Abstract
Background & Aims
In gastrointestinal muscles, KIT is predominantly expressed by interstitial cells of Cajal (ICC) and PDGFRA is expressed by so-called fibroblast-like cells. KIT and PDGFRA have been reported to be co-expressed in ICC precursors and gastrointestinal stromal tumors (GISTs), which originate from the ICC lineage. PDGFRA signaling has been proposed to stimulate growth of GISTs that express mutant KIT, but the effects and mechanisms of selective blockade of PDGFRA are unclear. We investigated whether inhibiting PDGFRA could reduce proliferation of GIST cells with mutant KIT via effects on the KIT-dependent transcription factor ETV1.
Methods
We studied 53 gastric, small intestinal, rectal, or abdominal GISTs collected immediately after surgery or archived as fixed blocks at the Mayo Clinic and University of California, San Diego. In human GIST cells carrying imatinib-sensitive and imatinib-resistant mutations in KIT, PDGFRA was reduced by RNA interference (knockdown) or inhibited with crenolanib besylate (a selective inhibitor of PDGFRA and PDGFRB). Mouse ICC precursors were retrovirally transduced to overexpress wild-type Kit. Cell proliferation was analyzed by methyltetrazolium, 5-ethynyl-2'-deoxyuridine incorporation, and Ki-67 immunofluorescence assays; we also analyzed growth of xenograft tumors in mice. Gastric ICC and ICC precursors, and their PDGFRA+ subsets, were analyzed by flow cytometry and immunohistochemistry in wild-type, Kit+/copGFP, Pdgfra+/eGFP and NOD/ShiLtJ mice. Immunoblots were used to quantify protein expression and phosphorylation.
Results
KIT and PDGFRA were co-expressed in 3%–5% of mouse ICC, 35%−44% of ICC precursors, and most human GIST samples and cell lines. PDGFRA knockdown or inhibition with crenolanib efficiently reduced proliferation of imatinib-sensitive and imatinib-resistant KIT+ETV1+PDGFRA+ GIST cells (half-maximal inhibitory concentration: IC50=5-32 nM), but not of cells lacking KIT, ETV1, or PDGFRA (IC50>230 nM). Crenolanib inhibited phosphorylation of PDGFRA and PDGFRB but not KIT. However, Kit overexpression sensitized mouse ICC precursors to crenolanib. ETV1 knockdown reduced KIT expression and GIST proliferation. Crenolanib downregulated ETV1 by inhibiting ERK-dependent stabilization of ETV1 protein and also reduced expression of KIT and PDGFRA.
Conclusions
In KIT-mutant GIST, inhibition of PDGFRA disrupts a KIT–ERK−ETV1−KIT signaling loop by inhibiting ERK activation. The PDGFRA inhibitor crenolanib might be used to treat patients with imatinib-resistant, KIT-mutant GIST.
Keywords: signal transduction, stem cells, receptor tyrosine kinase, cancer
INTRODUCTION
KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog), platelet-derived growth factor (PDGF) receptor-alpha (PDGFRA) and PDGFR-beta (PDGFRB) are closely related type III receptor tyrosine kinases (RTKs).1 The ligand for KIT is stem cell factor; PDGFRA, PDGFRB and their heterodimers bind PDGF polypeptides. Activation of these pathways promotes cell proliferation, migration, survival and, in differentiated cells, function.1
In gastrointestinal muscles, KIT and PDGFRA are mainly expressed in distinct mesenchymal cells: interstitial cells of Cajal (ICC)2 and ‘fibroblast-like cells’ (FLC),3,4 respectively. These cells perform critical functions in neuroregulation and motility: ICC generate electrical slow waves, mediate cholinergic excitatory and nitrergic inhibitory neuromuscular neurotransmission and set smooth muscle membrane potential,5,6 whereas FLC mediate purinergic inhibitory neuromuscular signaling.3 While previous studies did not indicate PDGFRA expression in mature ICC of adult tissues,2,4 both fetal and adult ICC precursors were found to co-express KIT and PDGFRA.7,8 Thus, KIT and PDGFRA expression may not be as completely compartmentalized as previously suggested.
Gastrointestinal stromal tumors (GIST), the most common human sarcoma,9 originate from the ICC lineage.8,10 Most GIST are driven by activating mutations in KIT (75-80%)10 or PDGFRA (<10%)11, which are mutually exclusive and most often heterozygous. Activating KIT mutations cooperate with ETV1 (ets variant 1), master regulator of the ICC transcription program whose cellular levels are controlled by the KIT–mitogen-activated protein kinase (MAPK) 3/1 (extracellular signal-regulated kinase; ERK1/2) cascade, to bring about GIST oncogenesis.9,12 The majority of GIST are initially responsive to KIT/PDGFRA RTK inhibitors such as imatinib, the first-line treatment of advanced GIST.13 However, medical therapy is rarely, if ever, curative due to disease persistence8,14,15 and the eventual emergence of drug-resistant mutations.13 Similarly to ICC precursors, GIST were found to co-express PDGFRA and KIT in a small cohort of patients.16 KIT/PDGFRA heterodimers containing wild-type receptors maintain signaling activation even after blocking oncogenic signaling by imatinib,16,17 suggesting that PDGFRA may contribute to the survival and proliferation of KIT-mutant GIST and could be considered a potential therapeutic target. However, the effects and mechanisms of selective PDGFRA blockade remain unclear.
Here, we quantified KIT/PDGFRA co-expression in ICC and GIST and investigated the role of PDGFRA–KIT cooperation in KIT-mutant GIST by selectively ablating PDGFRA by RNA interference (RNAi) or inhibiting PDGFRA signaling with crenolanib besylate (CP-868,596). Crenolanib is orally bioavailable and clinically safe inhibitor of both wild-type and constitutively active type III RTKs with low nanomolar-to-subnanomolar half-maximal inhibitory concentrations (IC50) toward PDGFRA/B and fms-related tyrosine kinase 3 (FLT3) and 25- to >100-fold lower potency toward other RTKs including KIT.18,19 We report that PDGFRA is expressed in 3-5% of ICC, 35-44% of ICC precursors and in most GIST. Selective ablation or inhibition of PDGFRA, but not PDGFRB, inhibited the proliferation of KIT+, KIT− mutant GIST cells regardless of their imatinib sensitivity by disrupting, in an ERK1/2 MAPK-dependent manner, a signaling loop between KIT and ETV1. Thus, crenolanib is a promising candidate for treatment of imatinib-resistant, KIT-mutant GIST.
METHODS
Standard methods {immunohistochemistry, Western blotting (WB), RNAi by small interfering RNA (siRNA), cell proliferation, viability and apoptosis assays, retroviral transduction, gene expression and xenograft studies, Chou-Talalay analysis20} and additional details are described in the Supplementary Methods.
Ethics statements
De-identified GIST tissues archived as formalin-fixed, paraffin-embedded (FFPE) blocks (Supplementary Tables S1-S2) or collected following surgery at the Mayo Clinic (Supplementary Table S3) or the University of California, San Diego (Supplementary Table S4) were obtained with the approval of the respective Institutional Review Boards (#10-001221 and #090401). Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocols were approved by the Institutional Animal Care and Use Committee of the Mayo Clinic (A50613, A64812).
