Abstract
Designing delivery agents for therapeutics is an ongoing challenge. As treatments and desired cargoes become more complex, the need for improved delivery vehicles becomes critical. Excellent delivery vehicles must ensure the stability of the cargo, maintain the cargo’s solubility, and promote efficient delivery and release. In order to address these issues, many research groups have looked to nature for design inspiration. Proteins, such as HIV-1 TAT and Antennapedia homeodomain protein, are capable of crossing cellular membranes. However, due to the complexities of their structures, they are synthetically challenging to reproduce in the laboratory setting. Being able to incorporate the key features of these proteins that enable cell entry into simpler scaffolds opens up a wide range of opportunities for the development of new delivery reagents with improved performance. This review charts the development of protein mimics based on cell-penetrating peptides and how structure-activity relationships with these molecules and their protein counterparts ultimately led to the use of polymeric scaffolds. These scaffolds deviate from the normal peptide backbone, allowing for simpler, synthetic procedures to make carriers and tune chemical compositions for application specific needs. Successful design of polymeric protein mimics would allow researchers to further understand the key features in proteins and peptides necessary for efficient delivery and to design the next generation of more efficient delivery reagents.
Keywords: Cell-penetrating peptides, polymers, guanidinium-rich molecular transporters, protein mimics, peptidomimetics, delivery
INTRODUCTION
Proteins are large, complex biomolecules that perform numerous biological functions.1 They contain both secondary and tertiary structure, which helps them arrange and fold into specific and functional conformations. While nature has developed efficient ways to generate correctly folded and functional proteins, it is substantially more difficult to recreate these structures synthetically. Many research groups have successfully developed peptidomimetics that mimic conformations of short peptide sequences, but to date, mimicking larger protein surfaces or entire protein functions represent more significant challenges.2 The field of proteomimetics looks to specifically address these challenges by moving away from naturally occurring amino acids and developing non-peptidic materials that can capture key secondary structures found in proteins.2
An elegant example of synthetic protein mimic development is from Andrew Hamilton and coworkers in which they were able to mimic part of the protein helix from the myosin light chain kinase using a terphenyl scaffold.3 These synthetic mimics operate on the premise that the critical residues needed for efficient protein-protein interactions lie along one face of the α-helix.3,4 Using this scaffold, they were also able to assess binding affinities for calmodulin, which is a calcium-binding messenger that aids in cell signaling.3 Specifically, they were able to prove that the synthetic α-helix had similar binding properties to the myosin light chain kinase, thus demonstrating the protein mimicking capacity of these materials.3 Other excellent work has also been published in this field.5–8
In addition, many researchers have explored foldamers, which are chains of molecules that can fold into organized structures, such as α-helices and β-sheets, in solution.9–22 Foldamers differ from other proteomimetics in that they require non-covalent interactions, such as hydrogen bonding, π-interactions, electrostatic interactions, van der Waal’s interactions, and solvophobic effects with non-adjacent surfaces.9,10,12,15,16,18,20,21 These molecules have been used to mimic the folding of proteins, polysaccharides, and nucleic acids.9,10,12,15,16,18,20,21 One specific subset of foldamers, referred to as abiotic foldamers, aim to capture key features of proteins, such as secondary structure, with non-natural materials.9–17,19–22
While mimicking protein secondary structure is an impressive feat and can aid the future development of entirely synthetic protein mimics, it remains difficult to generate these scaffolds and to predict the proper folding or assembly processes. Protein mimics that capture key features using simpler scaffolds without secondary structures can more easily be attained with tunable, synthetic platforms.23,24 One prominent example involves mimicking antimicrobial peptides (AMPs). AMPs are potent antibacterial agents that are part of the innate immune systems for many organisms.25–28 Magainin 2, which is one of the many AMPs currently found in nature, and other similar antimicrobial agents have been shown to be effective against both Gram positive and Gram negative bacteria.25–27,29,30 These peptides have facially amphiphilic topologies, enabling segregation of hydrophilic (cationic) and hydrophobic residues for improved membrane activity and antimicrobial properties.25,31 Much effort has been devoted to understanding these peptides and the mechanisms by which they kill bacteria, both experimentally and computationally.25,32–49 Despite the level of controversy surrounding their mode of action, hydrophobic and electrostatic interactions between these peptides and the bacterial cell wall play prominent roles in pore formation, which ultimately leads to bacterial cell death.44,45
Given the rise in antibacterial resistance, researchers have turned to AMPs as a source of inspiration. Incorporating key features of these peptides into synthetic scaffolds offer more structural options for tuning chemical compositions for improved performance.15,25,45,50–54 These molecules are often referred to as synthetic mimics of antimicrobial peptides (SMAMPs). One example from DeGrado and coworkers demonstrated the use of β-peptides, which mimic the α-helical structures and amphiphilicity of many AMPs.55 Although many of these molecules contained α-helical structures, linear β-peptides made by Gellman and coworkers and β- and ϒ-peptides made by Seebach and coworkers demonstrated that α-helical structures were not necessary for potent antimicrobial properties.56,57 These studies paved the way for the development of synthetic mimics with completely abiotic scaffolds. Such molecules, designed by Tew and coworkers, used facially amphiphilic triaryl scaffolds in which the hydrophobic content and cationic charge content (ammonium or guanidinium groups) could be tuned for improved antimicrobial properties and selectivities.58–61 By converting AMP designs to simple, synthetic scaffolds, production time and costs are considerably reduced.51 In addition, peptide in vivo limitations, such as proteolysis, tissue distribution, and toxicity, are overcome and robust in vivo antibacterial activity against drug resistant infections has been demonstrated, specifically with a compound in phase II clinical trials.51 An important new development was the demonstration that polymers could be designed with AMP-like biological activity.15,17,23,50,53,54,62–72 Unlike proteins and peptides, which typically have a single, exact molar mass, synthetic polymers, even when termed monodisperse, are characterized by a distribution of molecular weights. Although it is unclear how antimicrobial activity trends with dispersity, this opens a wider range of molecules and chemistries that can be used for the development of synthetic antimicrobial mimics.
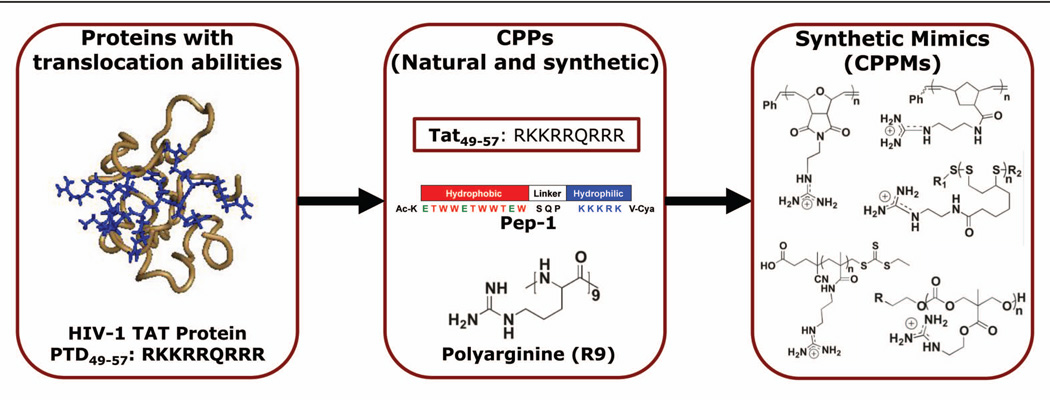
By using the same process that gave rise to SMAMPs, researchers have used proteins and peptide sequences as inspiration for the design of the next generation of delivery reagents (Figure 1).23 This is an area of particular interest since delivery of therapeutic agents is an ongoing challenge. Although there is an increasing demand for new treatments and treatment options, the field lacks a clear understanding of how to efficiently and reliably deliver bioactive molecules across cellular membranes, especially as therapies move increasingly toward more complex biologics.73–79 Nature, however, is already capable of designing proteins that can perform these functions. One example is HIV-1 TAT (trans-activator of transcription) protein, which is responsible for the spread of the virus80,81 This protein, along with others, contain a region referred to as a protein transduction domain (PTD) that is responsible for their abilities to enter cells.82–84 The study of TAT and other PTDs have subsequently led to the development of cell-penetrating peptides (CPPs), which are peptides that are capable of delivering cargo, such as small molecules, siRNA, pDNA, antibodies, and proteins, into cells via covalent or non-covalent interactions.85–90 Two examples of such molecules include TAT49–57, which is a guanidinium-rich sequence, and Pep-1, which has a segregated architecture similar to a block copolymer.83,84,90–92 While both peptides are cation-rich, Pep-1 also incorporates a hydrophobic segment that is thought to further aid in cellular uptake.
Figure 1.
The progression from proteins and peptides to guanidinium-rich CPPMs.
Despite the development of many CPPs such as Pep-1, which is now commercially available through Active Motif as Chariot™, moving away from the peptide scaffold offers distinct advantages. Most peptides are prepared by solid phase synthesis, which is both costly and time consuming because amino acids need to be sequentially added using a series of deprotection, addition, wash, and protection steps. Switching to a completely abiotic backbone allows delivery agents to be made cheaper and potentially in larger quantities. In addition, a non-peptide-based system offers many more structural options in terms of chemical compositions and molecular arrangements because it is not restricted to the incorporation of natural amino acids.23 This expanded chemistry toolset is expected to generate more efficient structures than their natural peptide analogs.23
This review aims to document the very early development of cell-penetrating peptide mimics (CPPMs), in particular ones based on polymeric scaffolds. A number of recent reports suggest this area will develop similarly to SMAMPs and will provide new tools for biology and perhaps new delivery opportunities for society.23,24 A summary of the early CPP work is highlighted followed by an overview of polymeric CPPMs developed to date.
