Abstract
A high-throughput method has been developed for the systematic mutagenesis of the Escherichia coli genome. The system is based on in vitro transposition of a modified Tn5 element, the Sce-poson, into linear fragments of each open reading frame. The transposon introduces both positive (kanamycin resistance) and negative (I-SceI recognition site) selectable markers for isolation of mutants and subsequent allele replacement, respectively. Reaction products are then introduced into the genome by homologous recombination via the λRed proteins. The method has yielded insertion alleles for 1976 genes during a first pass through the genome including, unexpectedly, a number of known and putative essential genes. Sce-poson insertions can be easily replaced by markerless mutations by using the I-SceI homing endonuclease to select against retention of the transposon as demonstrated by the substitution of amber and/or in-frame deletions in six different genes. This allows a Sce-poson-containing gene to be specifically targeted for either designed or random modifications, as well as permitting the stepwise engineering of strains with multiple mutations. The promiscuous nature of Tn5 transposition also enables a targeted gene to be dissected by using randomly inserted Sce-posons as shown by a lacZ allelic series. Finally, assessment of the insertion sites by an iterative weighted matrix algorithm reveals that these hyperactive Tn5 complexes generally recognize a highly degenerate asymmetric motif on one end of the target site helping to explain the randomness of Tn5 transposition.
Complete genomic sequences provide not only the opportunity to identify an organism's entire gene complement but also the challenge to understand the function of each gene. Initial annotation of the Escherichia coli genome suggested that there were 4,288 protein encoding genes, nearly 40% of which were of unknown function (4). Subsequent experimental and in silico analysis has elucidated the role of many of those genes, but the function of ca. 1,000 genes is still obscure. Furthermore, the pathway(s) each gene participates in and the interactions between their products remain open questions for the majority of genes. Mutant analysis remains the most powerful means of addressing these basic questions. A collection of mutants in every gene of Saccharomyces cerevisiae has greatly aided a global genetic understanding of the organism (1); however, a similar resource for E. coli is not yet available.
Several methods exist for generating large mutant sets in E. coli, including both random and directed approaches. Historically, mutants were generated randomly by using chemical reagents or transposons. Transposons offer the significant advantage of allowing useful features such as selectable markers to be included, and the efficiency of generating large mutant pools by transposition has been improved recently using modified Tn5-based reagents (13). Although random transposition can produce saturating numbers of mutants in a single reaction, it suffers both from the difficulty in identifying the insertion sites as well as in isolating mutants in every gene (i.e., closure). These limitations are minimized in directed approaches in which individual genes are mutagenized in vitro and then introduced into the genome by homologous recombination. However, unlike yeast cells, which readily recombine incoming linear DNA fragments (2, 31), the RecBCD nuclease rapidly degrades such molecules in E. coli. This problem has recently been overcome by using the bacteriophage λ homologous recombination system encoded by the three red genes: bet, gam, and exo (9, 22, 23, 32). The encoded proteins not only increase the homologous recombination efficiency by >70-fold (22), they require as little as 30 bp of homology on both ends of the targeted fragment to efficiently catalyze the reaction (32). These observations led to a general strategy in which linear fragments containing a selectable marker flanked by short regions of homology are generated by PCR and then recombined into the chromosome by using λRed (9, 23, 32). This approach does, however, require long primer pairs for each gene to provide the necessary homology for recombination and sufficient priming sequence for amplification of the selectable cassette.
In order to efficiently create a complete set of mutant strains for E. coli, several criteria must be met. First, the method must be sufficiently robust to allow the mutant to be isolated with minimal screening, and it must be scalable. Second, the mutations created should be easily convertible to other types of alleles in a single step. Third, it should be cost-effective. To this end, we have developed a flexible, high-throughput method for systematically creating insertion mutants in every nonessential gene. The procedure is based on in vitro transposition of modified Tn5 complexes into PCR products corresponding to each open reading frame (ORF) followed by λRed-mediated recombination into the chromosome in vivo. A description of our progress in constructing a complete mutant strain collection for E. coli, as well as additional applications of the system, is presented.
MATERIALS AND METHODS
Bacterial strains and preparation of electrocompetent cells.
MG1655 (4) cells containing pKD46 (9) were the transformation recipients for all transposition reactions except for replacement experiments where the appropriate Sce-poson containing insertion strain harboring pKD46 was used. Electrocompetent cells were made by growing cells at 30°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) to an optical density at 600 nm (OD600) of 0.1. Arabinose was then added to a final concentration of 0.2% (wt/vol) to induce λred expression and cultures grown until the OD600 reached 0.5. Cells were washed once with ice-cold water and twice with ice-cold 10% glycerol before being concentrated ∼100-fold in 10% glycerol.
In vitro transposition and isolation of insertion mutants.
The Sce-poson was created by PCR amplification of EZ::TN〈KAN-2〉 (Epicentre) with appropriate primers to introduce the 18-bp I-SceI recognition site downstream of the kanamycin resistance (Kanr) gene. The product was then blunt end cloned into PvuII-linearized LITMUS28 (New England Biolabs). The transposon was produced by PvuII digestion of the plasmid, agarose gel separation, and purification by using a QIAquick gel extraction kit (Qiagen) or purchased from the manufacturer (Epicentre). Transposition reactions were done in 96-well format, with each well containing a PCR product corresponding to an annotated ORF (∼200 ng), 200 ng of purified transposon, and 0.1 μg of transposase. Reactions were incubated in reaction buffer [50 mM Tris (pH 7.5), 150 mM CH3COOK, 10 mM (CH3COO)2Mg, 4 mM spermidine] at 37°C for 2 h and then stopped by the addition of 1% sodium dodecyl sulfate and heating to 70°C for 10 min. Reactions were desalted by Sephadex G-50 purification prior to being electroporated by using a GenePulser (Bio-Rad) according to the manufacturer's instructions. Cells were resuspended and grown in LB medium at 37°C for 1 h before being plated on medium containing ampicillin (100 μg/ml) and kanamycin (12.5 μg/ml) and then incubated at 30°C.