Animals and tissue preparation
BALB/c mice were from Harlan Laboratories (Madison, WI). Kit+/copGFP (B6.129S7-Kittm1Rosay/J) mice expressing copepod green fluorescent protein (copGFP) from the endogenous Kit locus,21 Pdgfra+/eGFP (B6.129S4-Pdgfratm11(EGFP)Sor/J) mice expressing histone H2B-enhanced GFP (eGFP) fusion protein from the endogenous Pdgfra locus22 and strain-matched wild-type mice were from The Jackson Laboratory (Bar Harbor, ME). C57BL/6J, WBB6F1/J, WCB6F1/J and NOD/ShiLtJ mice8 were also from The Jackson Laboratory. Athymic NCr-nu/nu mice8 were from NCI-Frederick. Mice were killed by decapitation performed under deep isoflurane inhalation anesthesia. Intact gastric tunica muscularis tissues were dissociated or used as organotypic cultures.23
Multi-parameter flow cytometry
Murine gastric Kit+Cd44+Cd34– ICC and KitlowCd44+Cd34+ ICC stem cells (ICC-SC)8,23 and their Pdgfra+ subsets were enumerated using previously published protocols8 with modifications (see Supplementary Methods, Supplementary Tables S5-S6 and Supplementary Figure S1). In experiments utilizing Kit and Pdgfra reporter mice, Kit+ and Pdgfra+ cells were identified by detecting GFP in addition to immunostaining.
Cell lines
KIT+ human GIST cells carrying imatinib-sensitive and imatinib-resistant KIT mutations and their KITlow/– derivatives were used as GIST models. The KIT+, imatinib-sensitive GIST-T1 cells contain a heterozygous in-frame deletion of 57 bases in the exon 11 juxtamembrane domain of KIT, the most common KIT mutation region in GIST.24 GIST-T1-5R cells carrying an additional missense mutation encoding KITT670I (exon14), which is frequently found in imatinib-resistant GIST patients, were derived from the GIST-T1 line by extended (~1 year) exposure to 5 μM imatinib.14 GIST-T1-10R cells were also derived from GIST-T1 cells as an imatinib-resistant clone that arose by short-term (~1 month) culturing with 10 μM imatinib (A.G. and B.P.R., unpublished). These cells do not contain an imatinib-resistant mutation but express very little or no KIT protein (KITlow/–). The KIT+, imatinib-sensitive GIST882 cells contain a homozygous KITK642E mutation affecting the first part of the split tyrosine kinase domain (exon 13).25 GIST48B cells expressing very little or no KIT (KITlow/–) were derived from KIT+ GIST48 cells containing KIT exon 11 (homozygous V560D: imatinib-sensitive) and exon 17 phosphotransferase domain (heterozygous D820A: imatinib-resistant) mutations by prolonged inhibition of the KIT chaperone heat shock protein 90.26 GIST882, GIST48(B), GIST-T1 and derivatives were maintained as described.27 GIST-T1-5R and GIST-T1-10R cells were grown in the presence of 1 μM imatinib mesylate (LC laboratories, Woburn, MA), which was removed from the media 4 days before experiments27.
The murine Kitlow/– ICC-SC line 2XSCS2F10 was derived from gastric tunica muscularis of 7-day-old C57BL6/J mice and maintained as previously described.8
Statistical analyses
“n” indicates the number of GIST samples, freshly isolated or cultured mouse stomachs or experiments performed in cultured cells. Data are expressed as the mean±standard error of the mean. Individual data points are shown when n=3. Mann-Whitney rank sum test was used for comparing two data sets. P<0.05 was considered statistically significant. Drug concentrations producing 50% and 90% inhibition (IC50 and IC90, respectively) were obtained from dose-response plots represented by spline curves (SigmaPlot 10; Systat Software, San Jose, CA).
RESULTS
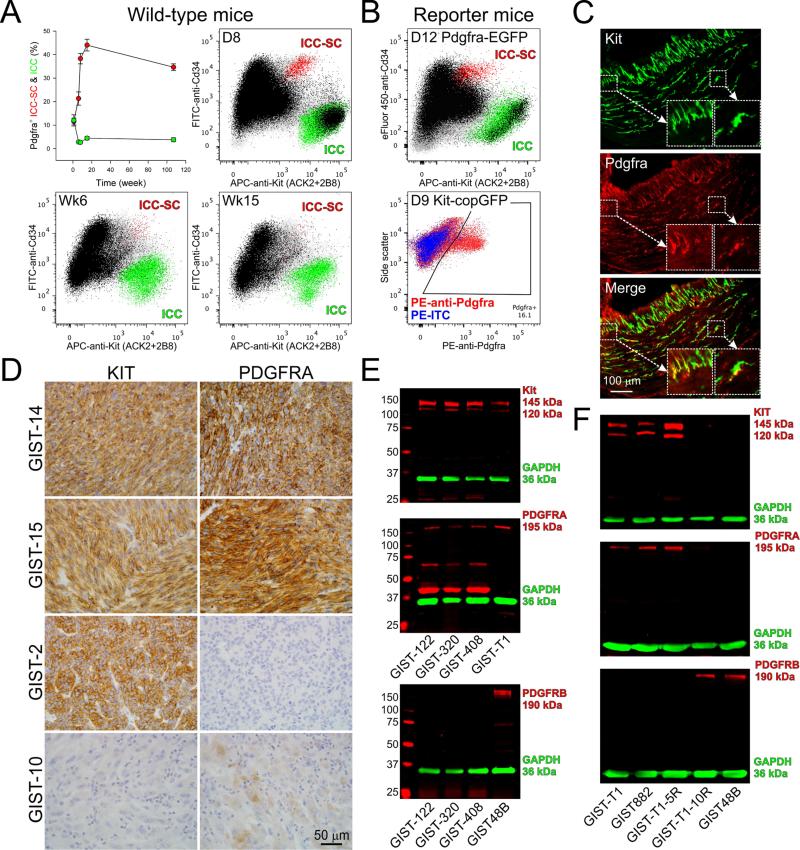
We first quantified Kit/Pdgfra co-expression in the ICC lineage by flow cytometry in the stomach of mixed-background wild-type mice (Figure 1A), as well as in Pdgfra+/eGFP and Kit+/copGFP mice expressing GFP from the endogenous Pdgfra and Kit loci, respectively (Figure 1B). At ~1 week of age, ICC and ICC-SC contained near-identical proportions of Pdgfra+ cells by live immunolabeling (12±2% and 12±1%, respectively) (Figure 1A). In 8-14-day-old Kit+/copGFP mice, copGFP+ cells of the ICC lineage contained 17±2% Pdgfra-expressing cells, whereas in 9-12-day-old Pdgfra+/eGFP mice, higher co-expression of eGFP and Kit was detected (ICC-SC: 30±3%; ICC: 23±1%) (Figure 1B), possibly reflecting the better signal-to-noise ratio obtained with the nucleus-targeted eGFP than by Pdgfra immunolabeling (Supplementary Figure S1). By 6 weeks of age, the frequency of Pdgfra+Kit+ ICC dropped to 3-5% and remained at this level for ~2 years (Figure 1A). Similarly, we detected 4.5±1% Pdgfra+ ICC in the stomach of adult, non-diabetic NOD/ShiLtJ mice by immunohistochemistry (Figure 1C). In contrast, the proportion of Pdgfra+ ICC-SC increased to 21% by 6 weeks of age and plateaued at 35-44% during adulthood (Figure 1A). These results indicate that Pdgfra expression is intrinsic to the ICC lineage, with Pdgfra+ ICC-SC and ICC becoming proportionally more and less frequent, respectively, with age.
Figure 1. Co-expression of KIT and PDGFRA/B in the ICC/GIST lineage.