CELL-PENETRATING PEPTIDES (CPPs)
CPPs are a class of peptides that can facilitate the delivery of various cargoes into cells.85–90 These peptides are generally 7–30 amino acid residues in length and cation-rich, usually containing multiple arginine and/or lysine residues.85 In general, CPPs can be broken down into three broad categories: protein-derived, chimera-derived, and synthetic. These classes of peptides are summarized in the following subsections and in Table 1.85
Table 1.
Summary of Cell Penetrating Peptides (CPPs) color-coded based on classification into three categories: protein-derived (top, pink), chimera (middle, blue), and synthetic (bottom, yellow).
| Peptide | Sequence | Derivation | Total # of charged residues |
Cargo | Alternative Classification |
|---|---|---|---|---|---|
| HIV-1 TAT49–5774,93 | RKKRRQRRR | Human Immunodeficiency Virus | 8 | protein, peptides, siRNA | Non-amphipathic |
| Penetratin94 | RQIKIWFQNRRMKWKK | Drosophila homeoprotein antennapedia | 7 | protein, PNA, siRNA | Secondary amphipathic |
| pVEC95 | LLIILRRRIRKQAHAHSK | Murine VE-cadherin | 6 | protein, peptides | Secondary amphipathic |
| VP2296 | NAKTRRHERRRKLAIER | Herpes simplex virus | 8 | protein | Secondary amphipathic |
| Pep-190–92,97 | KETWWETWWTEWSQPKKKRKV | NLS from SV40 T-antigen and a Tryptophan-rich domain | 5 | protein, peptides | Primary amphipathic |
| MPG91,98,99 | GALFLGFLGAAGSTMGAWSQPKKKRKV | Hydrophbic domain from fusion sequence of HIV gp41 and NLS of SV40 T-antigen | 5 | siRNA, plasmids | Primary amphipathic |
| Transportan (TP10)100,101 | AGYLLGKINLKALAALAKKIL | Galanin and mastoparan | 4 | protein, PNA, siRNA | Primary amphipathic |
| M918102 | MVTVLFRRLRIRRACGPPRVRV | Tumor suppressor protein p14ARF | 7 | Proteins, PNA | Secondary amphipathic |
| YTA2103 | YTAIAWVKAFIRKLRK | MMP cleavage site | 5 | small molecules | Secondary amphipathic |
| YTA4103 | IAWVKAFIRKLRKGPLG | MMP cleavage site | 5 | small molecules | Secondary amphipathic |
| MAP104,105 | KLALKLALKALKAALKLA | Synthetic | 5 | small molecules, plasmids | Secondary amphipathic |
| Polyarginine83,84,89,106 | R5–15 | HIV-1 TAT | 5–15 | protein, peptides, siRNA | Non-amphipathic |
| CADY107,108 | GLWRALWRLLRSLWRLLWRA | Derived from PPTG1 peptide | 5 | protein, peptides, siRNA | Secondary amphipathic |
PROTEIN-DERIVED CPPS
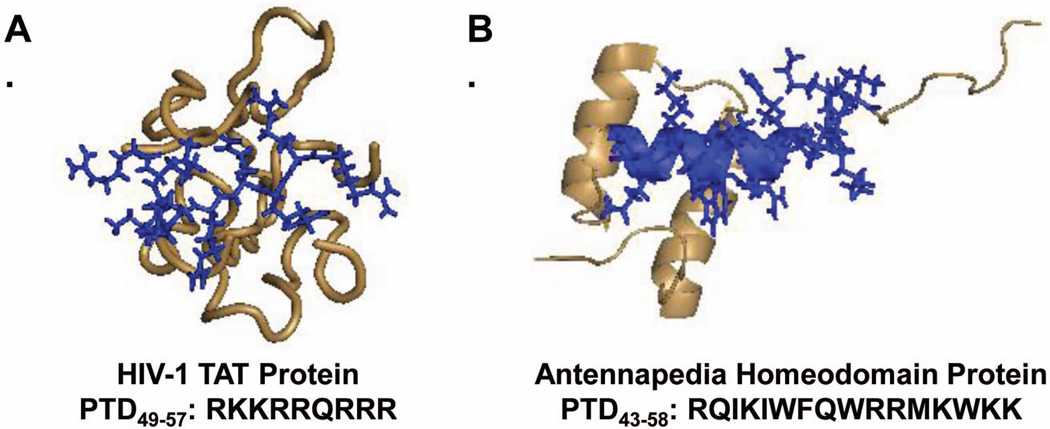
Protein-derived CPPs are based on known sequences from naturally occurring proteins.85 The specific sequences used for these CPPs are generally derived from the protein transduction domains (PTDs) of these molecules. Disruptions in these sequences often lead to partial or complete uptake inhibition.82,84,109 Two examples of protein-derived CPPs are TAT49–57 and the Antennapedia homeodomain protein derivative Penetratin. These proteins are modeled in Figure 2 with their PTDs highlighted in blue.
Figure 2.
Models of HIV-1 TAT and antennapedia homeodomain protein with their protein transduction domains modeled in blue.
In 1988, Green and Loewenstein, as well as Frankel and Pabo, independently reported that HIV-1 TAT had the unique ability to translocate into and out of cells.80,81 Then, in 1994, Fawell et al. tested the delivery efficiencies of two truncated HIV-1 TAT sequences: TAT1–72 and TAT38–72.110 The former was examined because it was thought to be involved in protein binding and cellular uptake and the latter was examined because it lacked the cysteine-rich region (residues 22–37).110 Both sequences were able to deliver proteins, demonstrating that the entire protein sequence was not required for efficient delivery.110 This study also compared these sequences to shorter peptides, Tat37–58 and Tat47–58, which were also able to deliver proteins to cells.110 Following this study, in 1997, Vives et al. studied four HIV-1 TAT-derived sequences to test the effects of altering the α-helical and basic regions on cellular uptake.82 Uptake data illustrated that the α-helix was not required for cellular uptake but that there could be no interruptions in the basic domain.82
In 2000, Wender et al. also studied the TAT peptide, as well as polyarginine. They confirmed that the basic region, amino acids 49–57, must be completely preserved in order for it to maintain its function.82,84 Truncating the sequence or substituting individual amino acids with alanine residues all resulted in a reduction in uptake efficiency. By studying Tat49–57 sequences that were synthesized with D-amino acids and/or in reverse, they also illustrated that backbone chirality, hydrogen bonding, and overall peptide backbone were not critical for efficacy.84
Similar studies were also performed for the antennapedia homeodomain protein. This protein contains approximately 60 amino acids, which fold to give three α-helices, and is a transcription factor that aids in DNA binding in Drosophila.111 In 1991, it was discovered that this antennapedia homeodomain protein could translocate into cells and that the third α-helix was important for the its cellular uptake ability.111 Similar studies to those performed on HIV-1 TAT, demonstrated that the entire protein sequence was not required for cellular uptake, just the 3rd α-helix and that backbone chirality, hydrogen bonding, and peptide secondary structure were not also required for uptake.109,112 This sequence of amino acids required for uptake has since been referred to as Penetratin.109 Other CPPs inspired by proteins include pVEC and VP22.95,96
Through studying HIV-1 TAT, antennapedia homeodomain protein, and their structural derivatives, it became apparent that full protein sequences, protein secondary structures, and peptide-based backbones are not necessarily required for efficient cellular uptake but that the cationic charge content was absolutely critical.82–84,94,109,110,112 These results opened up the possibility of designing synthetic mimics.
CHIMERA-DERIVED CPPS
As an alternative to using shortened TAT sequences to overcome some of the limitations of using the full TAT-protein, amphiphilic peptides with improved stability were developed based on a chimeric scaffold.90 Chimera-derived CPPs are fusions of two or more naturally occurring protein or peptide sequences.85 Most often, they are the combination of sequences that enable specific protein functions, such as nuclear localization sequences (NLSs) and signaling sequences.85 One of the first chimera-derived CPPs reported in the literature was Transportan (AGYLLGKINLKALAALAKKIL-NH2), which was a fusion of the neuropeptide galanin1–13 and the wasp venom peptide mastoparan.100,101,113,114
One of the most popular examples of a chimera-derived CPP is Pep-1.90–92 This peptide has a block copolymer-like structure with a tryptophan-rich hydrophobic domain that is segregated from a lysine-rich hydrophilic domain by a linker and is based on the NLS of the simian virus 40 (SV-40) large T antigen, as well as on the reverse transcriptase of HIV.90–92 Pep-1 is considered to be a primary amphipathic peptide because the hydrophilic/hydrophobic segregation is not dependent on the secondary structure of the peptide.90–92 Although stability is often an issue with peptide-based carriers, Pep-1 has an acetylated N-terminus and a cystamide C-terminus to improve shelf life. Since the development of Pep-1, structural variations have been made to try to enhance delivery of proteins, peptides, and peptide nucleic acids.91 In addition, Divita and coworkers have developed MPG for nucleic acid delivery, which is structurally similar CPP with a primary amphipathic and alpha-helical structure.91,98,99 This CPP is commercially available as DeliverX™ through Panomics.91,98,99
In addition to their amphiphilicity, what set Transportan, Pep-1, and MPG apart from others CPPs was their ability to deliver cargo using non-covalent attachment.90–92,99–101,113,114 Many CPPs, such TAT49–57 and Penetratin, require covalent attachment for efficient cargo delivery.81,82,110–112 By developing a non-covalent system, the carrier can simply be mixed with its cargo to form a transient complex that can dissociate upon entry into cells. Although this presents in vivo limitations because of the high risk of non-specific binding, it could mean less complex synthetic procedures and fewer experiments to ensure activity is not lost upon chemical conjugation.
