cell lines that retain virus-mediated type I IFN responses, T3M4, Capan-2, and ASPC-1 (Moerdyk-Schauwecker et al., 2013). Additionally, we chose one control cell line, a “normal” (non-malignant) human pancreatic ductal epithelia cell line, HPDE. Using genomic DNA, we analyzed the p53 status of each cell line with Ion Ampliseq Hotspot analysis. The TP53 mutations identified by Ion Ampliseq for all cell lines were independently confirmed using PCR amplification and sequencing of a region of genomic DNA between TP53 exons 5 and 9, where most p53 mutations occur (Table 3). As expected for most human PDACs, all PDAC cell lines contained mutations in the TP53 gene. The R273H mutation for Suit2 and Capan2 is a known dominant-negative mutation, while the Y220C mutation for T3M4 is shown only to have moderate dominant-negative activity. For ASPC-1, we identified an early frameshift mutation (C135fs) that results in a small truncated protein product that could not be detected by Western blot analysis. As expected for non-malignant cell line, HPDE had a wt TP53 gene sequence.
To investigate how VSV-encoded p53 expression affects antiviral signaling in these cell lines, cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 5 and total RNA was isolated 8 h p.i. and analyzed by RT-PCR for specific gene expression levels. As was seen previously in Suit2, there was p53-mediated downregulation of transcripts associated with type I IFN as well as NF-κB signaling in T3M4, Capan-2, and ASPC-1 (Fig. 9). In particular, both p53wt and p53-CC reduced expression of the IFN-β mRNA at 8 h p.i. This result suggests that the observation of virus-encoded p53 effect on type I IFN signaling is not limited only to Suit2 cell line or the R273H mutation in TP53. The results in Figure 9 confirm our previous findings that a non-malignant pancreatic cell line HPDE (resistant to VSV) constitutively expresses MxA, OAS, IFN-α, IFN-β (Moerdyk-Schauwecker et al., 2013). Importantly, expression of p53 encoded by VSV-p53wt or VSV-p53-CC did not inhibit type I IFN or NF-κB signaling in HPDE. This suggests that the observed p53-mediated inhibition of IFN antiviral signaling could be limited to malignant PDAC cells (Fig. 9).
Figure 9. RT-PCR analysis of selected mRNA transcripts.
Suit2, T3M4, Capan2, and ASPC-1 PDAC cells and HPDE cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs, at MOI 5. Total RNA was extracted, reverse transcribed and analyzed by PCR for the indicated genes.
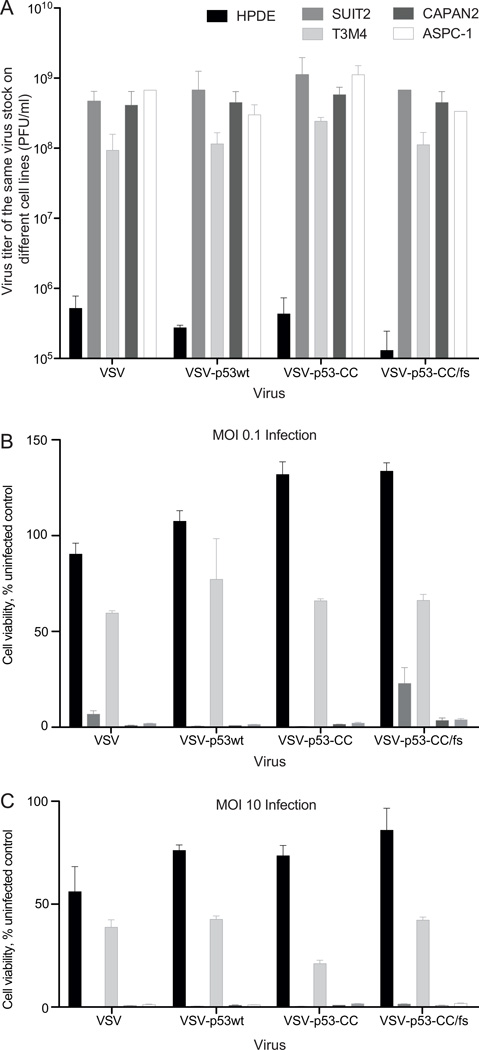
VSV-p53 recombinants retain their oncoselectivity and are attenuated in non-malignant pancreatic ductal cells
As patient safety is a major concern for treatment with replication-competent viruses, it is imperative that any virus-encoded transgene not cause the virus to lose oncoselectivity and attenuation in normal cell types. The data above suggests that VSV-p53wt or VSV-p53-CC do not inhibit IFN-β signaling in HPDE as they do in PDAC cells. To further examine whether p53 transgene expression may result in the loss of oncoselectivity of VSV recombinants, the same virus stocks of VSV, VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs were titrated on HPDE, Suit2, T3M4, Capan2, and ASPC-1 cells using plaque assay (Fig. 10A). All viruses showed cell-specific titers that were at least 2 logs lower in HPDE cells compared to PDAC cell lines, suggesting similar levels of attenuation in HPDE cells, regardless of p53 or p53-CC expression (Fig. 10A). Further, the cell lines were mock treated or infected at MOI 0.1 or 10, and cell viability was measured with MTT assay 5 days p.i. At MOI 0.1 (Fig. 10B), HPDE showed limited cell death with all tested viruses. Compared to the PDAC cell lines, where Capan2 and ASPC-1 showed almost no viable cells and T3M4 and Suit2 