(A) Age-dependent expression of Pdgfra detected by immunofluorescence (black) in murine ICC (Kit+Cd44+Cd34– cells; green) and ICC-SC (KitlowCd44+Cd34+ cells; red), identified in the hematopoietic marker-negative fraction (gray) of the dissociated gastric corpus+antrum tunica muscularis. Upper left panel shows time course data from 3-6 mixed-lineage wild-type mice/group; other panels are representative projections from mice of the ages indicated. APC, allophycocyanin; FITC, fluorescein isothiocyanate. Note age-related decline and increase in Pdgfra+ ICC and ICC-SC, respectively. (B) Upper panel: Pdgfra expression detected by eGFP fluorescence (black) in ICC (green) and ICC-SC (red) identified by immunolabeling in Pdgfra+/eGFP reporter mice. A representative projection from 6 mice is shown. Lower panel: Pdgfra expression detected by immunolabeling in the ICC lineage identified by copGFP fluorescence in Kit+/copGFP reporter mice. (Note that Kit+ and Kitlow cells could not be reliably distinguished in this line; see Supplementary Figure S1). Red and blue color indicate copGFP+ cells stained with phycoerythrin (PE)-anti-Pdgfra and PE-isotype control antibody (ITC), respectively. A representative projection from 9 mice is shown. Experiments in reporter mice confirmed expression of Pdgfra in the ICC lineage. (C) Representative immunofluorescent images demonstrating Pdgfra expression in a subset of gastric ICC of a non-diabetic NOD/ShiLtJ mouse (4.5±1% Pdgfra+ ICC out of 1,446 Kit+ ICC counted in 4 mice×3 sections; see Supplementary Table S7 for antibodies). Insets show enlarged regions identified by rectangles in the images. Yellow color signifies co-localization. (D) KIT and PDGFRA expression (brown color) detected by immunohistochemistry in representative FFPE GIST samples from the cohort described in Supplementary Table S1 (n=20 patients). Note samples with different expression profiles. (E) Profiling of KIT, PDGFRA and PDGFRB expression in unfixed GIST samples by WB (see Supplementary Tables S3 and S8 for details). Target proteins and GAPDH (loading control) were detected simultaneously using LI-COR Biosciences (Lincoln, NE) secondary antibodies tagged with near-infrared and infrared fluorescent dyes (IRDye700: red pseudocolor; IRDye800CW: green pseudocolor).27 GIST-T1 and GIST48B cells were included as positive controls. All GIST samples in this cohort were KIT+PDGFRA+PDGFRB–. (F) Profiling of KIT, PDGFRA and PDGFRB expression in GIST cell lines by WB. PDGFRA but not PDGFRB was consistently co-expressed with KIT; KIT– lines expressed PDGFRB but not PDGFRA.
We next examined KIT and PDGFRA expression in GIST by immunohistochemistry in 43 samples (14 imatinib-treated and 29 untreated samples in two cohorts; Figure 1D, Supplementary Figure S2A and Supplementary Tables S1-S2) and WB in 10 additional samples (3 treated and 7 untreated samples in two cohorts; Figure 1E, Supplementary Figure S2B and Supplementary Tables S3-S4). KIT protein was expressed in 50 of 53 tumors (94.3%; note that all 3 KIT– samples were from imatinib-treated patients: GIST-10, GIST-910 and GIST-914). PDGFRA immunoreactivity was detected in 48 of 53 samples (90.6%) and 45 of the 50 KIT+ GIST (90%). In most cases, PDGFRA immunostaining was strong and uniform; focal, low-level expression was only seen in 3 tissues {KIT–: GIST-10 (Figure 1D); KIT+: GIST-904, GIST-909}. PDGFRA immunostaining scores were not different between imatinib-treated and untreated samples (P=0.866). By WB, the KIT+PDGFRA+ GIST samples examined did not express PDGFRB (Figure 1E). These results extend previous findings of widespread KIT/PDGFRA co-expression in GIST.16
To facilitate mechanistic studies, we also examined KIT, PDGFRA and PDGFRB expression in 5 KIT-mutant GIST cell lines (Figure 1F). As reported previously,17 the imatinib-sensitive GIST882 cells co-expressed KIT and PDGFRA; however, we did not detect PDGFRB expression. The same constellation was observed in the imatinib-sensitive GIST-T1 cells and their GIST-T1-5R sub-line containing the imatinib-resistant mutation KITT670I. These cells therefore model the majority of GIST analyzed in this study. In contrast, GIST-T1-10R and GIST48B cells expressing very little or no KIT protein (KITlow/–) also did not contain PDGFRA but expressed PDGFRB. Therefore, these cells, which were generated by prolonged KIT inhibition or depletion,26 model long-term imatinib-treated GIST such as GIST-10, GIST-910 and GIST-914.
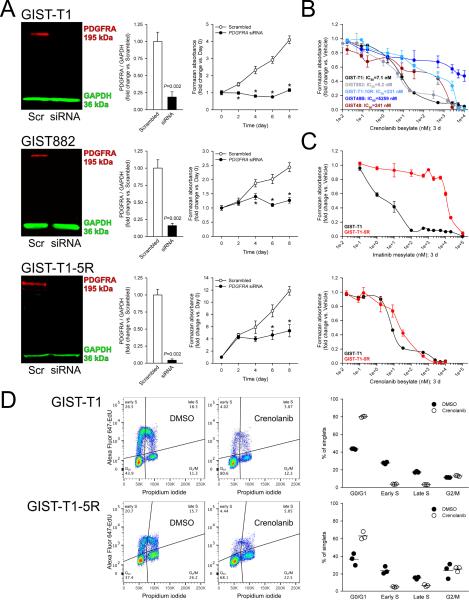
We first studied the functional significance of PDGFRA expression in KIT-mutant, KIT+ GIST cell models by RNAi-mediated knock-down of PDGFRA. PDGFRA-targeting siRNA effectively downregulated PDGFRA protein by WB (Figure 2A) without contemporaneously reducing KIT protein (Supplementary Figure S3), and robustly inhibited the growth of the imatinib-sensitive GIST-T1 and GIST882 cells and the imatinib-resistant GIST-T1-5R line by methyl-tetrazolium salt (MTS) assay (Figure 2A). These results indicate that wild-type PDGFRA is important for the viability of KIT-mutant GIST cells and thus its role in GIST biology is likely more significant than previously recognized.16,17
Figure 2. PDGFRA knock-down or inhibition suppresses cell proliferation of both imatinib-sensitive and imatinib-resistant, KIT+PDGFRA+, KIT-mutant GIST cells.
(A) siRNA-mediated PDGFRA knock-down inhibited the growth of the imatinib-sensitive GIST-T1 and GIST882 and the imatinib-resistant GIST-T1-5R cells (n=6/data point). Knock-down efficacy was verified by WB 3 days after siRNA transfection. Viable cell numbers determined by MTS assay were evaluated every other day. (B) Crenolanib applied for 72 hours inhibited the growth of KIT+PDGFRA+ lines (GIST-T1, GIST882) with IC50`s comparable to that of imatinib but had poor efficacy or efficiency in cells lacking KIT and/or PDGFRA (MTS assays; n=3/cell line). (C) Crenolanib inhibited the growth of the imatinib-resistant GIST-T1-5R cells and its imatinib-sensitive GIST-T1 parent line with comparable potency. Upper and lower panels show MTS dose-response curves obtained with imatinib and crenolanib, respectively (n=3/cell line/treatment). IC50/IC90 values are shown in Supplementary Figure S4A. (D) Crenolanib blocked G1/S transition and DNA synthesis in both the imatinib-sensitive GIST-T1 and the imatinib-resistant GIST-T1-5R cell lines. GIST-T1 and GIST-T1-5R cells maintained with complete growth media were treated for 24 h with 8.3 and 33.2 nM crenolanib, respectively and subjected to cell cycle analysis by combined Alexa Fluor 647-EdU incorporation and PI labeling. Left panels: representative Alexa Fluor 647-EdU vs. PI (area) projections of events gated for single cells with at least diploid DNA content. Right panels show the cell frequencies in the cell cycle phases identified in the quadrants (n=3/cell line/treatment). Horizontal lines indicate mean frequencies.