SYNTHETIC CPPS
Synthetic CPPs are based on a peptide backbone but are not derived from naturally occurring protein or peptide sequences.85 CPPs that fall into this category include polyarginine; Amphiphatic Model Peptide (MAP), which is a lysine-rich secondary amphiphatic peptide; and CADY, which is arginine-rich and self-assembles to yield a facially amphiphilic structure.104,115 While polyarginine does not require secondary structure or self assembly for delivery, the delivery activities of MAP and CADY are highly dependent on their secondary structures.104,115 Disruptions in the helices or self assembly lead to diminished delivery efficiencies.104,115 The primary focus will be placed on polyarginine due to its importance in the development of synthetic guanidinium-rich molecular transporters.
“Polyarginine” and “oligoarginine” broadly define a series of peptide sequences that only contain arginine residues and, depending on length, may be synthetically easier to prepare than TAT49–57.80–82,84,110 As part of Wender et al’s study in 2000, polyarginine (lengths = five to nine resides) was shown to outperform TAT49–57, with longer sequences performing better than shorter ones.84 Polyarginine sequences were also compared to their D enantiomers and their corresponding peptoids.84 Both sets led to better cellular uptake. This indicated that cellular uptake was not dependent on hydrogen bonding present in the peptide backbone and that the peptide backbone may not be necessary.84 Another study from Mitchell et al. in 2000 showed that guanidinium groups led to superior uptake efficiencies compared to other possible cationic residues.83 They also demonstrated that cellular uptake increased as the number of arginine residues increased up to 15 and that further increases in arginine content led to decreased cellular uptake.83 This suggested that there is a critical number of arginine residues required for efficient uptake/delivery.
Unlike other hydrophobic-containing CPPs such as Pep-1, MPG, MAP, and CADY, which contain hydrophobic components to aid in cellular uptake, polyarginine is purely hydrophilic. To further explore polyarginine’s internalization efficiencies, Matile and coworkers studied the effect counter ions have on cellular uptake.106 In 2005, they reported that pairing polyarginine with bulky, aromatic activators, such as pyrenebutyrate, led to better peptide activity. This study suggested that there was some intrinsic benefit to having a hydrophobic component in addition to cationic charge as opposed to just having cationic charge.106
Through systematic studies with polyarginine and other synthetic peptides, it became clear that efficient delivery could be achieved without restricting structures to naturally occurring sequences.83,84,106 Not only did structure-activity relationships (SARs) reveal that these molecules could be optimized for more efficient delivery through altering molecular weights and/or structural compositions, but also that the peptide backbone was not necessarily essential for successful delivery. This set the stage for the development of tunable delivery vehicles based on novel, abiotic scaffolds.
ALTERNATIVE CLASSIFICATION OF CPPs
As an alternative to the afore mentioned CPP classification system, CPPs can be classified based on their distribution of hydrophilic/cationic amino acids and hydrophobic amino acids.116 These categories include primary amphipathic, secondary amphipathic, and non-amphipathic CPPs.116,117 A summary of CPPs that fall into these categories can be found in the last column of Table 1. For primary amphipathic peptides, segregation of cationic and hydrophobic amino acids is based on the sequential order of amino acids. Examples of these peptides include Pep-1, MPG, and Transportan.90–92,98–100 Secondary amphipathic peptides, such as penetratin, MAP, pVEC, and M918, achieve the segregation of cationic and hydrophobic amino acids through their secondary structures, which enable them to either form α-helices or β-sheets.95,102,104,105,109,118 Lastly, non-amphipathic peptides contain mostly cationic residues, with no cationic/hydrophobic amino acid segregation including polyarginine and TAT49–57.82–84,89,110
SURVEY OF CELL PENETRATING PEPTIDE MIMCS
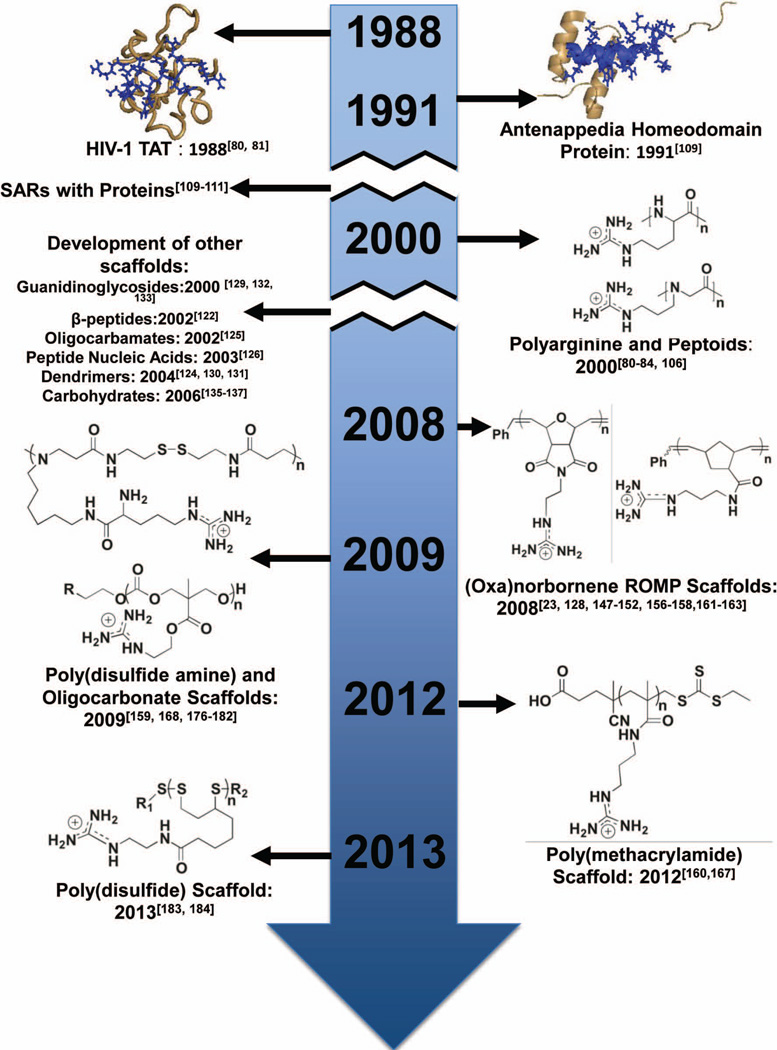
Although a large body of work focused on the development of peptide and peptidomimetic scaffolds for intracellular delivery,79,83,84,119–127 moving away from naturally occurring amino acid residues and peptide backbones offers many advantages. Solid phase peptide synthesis can be avoided, which saves both time and money in producing the desired delivery vehicles. Additionally, a non-peptidic system, such as a polymeric scaffold, permits the use of different chemistries and allows the chemical compositions and polymer architectures to be tailored for efficient cargo delivery.23 As a direct result of the easily tunable scaffolds, these materials are expected to be more potent and provide new models for fundamental studies. One prominent example of this is Tew and workers oxanorbornene-based mimics of R9 referred to as GMe9 and dG9.128 These polymeric-based mimics both contain nine repeat units, but dG9 has double the guanidine density (18 charges) than GMe9 (9 charges).128 Both of these mimics perform better than R9, with dG9 outperformed GMe9, suggesting that higher guanidinium density yields better cellular uptake.128 Branched peptide scaffolds have been developed127, which increase the guanidinium density. These molecules have similar uptake efficiencies to their linear analogs; however, they are synthetically more difficult to access and develop, making it harder to tune uptake and delivery efficiencies.127 Figure 3 charts the development and progress of guanidinium-rich molecular transporters. While this review highlights the use of guanidinium-rich, polymeric materials, other non-polymeric scaffolds have been developed and extensively studied, including guanidinioglycosides, dendrimers/branched materials, and carbohydrates.129–137
Figure 3.
Development timeline for key guanidinium-rich CPPM scaffolds.
Ring-opening Metathesis Polymerization-based Scaffold
In 2008, Tew and coworkers, as well as Kiessling and coworkers independently demonstrated that they could use ring-opening metathesis polymerization-based scaffolds to design materials with CPP-like activity. Tew and coworkers have used oxanorbornene-based systems for the design of their materials and Kiessling and coworkers have primarily used a norbornene-based system. Both groups selected ROMP because it offers fast, efficient, and functional group tolerant polymerizations.138–146 These polymerizations are often living, allowing for good control over molecular weights and polydispersities and allowing for the formation of more advanced architectures, such as block copolymers.23,129,147,148 Both groups’ work is summarized below.
Oxanorbornene-based CPPMs
The first ROMP-based CPPM developed by Tew and coworkers was polyguanidinium oxanorbornene (PGON).149,150 This molecule (Figure 4) was originally designed as an alternative to their amine-based antimicrobial agents.149 Although it had good antimicrobial activity against E. coli and S. aureus and was non-hemolytic, it was also not membrane-disruptive, suggesting it could also be used as a molecular transporter.149 Preliminary dye release experiments using model vesicle systems confirmed that PGON (degree of polymerization (DP) = 5–41) was able to induce dye release in a non-linear, molecular weight dependent fashion, further indicating it was membrane active and could be used as a molecular transporter.150
Figure 4.