show some resistance, the p53 transgene does not appear to increase cell killing of HPDE. Even more, at MOI 10 HPDE showed only 50 percent cell viability when infected with VSV, and 75 percent cell viability when infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs (Fig. 10C). Compared to the PDAC cell lines, where Suit2, Capan2, and ASPC-1 show no viable cells and T3M4 remains resistant, this data suggests that HPDE cells retain resistance to VSV regardless of p53 or p53-CC expression. Together, our data show that VSV recombinants encoding p53 retain their oncoselectivity and are attenuated in non-malignant cells.
Figure 10. Analysis of PDAC and HPDE cell infectivity and cell viability at multiple MOIs.
(A) To determine a relative infectivity of VSV recombinants on different cells lines (PDAC cells and non-malignant HPDE cells), the same virus stock of each virus was titrated on different cell lines using plaque assay. The Y-axis shows virus titer of the same stock for each cell lines, with higher titer indicating higher infectivity of VSV on the given cell line. (B) and (C) Cells were mock treated or infected with VSV-p53wt, VSV-p53-CC, or VSV-p53-CC/fs at MOI 0.1 (B) or 10 (C). Cell viability was analyzed 5 d p.i. by MTT cell viability assay and is expressed as a percentage of mock-treated cells. All MTT assays were done in triplicate, and the data represent the means ±SEM.
DISCUSSION
In the present study, novel VSVs encoding functional human p53 or the chimeric p53-CC have been generated. Our study is the first to date to conduct a global analysis of PDAC mRNA expression following infection with any VSV. Based on this analysis and other experiments, we report p53-mediated inhibition of antiviral signaling. Our results suggest that virus-encoded p53 expression can simultaneously produce anticancer activities while assisting, rather than inhibiting, viral replication in PDAC cells.
Our data is in direct contrast to previous reports that p53 enhances type I IFN signaling pathway in normal cells and some cancer cell lines (Dharel et al., 2008; Lazo and Santos, 2011; Munoz-Fontela et al., 2005; Munoz-Fontela et al., 2011; Munoz-Fontela et al., 2008; Shin-Ya et al., 2005; Su et al., 2009; Takaoka et al., 2003; Yonish-Rouach et al., 1991). There are several potential reasons for the discrepancy between our results in human PDAC cells and previous results in other cell types: 1) differences between normal and cancer cells (and between different cancer cell types) in antiviral signaling, 2) constitutive activation of NF-κB pathway in a majority of PDACs, and 3) different timing and level of expression for VSV-encoded p53 in our study, and constitutively expressed p53 transgenes in other studies.
The unexpected inhibition of antiviral signaling by p53 can be explained by dysregulation of type I IFN signaling in cancer cells. In many cancer cell types, specific genes associated with type I IFN signaling are transcriptionally downregulated or functionally inactive (Balachandran and Barber, 2004; Marozin et al., 2008; Marozin et al., 2010; Moussavi et al., 2010; Zhang et al., 2010). Also, type I IFN mediated responses can be inhibited by MEK/ERK signaling, which is often upregulated in cancer cells (Noser et al., 2007), or by epigenetic silencing of IFN responsive transcription factors IRF7 or IRF5 (Li and Tainsky, 2011). Although all tested PDAC cell lines retain at least partially functional type I IFN signaling (Moerdyk-Schauwecker et al., 2013), it is likely that they have some dysregulation of this pathway, as type I IFN responses are generally anti-proliferative and pro-apoptotic (Stojdl et al., 2000; Vitale et al., 2007). Such dysregulation may result in the inability of p53 to positively affect antiviral signaling in PDAC cells.