To identify disruptions, two ampicillin-and kanamycin-resistant (Ampr Kanr) colonies were streaked to single colonies, and individual colonies from each were grown in selective media overnight at 30°C. Cultures were diluted 1:50 with sterile water, and the targeted locus was PCR amplified with gene-specific primers (26). Reactions were separated on a 1% agarose gel, and those with a product ∼1.2 kb larger than that of the wild type were identified as potential insertion alleles. These were then confirmed by sequencing the products with the KAN-2-FP-1 primer (Epicentre).
To cure cells of the pKD46 plasmid, cultures were streaked to single colonies on LB medium supplemented with kanamycin (12.5 μg/ml) and incubated at 43°C overnight. Representative colonies were screened for sensitivity to ampicillin by replica plating, and the Kanr Amps phenotype was reverified by PCR and sequencing as described above.
Weighted matrix evaluation of Tn5 insertion sites.
Sequences from 1,960 confirmed mutant insertion sites were analyzed by using a weighted matrix algorithm (18) in an iterative manner as follows. The 9-bp target site with five bases on either side was extracted from the coding strand of each gene and aligned to generate the initial matrix. Scores for both the coding strand and its reverse complement were then determined, and the higher scoring sequence used in a new alignment from which the next matrix was built. Both strands were then reevaluated compared to the new matrix, and the best matches again used to generate a refined matrix. This process was done iteratively until no changes in the matrix occurred. From this, the five positions showing the greatest degree of sequence bias (positions 1, 4, 6, 9, and 10) were used to build a new matrix, and the same iterative process was used to refine the result. The output of the analysis was viewed by using WebLogo (weblogo.berkeley.edu).
Replacement with different alleles.
Megaprimer PCR was used to produce both the xylA amber and D1 deletion fragments with appropriate primers (Table 1). Strategy and construction of the amber fragment has been described previously (17). The D1 fragment was constructed in two amplification steps with Pfu polymerase (Stratagene). First, the 1,358-bp region immediately upstream of the xylA start codon was amplified with the Forward1 and Reverse1 primers, and the 1,526-bp segment downstream of the stop codon was generated in a separate reaction with the Forward2 and Reverse2 primers. Second, the two products were then combined in a second round of amplification with the overlapping extensions contained at the ends of Reverse1 and Forward2. This results in the replacement of the intervening xylA coding region with a barcode and primer-binding site cassette as described previously (12).
TABLE 1.
Primers for constructing replacement fragments
| Genea | Direction | Sequence |
|---|---|---|
| xylA (A)b | Forward | ttgctcttcgttatttgtcgaacagataatggt |
| Reverse | ttgctcttccatgcaagcctattttgaccagctc | |
| Mutagenic | catagaccgtcgccTAGtcgtaatcatattg | |
| xylA (D1)c | Forward1 | ttgctcttcgttacgccattaatggcagaagttg |
| Reverse1 | CACCCGAGTTACGATCAAATAGAGACCTCGTGGACATCcggctaactgtgcagtccgt | |
| Forward2 | ATTTGATCGTAACTCGGGTGCGTACGCTGCAGGTCGACattgaactccataatcaggt | |
| Reverse2 | ttgctcttcgttacagctcgctctctttgtgga | |
| xylA (D2)d | Forward | catccatcacccgcggcattacctgattatggagttcaatCGGCTAACTG |
| Reverse | cgctaccgataaccgggccaacggactgcacagttagccgATTGAACTCC | |
| crp (D)d | Forward | tctggctctggagaaagcttataacagaggataaccgcgcTCCCGTCGGA |
| Reverse | aaaatggcgcgctaccaggtaacgcgccactccgacgggaGCGCGGTTAT | |
| rpoS (D)d | Forward | ccagcctcgcttgagactggcctttctgacagatgcttacAAGGTGGCTC |
| Reverse | ttgaatgttccgtcaagggatcacgggtaggagccaccttGTAAGCATCT | |
| rseB (D)d | Forward | tgcccgttttgccaggagacgacggtagcccactctttgaTACTGCGATT |
| Reverse | aggtgccaggaattcaaactttaggaacgcaatcgcagtaTCAAAGAGTG | |
| yeiE (D)d | Forward | acgaaatcgacacagcaccagcatgttcttgtacagcaacAGTCGCTTAC |
| Reverse | ttatatatttataattaatctttataagtggtaagcgactGTTGCTGTAC | |
| ygaA (D)d | Forward | gcgcaataaacctgcaggatttctatcaggccgggattatTAATGGGCAT |
| Reverse | tctgattgtttttactctataaataaaattatgcccattaATAATCCCGG | |
| yohI (D)d | Forward | aaagagctaacacattgtcaaaaaacatcactatggttttATCATCACCC |
| Reverse | aggcgctaacatagcgcctcatttttttgcgggtgatgatAAAACCATAG | |
| metA (D)d | Forward | tgtctaaacgtataagcgtatgtagtgaggtaatcaggttTCTTCTGTGA |
| Reverse | taaggtgctgaatcgcttaacgatcgactatcacagaagaAACCTGATTA |
Mutation designations shown in parentheses: A, amber; D, deletion.