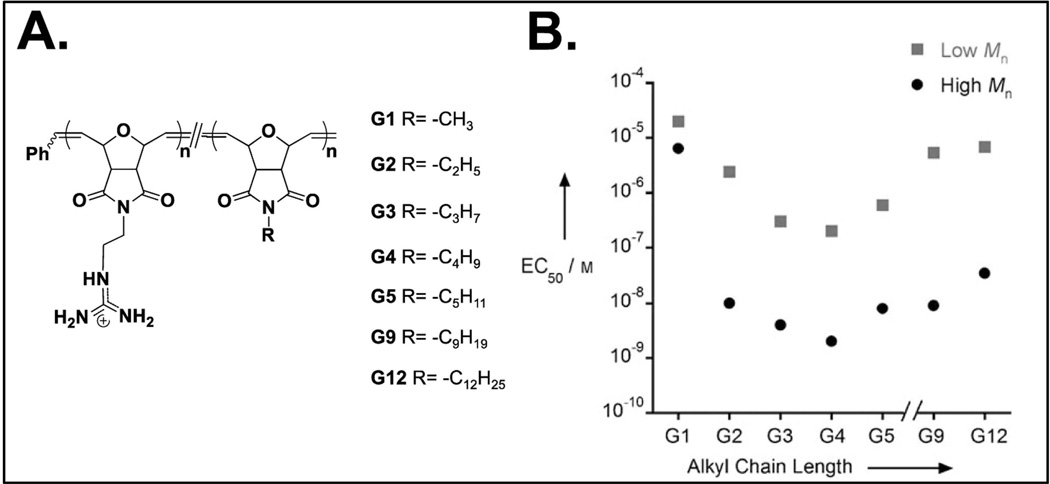
Oxanorbornene-based “self-activating” CPPMs. A) Molecular structures and B) Plot of EC50 vs. alkyl chain length.
In an effort to improve PGON’s activity, hydrophobic monomers, which contained alkyl chains ranging from methyl to dodecyl, were copolymerized with guanidinium-rich monomers to obtain a new series of CPPMs (Figure 4). Inspiration for this came from a Matile and coworkers study, which showed that hydrophobic counter ion activators improve cellular uptake of polyarginine.106 The goal was to develop CPPMs that were “self-activating”, meaning they did not need external activators for improved efficiency.151 CPPM activity improved with increasing alkyl chain length up until the incorporation of the butyl chain (Figure 4)151 Beyond this, CPPM insolubility led to a reduction in polymer activity.151 All CPPMs, however, performed better than PGON, indicating the importance of incorporating hydrophobic moieties.149–151 As a follow up to this study, CPPMs were designed in order to assess the role of aromaticity.152 Aromatic groups were selected for study because they are common structural features in CPPs, such as Pep-1 and Penetratin, and because some of the best CPP activators contain aromatic groups.90–92,106,109,112 Also, many cellular components contain aromatic groups, such as transmembrane proteins, which use aromatic amino acids to stabilize the interface between the hydrophilic and hydrophobic portions of the protein.153,154 In addition, it has been shown that the flat, rigid shape of aromatic rings along with their quadrupole moments can aid in membrane interactions.155 The CPPMs containing phenyl rings were the most active in the series, thus indicating the importance of aromaticity for CPPM activity.152 Further studies looked to probe the role of π-electronics in these phenyl ring systems through the incorporation of electron donating and electron withdrawing groups. All CPPMs, regardless of the electron rich or electron poor ring incorporated, maintained similar membrane activities.156 This demonstrated both the limits of the structural tuning that could be performed in this system and the number of structural options available without loss of membrane activity.156
Tew and coworkers also designed CPPMs using di-armed oxanorbornene monomers. This system added more synthetic versatility as each monomer contains two functionalities that can be independently tuned.23 Homopolymers containing one guanidine group and one hydrophobic group (aliphatic, aromatic, electron rich/electron poor aromatic systems) per monomer were designed as a direct comparison to their imide counterparts. These polymers had similar activities compared to the imide system.151,156 In addition, further studies were aimed at understanding the role of functional group segregation. The results indicated improved delivery efficiencies of a block copolymer scaffold without loss of activity in the presence of serum compared to its homopolymer counterparts (Figure 5A).157 This block copolymer was designed to capture the guanidinium-rich nature of TAT49–57 and the amphiphilicity of Pep-1.82–84,90–92 Follow-up studies explored the functional group distribution using constitutional macromolecular isomers, which are polymers of the same degree of polymerization but different arrangements of their pendent groups, ranging from completely segregated to completely mixed.158 These arrangements were accessed through block (completely segregated), gradient (partially segregated), and homopolymerizations (mixed distribution).158 Although studies indicated that the homopolymers enabled the best cellular uptake of the polymers, it is likely that block copolymers will deliver cargo more efficiently, as suggested by the literature.148,157,159,160
Figure 5.
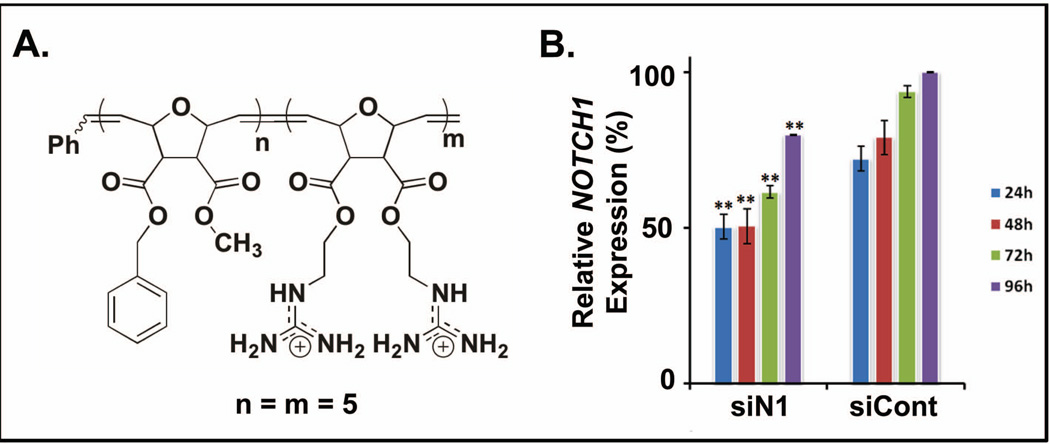
Delivery of siRNA to NOTCH1 into human peripheral blood mononuclear cells (PBMCs). A. Block copolymer structure used for delivery. B. Percent relative protein expression as a function of time for PBMCs that received siRNA to NOTCH1 (siN1) and PBMCs that received a scrambled, negative control (siCont).
While early studies with PGON strictly used model vesicle systems to assess membrane activity, di-armed homopolymers were used to assess the effect of guanidinium density on cellular uptake in three different cell lines: HEK293T, CHO, and Jurkat T cells.128 Although these polymers could enter all cell types with low cell death, uptake efficiencies were highly cell line dependent.128 In all cases, these CPPMs outperformed polyarginine (R9).128 These studies were extended to explore the constitutional macromolecular isomers, wherein cellular uptake was the best for the homopolymer tested and uptake efficiencies were found to be cell-type dependent. To further these studies and probe the effect of architecture when delivering biologically relevant cargo, a block copolymer was synthesized containing a 1:1 (n=m=5) ratio of hydrophobic to cationic monomers (Figure 4).157,161 This block copolymer was shown to deliver FITC-tagged siRNA and bioactive siRNA to NOTCH1 to Jurkat T cells and human peripheral blood mononuclear cells (hPBMCs), respectively, with > 90% viability.157 Delivery efficiencies were not altered by the presence of serum proteins in the media. The NOTCH1 knockdown results were noteworthy because 50% knockdown of an active gene was reported (Figure 5).157 This siRNA study represents proof of concept work that explored the delivery capabilities of this type of CPPM. Additional SARs studies are underway to develop design parameters for efficient siRNA delivery using these molecular transporters. These studies will also be expanded in order to develop design parameters for other biologically relevant cargo.23
Norbornene-based CPPMs
Kiessling and coworkers developed a norbornene-based scaffold.147,148 Single-armed norbornene monomers used for these materials initially contained succinimidyl ester moieties, which provided handles for post-polymerization functionalization. Polymeric materials could be reacted with N-(3-aminopropyl)guanidine in the presence of N-methyl morpholine to achieve near complete replacement of the succinimidyl ester pendent groups. This scaffold is based on previously published work.162 The guanidinium-rich scaffold builds on this initial work by terminating the polymers with an enol-ether, which allows for dye-labeling the polymer chains for easy tracking of the materials during cellular experiments.147 Uptake of these labeled molecules in HeLa cells was monitored using fluorescence imaging and results indicated that the polymer was trapped in endosomes, as noticed by punctate fluorescence, with some polymer dispersed in the cytosol.147 Greater than 95% viability was observed for all polymer concentrations tested (up to 5µM).147
Given that after post-polymerization functionalization only homopolymer and random copolymer architectures could be accessed, follow-up work looked at the formation of block copolymers.147,148 Two monomers were developed that could be modified post-polymerization by chemospecific methods, one succinimidyl ester-containing monomer and one alpha chloroacetamide-containing monomer.148 These monomers were polymerized sequentially in order to yield a block copolymer template. Based on the different chemistries involved, this template could be modified post-polymerization using separate reactions to yield segregation of attached functional groups.148 High degrees of conversion were demonstrated for both starting functional groups.148 Internalization of dye labeled block copolymers was demonstrated using HeLa cells, with cellular uptake following similar patterns to their previously reported homopolymer counterparts.148
More recently, Kiessling and coworkers developed a completely degradable ROMP scaffold using oxazinone-based monomers.163 Authors demonstrated that a wide range of functional groups could be incorporated into these scaffolds, making them potential candidates for drug delivery and biomaterials applications.163–166
Polymethacrylamides
In 2012, McCormick and coworkers reported the synthesis of guanidinium-rich polymethacrylamides using aqueous reversible addition fragmentation chain transfer (aRAFT).160 This synthetic method was advantageous for the synthesis of materials because polymerizations by this method are well-controlled and the guanidinium-containing monomers could be polymerized without protecting groups.160 Homopolymers were synthesized using a N-[3-(dimethylaminopropyl) methacrylamide monomer and copolymers were synthesized using a N-[3-(dimethylaminopropyl) methacrylamide and N-[2-(hydroxypropyl) methacrylamide monomer.160 Similar to polymers generated by the Tew and Wender groups, both sets of polymers were readily able to enter KB cells, with block copolymers out performing their homopolymer counterparts.148,157,159,160 Authors note that the good control over the polymerization technique could enable this platform to be used for bioactive cargo delivery.160 More recently, Peneva and coworkers developed a series of guanidinium-rich, statistical copolymers using aRAFT as potential siRNA delivery reagents.167 Binding strength and competitive binding were studied but, to date, no cellular uptake or viability results have been reported.167
Oligocarbonates
Wender/Hedrick/Waymouth and coworkers developed a series of CPPMs based on oligocarbonate polymers.159,168 Molecules were synthesized using metal-free, ring-opening polymerization of cyclic carbonates.159,168 Initiators for these materials were designed such that the drug molecule or molecular probe to be delivered could be attached at the beginning of the polymerization to allow for simpler conjugation to various cargoes.168 These molecules were shown to be biodegradable under physiological conditions, with half-lives around eight hours. Cellular uptake in Jurkat T cells revealed that these polymers entered in a charge-dependent manner, with longer polymers outperforming their shorter counterparts. To assess whether a biologically active cargo could be delivered, proof-of-concept experiments were performed in luciferin was successfully delivered and shown to luminesce in HepG2 cells.
