Abstract
Cryo-electron microscopy (cryo-EM) was used to solve the structures of human papillomavirus type 16 (HPV16) complexed with fragments of antibody (Fab) from three different neutralizing monoclonals (mAbs): H16.1A, H16.14J, and H263.A2. The structure-function analysis revealed predominantly monovalent binding of each Fab with capsid interactions that involved multiple loops from symmetry related copies of the major capsid protein. The residues identified in each Fab-virus interface map to a conformational groove on the surface of the capsomer. In addition to the known involvement of the FG and HI loops, the DE loop was also found to constitute the core of each epitope. Surprisingly, the epitope mapping also identified minor contributions by EF and BC loops. Complementary immunological assays included mAb and Fab neutralization. The specific binding characteristics of mAbs correlated with different neutralizing behaviors in pre- and post-attachment neutralization assays.
Keywords: Cryo-EM, virus Fab complex, HPV16, Neutralization, mAb, crosslinking, stabilization, epitope, human papillomavirus, conformation
INTRODUCTION
Human papillomaviruses (HPVs) cause epithelial tumors and are the etiologic agents of numerous anogenital and oropharyngeal cancers (1–3). Identification of neutralization-sensitive epitopes on the capsid protein structures (conformational epitopes) support investigations to develop improved recombinant vaccines that maximize effective and long-term antibody-mediated protection against multiple HPV types (4). As one of the major cancer-causing HPV types, HPV16 is extensively studied (1, 3, 5–7), and together with HPV18 comprises a major target for vaccine development (8, 9). Since the life cycle of HPVs rely on differentiation of basal cells into keratinocytes, purifying high titer virus stocks for structural studies is difficult. Therefore, other production methods have been developed as an alternative for studies of the native virions. Virus-like particles (VLPs) are comprised of only the major structural protein, L1, and are not infectious since they are devoid of viral genome (10). Quasivirions (QV16) and pseudovirions (PsV16) were used for our structural analysis and neutralization assays (11, 12) as both types of HPV 16 particles contain a mock genome.
Papillomaviruses form a T=7 icosahedral, non-enveloped ~55–60 nm diameter capsid containing a circular dsDNA genome of 8Kb. The capsid is comprised of 360 copies of the L1 structural protein and up to 72 copies of the L2 minor structural protein (12, 13). Five L1 proteins intertwine to form each capsomer, 72 of which make up one capsid. Twelve of the 72 capsomers lie on an icosahedral fivefold vertex and are described as pentavalent capsomers. The remaining 60 capsomers are each surrounded by six other capsomers and are consequently referred to as hexavalent capsomers. The C-terminus, or “C-terminal arm,” of each L1 protein extends along the capsid floor to interact with the neighboring capsomer and then returns to the original donor capsomer (14–16). Inter-capsomer disulfide bonds are formed between cysteine C428 and C175, which stabilize the capsid structure and play an important role in virus maturation (15, 17). The core of the capsomer is composed of the common viral structural motif, the antiparallel β-strands BIDG and CHEF (18), which are connected by surface loops of BC, DE, EF, FG, and HI. Nearly all conformational epitopes are located on one or more of these outwardly facing surface-exposed loops (19). Our knowledge of these epitopes has been largely obtained from mAb/Fab binding and neutralization assays (4, 20–22), hybrid virus loop exchange studies (23), and previous structural analysis (16, 24). These complementary studies represent an important approach to analyze the nature of conformational epitopes, neutralization mechanisms, and how the host immune system recognizes and responds to the virus.
H16.V5 is a well-characterized HPV16-specific neutralizing mAb induced by HPV16 L1 VLPs. This mAb has been extensively used in major HPV vaccination trials and is an especially important tool in inhibition-based HPV serological assays (8, 19, 20, 25–27). The neutralizing antibodies of H16.1A, H16.14J, and H263.A2 were raised against HPV16 L1 VLP (20) or hybrid capsids (39). Like H16.V5, based on previous immunological studies, all three antibodies were thought to recognize portions of the FG and HI loops. The H16.V5 neutralization mechanism has been shown to be one of capsid stabilization that consequently inhibits the conformational changes required during entry (8, 26–28). Although many immunological studies of H16.V5 neutralization have been published, no information on H16.V5 Fab has been recorded. For the three antibodies H16.1A, H16.14J, and H263.A2, details of neutralization are unknown.
Previously, two HPV16-H16.V5 complex cryo-EM maps of 20 Å (29) and 10 Å (16) resolution showed that H16.V5 Fab binding induced conformational changes and bound predominately to the hexavalent capsomers. Here we present three new cryo-EM structures of HPV16 complexed with the Fabs from the specific mAbs, H16.1A, H16.14J, and H263.A2 at ~12 Å resolution (Fig. 1). Atomic structures of the component parts, virus and Fab, were fitted into the cryo-EM complex maps using rigorous fitting algorithms developed for this purpose (30–32). The resulting pseudo-atomic model was used to define the Fab binding sites and identify the amino acids that likely comprise the complex conformational epitope (30, 32, 33). We found that besides the well-known FG and HI loops, the DE loop also composes the core of each footprint; however, additional participation by BC and EF loops vary between the antibodies. The structural results were complemented with immunological studies that showed Fabs from all four neutralizing antibodies are also neutralizing, albeit at higher molar concentrations. H16.1A and H16.14J Fabs can distinguish conformational changes of the capsid after host cell attachment, whereas H16.V5 and H263.A2 do not. Differences in the antibody footprints correlate with these immunological differences. Thus, the structure-function study predicts the neutralization mechanisms to be a combination of stabilization and cross-linking.
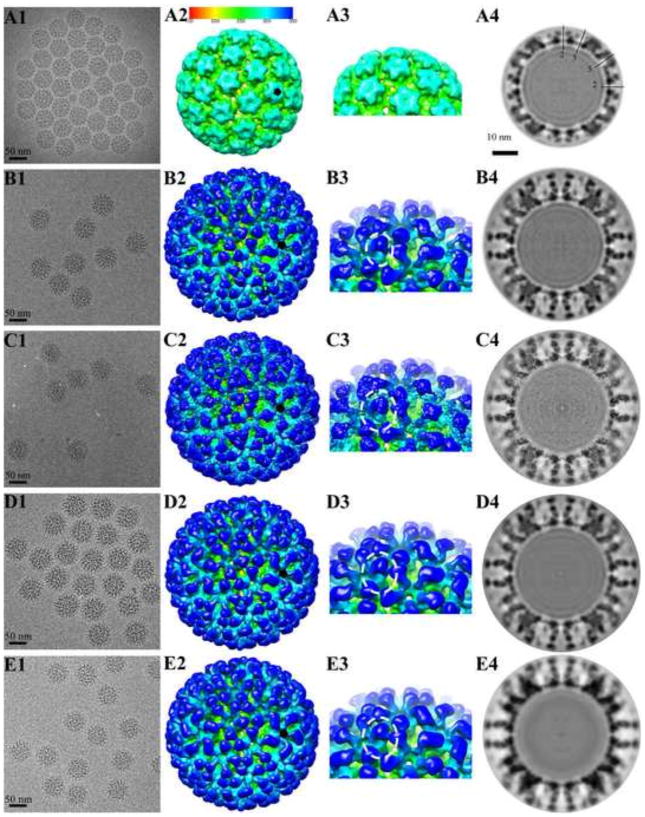
FIGURE 1. Cryo-EM reconstructions of HPV16 and HPV16-Fab complexes.