Capital letters in the mutagenic primer indicate the amber codon. SapI recognition sites are in boldface.
Barcode sequences are capitalized and primer-binding sites are italicized capitals. SapI restriction sites are in boldface.
For forward primers, the 10 bases adjacent to the start codon are in boldface and the 10 nucleotides flanking the stop are capitalized. For reverse primers, the reverse complement of the 10 bases immediately after the stop and preceding the start are indicated in boldface and capitals, respectively.
The xylA D2 deletion and all other deletions were created by overlapping oligonucleotide extension with primers listed in Table 1. In each case, two 50-nucleotide (nt) oligonucleotides were synthesized: (i) a forward primer containing 40 nt immediately 5′ of the start codon, followed by the 10-nt region downstream of the stop codon, and (ii) a reverse primer with the 40-nt downstream sequence joined to 10 nt of upstream DNA. Oligonucleotides were mixed in equimolar amounts, heated to 100°C for 5 min, and slow cooled to allow annealing via the 20-bp overlap. Annealed oligonucleotides were filled in by incubation with the Klenow fragment of DNA polymerase (New England Biolabs) in 1× reaction buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 7.5 mM dithiothreitol) and deoxynucleoside triphosphates at 37°C for 5 min. The enzyme was heat inactivated at 75°C for 20 min and desalted by using Sephadex G50 prior to electroporation.
Insertion strains expressing λRed functions were made electrocompetent as described above. pBC-I-SceI was constructed by excising the 795-bp XbaI-SalI fragment from pST76-ASceP (25) and ligating it with XbaI-SalI-digested pBCSK (Stratagene). Linear fragments and pBC-I-SceI were mixed at an approximate molar ratio of 40:1 and electroporated into the appropriate strain, and outgrowths were plated onto LB medium containing 25 μg of chloramphenicol/ml. Typically, 50 chloramphenicol-resistant (Camr) colonies were replica patched onto LB plates containing either chloramphenicol or kanamycin, and Camr Kans colonies were identified. Candidates were then assayed by colony PCR with flanking primers distal to the mutated gene. A representative clone with the predicted-size PCR product was then sequenced to verify the mutation was correct.
RESULTS
Strategy.
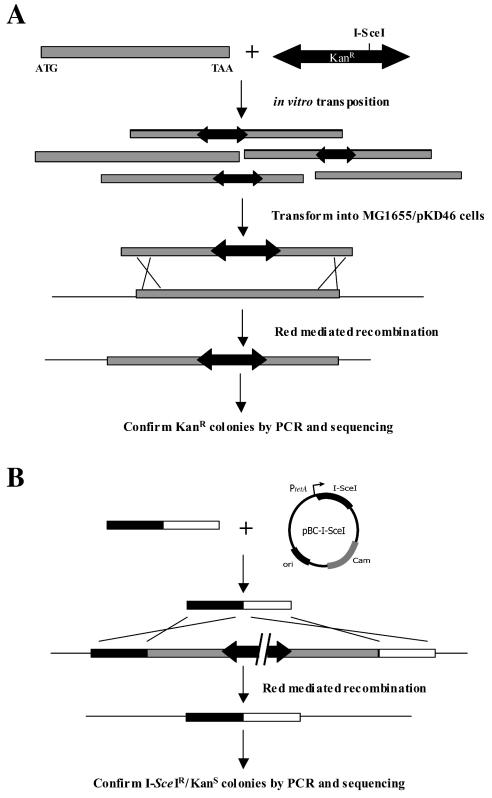
As a means of systematically mutating each annotated ORF, we developed a two-step method outlined in Fig. 1A. First, the 4,288 predicted ORFs (4) are PCR amplified individually by using a set of gene-specific primers (26). Each product is then subjected to an in vitro transposition reaction with a modified Tn5 transposon, Tn-Kan-I-SceI (referred to here as the Sce-poson). This transposon contains the Kanr gene and the recognition sequence for the yeast meganuclease, I-SceI. These elements are flanked by mosaic transposable ends that, in combination with a hyperactive transposase, efficiently catalyze transposition in vitro (15). Note, this reaction also results in a 9-bp duplication at the site of insertion (3). After inactivation of the transposase, the reaction mixture is electroporated into MG1655 cells expressing the λred genes from the pKD46 plasmid (9). Recombinants are selected on kanamycin-containing medium, and the targeted locus is amplified by PCR with the appropriate gene-specific primers. Products that increase by the size of the Sce-poson (1,239 bp) relative to the wild type are diagnostic of the desired insertion event. The precise site and orientation of the insert are determined by sequencing with a transposon-specific primer. The temperature-sensitive replicon of pKD46 allows plasmid curing by growth at 43°C (9).
FIG. 1.
General strategy for systematic creation of insertion alleles and replacements. (A) Insertion mutants are made by in vitro transposition of Tn-Kan-I-SceI (black double-headed arrows) into PCR products corresponding to each ORF (shaded bars). The mixture is then electroporated into wild-type cells expressing the λRed proteins and introduced into the chromosome via homologous recombination. (B) Replacement of insertion alleles with markerless mutations by using I-SceI counterselection. A gene deletion strategy is depicted. Linear fragments (bar) consisting of a 5′ homology (solid portion of the bar) fused to a 3′ homology (open portion of the bar) are coelectroporated with pBC-I-SceI into the appropriate insertion strain. I-SceI-mediated cleavage of the genome (gapped double-headed arrow) stimulates λRed-mediated recombination at the targeted locus.