In a follow-up study, Wender/Hedrick/Waymouth and coworkers showed that they could tune their oligocarbonate structures through the incorporation of hydrophobic moieties for more efficient siRNA complexation and release.159 Experiments with a dual-functional reporter HaCaT cell line that expressed both enhanced green fluorescent protein (EGFP) and Tomato fluorescent protein (tdTOM) was utilized to show specificity of knockdown.159 Delivery of siRNA to tdTOM was shown to yield efficient reduction in tdTOM protein levels but to have a negligible effect on EGFP protein levels, thus demonstrating good knockdown specificity.159 Polymers were shown to perform better in serum free media (86% knockdown) than in serum-containing media (64%).159 Oligomers that incorporate longer alkyl chains (up to dodecyl) were also shown to out perform their counterparts that contained shorter alkyl chains.159 Shorter oligomer lengths were also shown to out perform their longer counterparts.159 Cell viability studies using the 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) assay demonstrated that the most active polymers showed significant toxicity at the higher concentrations tested (100 nM) but could be greatly improved by cutting the polymer concentrations in half (50 nM).159 Lastly, it was demonstrated that knockdown efficiencies and complex sizes could be tuned by mixing different oligomers.159 Overall Wender/Hedrick/Waymouth and coworkers have developed a versatile, biodegradable platform for efficient delivery of biologically active siRNA. This platform has since been expanded by Wender et al. to include glycerol-based monomers to allow for better control over oligocarbonate/siRNA complex stability.169
Poly(disulfide)s
Poly(disulfide)s are polymers that contain at least one disulfide bond in the polymer repeat unit structure.170 These materials are distinctly different from proteins and vulcanized rubber, which both utilize disulfide bonds for crosslinking.170 Poly(disulfide)s can serve as efficient delivery reagents for nucleic acids and proteins, releasing their cargo through reductive de-polymerization in the presence of glutathione. Initially poly(amido amines) (PAAs) containing disulfide bonds were explored, which were synthesized by reacting cationic and/or hydrophobic monomers with cystaminebisacrylamide using Michael addition.171,172 This platform is very functional group tolerant, allowing for the incorporation of a wide range of functionalities, which enabled fine-tuning of PAA structures for efficient uptake and delivery.173–175 S. W. Kim and coworkers, utilized a similar platform to design poly(disulfide amines) and their guanidinium-rich counterparts, which are often referred to as arginine-grafted bioreducible polymers (ABPs) and guanidinylated bioreducible polymers (GBPs).176–182 The ABPs and GBPs molecules led to higher transfection efficiencies when compared to their amine counterparts.177,179–182 ABPs have been explored for RNAi applications related to anti-angiogenesis gene therapy of tumors as well as ex vivo pDNA delivery vehicles for treatment of ischemic heart diseases.177,179–182
While most poly(disulfide) delivery reagents utilize non-covalent delivery strategies, Matile and coworkers developed an efficient method of generating cell penetrating poly(disulfide)s that are covalently attached to their cargo utilizing surface-initiated ring-opening disulfide-exchange polymerization.183,184 Many probes, drugs, and bioactive cargoes contain or can be easily made to contain thiol moieties, which makes them convenient initiating species for this polymerization method. Molecule formation and subsequent depolymerization in the presence of dithiothreitol (DTT) were monitored using dye-loaded vesicles.183,184 Fast, efficient delivery (5 minutes), and subsequent fast depolymerization rates to release cargo into the cytosol (1 minute) were demonstrated in HeLa cells.184 Low toxicities were demonstrated for all molecules tested up to 10µM.184 Molecular uptake mechanism was independent of the cargo but could be altered based on the hydrophobic/cationic content of the materials.183 The authors also suggested a thiol/counterion-mediated uptake mechanism to explain cell entry and how chemical compositions of the delivery vehicles change their ability to enter cells efficiently.184
MODES OF INTERNALIZATION FOR CPP(M)s
The mechanisms of CPP(M) uptake remains highly debated in the literature.79,116,119,123 Early studies suggested direct translocation as the primary mode of internalization, which refers to molecules crossing membranes and directly entering the cytosol.185–187 However, many of these observations were shown to be erroneous, primarily due to cell fixation, which permeabilizes cell membranes and allows extracellular cargo to be internalized.185–187 Additional modes of uptake have been considered, including engulfment of particular molecules by the cell’s membrane through forms of endocytosis, such as Clathrin-mediated endocytosis, caveolae-mediated endocytosis, macropinocytosis, and receptor-mediated endocytosis.116,119,123,187 Glycosaminoglycans and lipid compositions are also thought to play a role in uptake mechanism.119 Complicating matters even further, experimental conditions including, but not limited to, composition, cargo, concentration, and cell types as well as incubation temperatures and times may play distinct roles in modes of internalization.79,116,119,123
Extensive effort has been made to understand the primary modes of internalization. One common way to do so is to inhibit certain entry methods and to compare uptake and delivery efficiencies to those cells under normal conditions.188–190 A summary of common methods can be found in Table 2. It is important to keep in mind that inhibiting specific modes of uptake may actually cause the cell to up-regulate other modes of entry or lead to off-target effects.188–190 It is also important to note that CPPs likely enter cells through multiple methods at the same time, with the different possible modes of cellular uptake being highly cell-type dependent.116,191,192
Table 2.
| Inhibitor | Affected Pathways | Blocking Mechanism |
|---|---|---|
| Lowered Temperature (4°C)193 | Energy dependent | Reduces cell metabolism, inhibiting energy-dependent pathways |
| Sodium azide and 2-deoxy-D-glucose194 | Energy dependent | Depletes ATP |
| Cytochalasin D119 | Macropinocytosis | Promotes disassembly of the actin cytoskeleton and blocks actin polymerization |
| Wortmannin195 | Macropinocytosis and Clathrin-mediated endocytosis | Inhibitor of phosphatidylinositol 3-kinase inhibitor |
| 5-(N-ethyl-N-isopropylamiloride)119 | Macropinocytosis | Inhibitor of sodium-proton pump exchange |
| Chloroquine196,197 | Endosomal Escape | Promoted endosomal escape |
| Nocodazole198,199 | Macropinocytosis | Blocks actin polymerization and disrupts actin cytoskeleton |
| Chlorpromazine and sucrose200 | Clathrin-mediated endocytosis | Depletes clathrin and AP2 adapter protein complex |
| Dynasore201 | Clathrin-mediated endocytosis | Dynamin inhibitor |
| Methyl-β-Cyclodextrin202 | Clathrin-independent endocytosis | Cholesterol extraction from membrane |
In addition, colocalization studies, which look at where polymer/cargo complexes end up in cells, and biophysical methods, which look at interactions between polymer/cargo complexes with lipid membranes, have also been utilized to elucidate uptake mechanism.106,151,152,203–211 To date, no one method has been shown to clearly document all possible and prominent methods of internalization.79,116 Often many methods need to be taken together in order to begin to understand what is happening in the cellular environment. This challenging problem is an area of active research.
PERSPECTIVES AND CONCLUSIONS
Structure-activity relationships with proteins led to essential design features for CPPs and enabled protein mimics to be realized. The extension to polymers represents another critical step in our fundamental understanding and is expected to produce some of the most useful CPPMs. Delivery of proteins and other biologics is going to be a central theme looking forward, which means delivery systems must be able to meet the challenges associated with these complex molecules. CPPMs appear to have an important role to play in this area, having already demonstrated their ability to perform in the arena of new immunological research and fundamental cell biology. Therapeutic delivery for the treatment of human disease, though a more difficult and complex issue, remains the ultimate goal. While the challenge is daunting, CPPMs are a promising technology undergoing continual refinement and offering many potential advantages.
ACKNOWLEDGEMENTS
The work was supported by the NIH (T32 GMO8515) and NSF (CHE-0910963). The authors would also like to thank Ms. Coralie Backlund, Ms. Kelly McLeod, and Mr. Nicholas Posey for their feedback on early drafts of this manuscript.