Representative regions of cryo-EM micrographs are shown for (A1) the HPV16 capsid (EM Database, EMD 5993) (16), (B1) HPV16 capsids complexed with H16.V5 (EM Database, EMD 5994) (16), (C1) H16.1A, (D1) H16.14J, and (E1) H16.263A2. (A2–E2) The 3-D complexes were radially colored according to the distance from the center of the capsid (color bar indicated) and surface rendered at 1σ. A small black pentagon indicates the position of a pentavalent capsomer. (A3–E3) In the zoomed in views of the complex maps the separate heavy chain and light chain Fab densities can be distinguished. (A4–E4) The central sections through the cryo-EM density maps were displayed in the same sequence as above. Capsids were cut vertically through the 2-, 3- and 5-fold icosahedral symmetry axes (black lines), with the central 2-fold axis appearing at the 12 o’clock position, and a scale bar equal to 10 nm.
EXPERIMENTAL PROCEDURES
Preparation of HPV16 quasivirus and pseudovirus
QV16 virions are comprised of HPV16 L1 and L2 proteins and encapsidate the cottontail rabbit papillomavirus genome (CRPV) containing the SV40 origin of replication. QV16 were prepared as described previously (34–36). PsV16 virions are comprised of the HPV16 L1 and L2 proteins and contain the pYSEAP (alkaline-phosphatase producing genome). Briefly, HPV16 sheLL plasmid (kindly provided by John Schiller, NIH) was transfected together with linear CRPV/SV40ori DNA (QV) or pYSEAP (PsV) into 293TT cells and prepared as described previously (12, 37). PsVs were purified by Optiprep gradient centrifugation as previously described (38). QVs were allowed to mature overnight and then pelleted by centrifugation. The centrifuged pellet was re-suspended in 1 M NaCl 0.2 M Tris, pH 7.4. After CsCl gradient purification, the lower band was collected, concentrated, and dialyzed against PBS, as described previously (16). The concentrated QVs were applied to Formavar-coated copper grids, stained with 2% phosphotungstic acid, and analyzed for integrity and concentration on a JEOL JEM 1400 electron microscope.
Preparation of Antibody and Fabs
Antibodies of H16.V5, H16.1A, H16.14J and H263.A2 were generated in Balb-C mice as described previously (20, 39). Hybridomas were acclimated to animal component free media (BD) and supernatant was purified on Protein A or G columns (Pierce). Fab was prepared by digestion with papain in the presence of cysteine (Pierce). Purity of the Fab was assessed by the lack of the fragment crystallizable (Fc) portion and integrity of the Fab was determined by ELISA. Antibody and Fab protein concentrations were determined by absorbance spectrometry at a wavelength of 280 nm.
Sequencing of antibody heavy and light chains
The hybridoma cells were pelleted by centrifugation and RNA was extracted using TRIzol® Reagent (Life Technologies). Total RNA was treated with DNase I (RNase-free) (New England Biolabs) to digest potentially contaminating DNA in the sample. cDNAs were synthesized from treated RNA with the RevertAid First Strand cDNA synthesis kit (Thermo Scientific). The cDNAs were used as a template for PCR and amplified using Pfu Turbo DNA Polymerase (Agilent) or Choice Taq DNA polymerase (Denville). PCR amplification used primers previously described by Wang et al (40). Immunoglobulin heavy chains were amplified using the isotype specific constant region 3′ primer and two highly degenerate 5′ primers. The light chains were amplified using the 3′ degenerate kappa chain constant region primer and the 5′ kappa chain framework one region universal degenerate primer. Prior to sequencing, PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). The same primers used for PCR amplification were also used as sequencing primers to obtain initial sequences. Resolution of the 5′ and 3′ ends of the sequence required sequence specific primers. The 263.A2 light chain required cloning into pUC19 to completely resolve the sequence.
Neutralization assays
The neutralization activity of mAbs and Fabs was investigated by pre- and post-virus attachment assays in 293TT cells. In the pre-attachment assay, PsVs were pre-incubated with mAb or Fabs at dilutions ranging from 66 nM-0.067 pM for 1 hour at 37 °C before adding to cells. For the post-attachment assay, PsVs were incubated with 293TT cells for 1 hour at 4 °C to allow for attachment. Cells were washed 1x with media prior to incubation with mAbs or Fabs. 72 hours later 30 ul of the cell culture supernatant was assayed with 4-Nitrophenyl phosphate disodium salt hexahydrate (pNPP) (Sigma) and the optical density was determined by absorbance spectrometry at OD405/490. The neutralization data was plotted at each antibody concentration and was used to determine the half-maximum neutralization titer.
Cryo-Electron Microscopy
To produce HPV16-H16.1A complexes, purified HPV16 virions and Fabs were incubated for one hour at room temperature and concentrated to 1.2 mg/ml in PBS buffer. An aliquot of 3 μl of complex was pipetted onto a Quantifoil R2/1 grid (Quantifoil Micro Tools GmbH, Jena, Germany) and blotted to remove excess sample before plunging into a liquid ethane-propane mixture using an Mk III Vitrobot (FEI, Hillsboro, OR) (41). Low-dose conditions were used to record images on Kodak SO-163 film (Kodak, Rochester, NY) in an FEI TF-20 cryo-electron microscope, which was operated at 200 kV at a nominal magnification of 50,000X. The microscope was equipped with a Gatan 626 cryo-holder (Gatan, Inc., Pleasanton, CA). Cryo-EM images were collected with a defocus range of 1.78–3.96 μm. Films were scanned using a Nikon Super Coolscan 9000 (Nikon, Melville, NY), giving a calibrated pixel size at the sample of 1.3 Å/pixel. A similar procedure was used to collect data for the HPV16-H16.14J and -H263.A2 complexes, which were vitrified on Quantifoil holey carbon support grids (Quantifoil, Jena, Germany) that were plunged into liquid ethane using a Cryoplunge 3 (Gatan, Pleasanton, CA). Low dose conditions were used to record digital images on an Ultrascan 4000 CCD camera (Gatan, Pleasanton, CA) in a JEOL 2100 LaB6 cryo-EM microscope (JEOL, Peabody, MA) operating at 200 kV and equipped with a Gatan 626 cryo-holder. The recorded CCD data for HPV16-H16.14J and H263.A2 complexes had calibrated pixel sizes of 2.33 Å/pixel, and 2.86 Å/pixel; nominal magnifications of 50,000X, 40,000X; and defocus ranges of 0.62–4.69 μm, and 1.49–5.52 μm (Table 1), respectively. AUTO3DEM and EMAN2 program suites were used for all image processing and 3D reconstructions (42, 43).