Once created, the insertion allele can be replaced with other mutations by using the I-SceI site as a negative selectable marker (Fig. 1B). Linear fragments containing the desired mutation (e.g., point mutations, deletions, other insertions) are generated by PCR and electroporated with the pBC-I-SceI plasmid into the corresponding insertion strain expressing λRed. pBC-I-SceI is a high copy number plasmid constitutively expressing the I-SceI gene from the tetA promoter and also contains the Camr gene for selection. I-SceI cleavage of the genome occurs only within the transposon sequences (there are no other sites in the MG1655 genome), producing a lethal double-strand break unless repaired or unless the recognition site is removed by recombination with the incoming fragment. A similar application of this counterselection has been shown to increase intramolecular recombination by 2 to 3 orders of magnitude (25). Transformants are selected as Camr (and therefore, I-SceIr) colonies and then screened for loss of the transposon by sensitivity to kanamycin. Candidates are verified by PCR and sequencing.
Results from the first round of mutagenesis.
To establish and refine the strategy outlined above, a first pass at mutating the 4,288 originally annotated ORFs has been completed, resulting in 1,976 characterized insertions (46% overall success rate). This includes 520 genes mutated with the EZ::TN〈KAN-2〉 transposon (Epicentre) and 1,456 genes constructed by using the Sce-poson. Most mutants are stored both with the pKD46 plasmid to facilitate additional manipulations and without it for phenotypic analysis. All strains are available through our website (www.genome.wisc.edu).
Mutants in ORFs ranging from 102 (tpr) to 4,617 (lhr) bp have been recovered and analysis of the lesion sites shows a clear bias toward insertion into the middle of genes (Fig. 2A). This is likely due to maximizing homology on both sides of the transposon and thereby increasing recombination efficiency, although preferential transposition into the middle of linear fragments could also contribute to this trend. Nevertheless, recombinants have been isolated with short homologies on either the 5′ end (e.g., yjiG, 13 bp) or the 3′ end (e.g., hflB, 22 bp). There is a slight but significant bias in the orientation of Kanr with respect to the direction of gene transcription. In 1,067 cases Kanr transcription is in the same direction as the affected gene (sense orientation), whereas 909 alleles have expression in the antisense orientation (P = 0.00041). This bias is especially pronounced with insertions near the 5′ end of a gene, where 70% of the inserts are oriented in the sense direction. Conversely, 62% of the 3′ insertions are in the antisense orientation.
FIG. 2.
Insertion site characteristics. (A) Histogram displaying the relative position and orientation of isolated insertion alleles. x-axis values refer to the position of transposon insertion as a percentage of gene length. The solid and open portions of each bar correspond to insertions in the sense and antisense orientations, respectively. (B) Sequence logo (28) representation of the Tn5 recognition motif obtained from coding strand only analysis. The sequence is oriented 5′ to 3′, and the target site refers to the region that is duplicated upon transposition. (C) Sequence logo representation of the motif derived from independent consideration of both strands by using a weighted matrix. Note that the y-axis scales differ between panels A and B.
Tn5 target site preferences.
To investigate the randomness of Tn5 transposition, we assessed 1,960 insertion sites for common sequence elements. The 9-bp target site that becomes duplicated upon transposition plus 20 nt from both flanks were extracted from the coding strand of each gene and aligned. No significant sequence information was detected outside the first 5 nt flanking the target site, and thus only the core 19 bp are considered below. Similar to previous reports (14, 29), an apparent symmetrical consensus emerged with the two most dominant features being a guanine at the beginning of the target site (Fig. 2B, position 1) and a cytosine at the end (position 9). It was noticed, however, that the incidence of symmetrical sequences at the target site was limited (436 GN7C cases of 1,020 GN8 and 611 cases of 1,960 overall). Further, there were 502 cases of HN7C (where H is not G), placing the guanine in the first 5′ position on the noncoding strand. These results raised the possibility that the recognition motif was asymmetric and could occur on either strand.
To evaluate the sequence information of both strands at the transposed sites, an iterative procedure was developed by using a weighted matrix algorithm (18) to identify the strand that most closely matched an emerging consensus (Materials and Methods). These sequences were then realigned and used to build a new matrix. This refinement process was repeated until no change in the consensus occurred. The resultant motif was indeed asymmetric, with only five positions showing significant sequence bias (Fig. 2C). The strongest signal was a guanine in the first 5′ duplicated position (position 1; 1,428 cases of 1,960 [72.8%]). In addition, a cytosine at position −2 is clearly preferred (1,092 of 1,960 being CN10, 857 of which are CNGN8). The three additional positions were more degenerate: G or C at position 1, 68% of all insertions; T or C at position 4, 81% of all insertions; and A or T at position 5, 71% of all insertions. No significant sequence preference was seen 3′ of the duplication center (position 5). Similar results were also obtained by using a heuristic sequence parsing approach (data not shown). Further, by using a set of 35 characterized Tn5-derivative insertions from an in vivo mutagenesis of the genome (K. M. Winterberg and W. Reznikoff, unpublished data), a nearly identical site preference was observed, although the T at position 4 was greatly enhanced (data not shown). Together, these results indicate that Tn5 preferentially recognizes a highly degenerate asymmetric sequence present in either 5′ half of the target site. This flexibility in target site recognition is clearly a critical facet in the observed randomness of Tn5 transposition.