REFERENCES
- 1.Jeffrey GA, Saenger W. Hydrogen Bonding in Biological Structures. Berlin: Springer-Verlag; 1991. [Google Scholar]
- 2.Fletcher S, Hamilton AD. Curr Opin Chem Biol. 2005;9:632–638. doi: 10.1016/j.cbpa.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Orner BP, Ernst JT, Hamilton AD. J Am Chem Soc. 2001;123:5382–5383. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 4.Fairlie DP, West ML, Wong AK. Curr Med Chem. 1998;5:29–62. [PubMed] [Google Scholar]
- 5.Weiser PT, Chang CY, McDonnell DP, Hanson RN. Bioorg Med Chem. 2014;22:917–926. doi: 10.1016/j.bmc.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayatunga MK, Thompson S, Hamilton AD. Bioorg Med Chem Lett. 2014;24:717–724. doi: 10.1016/j.bmcl.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Barnard A, Long K, Yeo DJ, Miles JA, Azzarito V, Burslem GM, Prabhakaran P, T AE, Wilson AJ. Org Biomol Chem. 2014;12:6794–6799. doi: 10.1039/c4ob00915k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard A, Long K, Martin HL, Miles JA, Edwards TA, Tomlinson DC, Macdonald A, Wilson AJ. Angew Chem Int Ed Engl. 2015;54:2960–2965. doi: 10.1002/anie.201410810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 10.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev. 2001;101:3893–4012. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 11.Ernst JT, Bercerril J, Park HS, Yin H, Hamilton AD. Angew Chem Int Ed Engl. 2003;42:535–539. doi: 10.1002/anie.200390154. [DOI] [PubMed] [Google Scholar]
- 12.Saraogi I, Hamilton AD. Chem Soc Rev. 2009;38:1726–1743. doi: 10.1039/b819597h. [DOI] [PubMed] [Google Scholar]
- 13.Jones TV, Blatchly RA, Tew GN. Org Lett. 2003;5:3297–3299. doi: 10.1021/ol0352254. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel GJ, Tew GN. Org Biomol Chem. 2008;6:417–423. doi: 10.1039/b714490n. [DOI] [PubMed] [Google Scholar]
- 15.Tew GN, Scott RW, Klein ML, Degrado WF. Acc Chem Res. 2010;43:30–39. doi: 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stigers KD, Soth MJ, Nowick JS. Curr Opin Chem Biol. 1999;3:714–723. doi: 10.1016/s1367-5931(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 17.Slutsky MM, Phillip JS, Tew GN. N J Chem. 2008;32:670–675. [Google Scholar]
- 18.Martinek TA, Fulop F. Chem Soc Rev. 2012;41:687–702. doi: 10.1039/c1cs15097a. [DOI] [PubMed] [Google Scholar]
- 19.Jones TV, Slutsky MM, Tew GN. N J Chem. 2008;32:676–679. [Google Scholar]
- 20.Horne WS, Gellman SH. Acc Chem Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnt L, Tew GN. Macromolecules. 2004;37:1283–1288. [Google Scholar]
- 23.Sgolastra F, deRonde BM, Sarapas JM, Som A, Tew GN. Acc Chem Res. 2013 doi: 10.1021/ar400066v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanzl EG, Trantow BM, Vargas JR, Wender PA. Acc Chem Res. 2013;46:2944–2954. doi: 10.1021/ar4000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brogden KA. Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 26.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 27.Hancock RE, Diamond G. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 28.Benko-Iseppon AM, Galdino SL, Calsa T, Jr, Kido EA, Tossi A, Belarmino LC, Crovella S. Curr Protein Pept Sci. 2010;11:181–188. doi: 10.2174/138920310791112075. [DOI] [PubMed] [Google Scholar]
- 29.Hancock RE, Sahl HG. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 30.Wilmes M, Sahl HG. Int J Med Microbiol. 2014;304:93–99. doi: 10.1016/j.ijmm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Som A, Vemparala S, Ivanov I, Tew GN. Biopolymers. 2008;90:83–93. doi: 10.1002/bip.20970. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Gordon VD, Trinkle DR, Schmidt NW, Davis MA, DeVries C, Som A, Cronan JE, Jr, Tew GN, Wong GC. Proc Natl Acad Sci U S A. 2008;105:20595–20600. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vriens K, Cammue BP, Thevissen K. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shai Y, Makovitzky A, Avrahami D. Curr Protein Pept Sci. 2006;7:479–486. doi: 10.2174/138920306779025620. [DOI] [PubMed] [Google Scholar]
- 35.Shai Y. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt NW, Tai KP, Kamdar K, Mishra A, Lai GH, Zhao K, Ouellette AJ, Wong GC. J Biol Chem. 2012;287:21866–21872. doi: 10.1074/jbc.M112.358721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GC. J Am Chem Soc. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira MD, Franco OL, Nascimento JM, de Melo CP, Andrade CA. Curr Protein Pept Sci. 2013;14:543–555. doi: 10.2174/13892037113149990070. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Tailhades J, O'Brien-Simpson NM, Separovic F, Otvos L, Jr, Hossain MA, Wade JD. Amino Acids. 2014;46:2287–2294. doi: 10.1007/s00726-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 40.Lemeshko VV. Arch Biochem Biophys. 2014;545:167–178. doi: 10.1016/j.abb.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Harrison PL, Abdel-Rahman MA, Miller K, Strong PN. Toxicon. 2014;88:115–137. doi: 10.1016/j.toxicon.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hancock RE, Rozek A. FEMS Microbiol Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 43.Bolintineanu DS, Kaznessis YN. Peptides. 2011;32:188–201. doi: 10.1016/j.peptides.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devine DA, Hancock RE. Curr Pharm Des. 2002;8:703–714. doi: 10.2174/1381612023395501. [DOI] [PubMed] [Google Scholar]
- 45.Yeaman MR, Yount NY. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 46.Polyansky AA, Chugunov AO, Vassilevski AA, Grishin EV, Efremov RG. Curr Protein Pept Sci. 2012;13:644–657. doi: 10.2174/138920312804142147. [DOI] [PubMed] [Google Scholar]
- 47.Mangoni ML, Shai Y. Cell Mol Life Sci. 2011;68:2267–2280. doi: 10.1007/s00018-011-0718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen LT, Haney EF, Vogel HJ. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Scocchi M, Tossi A, Gennaro R. Cell Mol Life Sci. 2011;68:2317–2330. doi: 10.1007/s00018-011-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi H, Palermo EF, Yasuhara K, Caputo GA, Kuroda K. Macromol Biosci. 2013;13:1285–1299. doi: 10.1002/mabi.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott RW, DeGrado WF, Tew GN. Curr Opin Biotechnol. 2008;19:620–627. doi: 10.1016/j.copbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotem S, Mor A. Biochim Biophys Acta. 2009;1788:1582–1592. doi: 10.1016/j.bbamem.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 53.Kuroda K, Caputo GA. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2013;5:49–66. doi: 10.1002/wnan.1199. [DOI] [PubMed] [Google Scholar]
- 54.Gabriel GJ, Som A, Madkour AE, Eren T, Tew GN. Mater Sci Eng R Rep. 2007;57:28–64. doi: 10.1016/j.mser.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamuro Y, Schneider JP, DeGrado WF. J Am Chem Soc. 1999;121:12200–12201. [Google Scholar]
- 56.Seebach D, Beck AK, Bierbaum DJ. Chem Biodivers. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 57.Porter EA, Weisblum B, Gellman SH. J Am Chem Soc. 2002;124:7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 58.Thaker HD, Cankaya A, Scott RW, Tew GN. ACS Med Chem Lett. 2013;4:481–485. doi: 10.1021/ml300307b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thaker HD, Sgolastra F, Clements D, Scott RW, Tew GN. J Med Chem. 2011;54:2241–2254. doi: 10.1021/jm101410t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thaker HD, Som A, Ayaz F, Lui DH, Pan WX, Scott RW, Anguita J, Tew GN. J Am Chem Soc. 2012;134:11088–11091. doi: 10.1021/ja303304j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Som A, Navasa N, Percher A, Scott RW, Tew GN, Anguita J. Clin Vaccine Immunol. 2012;19:1784–1791. doi: 10.1128/CVI.00291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lienkamp K, Madkour AE, Musante A, Nelson CF, Nusslein K, Tew GN. J Am Chem Soc. 2008;130:9836–9843. doi: 10.1021/ja801662y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Badri ZM, Som A, Lyon S, Nelson CF, Nusslein K, Tew GN. Biomacromolecules. 2008;9:2805–2810. doi: 10.1021/bm800569x. [DOI] [PubMed] [Google Scholar]
- 64.Arnt L, Nusslein K, Tew GN. J Polym Sci A1. 2004;42:3860–3864. [Google Scholar]
- 65.Colak S, Nelson CF, Nusslein K, Tew GN. Biomacromolecules. 2009;10:353–359. doi: 10.1021/bm801129y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ilker MF, Nusslein K, Tew GN, Coughlin EB. J Am Chem Soc. 2004;126:15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- 67.Lienkamp K, Kumar KN, Som A, Nusslein K, Tew GN. Chem Eur J. 2009;15:11710–11714. doi: 10.1002/chem.200802558. [DOI] [PubMed] [Google Scholar]
- 68.Lienkamp K, Madkour AE, Kumar KN, Nusslein K, Tew GN. Chem Eur J. 2009;15:11715–11722. doi: 10.1002/chem.200900606. [DOI] [PubMed] [Google Scholar]
- 69.Madkour AE, Dabkowski JM, Nusslein K, Tew GN. Langmuir. 2009;25:1060–1067. doi: 10.1021/la802953v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rennie J, Arnt L, Tang H, Nusslein K, Tew GN. J Ind Microbiol Biotechnol. 2005;32:296–300. doi: 10.1007/s10295-005-0219-0. [DOI] [PubMed] [Google Scholar]