TABLE 1. CryoEM image reconstruction data.
| HPV16-V5* | HPV16-1A | HPV16-14J | HPV16-263A2 | |
|---|---|---|---|---|
| Number of micrographs | 411 | 80 | 385 | 264 |
| Defocus level range (μm) | 0.69–3.99 | 1.78–3.96 | 0.62–4.69 | 1.49–5.52 |
| Number of particles selected from micrographs | 2306 | 2071 | 7074 | 8908 |
| Number of particles used for reconstruction | 2075 | 1657 | 5642 | 6231 |
| Final resolution (Angstrom) | 12.0 | 10.6 | 12.0 | 12.1 |
Icosahedral reconstruction
Virus-Fab complexes were selected from micrographs and used for calculating 3D reconstructions (Table 1). Semiautomatic particle selection was performed using EMAN2’s e2boxer.py to obtain the particle coordinates, followed by particle extraction, linearization, normalization, and apodization of the images using AUTO3DEM (42, 43). Each icosahedrally averaged reconstruction was initiated using a random model generated from the raw data (44) and the resolution was estimated where the Fourier Shell Correlation (FSC) dropped below 0.3 (Fig. 2). To correct for contrast transfer function, defocus and astigmatism values were assessed over the digitized images using ctffind3 and correction was applied in AUTO3DEM (42, 43).
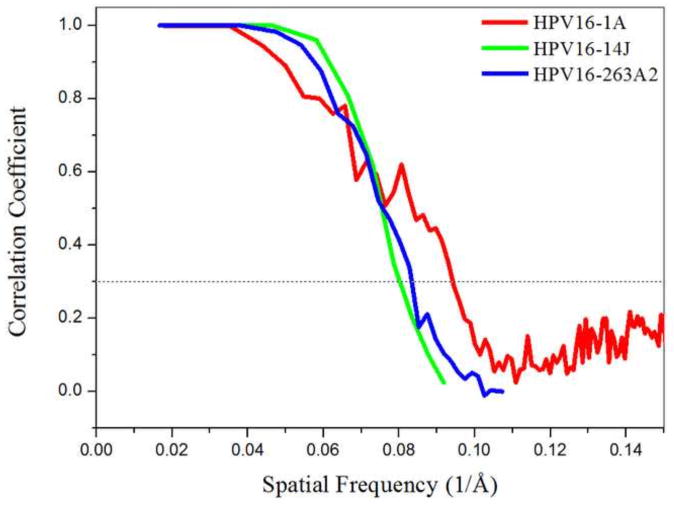
FIGURE 2. Plot showing Fourier shell correlation (FSC) versus spatial frequency of the icosahedrally averaged reconstructions for each of the virus-Fab complexes.
The resolutions of the reconstructions (10.6 Å for HPV16-1A, 12.0 Å for HPV16-14J, and 12.1 for HPV16-263.A2) were assessed where the FSC curve crossed a correlation value of 0.3. Due to different pixel sizes in the reconstructions, the FSC curves ended at different frequency values.
Fitting the Fab structures into corresponding cryo-EM density
Following a classic fitting protocol (30, 31, 45), the difference maps were calculated by scaling and subtracting the virus density from each HPV16-Fab complex map. Four Fab densities marked as 1 to 4 on the asymmetric unit of HPV16 capsid were used for fitting, excluding the two clashing Fab densities near the pentavalent capsomer (Fig. 3A). Fab structural models were predicted according to the amino acid sequence using the Rosetta Online Server, ROSIE (http://rosie.rosettacommons.org/antibody) (46). These ROSIE models were fitted into the related densities in each of the HPV16-Fab complexes with the crystal structure of a murine Fab used for fitting the constant domain (PDB ID 3GK8) (33). The Fabs were fitted stepwise, with the constant domain being fitted prior to placing the ROSIE model for each variable domain. Four of the six Fabs corresponding to one asymmetric unit were fitted into the difference map. The resulting fitted structure was then placed into the complex map and refined simultaneously with the crystal structure of the HPV16 L1 pentamer (Protein Data Bank PDB ID 3OAE) (47) using Situs (48). However, to fit successfully H263.A2, the approach had to be modified to include fitting of the variable domain simultaneously with constant domain of 3GK8 (48, 49). Contacts between the fitted crystal and ROSIE structures were identified using Chimera with the criteria for van der Waals overlap distances of −0.4 and 0.0 Å, respectively. All glycines in the interface were excluded from the epitope due to the limited likelihood of contributing to bonds or antigenicity.
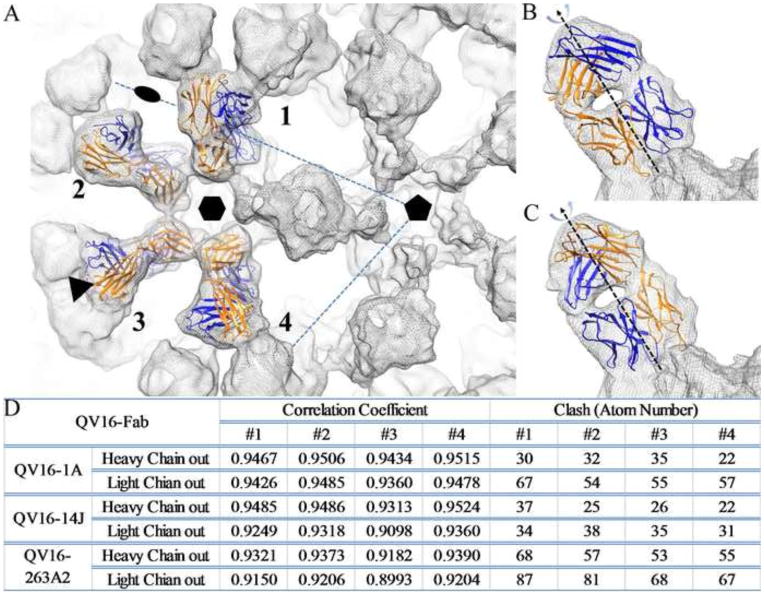
FIGURE 3. Fitting of four Fabs on one asymmetric unit of capsid.