The NF-κB pathway has been previously shown to be constitutively active in a majority of PDACs (Carbone and Melisi, 2012). Such abnormal activation of NF-κB plays a major role in tumor development maintaining expression of cellular genes that promote cell growth, proliferation and survival. Our data suggest that VSV-encoded p53wt or p53-CC downregulate antiviral responses via inhibition of NF-κB. As was shown previously, NF-κB is crucial for early IFN-β expression and resistance to virus replication (Wang et al., 2010). Our results are in agreement with previous studies demonstrating inhibition of NF-κB pathway by p53 via various mechanisms, including prevention of NF-κB nuclear translocation through upregulation of NF-κB inhibitor, IκBα (Shao et al., 2000), competition for common transcription cofactors (p300 and cAMP response element-binding protein (CBP) (Culmsee et al., 2003), and/or directly binding to NF-κB (RelA subunit p65) to disrupt its binding to DNA (Kawauchi et al., 2008). While beyond the scope of this study, the exact mechanism of p53-mediated NF-κB inhibition in the tested PDAC cell lines could be determined in the future.
Finally, the constitutively expressed p53 transgenes in other studies are likely stabilized and activated rapidly in response to viral infection, while VSV-mediated expression of p53 in our study probably takes longer. From our own observation, even following high MOI infection, VSV-encoded RFP was not visible until at least 5 h p.i. (data not shown), suggesting that expression of virus-encoded p53 and its effects occur after initial virus detection and stimulation of antiviral signaling.
Interestingly, VSV expressing p53 (wt or p53-CC) remained attenuated in non-malignant pancreatic ductal cell line HPDE, suggesting that these novel OVs would retain their safety in vivo. Future studies with VSVs expressing human p53 in animal models will determine if the oncoselective phenotype seen in PDAC cell lines is also observed in vivo. As beneficial p53 functions include promotion of enhanced antitumor immunity (Lowe et al., 2013), to fully examine the anticancer abilities of VSV-p53 or VSV-p53-CC, the viruses need to be tested in immunocompetent system. Unfortunately, the current design of our VSV-p53 recombinants complicates this task, as they encode human p53. Also, in our previous study all tested mouse PDAC cell lines had defective type I IFN signaling (Hastie et al., 2013), so our current in vivo mouse model of PDAC cannot be used to comprehensively test the novel VSV-p53 recombinants for their effect on antiviral signaling in those mouse PDAC cells. However, our data are in agreement with Heiber et al. (2011) who demonstrated that VSV encoding mouse p53 is not attenuated in vitro, but highly attenuated in normal tissues in vivo, and that in vivo treatment of mice results in enhanced tumor killing through stimulated antitumor immunity (Heiber and Barber, 2011). It should be noted that analysis of murine and human p53 response element consensus sequences show many differences between murine and human p53, suggesting that caution should be taken when generalizing about the similarity of regulation of the p53 pathway between humans and rodents (Horvath et al., 2007). Nevertheless, our present data and the previous study using VSV encoding mouse p53 in vivo (Heiber and Barber, 2011) both show VSV-p53 as a promising OV. Significantly, a recent clinical trial using vaccinia virus encoding p53 has also shown enhanced oncolytic efficacy in clinical trials through stimulation of an antitumor immune response (Hardwick et al., 2014). None of these previous studies has examined total gene expression in cancer cells in response to virus infection, but our data using VSV provides evidence of a new mechanism for enhanced oncolytic efficacy of viruses encoding p53 that will be important for future studies as OVs move forward in clinical application.