- 71.Ishitsuka Y, Arnt L, Majewski J, Frey S, Ratajczek M, Kjaer K, Tew GN, Lee KY. J Am Chem Soc. 2006;128:13123–13129. doi: 10.1021/ja061186q. [DOI] [PubMed] [Google Scholar]
- 72.Arnt L, Tew GN. J Am Chem Soc. 2002;124:7664–7665. doi: 10.1021/ja026607s. [DOI] [PubMed] [Google Scholar]
- 73.Park TG, Jeong JH, Kim SW. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Snyder EL, Dowdy SF. Pharm Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 75.Thomas M, Klibanov AM. Appl Microbiol Biotechnol. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 76.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deshayes S, Morris MC, Divita G, Heitz F. Cell Mol Life Sci. 2005;62:1839–1849. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fonseca SB, Pereira MP, Kelley SO. Adv Drug Deliv Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 79.Heitz F, Morris MC, Divita G. Brit J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Green M, Loewenstein PM. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 81.Frankel AD, Pabo CO. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 82.Vives E, Brodin P, Lebleu B. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 83.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 84.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lindgren M, Langel U. Cell-Penetrating Peptides: Methods and Protocols. 2011;683:3–19. doi: 10.1007/978-1-60761-919-2_1. [DOI] [PubMed] [Google Scholar]
- 86.Siprashvili Z, Reuter JA, Khavari PA. Mol Ther. 2004;9:721–728. doi: 10.1016/j.ymthe.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Opalinska JB, Gewirtz AM. Nat Rev Drug Discov. 2002;1:503–514. doi: 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- 88.Patel LN, Zaro JL, Shen WC. Pharm Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 89.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 90.Morris MC, Depollier J, Mery J, Heitz F, Divita G. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 91.Morris MC, Deshayes S, Heitz F, Divita G. Biol Cell. 2008;100:201–217. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- 92.Morris MC, Vidal P, Chaloin L, Heitz F, Divita G. Nucleic Acids Res. 1997;25:2730–2736. doi: 10.1093/nar/25.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 94.Joliot A, Prochiantz A. Nat Cell Biol. 2004;6:189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 95.Elmquist A, Lindgren M, Bartfai T, Langel U. Exp Cell Res. 2001;269:237–244. doi: 10.1006/excr.2001.5316. [DOI] [PubMed] [Google Scholar]
- 96.Elliott G, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 97.Gros E, Deshayes S, Morris MC, Aldrian-Herrada G, Depollier J, Heitz F, Divita G. Biochim Biophys Acta. 2006;1758:384–393. doi: 10.1016/j.bbamem.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 98.Simeoni F, Arvai A, Bello P, Gondeau C, Hopfner KP, Neyroz P, Heitz F, Tainer J, Divita G. Biochemistry. 2005;44:11997–12008. doi: 10.1021/bi050427o. [DOI] [PubMed] [Google Scholar]
- 99.Simeoni F, Morris MC, Heitz F, Divita G. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pooga M, Hallbrink M, Zorko M, Langel U. FASEB J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 101.Soomets U, Lindgren M, Gallet X, Hallbrink M, Elmquist A, Balaspiri L, Zorko M, Pooga M, Brasseur R, Langel U. BBA-Biomembranes. 2000;1467:165–176. doi: 10.1016/s0005-2736(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 102.El-Andaloussi S, Johansson HJ, Holm T, Langel U. Mol Ther. 2007;15:1820–1826. doi: 10.1038/sj.mt.6300255. [DOI] [PubMed] [Google Scholar]
- 103.Lindgren M, Rosenthal-Aizman K, Saar K, Eiriksdottir E, Jiang Y, Sassian M, Ostlund P, Hallbrink M, Langel U. Biochem Pharmacol. 2006;71:416–425. doi: 10.1016/j.bcp.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 104.Oehlke J, Scheller A, Wiesner B, Krause E, Beyermann M, Klauschenz E, Melzig M, Bienert M. BBA-Biomembranes. 1998;1414:127–139. doi: 10.1016/s0005-2736(98)00161-8. [DOI] [PubMed] [Google Scholar]
- 105.Wolf Y, Pritz S, Abes S, Bienert M, Lebleu B, Oehlke J. Biochemistry. 2006;45:14944–14954. doi: 10.1021/bi0606896. [DOI] [PubMed] [Google Scholar]
- 106.Nishihara M, Perret F, Takeuchi T, Futaki S, Lazar AN, Coleman AW, Sakai N, Matile S. Org Biomol Chem. 2005;3:1659–1669. doi: 10.1039/b501472g. [DOI] [PubMed] [Google Scholar]
- 107.Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R, Heitz F, Divita G. Mol Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurzawa L, Pellerano M, Morris MC. Biochim Biophys Acta. 2010;1798:2274–2285. doi: 10.1016/j.bbamem.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 109.Derossi D, Joliot AH, Chassaing G, Prochiantz A. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 110.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Proc Natl Acad Sci U S A. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Joliot A, Pernelle C, Deagostinibazin H, Prochiantz A. Proc Natl Acad Sci U S A. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Derossi D, Chassaing G, Prochiantz A. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 113.Deshayes S, Morris M, Heitz F, Divita G. Adv Drug Deliv Rev. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 114.Lindgren M, Hallbrink M, Elmquist A, Soomets U, Gallet X, Brasseur R, Zorko M, Langel U. Eur J Neurosci. 2000;12:48–48. [Google Scholar]
- 115.Crombez L, Morris MC, Deshayes S, Heitz F, Divita G. Curr Pharm Des. 2008;14:3656–3665. doi: 10.2174/138161208786898842. [DOI] [PubMed] [Google Scholar]
- 116.Madani F, Lindberg S, Langel U, Futaki S, Graslund A. J Biophys. 2011;2011:414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ziegler A. Adv Drug Deliv Rev. 2008;60:580–597. doi: 10.1016/j.addr.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 119.Copolovici DM, Langel K, Eriste E, Langel U. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 120.Fillon YA, Anderson JP, Chmielewski J. J Am Chem Soc. 2005;127:11798–11803. doi: 10.1021/ja052377g. [DOI] [PubMed] [Google Scholar]
- 121.Rothbard JB, Kreider E, Vandeusen CL, Wright L, Wylie BL, Wender PA. J Med Chem. 2002;45:3612–3618. doi: 10.1021/jm0105676. [DOI] [PubMed] [Google Scholar]
- 122.Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH. J Am Chem Soc. 2002;124:368–369. doi: 10.1021/ja017283v. [DOI] [PubMed] [Google Scholar]
- 123.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. Adv Drug Deliv Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wender PA, Kreider E, Pelkey ET, Rothbard J, Vandeusen CL. Org Lett. 2005;7:4815–4818. doi: 10.1021/ol051496y. [DOI] [PubMed] [Google Scholar]
- 125.Wender PA, Rothbard JB, Jessop TC, Kreider EL, Wylie BL. J Am Chem Soc. 2002;124:13382–13383. doi: 10.1021/ja0275109. [DOI] [PubMed] [Google Scholar]
- 126.Zhou P, Wang M, Du L, Fisher GW, Waggoner A, Ly DH. J Am Chem Soc. 2003;125:6878–6879. doi: 10.1021/ja029665m. [DOI] [PubMed] [Google Scholar]
- 127.Futaki S, Nakase I, Suzuki T, Youjun Z, Sugiura Y. Biochemistry. 2002;41:7925–7930. doi: 10.1021/bi0256173. [DOI] [PubMed] [Google Scholar]
- 128.Tezgel AO, Telfer JC, Tew GN. Biomacromolecules. 2011;12:3078–3083. doi: 10.1021/bm200694u. [DOI] [PubMed] [Google Scholar]
- 129.Baker TJ, Luedtke NW, Tor Y, Goodman M. J Org Chem. 2000;65:9054–9058. doi: 10.1021/jo001142e. [DOI] [PubMed] [Google Scholar]
- 130.Chung HH, Harms G, Seong CM, Choi BH, Min C, Taulane JP, Goodman M. Biopolymers. 2004;76:83–96. doi: 10.1002/bip.10597. [DOI] [PubMed] [Google Scholar]
- 131.Huang K, Voss B, Kumar D, Hamm HE, Harth E. Bioconjug Chem. 2007;18:403–409. doi: 10.1021/bc060287a. [DOI] [PubMed] [Google Scholar]
- 132.Luedtke NW, Baker TJ, Goodman M, Tor Y. J Am Chem Soc. 2000;122:12035–12036. [Google Scholar]
- 133.Luedtke NW, Carmichael P, Tor Y. J Am Chem Soc. 2003;125:12374–12375. doi: 10.1021/ja0360135. [DOI] [PubMed] [Google Scholar]
- 134.Moazed D, Noller HF. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 135.Maiti KK, Jeon OY, Lee WS, Chung SK. Chemistry. 2007;13:762–775. doi: 10.1002/chem.200600898. [DOI] [PubMed] [Google Scholar]
- 136.Maiti KK, Jeon OY, Lee WS, Kim DC, Kim KT, Takeuchi T, Futaki S, Chung SK. Angew Chem Int Ed Engl. 2006;45:2907–2912. doi: 10.1002/anie.200600312. [DOI] [PubMed] [Google Scholar]
- 137.Maiti KK, Lee WS, Takeuchi T, Watkins C, Fretz M, Kim DC, Futaki S, Jones A, Kim KT, Chung SK. Angew Chem Int Ed Engl. 2007;46:5880–5884. doi: 10.1002/anie.200701346. [DOI] [PubMed] [Google Scholar]
- 138.Bielawski CW, Benitez D, Grubbs RH. J Am Chem Soc. 2003;125:8424–8425. doi: 10.1021/ja034524l. [DOI] [PubMed] [Google Scholar]
- 139.Bielawski CW, Grubbs RH. Angew Chem Int Ed Engl. 2000;39:2903–2906. doi: 10.1002/1521-3773(20000818)39:16<2903::aid-anie2903>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 140.Bielawski CW, Grubbs RH. Prog. Polym. Sci. 2007;32:1–29. [Google Scholar]
- 141.Cannizzo LF, Grubbs RH. Macromolecules. 21:1961–1967. [Google Scholar]