(A) Four Fab densities marked as 1 to 4 on an asymmetric unit of the HPV16 capsid were used for fitting. The icosahedral 2-fold, 3-fold and 5-fold symmetry axes are indicated and the hexavalent capsomer is marked on the virus-Fab complex density map (grey mesh). Due to the pseudo-two-fold symmetry axis between Fab heavy (Blue) and light chains (Orange), two fitting modes were used for refinement of fit: the Fab heavy chain facing outwards from the center of the capsomer (B) and Fab light chain facing outwards (C). Statistics for the fitted Fab structures into corresponding cryo-EM density are reported for both fitting modes (D) according to the correlation coefficient and the number of atomic clashes with the fitted virus structure (crystal structure of the HPV16 L1 pentamer, Protein Data Bank, PDB ID 3OAE) (47)).
To identify the correct Fab placement, the Fab was fitted and refined in two orientations according to its pseudo-two-fold symmetry axis between the heavy and light chain (Fig. 3B and C). The fitting quality was evaluated by four structural criteria: correlation coefficient number, number of clashes (Fig. 3D), elbow angle, and surface charge (49). Stereographic projections were made using the program RIVEM (50). The buried surface area was calculated using a 1.70 Å probe through the CCP4 program AREAIMOL (51).
Cryo-EM maps for the capsid and capsid-Fab complexes are deposited in the EM database (www.emdatabank.org/) with accession numbers EMD-5990 (capsid-H16.1A), - 6121 (capsid-H16.14J), and -6184 (capsid-H263.A2). Fitted structures of Fab (ROSIE) and PDB ID 3OAE are deposited in the PDB: 3J8Z (H16.1A), 3J8V (H16.14J) and 3J8W (H263.A2) respectively.
RESULTS
The Cryo-EM image reconstructions for each of the three Fab-labeled HPV16 complexes showed strong Fab densities
For each complex, HPV16 virus was incubated with excess Fab and the quality of the HPV16-Fab preparation was confirmed by negative stain transmission electron microscopy (TEM) before proceeding to cryo-EM data collection. The icosahedral symmetry of the HPV16-Fab complexes was readily apparent in low-dose cryo-EM images (Fig. 1B1 to E1). All the Fab-complexed particles displayed nearly spherical outer profiles with a highly uniform diameter around 71–72 nm, which was significantly larger than the 59 nm diameter of the virus alone (Fig. 1A1) and consistent for HPV with Fab bound (16). Uniformly, the virus-Fab complex maps clearly showed bulbs of Fab density bound in nearly saturating amounts.
The three dimensional (3D) reconstructions (Fig. 1A2 to E2) were comparable with resolutions of 11–12 Å, as estimated where the Fourier shell correlation (FSC) dropped below 0.3 (Fig. 2). Fab density was located on the distal-most surface of each L1 capsomer unit bound at ~40 degree angle relative to the virus capsid. The heavy and light chain domains were clearly identified in the Fab densities which covered the top of the star-shaped capsomers, leaving only the small central dimple free of Fab (Fig. 1A3 to E3). Central sections through the cryo-EM density maps of capsids and Fab-labeled capsids illustrated the magnitude of the protein density (Fig. 1A4 and E4) and the quality of the cryo-EM maps. The Fab densities with clear constant and variable domains were identified near the edge of the capsid shell and it was evident that the Fab fragments bound to the distal tips of the capsomers.
Most of the Fabs from adjacent capsomers were in close proximity to one another and radiated outwardly from the intra-capsomer regions without steric interference. This mode of binding allowed near saturation of binding sites as attested by the Fab density being nearly equal in magnitude to that of the capsid. For every Fab binding to a pentavalent capsomer site there was a clash with the nearest Fab bound to the neighboring hexavalent capsomer (Fig. 3A). Thus due to these steric limitations only one Fab was bound to either the pentavalent or hexavalent site, which made the total occupancy decrease from 360 to 300.
Atomic models were generated for Fabs based on sequence
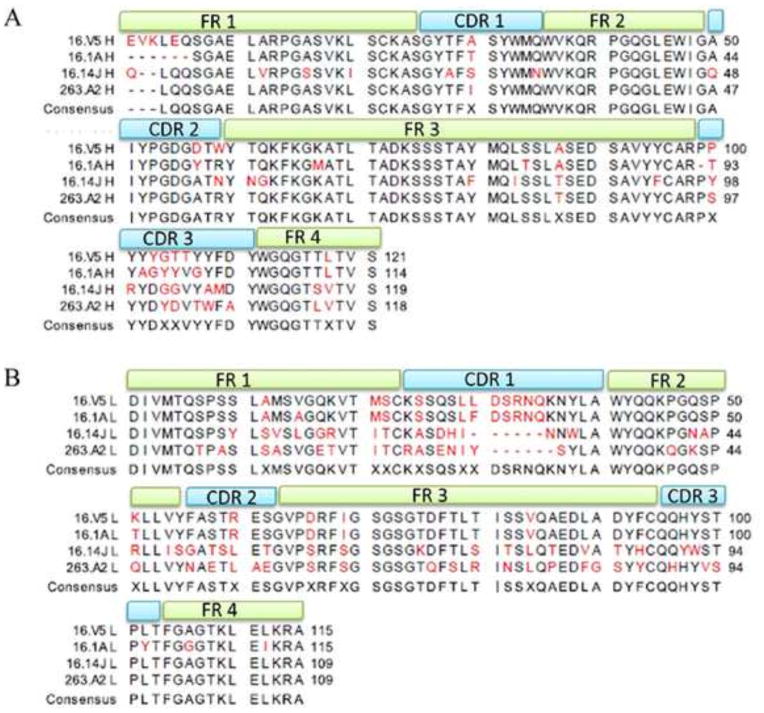
The sequence of the variable regions of H16.1A, H16.14J, and H263.A2 were aligned and annotated so that the complementarity determining regions (CDRs) of both the antibody heavy and light chains could be assigned using the Rosetta online server (ROSIE) (Methods). Because the atomic structures of the Fabs were unknown, a ROSIE model was predicted for each based on the high structural conservation of the constant domains and the sequence of the variable domains (46). Although the virus-H16.V5 cryo-EM structure was described elsewhere (16), the sequence of H16.V5 is reported for the first time here. Most of the differences between Fabs were found in the CDRs (Fig. 4). Among the heavy chain sequences, H16.14J had the most variability within the framework region. However, compared to the light chain sequences, the heavy chains are largely conserved. Among the differences between the light chains, H263.A2 and H16.14J appear to be the most divergent from H16.1A and H16.V5. Additionally, H263.A2 and H16.14J have the most differences within the framework region compared to the H16.V5 and H16.1A sequences.
FIGURE 4. Sequence alignment of the heavy chain (A) and light chain (B) variable regions of H16.1A, H16.14J, H16.V5, and H263.A2.
Complementarity determining regions (CDR 1–3) were annotated at the top of the alignment with each framework region (FR 1–4) annotated in the same manner. The consensus sequence was displayed as the last sequence and red residues indicate differences among the sequences.