Isolation of insertion alleles in essential genes.
One expected limitation of this method is the inability to mutate essential genes (defined here as required for growth on LB medium at 30°C). However, inspection of the current collection indicates that insertions in 187 of the 620 known and putative essential genes (10) have been isolated (for a detailed list, see Table S1 in the supplemental data [see also http://www.genome.wisc.edu/resources/cloneandmut/TableS1.txt]). Among the group mutated, 50 represent “y” genes and 40 are only designated by a b-number. Most of these discrepancies are likely due to mutants in the footprinting experiment that affect competitive fitness rather than actual essential functions, as discussed by the authors (10). However, three scenarios can be envisioned whereby viable insertions could be recovered in truly essential genes, and examples of at least two are present in this collection.
First, transposition could occur in a nonessential 5′ ORF segment with transcription initiating from within the transposon producing a translatable message encoding the essential portion of the protein. In addition to possible readthrough from the Kanr gene, EZ::TN〈KAN-2〉 sequence and insertion analysis indicates cryptic promoters likely exist on both ends of the element, explaining the relatively nonpolar nature of this transposon (11). Further, inspection of the transposon ends shows that initiation codons in two frames exist reasonably spaced downstream of a Shine-Dalgarno-like sequence (Fig. 3A). Mutants with this predicted structure were isolated in two ribosomal genes, rplI and rplU (Fig. 3B). Both genes are presumed to be essential since ribosomes lacking these components have not been isolated (8), although subtle mutations in each gene have been obtained (7). In both cases, the transposon is located at the 5′ end of the ORF (nt 60 and 13 of the rplI and rplU coding sequences, respectively) and is oriented in the sense direction. For rplI, the putative fusion protein contains six residues from the end of the transposon and the carboxy-terminal 129 amino acids (i.e., amino acids 21 to 149) of the gene. The Tn-RplU chimera is predicted to consist of 5 amino acids from the transposon, initiating at a GTG codon, followed by the region of RplU from amino acids 5 to 103. These results imply that the amino-terminal 20 and 4 amino acids of RplI and RplU, respectively, are not required for protein function.
FIG. 3.
Viable insertion mutants in essential genes. (A) Sequence and reading frames at the transposon end downstream of the Kanr gene. The Shine-Dalgarno sequence is underlined, and bars above the sequence indicate initiation codons. Single-letter designations are used for amino acids, and the stop codon is denoted by an asterisk. (B) Insertions into 5′ ends of essential ORFs. Schematic shows orientation of transposon insertion relative to the Kanr gene and gene transcription for two isolated insertion alleles. Sequences flanking the putative fusion protein start point show transposon (lowercase) and gene (capitals) segments with the predicted amino acid sequence shown below. Underlined nucleotides denote potential Shine-Dalgarno sites, and the position of first ORF nucleotide downstream of the insertion is marked. (B) Insertions into the 3′ ends of essential genes. Schematics show the insertion points and orientation of transposon insertions into indicated essential genes. Numbering refers to amino acid positions corresponding to the upstream insertion site and the wild-type length of the protein.
Second, insertions into nonessential carboxy-terminal regions can be recovered (Fig. 3C). For example, an insertion located at nt 1637 of the methionyl-tRNA synthetase encoding gene, metG, was isolated, leading to the truncation of amino acids 546 to 676. This is almost precisely the same region previously identified to be dispensable for MetG function (21). Also, the nusA gene has been shown to be essential (5, 24, 27), and yet an insertion corresponding to amino acid position 417 was isolated, suggesting that the carboxy-terminal 79 residues are not necessary for function.
Third, in cases in which genes are capable of intracistronic complementation, such as lacZ, properly positioned transposons could yield viable cells by a mechanism postulated below. Although this is expected to be relatively uncommon, the diversity of transposed positions across genes suggests it may occur more frequently than anticipated.
These results demonstrate that a subset of essential genes can be mutated by this method and provide information on dispensable domains as well as starting material for further dissection using a simple replacement strategy (Fig. 1B). Combining data from both footprinting and systematic mutagenesis will also more accurately define the set of essential genes in bacteria.
Replacement of insertion alleles with other mutations.
The limitations intrinsic to insertion mutants, as well as the breadth of information available from other types of mutations, make it important that these alleles be easily convertible. As an initial test of the strategy outlined above (Fig. 1B), we constructed three linear fragments carrying markerless mutations in xylA. The first consisted of the entire ORF with a nonsense mutation at position 1065 converting the alanine residue to an amber stop. The other two products were designed to precisely remove the entire xylA ORF. One contained 1,358 bp of 5′ homology starting immediately upstream of the start codon fused to a 1,526-bp segment 3′ of the translational stop. The second consisted of 40-bp homologies directly adjacent to either end of the ORF. Fragments were electroporated with the I-SceI-expressing plasmid, pBC-I-SceI, into the xylA::Tn insertion strain producing λRed from pKD46. Camr transformants (pBC-I-SceI+) were then screened for sensitivity to kanamycin, and Camr Kans colonies were considered putative replacements. These candidates were verified by PCR and sequencing. Transformation with any of these three fragments resulted in ca. 25% Kans colonies, a finding indicative of transposon loss (Table 2). PCR established that all 18 Kans clones from the two deletion fragment tests had completely removed xylA, whereas two of seven colonies transformed with the nonsense-harboring segment were confirmed as amber alleles (Table 2). In the other five cases, a wild-type xylA gene was recovered, a finding consistent with crossover events that occurred between the amber and the transposon. Control transformations with an empty pBCSK vector, instead of pBC-I-SceI, yielded no Kans colonies, demonstrating that the I-SceI enzyme is necessary for efficient replacement (data not shown). The number of Camr transformants obtained with equal amounts pBC-I-SceI was at least 2 orders of magnitude lower than with pBCSK (data not shown), indicating the effectiveness of the counterselection and similar to previous estimates of I-SceI killing (16). Finally, cotransformations into xylA::Tn cells cured of the pKD46 plasmid produced no replacements, indicating that λRed is also required (data not shown).