- 142.Love JA, Morgan JP, Trnka TM, Grubbs RH. Angew Chem Int Ed Engl. 2002;41:4035–4037. doi: 10.1002/1521-3773(20021104)41:21<4035::AID-ANIE4035>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 143.Schrock RR, Hoveyda AH. Angew Chem Int Ed Engl. 2003;42:4592–4633. doi: 10.1002/anie.200300576. [DOI] [PubMed] [Google Scholar]
- 144.Schwab P, France MB, Ziller JW, Grubbs RH. Angew Chem Int Ed Engl. 1995;34:2039–2041. [Google Scholar]
- 145.Singh R, Czekelius C, Schrock RR. Macromolecules. 2006;39:1316–1317. [Google Scholar]
- 146.Trnka TM, Grubbs RH. Acc Chem Res. 2001;34:18–29. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]
- 147.Kolonko EM, Kiessling LL. J Am Chem Soc. 2008;130:5626-+. doi: 10.1021/ja8001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kolonko EM, Pontrello JK, Mangold SL, Kiessling LL. J Am Chem Soc. 2009;131:7327–7333. doi: 10.1021/ja809284s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gabriel GJ, Madkour AE, Dabkowski JM, Nelson CF, Nusslein K, Tew GN. Biomacromolecules. 2008;9:2980–2983. doi: 10.1021/bm800855t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hennig A, Gabriel GJ, Tew GN, Matile S. J Am Chem Soc. 2008;130:10338–10344. doi: 10.1021/ja802587j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Som A, Tezgel AO, Gabriel GJ, Tew GN. Angew Chem Int Ed Engl. 2011;50:6147–6150. doi: 10.1002/anie.201101535. [DOI] [PubMed] [Google Scholar]
- 152.Som A, Reuter A, Tew GN. Angew Chem Int Ed Engl. 2012;51:980–983. doi: 10.1002/anie.201104624. [DOI] [PubMed] [Google Scholar]
- 153.de Planque MR, Bonev BB, Demmers JA, Greathouse DV, Koeppe RE, 2nd, Separovic F, Watts A, Killian JA. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 154.Yau WM, Wimley WC, Gawrisch K, White SH. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 155.White SH, Wimley WC. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 156.deRonde BM, Birke A, Tew GN. Chem Eur J. 2015;21:3013–3019. doi: 10.1002/chem.201405381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tezgel AO, Gonzalez-Perez G, Telfer JC, Osborne BA, Minter LM, Tew GN. Mol Ther. 2013;21:201–209. doi: 10.1038/mt.2012.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sgolastra F, Minter LM, Osborne BA, Tew GN. Biomacromolecules. 2014;15:812–820. doi: 10.1021/bm401634r. [DOI] [PubMed] [Google Scholar]
- 159.Geihe EI, Cooley CB, Simon JR, Kiesewetter MK, Edward JA, Hickerson RP, Kaspar RL, Hedrick JL, Waymouth RM, Wender PA. Proc Natl Acad Sci U S A. 2012;109:13171–13176. doi: 10.1073/pnas.1211361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Treat NJ, Smith D, Teng C, Flores JD, Abel BA, York AW, Huang F, McCormick CL. ACS Macro Lett. 2012;1:100–104. doi: 10.1021/mz200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tezgel AO, Jacobs P, Telfer JC, Tew GN. 2015 Submitted. [Google Scholar]
- 162.Strong LE, Kiessling LL. J Am Chem Soc. 1999;121:6193–6196. [Google Scholar]
- 163.Fishman JM, Kiessling LL. Angew Chem Int Ed Engl. 2013;52:5061–5064. doi: 10.1002/anie.201300293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Smith D, Pentzer EB, Nguyen ST. Polym Rev. 2007;47:419–459. [Google Scholar]
- 165.Shoichet MS. Macromolecules. 2010;43:581–591. [Google Scholar]
- 166.Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Annu Rev Chem Biomol. 2010;1:149–173. doi: 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tabujew I, Freidel C, Krieg B, Helm M, Koynov K, Mullen K, Peneva K. Macromol Rapid Commun. 2014;35:1191–1197. doi: 10.1002/marc.201400120. [DOI] [PubMed] [Google Scholar]
- 168.Cooley CB, Trantow BM, Nederberg F, Kiesewetter MK, Hedrick JL, Waymouth RM, Wender PA. J Am Chem Soc. 2009;131:16401–16403. doi: 10.1021/ja907363k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wender PA, Huttner MA, Staveness D, Vargas JR, Xu AF. Mol Pharm. 2015;12:742–750. doi: 10.1021/mp500581r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bang EK, Lista M, Sforazzini G, Sakai N, Matile S. Chem Sci. 2012;3:1752–1763. [Google Scholar]
- 171.Hartmann L, Hafele S, Peschka-Suss R, Antonietti M, Borner HG. Macromolecules. 2007;40:7771–7776. [Google Scholar]
- 172.Hartmann L, Krause E, Antonietti M, Borner HG. Biomacromolecules. 2006;7:1239–1244. doi: 10.1021/bm050884k. [DOI] [PubMed] [Google Scholar]
- 173.Lin C, Blaauboer CJ, Timoneda MM, Lok MC, van Steenbergen M, Hennink WE, Zhong Z, Feijen J, Engbersen JF. J Control Release. 2008;126:166–174. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 174.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. J Control Release. 2007;123:67–75. doi: 10.1016/j.jconrel.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 175.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. Bioconjug Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 176.Kim SH, Jeong JH, Ou M, Yockman JW, Kim SW, Bull DA. Biomaterials. 2008;29:4439–4446. doi: 10.1016/j.biomaterials.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim TI, Kim SW. React Funct Polym. 2011;71:344–349. doi: 10.1016/j.reactfunctpolym.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Bioconjug Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kim SH, Jeong JH, Kim TI, Kim SW, Bull DA. Mol Pharm. 2009;6:718–726. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kim TI, Lee M, Kim SW. Biomaterials. 2010;31:1798–1804. doi: 10.1016/j.biomaterials.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim TI, Ou M, Lee M, Kim SW. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ou M, Kim TI, Yockman JW, Borden BA, Bull DA, Kim SW. J Control Release. 2010;142:61–69. doi: 10.1016/j.jconrel.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bang EK, Gasparini G, Molinard G, Roux A, Sakai N, Matile S. J Am Chem Soc. 2013;135:2088–2091. doi: 10.1021/ja311961k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gasparini G, Bang EK, Molinard G, Tulumello DV, Ward S, Kelley SO, Roux A, Sakai N, Matile S. J Am Chem Soc. 2014;136:6069–6074. doi: 10.1021/ja501581b. [DOI] [PubMed] [Google Scholar]
- 185.Lundberg M, Wikstrom S, Johansson M. Mol Ther. 2003;8:143–150. doi: 10.1016/s1525-0016(03)00135-7. [DOI] [PubMed] [Google Scholar]
- 186.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 187.Trabulo S, Cardoso AL, Mano M, Pedroso de Lima M. Pharmaceuticals. 2010;3:961–993. doi: 10.3390/ph3040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ivanov AI. Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 189.Ivanov AI. Methods Mol Biol. 2014;1174:3–18. doi: 10.1007/978-1-4939-0944-5_1. [DOI] [PubMed] [Google Scholar]
- 190.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC, Sanders NN, Braeckmans K. Mol Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Maiolo JR, Ferrer M, Ottinger EA. Biochim Biophys Acta. 2005;1712:161–172. doi: 10.1016/j.bbamem.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 192.Manceur A, Wu A, Audet J. Anal Biochem. 2007;364:51–59. doi: 10.1016/j.ab.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 193.Silverstein SC, Steinman RM, Cohn ZA. Annu Rev Biochem. 1977;46:669–722. doi: 10.1146/annurev.bi.46.070177.003321. [DOI] [PubMed] [Google Scholar]
- 194.Schmid SL, Carter LL. J Cell Biol. 1990;111:2307–2318. doi: 10.1083/jcb.111.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ohkuma S, Poole B. Proc Natl Acad Sci U S A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wibo M, Poole B. J Cell Biol. 1974;63:430–440. doi: 10.1083/jcb.63.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Samson F, Donoso JA, Heller-Bettinger I, Watson D, Himes RH. J Pharmacol Exp Ther. 1979;208:411–417. [PubMed] [Google Scholar]
- 199.Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. Mol Biol Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang LH, Rothberg KG, Anderson RG. J Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 202.Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Mol Biol Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sandgren S, Cheng F, Belting M. J Biol Chem. 2002;277:38877–38883. doi: 10.1074/jbc.M205395200. [DOI] [PubMed] [Google Scholar]
- 204.Schmidt NW, Lis M, Zhao K, Lai GH, Alexandrova AN, Tew GN, Wong GC. J Am Chem Soc. 2012;134:19207–19216. doi: 10.1021/ja308459j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sieczkarski SB, Whittaker GR. J Gen Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 206.Almeida PF, Pokorny A. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Almeida PF, Pokorny A. Methods Mol Biol. 2010;618:155–169. doi: 10.1007/978-1-60761-594-1_11. [DOI] [PubMed] [Google Scholar]
- 208.Perret F, Nishihara M, Takeuchi T, Futaki S, Lazar AN, Coleman AW, Sakai N, Matile S. J Am Chem Soc. 2005;127:1114–1115. doi: 10.1021/ja043633c. [DOI] [PubMed] [Google Scholar]
- 209.Sakai N, Futaki S, Matile S. Soft Matter. 2006;2:636–641. doi: 10.1039/b606955j. [DOI] [PubMed] [Google Scholar]
- 210.Sakai N, Matile S. J Am Chem Soc. 2003;125:14348–14356. doi: 10.1021/ja037601l. [DOI] [PubMed] [Google Scholar]
- 211.Sakai N, Takeuchi T, Futaki S, Matile S. Chembiochem. 2005;6:114–122. doi: 10.1002/cbic.200400256. [DOI] [PubMed] [Google Scholar]