Supplementary Material
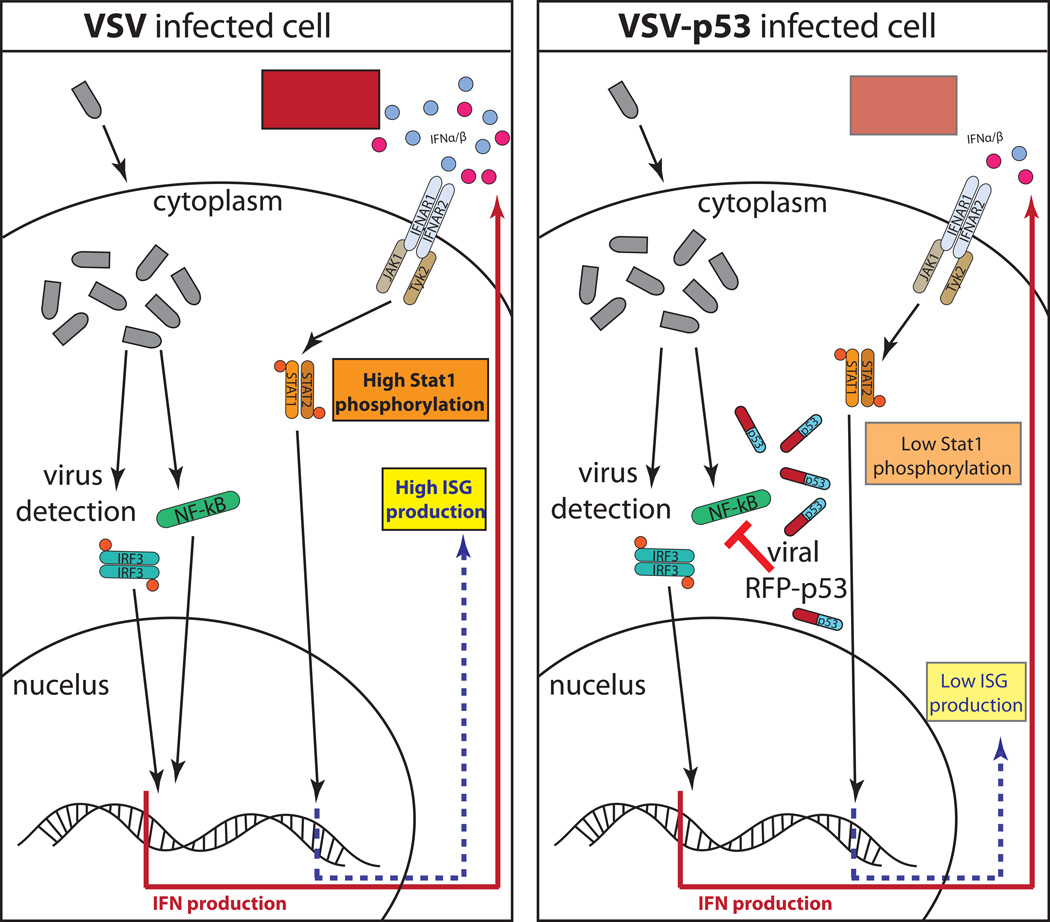
Figure 11.
The hypothetical model illustrating p53 transgene mediated inhibition of Type I IFN signaling during VSV infection. (Left) In the absence of a functional p53, VSV infection is detected through IRF-3 and NF-kB mediated signaling pathways and results in production of high levels of Type I IFNs and subsequent IFN signaling that stimulates high levels STAT1 phosphorylation and ISG expression. (Right) VSV-p53 infection is detected through IRF-3 and NF-kB mediated antiviral signaling pathways, however p53 transgene expression inhibits the NF-kB antiviral signaling pathway and results in reduced IFN production and reduced STAT1 phosphorylation and ISG expression.
HIGHLIGHTS.
p53 transgene expression enhances anticancer activities of oncolytic viruses
However, p53 is also known to enhance antiviral interferon-mediated responses
We examined effect of p53 transgenes on antiviral signaling in cancer cells
We show that p53 expression dramatically inhibited type I interferon responses
This unexpected inhibition occurs through inhibition of the NF-κB pathway by p53
ACKNOWLEDGEMENTS
We are grateful to Jack Rose (Yale University) for providing VSV-XN2 and VSV-XN2-ΔM51 plasmids, Randall Kimple (University of North Carolina at Chapel Hill) for Capan-2 and T3M4 cells, Timothy Wang (Columbia University) for AsPC-1 cells, and Michael Hollingsworth (University of Nebraska) for Suit2 cells. We thank Megan Moerdyk-Schauwecker, Sébastien Felt, and David Gray for technical assistance, and Sébastien Felt, Travis Porter and Sarah Greene for critical comments on the manuscript. This work was supported by NIH grant 1R15CA167517-01 (to V.Z.G.) from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 2003;77:4646–4657. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ak P, Levine AJ. p53 and NF-kappaB: different strategies for responding to stress lead to a functional antagonism. FASEB J. 2010;24:3643–3652. doi: 10.1096/fj.10-160549. [DOI] [PubMed] [Google Scholar]
- Azmi AS, Philip PA, Aboukameel A, Wang Z, Banerjee S, Zafar SF, Goustin AS, Almhanna K, Yang D, Sarkar FH, Mohammad RM. Reactivation of p53 by novel MDM2 inhibitors: implications for pancreatic cancer therapy. Curr. Cancer Drug Targets. 2010;10:319–331. doi: 10.2174/156800910791190229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Barber GN. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell. 2004;5:51–65. doi: 10.1016/s1535-6108(03)00330-1. [DOI] [PubMed] [Google Scholar]
- Bauzon M, Hermiston T. Armed therapeutic viruses - a disruptive therapy on the horizon of cancer immunotherapy. Front. Immunol. 2014;5:74. doi: 10.3389/fimmu.2014.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Brown CW, Stephenson KB, Hanson S, Kucharczyk M, Duncan R, Bell JC, Lichty BD. The p14 FAST protein of reptilian reovirus increases vesicular stomatitis virus neuropathogenesis. J. Virol. 2009;83:552–561. doi: 10.1128/JVI.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp ER, Wang C, Little EC, Watson PM, Pirollo KF, Rait A, Cole DJ, Chang EH, Watson DK. Transferrin receptor targeting nanomedicine delivering wild-type p53 gene sensitizes pancreatic cancer to gemcitabine therapy. Cancer Gene Ther. 2013;20:222–228. doi: 10.1038/cgt.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone C, Melisi D. NF-kappaB as a target for pancreatic cancer therapy. Expert Opin. Therap. Targets. 2012;16(Suppl 2):S1–S10. doi: 10.1517/14728222.2011.645806. [DOI] [PubMed] [Google Scholar]
- Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat. Rev. Clin. Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Siewe J, Junker V, Retiounskaia M, Schwarz S, Camandola S, El-Metainy S, Behnke H, Mattson MP, Krieglstein J. Reciprocal inhibition of p53 and nuclear factor-kappaB transcriptional activities determines cell survival or death in neurons. J. Neurosci. 2003;23:8586–8595. doi: 10.1523/JNEUROSCI.23-24-08586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharel N, Kato N, Muroyama R, Taniguchi H, Otsuka M, Wang Y, Jazag A, Shao RX, Chang JH, Adler MK, Kawabe T, Omata M. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology. 2008;47:1136–1149. doi: 10.1002/hep.22176. [DOI] [PubMed] [Google Scholar]
- Ebert O, Harbaran S, Shinozaki K, Woo SL. Systemic therapy of experimental breast cancer metastases by mutant vesicular stomatitis virus in immune-competent mice. Cancer Gene Therapy. 2005;12:350–358. doi: 10.1038/sj.cgt.7700794. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am. J. Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- Geoerger B, Vassal G, Opolon P, Dirven CM, Morizet J, Laudani L, Grill J, Giaccone G, Vandertop WP, Gerritsen WR, van Beusechem VW. Oncolytic activity of p53-expressing conditionally replicative adenovirus AdDelta24-p53 against human malignant glioma. Cancer Res. 2004;64:5753–5759. doi: 10.1158/0008-5472.CAN-04-0499. [DOI] [PubMed] [Google Scholar]
- Grdzelishvili VZ, Smallwood S, Tower D, Hall RL, Hunt DM, Moyer SA. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 2005;79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick NR, Carroll M, Kaltcheva TI, Qian D, Lim D, Leong L, Chu P, Kim J, Joseph C, Fakih MG, Yen Y, Espenschied J, Ellenhorn JD, Diamond DJ, Chung V. p53MVA therapy in patients with refractory gastrointestinal malignancies elevates p53-specific CD8+ T cell responses. Clin. Cancer. Res. 2014:4459–4470. doi: 10.1158/1078-0432.CCR-13-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie E, Besmer DM, Shah NR, Murphy AM, Moerdyk-Schauwecker M, Molestina C, Roy LD, Curry JM, Mukherjee P, Grdzelishvili VZ. Oncolytic Vesicular Stomatitis Virus in an Immunocompetent Model of MUC1-Positive or MUC1-Null Pancreatic Ductal Adenocarcinoma. J..Virol. 2013;87:10283–10294. doi: 10.1128/JVI.01412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie E, Grdzelishvili VZ. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J. Gen. Virol. 2012;93:2529–2545. doi: 10.1099/vir.0.046672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiber JF, Barber GN. Vesicular stomatitis virus expressing tumor suppressor p53 is a highly attenuated, potent oncolytic agent. J. Virol. 2011;85:10440–10450. doi: 10.1128/JVI.05408-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman DA, Steenbergen RD, van der Torre J, Scheffner M, Alemany R, Gerritsen WR, Meijer CJ, Snijders PJ, van Beusechem VW. Oncolytic adenovirus expressing a p53 variant resistant to degradation by HPV E6 protein exhibits potent and selective replication in cervical cancer. Mol. Ther. 2005;12:1083–1090. doi: 10.1016/j.ymthe.2005.06.443. [DOI] [PubMed] [Google Scholar]
- Heyne K, Winter C, Gerten F, Schmidt C, Roemer K. A novel mechanism of crosstalk between the p53 and NFkappaB pathways: MDM2 binds and inhibits p65RelA. Cell Cycle. 2013;12:2479–2492. doi: 10.4161/cc.25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Grandvaux N, Sharma S, Tenoever BR, Servant MJ, Lin R. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. NY Acad. Sci. 2003;1010:237–248. doi: 10.1196/annals.1299.042. [DOI] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genetics. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N. Engl. J. Med. 2014;370:1352–1353. doi: 10.1056/NEJMcibr1400189. [DOI] [PubMed] [Google Scholar]
- Idema S, Lamfers ML, van Beusechem VW, Noske DP, Heukelom S, Moeniralm S, Gerritsen WR, Vandertop WP, Dirven CM. AdDelta24 and the p53-expressing variant AdDelta24-p53 achieve potent anti-tumor activity in glioma when combined with radiotherapy. J. Gene Med. 2007;9:1046–1056. doi: 10.1002/jgm.1113. [DOI] [PubMed] [Google Scholar]