To investigate whether the inhibition of PDGFRA signaling could also reduce GIST cell viability, we utilized crenolanib besylate, a potent inhibitor of wild-type and constitutively active PDGFRA/B with 25- to >100-fold lesser potency toward KIT.18,19 Crenolanib also inhibits FLT3 but none of the GIST lines studied expressed FLT3 (not shown). Crenolanib potently inhibited the growth of both the imatinib-sensitive GIST-T1 and GIST882 cells (IC50≤8 nM) and the imatinib-resistant GIST-T1-5R cells (IC50=23±6 nM) (Figure 2B-C) by MTS assay. Furthermore, crenolanib had lower 90% inhibitory concentration in GIST-T1-5R than GIST-T1 cells (IC90=518±90 nM and 1340±57 nM, respectively; Supplementary Figure S4A). In GIST-T1 and GIST882 cells, 16.6 nM crenolanib only modestly increased apoptosis by Caspase Glo-3/7 assay (GIST-T1: 1.38±0.04-fold vs. vehicle; GIST882: 1.55±0.02-fold vs. vehicle) and we observed no significant accumulation of sub-G0/1 cells by propidium iodide (PI) labeling and flow cytometry in GIST-T1 and GIST-T1-5R cells (not shown). In contrast, 8.3 and 33.2 nM crenolanib applied for 24 or 72 h significantly inhibited GIST-T1 and GIST-T1-5R cell proliferation detected by Ki-67 immunofluorescence (Supplementary Figure S4B). In both cell lines, cell cycle analysis by flow cytometry utilizing Alexa Fluor 647-tagged 5-ethynyl-2'-deoxyuridine (EdU) incorporation and PI labeling indicated that the reduced cell proliferation was due to the blockade of G1/S transition and DNA synthesis (Figure 2D).
Surprisingly, crenolanib failed to efficiently inhibit the growth of the KIT–PDGFRA– cell lines GIST48B and GIST-T1-10R (IC50>230 nM; Figure 2B) despite their expression of PDGFRB and crenolanib's ability to potently inhibit PDGFRB phosphorylation in GIST48B cells (see below). This result indicates that PDGFRB is unlikely to contribute to GIST proliferation despite complexing with, and being cross-phosphorylated by, oncogenic KIT.17 In contrast, and consistent with the RNAi results above, PDGFRA appeared to be required for crenolanib to efficiently block GIST cell proliferation as indicated by the ~240 nM IC50 obtained in the KIT+PDGFRA–PDGFRB+ GIST48 cells, the parent line of GIST48B cells17 (Figure 2B).
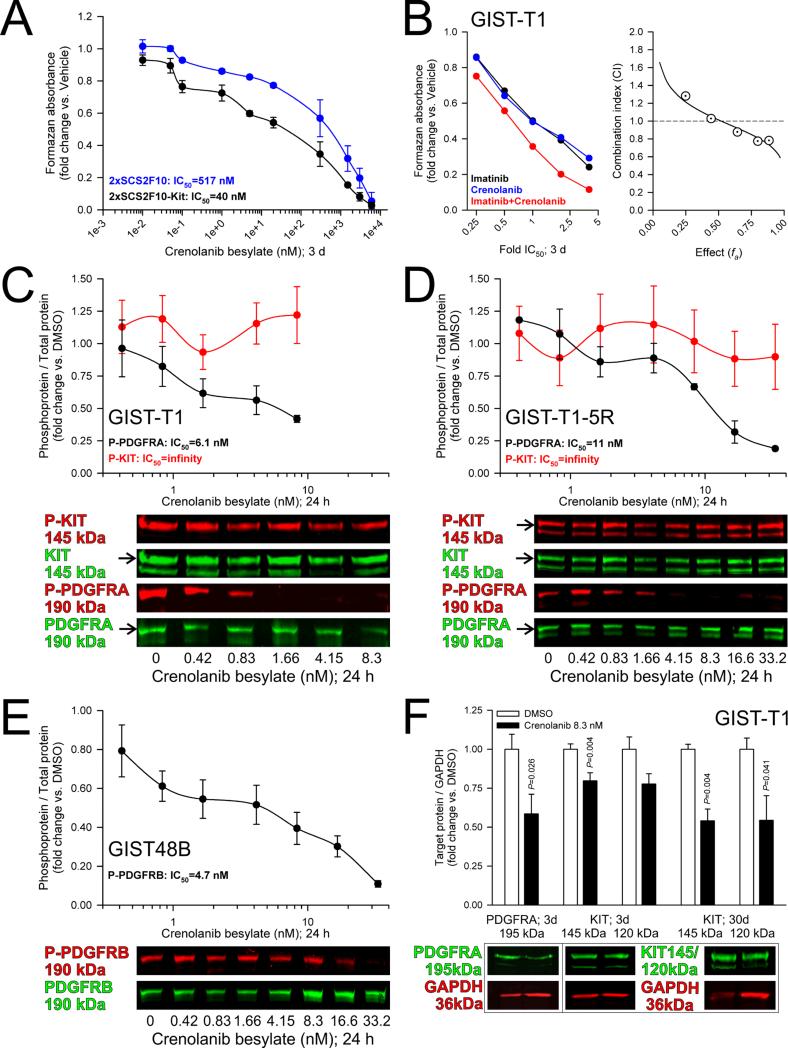
Crenolanib also poorly inhibited cell proliferation in the murine wild-type ICC-SC line 2xSCS2F108 (IC50=517 nM; Figure 3A), which expresses both Pdgfra and Pdgfrb but lacks significant Kit protein (Supplementary Figure S5A-B). Stable retroviral transduction with wild-type murine Kit (Supplementary Figure S5B-C) shifted the IC50 to 40 nM (Figure 3A). Thus, in the absence of oncogenic Pdgfra mutation (verified by RNA-sequencing; not shown), not only Pdgfra but also Kit is required for cytostatic effects of crenolanib. We then investigated the nature of KIT–PDGFRA cooperation by probing the interaction between crenolanib and imatinib in GIST-T1 cells using the Chou-Talalay combination index (CI) method.20 Although these drugs have been reported to have near-identical IC50 values for wild-type PDGFRA in heterologous expression systems,19 wild-type PDGFRA has been shown to maintain signaling activation in imatinib-treated, imatinib-responsive GIST.16 Consistent with different modes-of-action and indicating cooperation between KIT- and PDGFRA-activated pathways, these experiments revealed slight-to-moderate synergism for concentrations ranging from 1× to 4×IC50 (CI: 0.88-0.77; moderate synergism at fa>0.75; Figure 3B).
Figure 3. KIT expression is important for crenolanib responsiveness without being a direct pharmacologic target.
(A) Expression of wild-type murine Kit from a retroviral construct sensitized the Kitlow/– ICC-SC line 2xSCS2F10 to crenolanib cytostatic effect (n=2/cell line). Note reduction of crenolanib IC50 from 517 to 40 nM. (B) Evaluation of the interaction between crenolanib and imatinib by the Chou-Talalay CI method. Left panel: MTS dose-response curves of drugs applied alone and in combination in 3 technical replicates. Right panel: CI vs. effect plot. Synergy, additivity and antagonism are defined as CI<1, CI=1, and CI>1, respectively. Note slight-to-moderate synergism for concentrations ranging from 1-4x IC50. (C-E) Effects of crenolanib on PDGFRA and KIT phosphorylation in GIST-T1 (C; n=3/dose/target) and GIST-T1-5R cells (D; n=3/dose/target) and on PDGFRB phosphorylation in GIST48B cells (E; n=7/dose). Crenolanib applied under full growth conditions for 24 hours dose-dependently inhibited PDGFRA and PDGFRB phosphorylation but not KIT phosphorylation. Note PDGFRA/B IC50 values in the nanomolar range. (F) Effects of crenolanib on PDGFRA and KIT protein expression in GIST-T1 cells (n=6/group). 8.3 nM crenolanib applied for 3-30 days reduced PDGFRA and KIT protein expression. Media were changed twice per week.