TABLE 2.
Insertion allele replacement
| Gene | A/Da | homology (bp)b
|
No. of colonies
|
|||
|---|---|---|---|---|---|---|
| 5′ | 3′ | Camr | Kans | Confirmedc | ||
| xylA | A | 1,064 | 259 | 24 | 7 | 2/7 |
| D1 | 1,358 | 1,526 | 40 | 9 | 9/9 | |
| D2 | 40 | 40 | 24 | 9 | 9/9 | |
| crp | D | 40 | 40 | 50 | 0 | |
| rpoS | D | 40 | 40 | 50 | 2 | 0/2 |
| rseB | D | 40 | 40 | 50 | 1 | 1/1 |
| yeiE | D | 40 | 40 | 50 | 10 | 8/8 |
| ygaA | D | 40 | 40 | 50 | 3 | 3/3 |
| yohI | D | 40 | 40 | 50 | 4 | 4/4 |
| metA | D | 40 | 40 | 50 | 16 | 9/10 |
A, amber-allele; D, deletion.
That is, the length of the sequence homology on either side of the mutation.
That is, the number of Kans colonies verified as replacements by PCR and sequencing of one representative clone.
To test the general utility of this procedure, linear fragments for deletions of seven additional genes were produced by extension of overlapping oligonucleotides (Table 2). These fragments were coelectroporated with pBC-I-SceI into the corresponding insertion strain expressing λRed and processed as described above. For four genes—yeiE, ygaA, yohI, and metA—Kans colonies were recovered at frequencies of 6 to 32%, and all but one of the candidates tested contained the expected deletion. The single Kans isolate of rseB was also shown to have the correct deletion. However, replacements were not obtained for either rpoS or crp. In the case of crp::Tn, this is almost certainly due to the requirement of active Crp for PBAD induction (34) and thereby λred expression. This further confirms the essentiality of Red functions for replacement. The reasons for the rpoS::Tn failures remain enigmatic, since deletion of this gene has been achieved (33). Nevertheless, the results demonstrate that replacing insertion alleles with markerless mutations by using I-SceI counterselection is straightforward in most cases.
Allelic series in lacZ.
To assess the diversity of alleles that can be generated in a given gene, 43 insertions in lacZ were isolated and sequenced (Fig. 4 and Table 3). There was again a slight bias toward insertions that occurred in the sense orientation (24 of 43); however, these were largely clustered in the 3′ third of the gene. In contrast, transposons in the antisense direction were more uniformly distributed. Two identical insertions were found at each of four positions; whether these represent true independent events or the result of division during the transformation outgrowth is not known.
FIG. 4.
Distribution of lacZ insertion alleles. The gene is depicted as an elongated box, and numbers refer to the nucleotide position within the ORF. Bars indicate insertion site locations, with those on top or below the gene denoting transposons oriented in the sense or the antisense direction, respectively. The four longer bars mark positions where two insertions were found. Asterisks denote alleles with partial function, and increasing font size corresponds to increasing enzymatic activity.
TABLE 3.
lacZ::Tn allelic series
| Allele no. | Orientationa | Insertion siteb | N fragmentc | Activityd |
|---|---|---|---|---|
| lacZ4106 | AS | 495 | 165 | − |
| lacZ4107 | AS | 726 | 242 | − |
| lacZ4108 | AS | 787 | 262 | − |
| lacZ4109 | AS | 1132 | 377 | − |
| lacZ4110 | AS | 1204 | 401 | − |
| lacZ4111 | AS | 1351 | 450 | − |
| lacZ4112 | AS | 1475 | 491 | − |
| lacZ4113 | AS | 1544 | 514 | − |
| lacZ4114 | AS | 1710 | 570 | − |
| lacZ4115 | AS | 1710 | 570 | − |
| lacZ4116 | AS | 1714 | 571 | − |
| lacZ4117 | AS | 1806 | 602 | − |
| lacZ4118 | AS | 1825 | 608 | − |
| lacZ4119 | AS | 2031 | 677 | − |
| lacZ4120 | AS | 2037 | 679 | − |
| lacZ4121 | AS | 2360 | 786 | − |
| lacZ4122 | AS | 2530 | 843 | − |
| lacZ4123 | AS | 2553 | 851 | − |
| lacZ4124 | AS | 2663 | 887 | − |
| lacZ4125 | S | 116 | 38 | + |
| lacZ4126 | S | 726 | 242 | + |
| lacZ4127 | S | 726 | 242 | + |
| lacZ4128 | S | 948 | 316 | − |
| lacZ4129 | S | 1341 | 447 | − |
| lacZ4130 | S | 1400 | 466 | − |
| lacZ4131 | S | 2107 | 702 | − |
| lacZ4132 | S | 2134 | 711 | − |
| lacZ4133 | S | 2259 | 753 | +/− |
| lacZ4134 | S | 2358 | 786 | − |
| lacZ4135 | S | 2378 | 792 | ++ |
| lacZ4136 | S | 2452 | 817 | − |
| lacZ4137 | S | 2483 | 827 | + |
| lacZ4138 | S | 2516 | 838 | − |
| lacZ4139 | S | 2628 | 876 | − |
| lacZ4140 | S | 2683 | 894 | − |
| lacZ4141 | S | 2683 | 894 | − |
| lacZ4142 | S | 2707 | 902 | − |
| lacZ4143 | S | 2707 | 902 | − |
| lacZ4144 | S | 2706 | 902 | − |
| lacZ4145 | S | 2744 | 914 | − |
| lacZ4146 | S | 2851 | 950 | − |
| lacZ4147 | S | 2929 | 976 | − |
| lacZ4148 | S | 2942 | 980 | − |
Orientation of transposon relative to Kanr transcription. AS, antisense; S, sense.