Fitting of Fab models and capsid atomic structures into the cryo-EM density maps
For fitting experiments, each of the three complex maps was used to calculate a difference map by subtracting a scaled virus map of similar quality. Into each difference map, the correct ROSIE Fab model was fitted and refined within each of the three complete Fab difference densities in the asymmetric unit of the capsid (Fab1-4) (Fig. 3A). The fitted Fabs were then placed into the corresponding complex map where they were simultaneously refined with the virus pentamer structure (PDB ID: 3OAE) (47) using the program Situs (48). Because of the Fab pseudo-2-fold symmetry axis, the heavy and light chains could be fitted into the same density in two orientations related by a 180-degree rotation (Fig. 3B and C). Each Fab was fitted and refined in both possible orientations, and the quality of the different docking modes was quantified by different fitting criteria (Fig. 3D). The correlation coefficient (cc) numbers for all fittings were high (>0.9); the difference (~ 1%) between the two orientations was too slight to use the cc alone to assign correct placement of the Fab. For each, the angle between the two pseudo-dyad rotation axes of the variable heavy and light chain and constant heavy and light chain (elbow angle) of the Fab was in the range of 140° to 180° which was within the observed range for Fab elbow angles of 127° to 225° (52). Thus the correct orientations of the Fabs could also not be identified by elbow angle alone.
The number of atoms between the fitted Fab and pentamer structures that were identified to be in positions of steric collision was then used to evaluate the correct binding mode. An orientation with the heavy chain facing away from the center of the capsomer (outward) resulted in fewer clashing atoms compared to the light chain facing outward mode for two of the three Fabs: H16.1A, and H263.A2. However, this approach was inconclusive for H16.14J because a similar number of clashes were defined in both orientations. To assess further the binding orientation for H16.14J, a Coulombic Surface Color model was generated for the fitted Fab and capsomer (data not shown). Based on the predicted charge interactions, the heavy chain facing-out model was proposed for H16.14J binding. The fitting of each Fab was refined using the heavy chain outwards orientation and the buried surface area was calculated. H16.V5 had a buried surface of 1467.7 Å2 calculated on the previously published binding mode (16). H263.A2 had a similar area of 1402.5 Å2; however both H16.1A (1159.8 Å2) and H16.14J (927.7 Å2) had significantly smaller buried surface.
Epitope analysis showed multiple L1-loop participation
The refined fitting of the Fab and capsomer structures produced a pseudo atomic model that was used for epitope analysis. Based on distance and geometry, all potential interactions between Fab and virus were identified (Table 2). Each Fab, including H16.V5, mapped to the surface of the virus with a distinct and different footprint. However, each of the four footprints involved multiple loops from more than one L1 protein. The epitope for H263.A2 included loops from three of the L1 proteins comprising the capsomer whereas H16.V5 (16), H16.1A, and H16.14J involved two neighboring copies of L1. Other specific differences included that H16.V5 and H16.1A did not have any BC loop interactions and H16.14J had the fewest DE and FG loop interactions.
TABLE 2. The pseudo-atomic models identified residues of HPV16 predicted to interact with Fab and define the antigenic epitopes.
In the contact region, two or three L1 proteins of the capsomer interacted with each Fab. For each footprint the copy of the L1 protein that made the major contributions is designated with a blue X, and additional minor interactions were made by a second neighboring L1 (indicated by red X′). In only one case, mAb HPV.263.A2, there was a third copy of L1 involved in the conformational epitope (green X″). The H16.V5 result included here was from our previously published paper (16). The alternative mode of binding for the light chain facing out has been included as supplemental material.
| L1 Protein Surface Loop | Fab | |||||
|---|---|---|---|---|---|---|
| V5* | 1A | 14J | 263A2 | |||
| BC | 54 | LYS | X′ | |||
| 55 | PRO | X′ | X′ | |||
| DE | 135 | TYR | X | X | ||
| 136 | ALA | X | X | |||
| 137 | ALA | X | X | |||
| 138 | ASN | X | X | X | X | |
| 139 | ALA | X | X | X | X | |
| 141 | VAL | X | X′ | |||
| 142 | ASP | X | X′ | |||
| 143 | ASN | X | ||||
| EF | 181 | GLN | X | X | X | X |
| 182 | PRO | X | X | |||
| 184 | ASP | X | ||||
| FG | 267 | VAL | X | |||
| 269 | GLN | X | ||||
| 270 | ASN | X | X | |||
| 273 | ASP | X | ||||
| 278 | LYS | X | X | |||
| 280 | SER | X | X | X | ||
| 282 | SER | X | X | X | X | |
| 283 | THR | X | X | |||
| 284 | ALA | X | X | |||
| 285 | ASN | X | X | X | X | |
| 286 | LEU | X | X | |||
| 287 | ALA | X | ||||
| HI | 348 | ILE | X′ | X′ | X′ | |
| 354 | THR | X′ | X″ | |||
| 358 | THR | X′ | X′ | X′ | X′ | |
| 361 | LYS | X′ | X′ | X′ | X′ | |
Distances between fitted Fabs indicate monovalent or bivalent binding modes
For an antibody to be able to bind bivalently, the distance between the C-terminal Cα atoms of adjacent heavy chains is typically in the range of 25–29 Å (52–54). For the fitted Fabs, the distance was measured across the two-, three- and five-fold symmetry axes (Table 3) between the closest adjacently bound Fabs. H16.V5, H16.14J, and H263.A2 bound to the capsid at distances ranging from 35–154Å, depending on the orientation, suggesting they are incapable of bivalent binding. However, at the three-fold symmetry axis, H16.1A Fab molecules were 27.5Å apart, indicating that bivalent binding might be possible.
TABLE 3. The distance between C-terminal heavy chain Cα atoms between the nearest icosahedrally related adjacently bound Fabs.
Since there were three pairs of Fabs related by icosahedral two-fold symmetry, three distances were measured separately and listed in the table. Calculation of H16.V5 was included for comparison and was based on our previously published fitting result (16).