Therefore, we next investigated the molecular mechanisms of KIT involvement by examining the effects of crenolanib on PDGFRA/B and KIT phosphorylation and expression. In GIST-T1 and GIST-T1-5R cells cultured in complete media, KIT and PDGFRA displayed activating phosphorylation on Y721 and Y742, respectively; and PDGFRB was phosphorylated on Y1021 in the KITlow/–PDGFRA–PDGFRB+ GIST48B cells (Figure 3B-E). Crenolanib inhibited PDGFRA phosphorylation in both the imatinib-sensitive GIST-T1 and the imatinib-resistant GIST-T1-5R cells with IC50 values closely resembling those obtained by MTS assays (see above) and, as shown previously in GIST-T1 cells,19 without inhibiting KIT phosphorylation (Figure 3B-D). As expected, crenolanib equally potently inhibited PDGFRB phosphorylation in GIST48B cells (Figure 3E). Together with the MTS results presented above (Figure 2B-C), these findings indicate that the KIT-dependency of the crenolanib effects is unlikely to reflect direct KIT inhibition at pharmacologically relevant concentrations.18 In contrast, 8.3 nM crenolanib applied for 3-30 days significantly reduced both PDGFRA and KIT protein expression in GIST-T1 cells (Figure 3F). KIT expression was also inhibited by 10-day siRNA-mediated knock-down of PDGFRA in GIST-T1 cells (Supplementary Figure S6) and by 30-day crenolanib treatment (16.6 nM) in murine gastric corpus+antrum tunica muscularis organotypic cultures (Supplementary Figure S7). Thus, downregulation of KIT and PDGFRA expression consistently occurs in response to longer-term inhibition of PDGFRA signaling and may contribute to its effects.
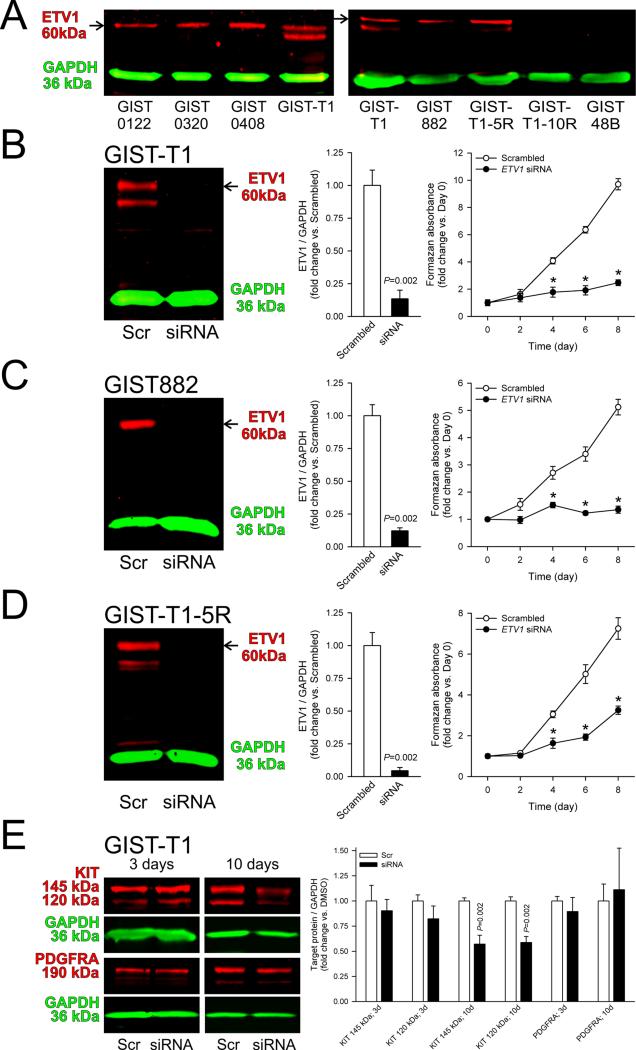
Based on the KIT-dependent effect of crenolanib we hypothesized that PDGFRA signaling could support GIST proliferation by modulating ETV1, a KIT-dependent transcription factor required for GIST oncogenesis.9,12 Indeed, we detected ETV1 protein expression only in the KIT+PDGFRA+, crenolanib-sensitive cell lines GIST-T1, GIST882 and GIST-T1-5R, but not in the KIT–, crenolanib-resistant GIST-T1-10R, GIST48B (Figure 4A) and 2xSCS2F10 cells (Supplementary Figure S5D). Consistent with previous reports,9,12 RNAi-mediated ETV1 knock-down inhibited the growth of the GIST882 line (Figure 4C) and we also found similar anti-proliferative effects in GIST-T1 and GIST-T1-5R cells (Figure 4B and D). In GIST-T1 cells, similarly to crenolanib and PDGFRA-targeting siRNA, 10-day ETV1 knock-down inhibited the expression of both mature and immature forms of KIT (~145 and ~120 kDa, respectively), although this effect occurred without a reduction in PDGFRA (Figure 4E). Besides suggesting a link between PDGFRA signaling and ETV1, these results support a role for ETV1 in controlling KIT expression.12
Figure 4. ETV1 regulates cell proliferation and KIT expression in crenolanib-sensitive GIST cells.
(A) Protein bands corresponding to ETV1 protein (arrows) were only detectable in crenolanib-sensitive, KIT+PDGFRA+ cells (GIST-T1, GIST882, GIST-T1-5R) but not in crenolanib-insensitive, KIT–PDGFRA– cells (GIST-T1-10R, GIST48B). Left panel shows ETV1 expression in unfixed, KIT+PDGFRA+ GIST samples for reference. (B-D) siRNA-mediated ETV1 knock-down inhibited the growth of the imatinib- and crenolanib-sensitive GIST-T1 and GIST882 and the imatinib-resistant, crenolanib-sensitive GIST-T1-5R cells (n=6/data point). Knock-down efficacy was verified by WB 3 days after siRNA transfection. Viable cell numbers determined by MTS assay were evaluated every other day. (E) Effects of siRNA-mediated ETV1 knock-down on PDGFRA and KIT protein expression 3 and 10 days after transfection (n=6/group/target). ETV1 knock-down inhibited KIT expression after 10 days.