- Iwamura T, Katsuki T, Ide K. Establishment and characterization of a human pancreatic cancer cell line (SUIT-2) producing carcinoembryonic antigen and carbohydrate antigen 19-9. Jpn. J. Cancer Res. 1987;78:54–62. [PubMed] [Google Scholar]
- Jenks N, Myers R, Greiner SM, Thompson J, Mader EK, Greenslade A, Griesmann GE, Federspiel MJ, Rakela J, Borad MJ, Vile RG, Barber GN, Meier TR, Blanco MC, Carlson SK, Russell SJ, Peng KW. Safety studies on intrahepatic or intratumoral injection of oncolytic vesicular stomatitis virus expressing interferon-beta in rodents and nonhuman primates. Hum. Gene Ther. 2010;21:451–462. doi: 10.1089/hum.2009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Nasar F, Coleman JW, Price RE, Javadian A, Draper K, Lee M, Reilly PA, Clarke DK, Hendry RM, Udem SA. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology. 2007;360:36–49. doi: 10.1016/j.virol.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 2009;83:7996–8003. doi: 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. Activated p53 induces NF-kappaB DNA binding but suppresses its transcriptional activation. Biochem. Biophys. Res. Commun. 2008;372:137–141. doi: 10.1016/j.bbrc.2008.05.021. [DOI] [PubMed] [Google Scholar]
- King JC, Lu QY, Li G, Moro A, Takahashi H, Chen M, Go VL, Reber HA, Eibl G, Hines OJ. Evidence for activation of mutated p53 by apigenin in human pancreatic cancer. Biochim. Biophys. Acta. 2012;1823:593–604. doi: 10.1016/j.bbamcr.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo T, Choi IK, Kim M, Lee JS, Oh E, Kim J, Yun CO. Negative regulation-resistant p53 variant enhances oncolytic adenoviral gene therapy. Hum. Gene Ther. 2012;23:609–622. doi: 10.1089/hum.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky SA, Willingham MC, Lyles DS. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 2001;75:12169–12181. doi: 10.1128/JVI.75.24.12169-12181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb. Perspect. Biol. 2010;2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo PA, Santos CR. Interference with p53 functions in human viral infections, a target for novel antiviral strategies? Rev. Med. Virol. 2011;21:285–300. doi: 10.1002/rmv.696. [DOI] [PubMed] [Google Scholar]
- Li J, Pan J, Zhu X, Su Y, Bao L, Qiu S, Zou C, Cai Y, Wu J, Tham IW. Recombinant adenovirus-p53 (Gendicine) sensitizes a pancreatic carcinoma cell line to radiation. Chin. J. Cancer Res. 2013;25:715–721. doi: 10.3978/j.issn.1000-9604.2013.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Tainsky MA. Epigenetic silencing of IRF7 and/or IRF5 in lung cancer cells leads to increased sensitivity to oncolytic viruses. PLoS One. 2011;6:e28683. doi: 10.1371/journal.pone.0028683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Shatz M, Resnick MA, Mendez D. Modulation of immune responses by the tumor suppressor p53. BioDiscovery. 2013;8 [Google Scholar]
- Marozin S, Altomonte J, Stadler F, Thasler WE, Schmid RM, Ebert O. Inhibition of the IFN-beta response in hepatocellular carcinoma by alternative spliced isoform of IFN regulatory factor-3. Mol. Ther. 2008;16:1789–1797. doi: 10.1038/mt.2008.201. [DOI] [PubMed] [Google Scholar]
- Marozin S, De Toni EN, Rizzani A, Altomonte J, Junger A, Schneider G, Thasler WE, Kato N, Schmid RM, Ebert O. Cell cycle progression or translation control is not essential for vesicular stomatitis virus oncolysis of hepatocellular carcinoma. PLoS One. 2010;5:e10988. doi: 10.1371/journal.pone.0010988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitlianga PG, Sioka C, Vartholomatos G, Goussia A, Polyzoidis K, Rao JS, Kyritsis AP. p53 enhances the Delta-24 conditionally replicative adenovirus anti-glioma effect. Oncol. Rep. 2006;15:149–153. [PubMed] [Google Scholar]