| Fab Binding with Heavy Chain Outwards | Distance (Å) to nearest symmetry-related Fab Heavy Chain Outwards | ||
|---|---|---|---|
| Two-Fold | Three-Fold | Five-Fold | |
| V5 | 93.8/97.2/154.8 | 36.6 | 81.2 |
| 1A | 67.9/108.3/139.5 | 27.5 | 76.4 |
| 14J | 94.8/95.5/152.3 | 35.5 | 79.9 |
| 263A2 | 92.9/95.8/154.8 | 35.3 | 78.9 |
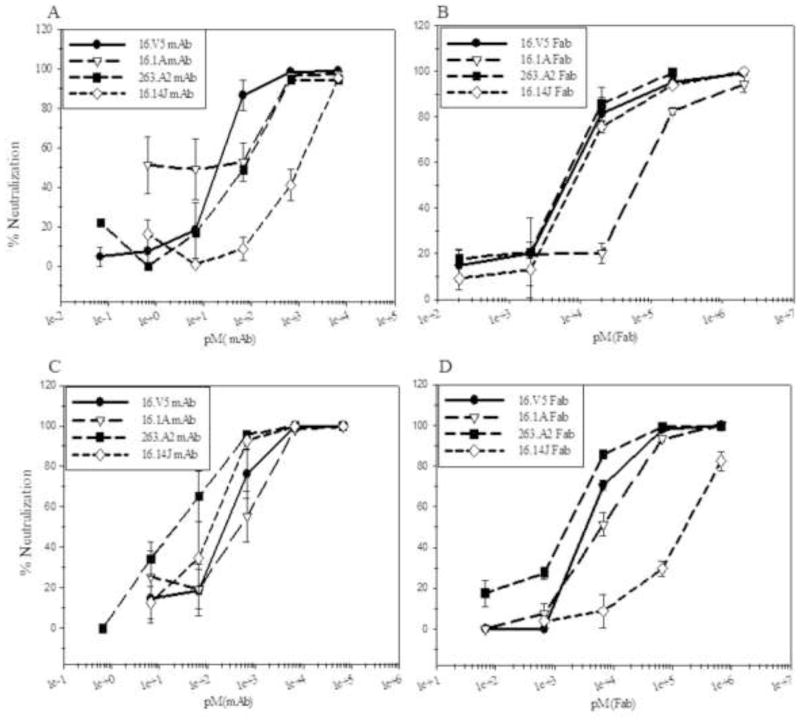
All four mAbs, H16.V5, H16.1A, H16.14J, and H263.A2 neutralize better than their Fabs
Intact mAb or Fab was pre-incubated with virus and applied to host cells in pre-attachment neutralization assays. Additionally, virus was allowed to attach to the cell surface first, followed by the addition of mAb or Fab in post-attachment neutralization assays. The mAb neutralization ability was similarly efficient between pre- and post- attachment assays for all of the four antibodies (Fig. 5A and C). Fab from each of the mAbs tested was capable of neutralization; however, at a much reduced efficiency (Fig. 5B and D). H16.1A Fab neutralized better in post-attachment than pre-attachment assays, whereas the reverse was true of H16.14J that neutralized better in pre-attachment assays. In each case, the intact antibody was more efficient at neutralization than the respective Fab, suggesting the bivalency of the mAbs may provide an enhancement in neutralization.
FIGURE 5. Neutralization was assessed pre- and post-attachment to host cells.
PsVs were pre-incubated with mAb (A) or Fab (B) at the indicated dilutions at physiological temperature prior to adding to cells. Alternatively, PsVs were incubated with cells for 1 hour for attachment before applying mAb (C) or Fab (D). The neutralization titers were graphed as % neutralization (Methods).
DISCUSSION
By fitting the atomic structures of Fab and virus into the ~12 Å cryo-EM complex density maps, the structures could be analyzed at near-atomic resolution (30, 31, 45). Because a Fab structure has a pseudo-dyad axis, two orientations related by a 180° rotation can be generated when fitting a Fab structure into density. The correct orientation for the fitted Fabs was identified according to multiple criteria, including correlation coefficient, steric interference between fitted molecules, Fab elbow angle, and surface charge compatibility (Fig. 3D). The best binding mode was determined to be with the heavy chain facing out, away from the center of the capsomer. Results from previous immunological studies (23, 26) corroborate this mode of binding. For example, BC loop interactions have been shown for H16.14J using hybrid loop exchange experiments (23). For the fitted H16.14J Fab, interactions with the BC loop were only detected when the Fab was placed into the density with the heavy chain facing outward. For H16.1A and H263.A2, EF loop participation was demonstrated previously (23) and EF interactions were only identified in the Fab fitting with the heavy chain facing outward. Nevertheless, the interactions predicted for fitted Fabs with the light chain facing outward have been included as supplementary material.
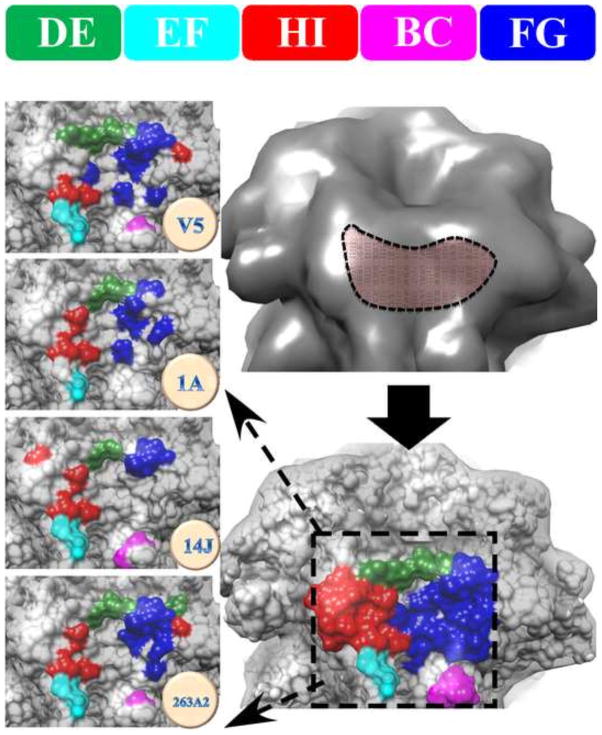
All four antibody footprints involved multiple loops from two or three L1 capsid proteins (Table 2, Fig. 6 and 7). Collectively the Fab footprints on the surface of the virus mapped to a topographical “groove” in the capsomer (Fig. 6). Most of the FG, HI and DE loops contributed to this groove whereas only smaller portions of the EF and BC loops were included. The Fab molecules filled the grooves on the capsomer crown, perhaps serving as a physical obstacle to block the loop movements necessary during virus conformational changes. Due to this mode of Fab binding and the interaction with multiple loops from more than one L1 molecule, the Fabs likely neutralize by stabilizing the capsomer and locking it into one conformation. As was shown previously with H16.V5, loop stabilization at the binding site was propagated throughout the capsid leading to hyperstabilization (16).
FIGURE 6. The location of the antibody binding groove on the surface of capsomer was made up of FG and DE loops with lesser contributions by the BC, EF, and HI loops.
All four epitopes map to a “groove” feature on the surface of the capsomer (upper right, dashed outline). The combined four footprints are color coded according to loop identity (lower right, dashed box). The left column shows the zoomed view of the individual epitopes of H16.V5, H16.1A, H16.14J and H263.A2 with specific loop contributions indicated by color (Top: color code bar for DE, EF, HI, BC, and FG).