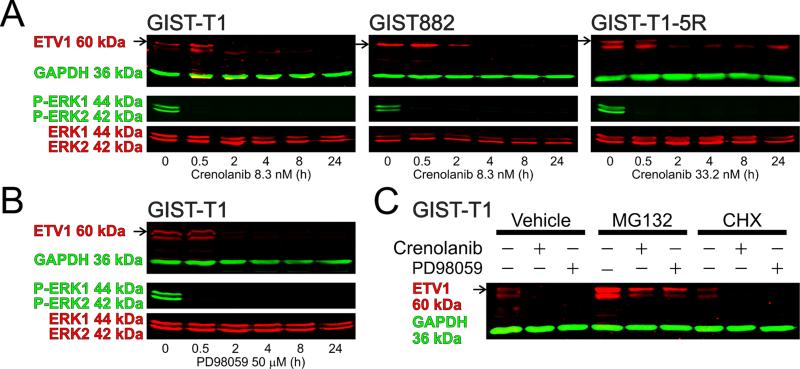
To obtain further evidence of an ETV1 role in mediating PDGFRA signaling in GIST, we analyzed ETV1 protein expression following 10-day PDGFRA knock-down in GIST-T1 cells. Similarly to KIT, ETV1 protein expression was significantly reduced (Supplementary Figure S6). Next, we measured ETV1 protein expression and ERK1/2 MAPK phosphorylation in GIST cells treated with crenolanib. In GIST-T1, GIST882 and GIST-T1-5R cells, 8.3-33.2 nM crenolanib profoundly inhibited ERK1/2 phosphorylation within 30 minutes and ETV1 protein expression in 2 hours (Figure 5A). The MAPK kinase inhibitor PD98059 (50 μM) fully reproduced crenolanib's effects both on ERK1/2 phosphorylation and ETV1 protein expression (Figure 5B), indicating that the crenolanib-induced inhibition of ERK1/2 activation was sufficient for ETV1 downregulation. Despite its rapid effect on ETV1, PD98059 only reduced KIT protein expression after 10 days (Supplementary Figure S8). These results are consistent with the time-course of KIT downregulation in response to ETV1 knock-down (Figure 4E).
Figure 5. Crenolanib induces ETV1 protein degradation by inhibiting ERK1/2 MAPK activation.
(A) Time course of crenolanib effects on ETV1 protein expression and ERK1/2 phosphorylation (n=3-6/cell/target). Crenolanib inhibited ERK1/2 activation in 30 min and ETV1 expression within 2 h. (B) Reproduction of the crenolanib effects by the MAPK kinase inhibitor PD98059 (50μM) in GIST-T1 cells (n=5-8/target). (C) Effects on ETV1protein of crenolanib (8.3 nM) and PD98059 (50μM) applied for 4 h in the absence or presence of the proteasome inhibitor MG132 (10μM) or the protein synthesis inhibitor CHX (10μg/ml) (n=6/group). MG132 reversed and CHX accentuated the effects of crenolanib and PD98059.
Previously, imatinib was shown to downregulate ETV1 protein levels by facilitating its proteasome-mediated degradation.9 Similarly, in GIST-T1 cells we found that the degree of ETV1 downregulation induced by 8.3 nM crenolanib or 50 μM PD98059 applied for 4 h exceeded the rate of ETV1's natural degradation revealed by treatment with the general protein synthesis inhibitor cycloheximide (CHX; 10 μg/mL) (Figure 5C). Furthermore, CHX accentuated both crenolanib- and PD98059-induced inhibition (from 50±14% of control to 27±6% and from 51±18% to 14±3%, respectively). The proteasome inhibitor MG132 (10 μM) increased baseline ETV1 levels and completely reversed both the crenolanib- and PD98059-induced downregulation (Figure 5C). These data indicate that, similarly to imatinib,9 crenolanib reduces cellular levels of ETV1 by facilitating its proteasome-mediated degradation via ERK1/2 inhibition.
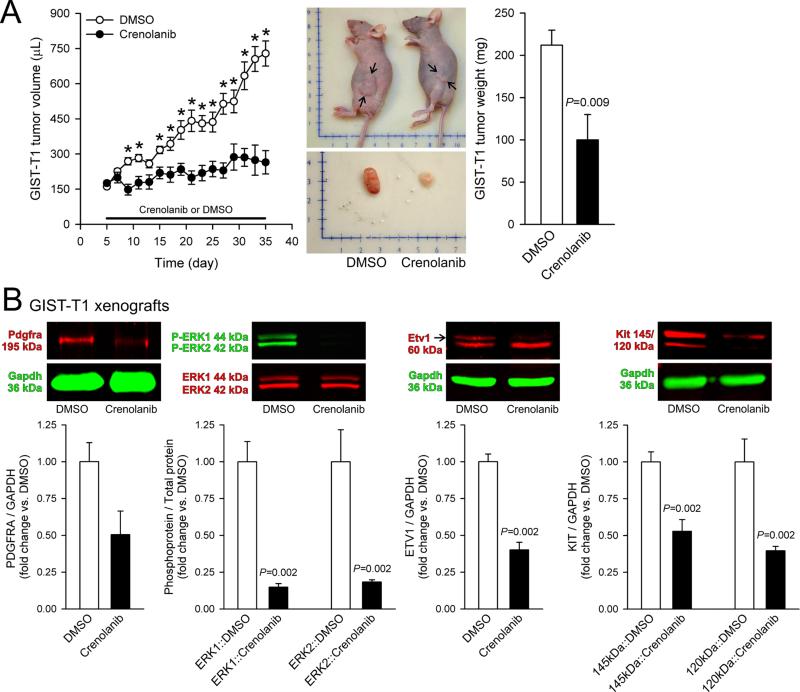
Finally, we investigated the in-vivo effects of crenolanib on GIST-T1 cells in a nude mouse xenograft model. Crenolanib administered intraperitoneally at 12.5 mg/kg BID for 30 days blocked the growth of tumors developed from transplanted GIST-T1 cells (Figure 6A) without reducing body weights (Supplementary Figure S9A), inhibited ERK1/2 phosphorylation and the expression of PDGFRA, KIT and ETV1 in the excised xenografts (Figure 6B) and also in the gastric corpus+antrum tunica muscularis of the host mice (Supplementary Figure 9B). These findings indicate that the crenolanib inhibitory effects seen in GIST models in vitro are also attainable in vivo.
Figure 6. Crenolanib inhibits the growth of tumors arising from xenotransplanted GIST-T1 cells and downregulates ETV1 expression in vivo.
(A) Crenolanib administered intraperitoneally at 12.5 mg/kg BID for 30 days inhibited the growth of GIST-T1 cells transplanted into NCr-nu/nu mice (n=6/group). Left panel: time course of changes in tumor volumes. Middle panel: representative tumor-bearing animals and excised tumors. Scales are in centimeters. The arrows in the upper panel mark the greatest extent of the tumors. Right panel: tumor weights at euthanasia. (B) Inhibition of PDGFRA, ETV1 and KIT protein expression and ERK1/2 phosphorylation in the excised GIST-T1 xenografts (n=6/group).
DISCUSSION
Expression of wild-type PDGFRA has been reported in GIST and proposed to contribute to GIST RTK signaling through cross-phosphorylation by constitutively active KIT and also via a ligand-activated autocrine/paracrine loop, which may remain active even after the blockade of oncogenic signaling by imatinib.16,17 Regulation of PDGFRA may have therapeutic implications in GIST, however, the consequences and mechanisms of specific PDGFRA blockade were not known. Here, we used RNAi and crenolanib to evaluate PDGFRA-mediated signaling and PDGFRA/KIT interactions in KIT-mutant GIST. Crenolanib is a novel potent and reported clinically safe pharmacological inhibitor of wild-type and oncogenic type III RTKs with high selectivity for PDGFRA/B relative to KIT.18,19 We report that selective depletion or inhibition of PDGFRA inhibited the growth of KIT+, KIT-mutant GIST cells, including cells with imatinib-resistant mutations, with surprising efficacy and efficiency (e.g., low nanomolar IC50 values for crenolanib). The crenolanib effect mainly reflected inhibition of G1/S transition and DNA synthesis but also included a modest increase in apoptosis. Drug interaction experiments demonstrated synergism between crenolanib and imatinib in imatinib-sensitive GIST cells. This finding suggests that imatinib and crenolanib, despite their near-identical IC50 for wild-type PDGFRA in heterologous expression systems,19 do not inhibit identical mechanisms in GIST cells, possibly reflecting crenolanib's predicted ability to bind to activated PDGFRA.19 In contrast to PDGFRA blockade, PDGFRB inhibition alone was not associated with reduced GIST proliferation. Crenolanib also profoundly inhibited the growth of tumors from xenografted GIST cells, demonstrating that the effects of PDGFRA blockade are also attainable in vivo. These results indicate that wild-type PDGFRA plays a more significant role in GIST pathogenesis than previously recognized and suggest that pharmacological targeting of PDGFRA could be beneficial not only in imatinib-resistant, PDGFRA-mutant GIST19 but also in KIT-mutant GIST including tumors harboring imatinib-resistant KIT.
