Abstract
A second lysyl endopeptidase gene (lepB) was found immediately upstream of the previously isolated lepA gene encoding a highly active lysyl endopeptidase in Lysobacter genomic DNA. The lepB gene consists of 2,034 nucleotides coding for a protein of 678 amino acids. Amino acid sequence alignment between the lepA and lepB gene products (LepA and LepB) revealed that the LepB precursor protein is composed of a prepeptide (20 amino acids [aa]), a propeptide (184 aa), a mature enzyme (274 aa), and a C-terminal extension peptide (200 aa). The mature enzyme region exhibited 72% sequence identity to its LepA counterpart and conserved all essential amino acids constituting the catalytic triad and the primary determining site for lysine specificity. The lepB gene encoding the propeptide and mature-enzyme portions was overexpressed in Escherichia coli, and the inclusion body produced generated active LepB through appropriate refolding and processing. The purified enzyme, a mature 274-aa lysine-specific endopeptidase, was less active and more sensitive to both temperature and denaturation with urea, guanidine hydrochloride, or sodium dodecyl sulfate than LepA. LepA-based modeling implies that LepB can fold into essentially the same three-dimensional structure as LepA by placing a peptide segment, composed of several inserted amino acids found only in LepB, outside the molecule and that the Tyr169 side chain occupies the site in which the indole ring of Trp169, a built-in modulator for unique peptidase functions of LepA, resides. The results suggest that LepB is an isozyme of LepA and probably has a tertiary structure quite similar to it.
Lysyl endopeptidases (EC 3.4.21.50), which hydrolyze lysyl bonds, have been found in the culture broths of Achromobacter lyticus M497-1 (9, 11, 13), Lysobacter enzymogenes (5), and Pseudomonas aeruginosa (4). In addition to narrow lysine specificity, Achromobacter lysyl endopeptidase (API) possesses unique characteristics: 10-fold higher peptidase activity than bovine trypsin (9), a broad pH optimum (pH 8.5 to 10.5) (13), and high stability against 4 M urea or 0.1% sodium dodecyl sulfate (SDS) (10). The endopeptidase has been successfully used to fragment polypeptide chains in protein sequence analysis (6, 12, 17, 27) and is available commercially. API consists of a single polypeptide chain of 268 amino acids (aa) (26) that is synthesized as an inactive precursor placing a signal peptide (20 aa), a propeptide (185 aa), a mature enzyme, and a C-terminal extension peptide (180 aa) from the N to the C terminus (16). The propeptide region is necessary for correct folding of the active peptidase, while the C-terminal extension peptide is unnecessary in the expression of the cloned gene in Escherichia coli (14). Analyses of API mutants, subsite mapping, and tertiary structure revealed that (i) His57, Asp113, and Ser194 are the three constituents of the catalytic triad; (ii) Asp225 in the S1 pocket is responsible for the lysine specificity; (iii) His210, Gly211, and Gly212 are substrate binding subsites S1, S2, and S3; and (iv) Trp169, stacking with His210 through the aromatic ring π-π interaction, contributes to broaden the optimal pH and to remarkably enhance catalytic activity (14, 15, 17, 18, 23-25).
Recently, a new lysyl endopeptidase-producing strain, Lysobacter sp. strain IB-9374, which has sixfold higher productivity than A. lyticus (3), was isolated. The endopeptidase (LepA; called Ls-LEP in the preceding paper [3]) was isolated from the Lysobacter strain, and it was found that the peptidase is synthesized as an inactive precursor protein including pre-, pro-, and C-terminal peptides like those of the API precursor. Differences between the deduced amino acid sequences of API and LepA are limited to two single substitutions at positions 321 and 425, nine consecutive substitutions at positions 440 to 448, and an 18-aa peptide chain elongation at the C-terminal end. All these differences occur far from the mature 268-aa endopeptidase portion. Eventually, mature LepA was identified as API, and then Lysobacter sp. strain IB-9374 was concluded to be a bacterial strain with higher LepA productivity than A. lyticus. In the course of this investigation, we have detected a new lysyl endopeptidase gene similar to the lepA gene on Lysobacter genomic DNA and cloned it. The cloned gene is 2,034 nucleotides long, and the deduced amino acid sequence is quite similar to the LepA sequence as well. To identify this new LepA-like protein, the cloned gene was expressed in E. coli cells using a glutathione-S-transferase (GST)-fused protein, and a mature recombinant protein, named LepB, was characterized as an isozyme of LepA. The present paper reports the primary structure, enzymatic properties, and a LepA-based three-dimensional model of this new lysyl endopeptidase of Lysobacter sp. strain IB-9374.