Last position in the gene from the translation start before the insertion.
Last amino acid residue of predicted C terminally truncated protein.
Relative enzymatic activity as judged on X-Gal-containing medium. Activity: −, none; +/−, very weak; +, weak; ++, intermediate; +++, wild type.
Most mutants are enzymatically inactive as assessed by colony color on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing medium. However, six alleles retain partial activity, apparently as the result of either α- or Ω-complementation (Fig. 4). Importantly, in all six cases the transposon is oriented in the sense direction and positioned so that the C-terminal fragment is in frame with one of the two translatable reading frames (Fig. 3A). Note that the first base of the gene downstream of the transposon is in a +1 codon position relative to the nucleotide listed in Table 3 due to the 9-bp duplication. Of the three putative α-complementing alleles, lacZ4135 disrupts codon 39, whereas lacZ4126 and lacZ4127 both have identical inserts in codon 243. These presumably produce a partially active enzyme by expressing the N-terminal region from the native promoter (an “α-donor”) and the remainder of the gene (corresponding to an “α-acceptor”) from a transcript originating within the transposon. The weak activity observed could result from a suboptimal α-complementing fragment and/or poor expression of the α-acceptor. The putative lacZ4135 α-donor (amino acids 1 to 38) is shorter than the minimal α-domain defined by the M15 deletion (amino acids 3 to 41) (20), whereas the two isolates expressing the N-terminal 242 amino acids are substantially larger and may be at the limit of an active α-domain (30). Similarly, three insertions were isolated near the presumed start of the ω-complementing region (6, 19). The order of apparent activity, lacZ4135 > 4137 > 4133, suggests that the optimal Ω-domain begins near position 790, which lies near the beginning of domain 5 (19). The adjacent upstream allele, lacZ4134, occurs in the reading frame containing a stop codon (Fig. 3A) and is not expected to be translated. Although lacZ4133 maintains almost all of domain 5 intact, it has extremely weak enzymatic activity and thus may affect proper folding of either the ω-donor and/or -acceptor.
These results demonstrate that a diverse array of alleles can be generated by this procedure, making this targeted approach a useful first step for structure-function studies. The sense orientation of all six partially active insertions also suggests that transcription readthrough from the Kanr gene is the major cause of the limited polarity observed with these transposons (11).
DISCUSSION
We describe here a high-throughput method for the systematic mutagenesis of the E. coli genome. The procedure takes advantage of an efficient Tn5-based in vitro transposition system to generate mutagenic linear fragments of each ORF and the λRed proteins to introduce those fragments into the chromosome via homologous recombination. Analysis of insertion sites from the collection defines several important characteristics of the system. First, it confirms that Tn5 transposition is largely random, although a degenerate asymmetric recognition motif was detected. Next, insertions are generally found near the gene center, presumably because of maximizing homology on both sides of the transposon and thus increasing recombination efficiency. Finally, evidence from both the putative α- and Ω-complementing lacZ alleles and two ribosomal mutants supports the idea that transcription readthrough from the Kanr gene is the key factor limiting polarity (11). This may also explain the slight overall bias toward sense-oriented insertions.
Although more laborious than random approaches (11, 13) in creating saturating insertion mutant populations, this directed method has several advantages during individual strain characterization and in closure of a complete set. Technically, PCR amplification and sequencing of a targeted locus is simple and fast compared to identifying the location of a random insertion. The directed method should only yield strains with one insertion, an important consideration for subsequent analysis and manipulation. The potential for losing mutants due to decreased competitive fitness during random mutagenesis is also minimized by mutating each gene separately. The significant number of putative essential genes (10) for which insertion alleles were isolated indicates the importance of this issue. Another potential problem overcome by this directed approach is the mutation of small genes that may be difficult to isolate in a random approach because of their limited target size. Mutations in genes as small as 102 bp (tpr) have been recovered during the first pass, and the observation that 35 bp of homology or less is sufficient for Red-mediated recombination (32) suggests that genes of any size should be amenable to this method. Whether Tn5 transposition has any target DNA size constraints, however, is not known.