We also found that the effect of crenolanib on KIT-mutant GIST cells depended on KIT expression, although crenolanib only inhibited PDGFRA but not KIT phosphorylation. This reflected the involvement of ETV1, a KIT-dependent transcription factor important for ICC development and GIST oncogenesis:9,12 Crenolanib promoted proteasomal degradation of ETV1 via reduced ERK MAPK phosphorylation. Consistent with these and previous findings,9,12 ETV1 knock-down reproduced the effects of PDGFRA knock-down or inhibition. Collectively, these findings indicate that PDGFRA cooperates with KIT in ETV1 stabilization; and inhibition of wild-type PDGFRA signaling reduces GIST proliferation to a great extent by intercepting this pathway (Figure 7). Furthermore, our results obtained in mouse gastric muscles indicate that KIT–PDGFRA cooperation also occurs in KIT+PDGFRA+ cells of the normal ICC lineage, where it may support the physiological, ETV1-dependent ICC transcriptional network.9
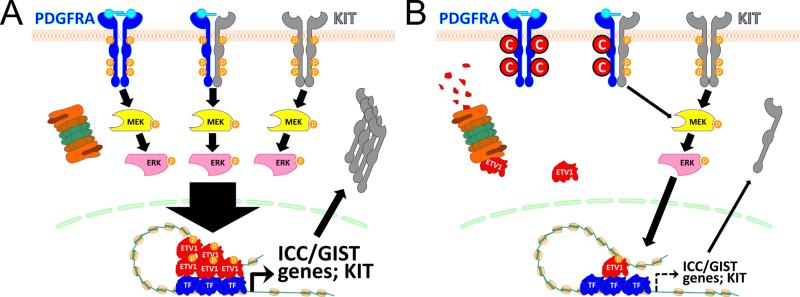
Figure 7. Proposed mechanism of PDGFRA inhibition-mediated cytostasis in KIT+PDGFRA+, KIT-mutant GIST cells.
(A) Under basal conditions, oncogenic and/or wild-type KIT and wild-type PDGFRA cooperatively activate ERK thereby preventing ETV1 degradation. High levels of ETV1 stimulate cell proliferation and tumorigenesis by hyperactivating ICC/GIST-specific transcriptional output including KIT expression. (B) PDGFRA knock-down or inhibition e.g. with crenolanib (C in red circle) facilitates ETV1 degradation by reducing ERK activation. Reduced ETV1 levels limit cell proliferation via reduced transcriptional activation of target genes including KIT. TF, other transcription factor. Assuming ligand-induced activation only, this model also applies to KIT–PGDFRA cooperation in the ICC lineage.
This transcriptional network was described by Chi et al. as a set of 38 genes.9 KIT has recently been added to these ETV1 targets.12 We detected reduced KIT protein in response to PDGFRA knock-down and inhibition, MAPK kinase blockade and ETV1 knock-down in GIST cells and in murine gastric muscles in response to PDGFRA inhibition. KIT downregulation can be expected to contribute to the effects of crenolanib as it would at least partially deprive KIT-mutant GIST cells of their principal driver oncogene.13 Robust effects on KIT required longer-term (>3-day) applications despite the rapid loss of ETV1 protein; a finding consistent with the results presented in refs.9,12 In light of the reported binding of ETV1 to KIT enhancers but not directly to the KIT promoter,12 this observation may reflect the role of additional factors in the maintenance of KIT expression. Furthermore, the robust inhibition of GIST growth even in the absence of complete KIT loss indicates that the effects of blocking the PDGFRA–ERK MAPK–ETV1 axis may have in part been mediated by other ETV1 target genes whose specific roles remain to be determined.
Our data indicate that PDGFRA but not PDGFRB expression confers significant growth advantage to KIT+ETV1+ cells. Together with previous reports,16,17 the present results also indicate that KIT/PDGFRA co-expression occurs in the majority of GIST. In the adult murine ICC lineage we found Pdgfra protein in a small subset of mature ICC and 35-44% of KitlowCd34+ ICC-SC, raising the intriguing possibility that the corresponding subsets in human tissues may be particularly susceptible to GIST oncogenesis.
The convergent effects of KIT and PDGFRA signaling on ETV1 protein stabilization further support the potential utility of pharmacological targeting of ETV1in GIST.9,12 Currently, inhibition of the ERK MAPK pathway with MEK162 is under investigation for this purpose in a phase 1b/2 clinical trial (NCT01991379). We propose that PDGFRA inhibition e.g. with crenolanib should also be explored as a tool for ETV1 downregulation. This approach, while sharing key benefits with MAPK kinase inhibition such as efficacy in imatinib-resistant GIST, has the added advantage of being able to robustly downregulate PDGFRA phosphorylation-dependent signaling cascades, such as PDGFRA Y742 phosphorylation, which regulates activation of the phosphatidylinositol-4,5-bisphosphate 3-kinase pathway,1 an effect that may have contributed to crenolanib synergism with imatinib detected in our study. Crenolanib has so far been found to have potent effects and a safe toxicity profile18 and is undergoing clinical evaluation including in advanced GIST patients with PDGFRA D842 mutations and deletions. Thus, crenolanib could be considered as a novel pharmacological for individualized treatment of ETV1+ GIST, particularly GIST harboring imatinib-resistant mutations.
Supplementary Material
Acknowledgements
We thank Dr. Gregory J. Gores (Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN) for granting us access to the LI-COR Odyssey Scanner and Dr. Scott W. Lowe (Memorial Sloan Kettering Cancer Center, New York, NY) for the pMSCV-PIG retroviral plasmid. We also thank Dr. Angela J. Mathison (Gastroenterology Research Unit, Mayo Clinic, Rochester, MN) for her aid in the retroviral transduction, Dr. Jason T. Lewis (Department of Pathology and Laboratory Medicine, Mayo Clinic, Rochester, MN) for providing blind scores of PDGFRA expression in GIST samples, and Dr. Felicity T. Enders (Division of Biostatistics and Informatics, Mayo Clinic, Rochester, MN) for advising the authors on statistical analysis. KIT/PDGFRA staining and genotyping of GIST samples were performed at the Mayo Clinic Center for Individualized Medicine Pathology Research Core (Director: Dr. Thomas J. Flotte) and in the Cytopathology and Molecular Anatomic Pathology Laboratory (Director: Dr. Andre M. Oliveira), respectively.
Grant support: Supported in part by NIH grants R01DK058185, P01DK068055, R01DK057061, P30DK084567, P30CA015083, P50CA127003, U54CA168512, K08CA168999, The Life Raft Group, the Virginia and Daniel K. Ludwig Trust for Cancer Research and the Mayo Clinic Center for Individualized Medicine (http://mayoresearch.mayo.edu/center-for-individualized-medicine). The funding agencies had no role in the study analysis or writing of the manuscript. Its contents are solely the responsibility of the authors.