- Moerdyk-Schauwecker M, Shah NR, Murphy AM, Hastie E, Mukherjee P, Grdzelishvili VZ. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: role of type I interferon signaling. Virology. 2013;436:221–234. doi: 10.1016/j.virol.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi M, Fazli L, Tearle H, Guo Y, Cox M, Bell J, Ong C, Jia W, Rennie PS. Oncolysis of prostate cancers induced by vesicular stomatitis virus in PTEN knockout mice. Cancer Res. 2010;70:1367–1376. doi: 10.1158/0008-5472.CAN-09-2377. [DOI] [PubMed] [Google Scholar]
- Munoz-Fontela C, Garcia MA, Garcia-Cao I, Collado M, Arroyo J, Esteban M, Serrano M, Rivas C. Resistance to viral infection of super p53 mice. Oncogene. 2005;24:3059–3062. doi: 10.1038/sj.onc.1208477. [DOI] [PubMed] [Google Scholar]
- Munoz-Fontela C, Gonzalez D, Marcos-Villar L, Campagna M, Gallego P, Gonzalez-Santamaria J, Herranz D, Gu W, Serrano M, Aaronson SA, Rivas C. Acetylation is indispensable for p53 antiviral activity. Cell Cycle. 2011;10:3701–3705. doi: 10.4161/cc.10.21.17899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fontela C, Macip S, Martinez-Sobrido L, Brown L, Ashour J, Garcia-Sastre A, Lee SW, Aaronson SA. Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 2008;205:1929–1938. doi: 10.1084/jem.20080383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Besmer DM, Moerdyk-Schauwecker M, Moestl N, Ornelles DA, Mukherjee P, Grdzelishvili VZ. Vesicular stomatitis virus as an oncolytic agent against pancreatic ductal adenocarcinoma. J. Virol. 2012;86:3073–3087. doi: 10.1128/JVI.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SH, Suzuki K, Downes M, Welch GL, De Jesus P, Miraglia LJ, Orth AP, Chanda SK, Evans RM, Verma IM. Tumor suppressor protein (p)53, is a regulator of NF-kappaB repression by the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA. 2011;108:17117–17122. doi: 10.1073/pnas.1114420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noser JA, Mael AA, Sakuma R, Ohmine S, Marcato P, Lee PW, Ikeda Y. The RAS/Raf1/MEK/ERK signaling pathway facilitates VSV-mediated oncolysis: implication for the defective interferon response in cancer cells. Mol. Ther. 2007;15:1531–1536. doi: 10.1038/sj.mt.6300193. [DOI] [PubMed] [Google Scholar]
- Odin L, Favrot M, Poujol D, Michot JP, Moingeon P, Tartaglia J, Puisieux I. Canarypox virus expressing wild type p53 for gene therapy in murine tumors mutated in p53. Cancer Gene Ther. 2001;8:87–98. doi: 10.1038/sj.cgt.7700279. [DOI] [PubMed] [Google Scholar]
- Okabe T, Yamaguchi NNO. Establishment and characterization of a carcinoembryonic antigen (CEA)-producing cell line from a human carcinoma of the exocrine pancreas. Cancer. 1983;51:662–668. doi: 10.1002/1097-0142(19830215)51:4<662::aid-cncr2820510419>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Okal A, Mossalam M, Matissek KJ, Dixon AS, Moos PJ, Lim CS. A chimeric p53 evades mutant p53 transdominant inhibition in cancer cells. Mol. Pharm. 2013;10:3922–3933. doi: 10.1021/mp400379c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Peng KW. Viruses as anticancer drugs. Trends Pharmacol. Sci. 2007;28:326–333. doi: 10.1016/j.tips.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Tsurumi T. Genome guardian p53 and viral infections. Rev. Med. Virol. 2013;23:213–220. doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- Shao J, Fujiwara T, Kadowaki Y, Fukazawa T, Waku T, Itoshima T, Yamatsuji T, Nishizaki M, Roth JA, Tanaka N. Overexpression of the wild-type p53 gene inhibits NF-kappaB activity and synergizes with aspirin to induce apoptosis in human colon cancer cells. Oncogene. 2000;19:726–736. doi: 10.1038/sj.onc.1203383. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nature Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Shemiakina II, Ryabova AV, Luker KE, Schmidt BT, Souslova EA, Gorodnicheva TV, Strukova L, Shidlovskiy KM, Britanova OV, Zaraisky AG, Lukyanov KA, Loschenov VB, Luker GD, Chudakov DM. Near-infrared fluorescent proteins. Nature Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin-Ya M, Hirai H, Satoh E, Kishida T, Asada H, Aoki F, Tsukamoto M, Imanishi J, Mazda O. Intracellular interferon triggers Jak/Stat signaling cascade and induces p53-dependent antiviral protection. Biochem. Biophys. Res. Commun. 2005;329:1139–1146. doi: 10.1016/j.bbrc.2005.02.088. [DOI] [PubMed] [Google Scholar]
- Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J. Biol. Chem. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, Knowles S, Marius R, Atkins H, Sonenberg N, Bell JC. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- Su WC, Liu WL, Cheng CW, Chou YB, Hung KH, Huang WH, Wu CL, Li YT, Shiau AL, Lai MY. Ribavirin enhances interferon signaling via stimulation of mTOR and p53 activities. FEBS Lett. 2009;583:2793–2798. doi: 10.1016/j.febslet.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS) J. Consum. Health. Internet. 2012;16:366–367. [Google Scholar]
- Trottier MD, Lyles DS, Reiss CS. Peripheral, but not central nervous system, type I interferon expression in mice in response to intranasal vesicular stomatitis virus infection. J. Neurovirol. 2007;13:433–445. doi: 10.1080/13550280701460565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G, van Eijck CH, van Koetsveld Ing PM, Erdmann JI, Speel EJ, van der Wansem Ing K, Mooij DM, Colao A, Lombardi G, Croze E, Lamberts SW, Hofland LJ. Type I interferons in the treatment of pancreatic cancer: mechanisms of action and role of related receptors. Annals Surgery. 2007;246:259–268. doi: 10.1097/01.sla.0000261460.07110.f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A, Balachandran S, Beg AA. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J. Immunol. 2010;185:1720–1729. doi: 10.4049/jimmunol.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su C, Cao H, Li K, Chen J, Jiang L, Zhang Q, Wu X, Jia X, Liu Y, Wang W, Liu X, Wu M, Qian Q. A novel triple-regulated oncolytic adenovirus carrying p53 gene exerts potent antitumor efficacy on common human solid cancers. Mol. Cancer Ther. 2008;7:1598–1603. doi: 10.1158/1535-7163.MCT-07-2429. [DOI] [PubMed] [Google Scholar]
- Webster GA, Perkins ND. Transcriptional cross talk between NF-kappaB and p53. Mol. Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, Aust D, Markert EK, Wu J, Grimmond SM, Pilarsky C, Prives C, Biankin AV, Lowe SW. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann G, Rogulin V, Simon I, Rose JK, van den Pol AN. Some attenuated variants of vesicular stomatitis virus show enhanced oncolytic activity against human glioblastoma cells relative to normal brain cells. J. Virol. 2010;84:1563–1573. doi: 10.1128/JVI.02040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki Y, Tazawa H, Hashimoto Y, Kojima T, Kuroda S, Yano S, Yoshida R, Uno F, Mizuguchi H, Ohtsuru A, Urata Y, Kagawa S, Fujiwara T. A novel apoptotic mechanism of genetically engineered adenovirus-mediated tumour-specific p53 overexpression through E1A-dependent p21 and MDM2 suppression. Eur. J. Cancer. 2012;48:2282–2291. doi: 10.1016/j.ejca.2011.12.020. [DOI] [PubMed] [Google Scholar]
- Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- Zhang KX, Matsui Y, Hadaschik BA, Lee C, Jia W, Bell JC, Fazli L, So AI, Rennie PS. Down-regulation of type I interferon receptor sensitizes bladder cancer cells to vesicular stomatitis virus-induced cell death. Int. J. Cancer. 2010;127:830–838. doi: 10.1002/ijc.25088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.