Given the advantages of directed approaches, several efficient λRed-based strategies have been developed for targeted mutagenesis of the genome (9, 23, 32). The method of Datsenko and Wanner (9) is being used to construct an in-frame deletion set that will provide a null allele for any nonessential genes (T. Baba, B. Wanner, and H. Mori, unpublished data). The ability of the Sce-poson system to generate a spectrum of alleles in a given gene, as exemplified by the lacZ allelic series, allows for both null alleles and partial loss-of-function mutations to be recovered, both of which are useful in structure-function analysis. Thus, in combination these collections provide a complementary set of reagents for the study of nonessential genes.
Inclusion of the 18-bp I-SceI recognition site as a negative selectable marker allows the Sce-poson alleles to be easily replaced by markerless mutations of virtually any type. This feature also permits the strains to be used as starting material for expanded studies on the targeted gene. For example, in addition to generating an allelic series of Tn5 insertions as demonstrated here with lacZ, higher-resolution analysis could be done by using random point mutants. For this purpose, a gene of interest would be PCR amplified under error-prone conditions and that reaction mixture used to replace the corresponding insertion allele. Kans colonies would contain a diverse array of substitutions in that gene that could be screened for a desired phenotype such as temperature sensitivity. Kans replacements can also be used to construct strains with multiple mutations at distinct loci in a stepwise fashion, either by P1 transduction of an existing Kanr marked allele or by repeating the Sce-poson mutagenesis at the next specified locus. Note that alleles constructed by the method of Datsenko and Wanner can also be subsequently retrofitted by using FLP recombinase, although a small scar corresponding to the FRT site is retained (9).
The methods described here are intended to create a flexible genomic resource available to all researchers. Together with an in-frame deletion collection and a variety of tools to further manipulate these strains, precise genotypes can now be more easily engineered to the specifications of any investigator.
Supplementary Material
Acknowledgments
We thank Sean Phillips, Nick Hermersmann, Patricia Borcelli, Buffy Spink, Nicole Zimmerman, and Jeff Laufenberg for outstanding technical support. Sarah Fendrick, Theo Ehlert, and Colette Johnston also provided excellent technical assistance. We also thank William Reznikoff for generously providing transposase and Mingzhu Liu for sequence parsing of the Tn5 insertion sites.
This study was supported by NIH grant GM35682 to F.R.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bader, G. D., A. Heilbut, B. Andrews, M. Tyers, T. Hughes, and C. Boone. 2003. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 13:344-356. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, D. E. 1989. Transposon Tn5, p. 185-216. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bylund, G. O., J. M. Lovgren, and P. M. Wikstrom. 2001. Characterization of mutations in the metY-nusA-infB operon that suppress the slow-growth of a ΔrimM mutant. J. Bacteriol. 183:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celada, F., A. Ullmann, and J. Monod. 1974. An immunological study of complementary fragments of β-galactosidase. Biochemistry 13:5543-5547. [DOI] [PubMed] [Google Scholar]
- 7.Dabbs, E. R. 1978. Mutational alterations in 50 proteins of the Escherichia coli ribosome. Mol. Gen. Genet. 165:73-78. [DOI] [PubMed] [Google Scholar]
- 8.Dabbs, E. R. 1979. Selection for Escherichia coli mutants with proteins missing from the ribosome. J. Bacteriol. 140:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 13.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 14.Goryshin, I. Y., J. A. Miller, Y. V. Kil, V. A. Lanzov, and W. S. Reznikoff. 1998. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95:10716-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goryshin, I. Y., and W. S. Reznikoff. 1998. Tn5 in vitro transposition. J. Biol. Chem. 273:7367-7374. [DOI] [PubMed] [Google Scholar]
- 16.Herring, C. D., and F. R. Blattner. 2004.. Conditional lethal amber mutations in essential Escherichia coli genes. J. Bacteriol. 186:2673-2681. [DOI] [PMC free article] [PubMed]
- 17.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311:153-163. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 19.Jacobson, R. H., X. J. Zhang, R. F. DuBose, and B. W. Matthews. 1994. Three-dimensional structure of β-galactosidase from E. coli. Nature 369:761-766. [DOI] [PubMed] [Google Scholar]
- 20.Langley, K. E., M. R. Villarejo, A. V. Fowler, P. J. Zamenhof, and I. Zabin. 1975. Molecular basis of beta-galactosidase alpha-complementation. Proc. Natl. Acad. Sci. USA 72:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellot, P., Y. Mechulam, D. Le Corre, S. Blanquet, and G. Fayat. 1989. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J. Mol. Biol. 208:429-443. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, Y., and H. Uchida. 1983. Isolation of conditionally lethal amber mutations affecting synthesis of the NusA protein of Escherichia coli. Mol. Gen. Genet. 190:196-203. [DOI] [PubMed] [Google Scholar]
- 25.Posfai, G., V. Kolisnychenko, Z. Bereczki, and F. R. Blattner. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schauer, A. T., D. L. Carver, B. Bigelow, L. S. Baron, and D. I. Friedman. 1987. lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J. Mol. Biol. 194:679-690. [DOI] [PubMed] [Google Scholar]
- 28.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko, Y., G. G. Bouffard, Y. S. Butterfield, R. W. Blakesley, J. L. Hartley, A. C. Young, M. A. Marra, S. J. Jones, J. W. Touchman, and E. D. Green. 2002. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ullmann, A., F. Jacob, and J. Monod. 1967. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J. Mol. Biol. 24:339-343. [DOI] [PubMed] [Google Scholar]
- 31.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 32.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zubay, G., D. Schwartz, and J. Beckwith. 1970. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. USA 66:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.