Abbreviations
- APC
allophycocyanin
- C+A
corpus plus antrum
- ChIP-seq
chromatin immunoprecipitation-sequencing
- CHX
cycloheximide
- copGFP
copepod (Pontellina plumata) green fluorescent protein
- CI
Chou-Talalay combination index
- CSF1R
colony stimulating factor 1 receptor
- DMSO
dimethyl sulfoxide
- EdU
5-ethynyl-2'-deoxyuridine
- eGFP
enhanced GFP
- ERK
extracellular signal-regulated kinase
- ETV1
ets variant 1
- FFPE
formalin-fixed, paraffin-embedded
- FITC
fluorescein isothiocyanate
- FLC
fibroblast-like cell(s)
- FLT3
fms-related tyrosine kinase 3
- GIST
gastrointestinal stromal tumor
- IC50
half-maximal inhibitory concentration
- IC90
concentration required for 90% inhibition
- ICC
interstitial cell(s) of Cajal
- ICC-SC
ICC stem cell(s)
- ITC
isotype control
- KIT
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- MAPK
mitogen-activated protein kinase
- MTS
methyl-tetrazolium salt
- PDGF
platelet-derived growth factor
- PDGFRA
platelet-derived growth factor receptor, alpha polypeptide
- PDGFRB
platelet-derived growth factor receptor, beta polypeptide
- PE
phycoerythrin
- PI
propidium iodide
- RNAi
RNA interference
- RTK
receptor tyrosine kinase
- siRNA
small interfering RNA
- WB
western immunoblotting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Jason K. Sicklick and Brian P. Rubin were members of the Speakers Bureau and the Advisory Board of Novartis Pharmaceuticals Corp. Simon J. Gibbons is a consultant to Millennium Pharmaceuticals Inc. Jonathan Fletcher is a member of the Advisory Boards of Novartis Pharmaceuticals Corporation, Bayer HealthCare AG and Ariad Pharmaceuticals, Inc. Abhijit Ramachandran is an employee of AROG Pharmaceuticals, the manufacturer of the drug crenolanib besylate utilized in this study. The other authors have nothing to disclose.
Author contributions: T.O. supervised the study. Y.H. and T.O. developed the study hypothesis and designed the experiments. Y.H., M.R.B., Y.T., S.M., G.B.G. and K.M.C. performed the experiments. Y.H., S.J.G., G.F. and T.O. analyzed and interpreted the data. K.M.R.-L., M.L.K., J.B-C., C.-H.T., J.K.S., T.T., A.G., B.P.R., J.A.F. and A.R. provided critical research resources and consultation. G.F., B.P.R., J.A.F. and A.R. provided scientific advice and helpful comments on the project. Y.H. and T.O. wrote the manuscript. All authors have read and approved the final manuscript.
REFERENCES
- 1.Heldin CH, Lennartsson J. Structural and functional properties of platelet-derived growth factor and stem cell factor receptors. Cold Spring Harb Perspect Biol. 2013;5:a009100. doi: 10.1101/cshperspect.a009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Ordog T, Chen J, et al. Differential gene expression in functional classes of interstitial cells of Cajal in murine small intestine. Physiol Genomics. 2007;31:492–509. doi: 10.1152/physiolgenomics.00113.2007. [DOI] [PubMed] [Google Scholar]
- 3.Kurahashi M, Zheng H, Dwyer L, et al. A functional role for the 'fibroblast-like cells' in gastrointestinal smooth muscles. J Physiol. 2011;589:697–710. doi: 10.1113/jphysiol.2010.201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iino S, Nojyo Y. Immunohistochemical demonstration of c-Kit-negative fibroblast-like cells in murine gastrointestinal musculature. Arch Histol Cytol. 2009;72:107–115. doi: 10.1679/aohc.72.107. [DOI] [PubMed] [Google Scholar]
- 5.Klein S, Seidler B, Kettenberger A, et al. Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat Commun. 2013;4:1630. doi: 10.1038/ncomms2626. [DOI] [PubMed] [Google Scholar]
- 6.Groneberg D, Lies B, Konig P, et al. Cell-specific deletion of nitric oxide-sensitive guanylyl cyclase reveals a dual pathway for nitrergic neuromuscular transmission in the murine fundus. Gastroenterology. 2013;145:188–196. doi: 10.1053/j.gastro.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 7.Kurahashi M, Niwa Y, Cheng J, et al. Platelet-derived growth factor signals play critical roles in differentiation of longitudinal smooth muscle cells in mouse embryonic gut. Neurogastroenterol Motil. 2008;20:521–531. doi: 10.1111/j.1365-2982.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- 8.Bardsley MR, Horvath VJ, Asuzu DT, et al. Kitlow stem cells cause resistance to Kit/platelet-derived growth factor alpha inhibitors in murine gastrointestinal stromal tumors. Gastroenterology. 2010;139:942–952. doi: 10.1053/j.gastro.2010.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi P, Chen Y, Zhang L, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 12.Ran L, Sirota I, Cao Z, et al. Combined inhibition of MAP kinase and KIT signaling effectively destabilizes the ETV1 protein and synergistically suppresses GIST tumorigenesis. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-14-0985. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, Roy S, Lazar AJ, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proc Natl Acad Sci U S A. 2010;107:14333–14338. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCaprio JA, Duensing A. The DREAM complex in antitumor activity of imatinib mesylate in gastrointestinal stromal tumors. Curr Opin Oncol. 2014;26:415–421. doi: 10.1097/CCO.0000000000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negri T, Bozzi F, Conca E, et al. Oncogenic and ligand-dependent activation of KIT/PDGFRA in surgical samples of imatinib-treated gastrointestinal stromal tumours (GISTs). J Pathol. 2009;217:103–112. doi: 10.1002/path.2450. [DOI] [PubMed] [Google Scholar]
- 17.Zhu MJ, Ou WB, Fletcher CD, et al. KIT oncoprotein interactions in gastrointestinal stromal tumors: therapeutic relevance. Oncogene. 2007;26:6386–6395. doi: 10.1038/sj.onc.1210464. [DOI] [PubMed] [Google Scholar]
- 18.Lewis NL, Lewis LD, Eder JP, et al. Phase I study of the safety, tolerability, and pharmacokinetics of oral CP-868,596, a highly specific platelet-derived growth factor receptor tyrosine kinase inhibitor in patients with advanced cancers. J Clin Oncol. 2009;27:5262–5269. doi: 10.1200/JCO.2009.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich MC, Griffith D, McKinley A, et al. Crenolanib inhibits the drug-resistant PDGFRA D842V mutation associated with imatinib-resistant gastrointestinal stromal tumors. Clin Cancer Res. 2012;18:4375–4384. doi: 10.1158/1078-0432.CCR-12-0625. [DOI] [PubMed] [Google Scholar]
- 20.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 21.Ro S, Park C, Jin J, et al. A model to study the phenotypic changes of interstitial cells of Cajal in gastrointestinal diseases. Gastroenterology. 2010;138:1068–1078. e1061–1062. doi: 10.1053/j.gastro.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton TG, Klinghoffer RA, Corrin PD, et al. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorincz A, Redelman D, Horvath VJ, et al. Progenitors of interstitial cells of cajal in the postnatal murine stomach. Gastroenterology. 2008;134:1083–1093. doi: 10.1053/j.gastro.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taguchi T, Sonobe H, Toyonaga S, et al. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest. 2002;82:663–665. doi: 10.1038/labinvest.3780461. [DOI] [PubMed] [Google Scholar]
- 25.Lux ML, Rubin BP, Biase TL, et al. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–795. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer S, Yu LK, Demetri GD, et al. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer Res. 2006;66:9153–9161. doi: 10.1158/0008-5472.CAN-06-0165. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi Y, Asuzu DT, Gibbons SJ, et al. Membrane-to-nucleus signaling links insulin-like growth factor-1- and stem cell factor-activated pathways. PLoS One. 2013;8:e76822. doi: 10.1371/journal.pone.0076822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.