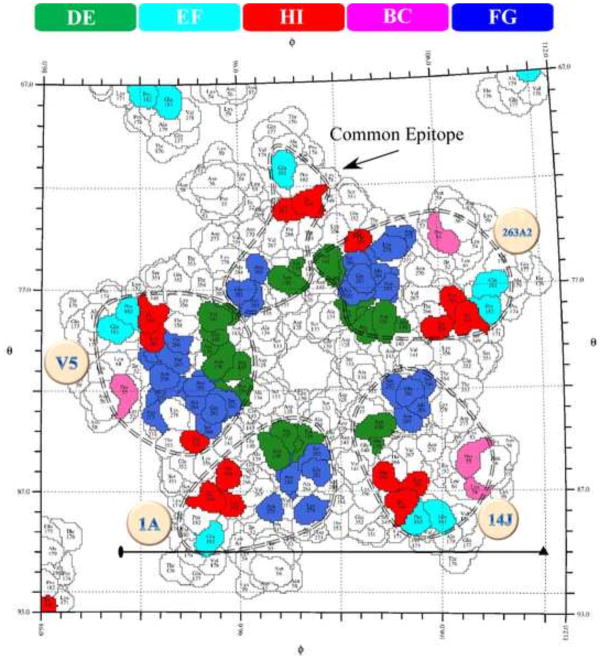
FIGURE 7. The roadmap shows the footprint of each antibody mapped to the stereographic projection of a capsomer.
The virus surface was represented as a quilt of amino acids, shown as a projection, for the icosahedral asymmetric unit, with the polar angles φ and θ representing the latitude and longitude (58). As in figure 6, the color bar identifies the contribution of the different antigenic L1 loops to each mAb epitope (dashed circle). The common epitope (arrow) includes all residues identified in each mAb footprint.
Since the internalization of bound virions is a slow process with a half-life of hours (55, 56), the virions remain accessible on the cell surface. However, virus binding to cell receptors triggers conformational changes to the capsid (29, 66, 67). Thus, during the post attachment assays, conformational changes of the capsid triggered by attaching to the cell may have affected binding and neutralization. H16.V5 and H236.A2 Fabs showed only slight differences of neutralization ability between pre- and post-attachment (Fig. 5), which suggests the capsid conformational changes had little or no effect on the antibody footprints for these Fabs. There are significant differences in pre- and post- attachment neutralization by H16.1A and H16.14J Fabs (Fig. 5) suggesting that capsid conformational changes overlap with the epitopes of these two Fabs.
Footprints for H16.1A and H16.14J involved fewer residues on the virus surface as was seen by the significantly smaller buried surface compared to H16.V5 and H263.A2 (around 20% to 35% smaller area). Specifically, our results predict H16.14J will make fewer interactions with DE and FG loops whereas the epitope of H16.V5, H16.1A and H263.A2 showed larger “core areas” formed by FG and DE loops with less participation of BC, EF, and HI loops (Table 2). Thus H16.14J, with fewer key interactions and less buried surface, must rely on the coordination of all five loops for an interaction and may have been more affected by the virus changing conformation during capsid attachment. Whilst for H16.1A, an enlarged or improved epitope may have been formed during L1 conformational changes during cell attachment, which enhanced the neutralizing ability of H16.1A. H16.V5 and H263.A2 were not affected by virus attachment, probably because these two antibodies have more DE and FG loop interactions and larger buried surface.
Mapping each epitope onto the roadmap of a single capsomer showed the similarities and differences (Fig. 7). The virus residues that interact with all four mAbs were mapped as a common epitope that consisted of ASN138.DE, ALA139.DE, GLN181.EF, SER282.FG, ASN285.FG, ILE348.HI, and LYS361.HI (Fig. 6). Among these seven residues may be the essential interactions for effective neutralization.
Participation of multiple loops from different L1 proteins for the antibodies agrees with previous studies suggesting a stabilization mechanism of neutralization (16, 21, 22, 27). However, mAbs showed significantly higher neutralization efficiency than Fabs, suggesting that the bivalency of the antibody may have a role to play in addition to the hyper-stabilization mechanism induced by the Fab-capsid interaction. The intact antibody can bind bivalently to one capsid surface to stabilize or bind monovalently with two capsids (54) to induce aggregation through cross-linking of viruses. Three of the four antibodies, H16.V5, H16.14J, and 263.A2, were clearly capable of monovalent binding (crosslinking) and incapable of bivalent binding (Table 3) as neighboring Fabs are too far apart. H16.1A is capable of bivalent binding at the three-fold, and monovalent binding at other sites. This complexity of binding suggests the process of mAb neutralization was more complicated than that for the corresponding Fab.
Here, we propose a model of neutralization by four HPV16 specific mAbs, H16.V5, H16.1A, H16.14J and H263.A2. These antibodies and their Fabs are likely capable of neutralizing by hyperstabilizing the capsid through interactions involving multiple loops from different L1 proteins. However, the structural analysis and functional assays indicate that bivalency of the intact antibody enhances or complements this type of neutralization. Likely mAbs neutralize by multiple mechanisms, including stabilization, crosslinking, and bivalent binding.
Supplementary Material
In the contact region, two L1 proteins of the capsomer interacted with each Fab. For each footprint the copy of the L1 protein that made the major contributions is designated with a blue X, and additional minor interactions were made by a second neighboring L1 (indicated by red X′).
Acknowledgments
We thank Dr. John Schiller for kindly providing HPV16 sheLL and pYSEAP plasmids. This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco CURE Funds. We thank the Microscopy Imaging Core Facility at the Pennsylvania State University College of Medicine. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. The work was supported by Penn State COM Institutional Research Grant #124171-IRG-13-042-01-IRG (SH), NIH SIG 1S10RR031780-01A1 (SH), and the Jake Gittlen Memorial Golf Tournament (NC).
Footnotes
Author contributions: JG, HL, SMB, and SAB performed research; REA performed microscopy and collected cryo-EM data; AMM and JFC collected cryo-EM data; JG and HL processed and analyzed cryo-EM data; JG, NDC, and SH wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shan KV International Biological Study on Cervical Cancer (IBSCC) Study Group. Prevalence of Human Papillomavirus in Cervical Cancer: a Worldwide Perspective. JNCI J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJF, Peto J, Meijer CJLM, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Crow JM. HPV: The global burden. Nature. 2012;488:S2–S3. doi: 10.1038/488S2a. [DOI] [PubMed] [Google Scholar]
- 4.Culp TD, Spatz CM, Reed CA, Christensen ND. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments, and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology. 2007;361:435–446. doi: 10.1016/j.virol.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castillo A. Human Papillomavirus and Carcinogenesis in the Upper Aero-Digestive Tract. In: Tonissen K, editor. Carcinogenesis. InTech; 2013. [Accessed April 23, 2014]. online http://www.intechopen.com/books/carcinogenesis/human-papillomavirus-and-carcinogenesis-in-the-upper-aero-digestive-tract. [Google Scholar]
- 6.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk RD, Chen Z, Van Doorslaer K. Human Papillomaviruses: Genetic Basis of Carcinogenicity. Public Health Genomics. 2009;12:281–290. doi: 10.1159/000214919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Q, Modis Y, High K, Towne V, Meng Y, Wang Y, Alexandroff J, Brown M, Carragher B, Potter CS, Abraham D, Wohlpart D, Kosinski M, Washabaugh MW, Sitrin RD. Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. Virol J. 2012;9:52. doi: 10.1186/1743-422X-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monie A, Tsen SWD, Hung CF, Wu TC. Therapeutic HPV DNA vaccines. Expert Rev Vaccines. 2009;8:1221–1235. doi: 10.1586/erv.09.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen ND. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir Chem Chemother. 2005;16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- 12.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med. 2005;119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 13.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. Maturation of the Human Papillomavirus 16 Capsid. mBio. 2014;5:e01104-14–e01104–14. doi: 10.1128/mBio.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. A CryoEM study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol. 2014 doi: 10.1128/JVI.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck CB, Thompson CD, Pang YYS, Lowy DR, Schiller JT. Maturation of Papillomavirus Capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossmann MG, Johnson JE. Icosahedral RNA Virus Structure. Annu Rev Biochem. 1989;58:533–569. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 19.Carter JJ, Wipf GC, Benki SF, Christensen ND, Galloway DA. Identification of a Human Papillomavirus Type 16-Specific Epitope on the C-Terminal Arm of the Major Capsid Protein L1. J Virol. 2003;77:11625–11632. doi: 10.1128/JVI.77.21.11625-11632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology. 1996;223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 21.Day PM, Thompson CD, Buck CB, Pang YYS, Lowy DR, Schiller JT. Neutralization of Human Papillomavirus with Monoclonal Antibodies Reveals Different Mechanisms of Inhibition. J Virol. 2007;81:8784–8792. doi: 10.1128/JVI.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day PM, Gambhira R, Roden RBS, Lowy DR, Schiller JT. Mechanisms of Human Papillomavirus Type 16 Neutralization by L2 Cross-Neutralizing and L1 Type-Specific Antibodies. J Virol. 2008;82:4638–4646. doi: 10.1128/JVI.00143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. Hybrid Papillomavirus L1 Molecules Assemble into Virus-like Particles That Reconstitute Conformational Epitopes and Induce Neutralizing Antibodies to Distinct HPV Types. Virology. 2001;291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 24.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Christensen N, Schiller JT, Dillner J. A monoclonal antibody against intact human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J Gen Virol. 1997;78 (Pt 9):2209–2215. doi: 10.1099/0022-1317-78-9-2209. [DOI] [PubMed] [Google Scholar]
- 26.Rizk RZ, Christensen ND, Michael KM, Muller M, Sehr P, Waterboer T, Pawlita M. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol. 2008;89:117–129. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- 27.Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in Cervarix™, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccin. 2010;6:407–419. doi: 10.4161/hv.6.5.11023. [DOI] [PubMed] [Google Scholar]
- 28.Chen HS, Conway MJ, Christensen ND, Alam S, Meyers C. Papillomavirus capsid proteins mutually impact structure. Virology. 2011;412:378–383. doi: 10.1016/j.virol.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q, Potter CS, Carragher B, Lander G, Sworen J, Towne V, Abraham D, Duncan P, Washabaugh MW, Sitrin RD. Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. Hum Vaccines Immunother. 2014;10:734–739. doi: 10.4161/hv.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-Ray Crystallography and Electron Microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmann MG, Bernal R, Pletnev SV. Combining Electron Microscopic with X-Ray Crystallographic Structures. J Struct Biol. 2001;136:190–200. doi: 10.1006/jsbi.2002.4435. [DOI] [PubMed] [Google Scholar]
- 32.Fabiola F, Chapman MS. Fitting of high-resolution structures into electron microscopy reconstruction images. Struct Lond Engl 1993. 2005;13:389–400. doi: 10.1016/j.str.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. Structural Comparison of Different Antibodies Interacting with Parvovirus Capsids. J Virol. 2009;83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brendle SA, Culp TD, Broutian TR, Christensen ND. Binding and neutralization characteristics of a panel of monoclonal antibodies to human papillomavirus 58. J Gen Virol. 2010;91:1834–1839. doi: 10.1099/vir.0.017228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mejia AF, Culp TD, Cladel NM, Balogh KK, Budgeon LR, Buck CB, Christensen ND. Preclinical Model To Test Human Papillomavirus Virus (HPV) Capsid Vaccines In Vivo Using Infectious HPV/Cottontail Rabbit Papillomavirus Chimeric Papillomavirus Particles. J Virol. 2006;80:12393–12397. doi: 10.1128/JVI.01583-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyeon D, Lambert PF, Ahlquist P. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc Natl Acad Sci U S A. 2005;102:9311–9316. doi: 10.1073/pnas.0504020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastrana DV, Buck CB, Pang YYS, Thompson CD, Castle PE, FitzGerald PC, Krüger Kjaer S, Lowy DR, Schiller JT. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology. 2004;321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 38.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient Intracellular Assembly of Papillomaviral Vectors. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christensen ND, Kreider JW, Kan NC, DiAngelo SL. The open reading frame L2 of cottontail rabbit papillomavirus contains antibody-inducing neutralizing epitopes. Virology. 1991;181:572–579. doi: 10.1016/0042-6822(91)90890-n. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Raifu M, Howard M, Smith L, Hansen D, Goldsby R, Ratner D. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J Immunol Methods. 2000;233:167–177. doi: 10.1016/s0022-1759(99)00184-2. [DOI] [PubMed] [Google Scholar]
- 41.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal Off J Microsc Soc Am Microbeam Anal Soc Microsc Soc Can. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan X, Sinkovits RS, Baker TS. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossmann MG. Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- 46.Lyskov S, Chou FC, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE) PloS One. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dasgupta J, Bienkowska-Haba M, Ortega ME, Patel HD, Bodevin S, Spillmann D, Bishop B, Sapp M, Chen XS. Structural Basis of Oligosaccharide Receptor Recognition by Human Papillomavirus. J Biol Chem. 2011;286:2617–2624. doi: 10.1074/jbc.M110.160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J Struct Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 53.Smith TJ, Olson NH, Cheng RH, Chase ES, Baker TS. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc Natl Acad Sci U S A. 1993;90:7015–7018. doi: 10.1073/pnas.90.15.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hewat EA, Blaas D. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 1996;15:1515–1523. [PMC free article] [PubMed] [Google Scholar]
- 55.Day PM, Schelhaas M. Concepts of papillomavirus entry into host cells. Curr Opin Virol. 2014;4:24–31. doi: 10.1016/j.coviro.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, Schelhaas M, Kast WM. The Evolving Field of Human Papillomavirus Receptor Research: a Review of Binding and Entry. J Virol. 2013;87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the contact region, two L1 proteins of the capsomer interacted with each Fab. For each footprint the copy of the L1 protein that made the major contributions is designated with a blue X, and additional minor interactions were made by a second neighboring L1 (indicated by red X′).