MATERIALS AND METHODS
Materials.
N-Benzoyl-dl-lysine-p-nitroanilide (Bz-Lys-pNA) was purchased from Wako Pure Chemical Industries, Ltd., Osaka, Japan. N-Benzoyl-l-arginine-p-nitroanilide, N-succinyl-l-alanine-p-nitroanilide, N-benzoyl-l-tyrosine-p-nitroanilide, t-butoxycarbonyl-valyl-leucyl-lysine-4-methylcoumaryl-7-amide (Boc-Val-Leu-Lys-MCA), and glucagon (human) were from Peptide Institute, Inc., Osaka, Japan. N-Acetyl-l-lysine-p-nitroanilide (Ac-Lys-pNA), l-arginine-p-nitroanilide, N-acetyl-l-leucine-p-nitroanilide, and N-acetyl-l-phenylalanine-p-nitroanilide were from Bachem AG, Bubendorf, Switzerland. t-Butoxycarbonyl-l-histidine-p-nitroanilide was from Vega Biochemicals, Tucson, Ariz. l-Lysine-p-nitroanilide (Lys-pNA) was from Merck KGa, Darmtadt, Germany. DEAE-Toyopearl 650 M was from Tosoh Co., Tokyo, Japan. AH-Sepharose 4B, Sephacryl S-200, and an electrophoresis calibration kit for molecular mass determination were from Amersham Pharmacia Biotech AB, Uppsala, Sweden. Other chemicals used were of reagent grade.
Enzyme assays.
The amidolytic activities of LepB were usually measured using Bz-Lys-pNA (3). The assay mixture (1.45 ml) containing 180 mM Tris-HCl buffer (pH 9.2) and 0.25 mM Bz-Lys-pNA was incubated at 30°C for 5 min. The reaction was initiated by the addition of 50 μl of appropriately diluted enzyme solution. After incubation for 5 min at 30°C, the reaction was terminated by adding 0.5 ml of 45% (vol/vol) acetic acid, and the absorbance at 405 nm was measured with an extinction coefficient of 9,620 M−1 cm−1 for the p-nitroaniline (28). One unit of amidolytic activity was defined as the amount of the enzyme that hydrolyzed 1 μmol of substrate in 1 min at 30°C.
pH-activity profiles for LepB and LepA were determined as follows. The reaction mixture contained 100 μM Boc-Val-Leu-Lys-MCA, 20 mM Tris-HCl, and 1 pM LepB or LepA in a total volume of 3 ml (24). After incubation for 5 min at 30°C, the reaction was initiated by the addition of enzyme solution. Increased fluorescence of liberated 7-amino-4-methylcoumarin was monitored at 440 nm upon excitation at 380 nm with a Shimadzu fluorescence spectrometer (RF-5000).
Nucleotide sequencing analysis.
DNA manipulation was performed according to the methods described by Sambrook and Russell (19). The nucleotide sequence of the lepB gene was determined on both strands by the dideoxy chain termination method of Sanger et al. (20) using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems Japan Ltd., Tokyo, Japan). The DNA sequence and the deduced amino acid sequence were examined with the sequence analysis programs of GENETYX (Software Development Co., Tokyo, Japan).
Construction of expression plasmid with LepB.
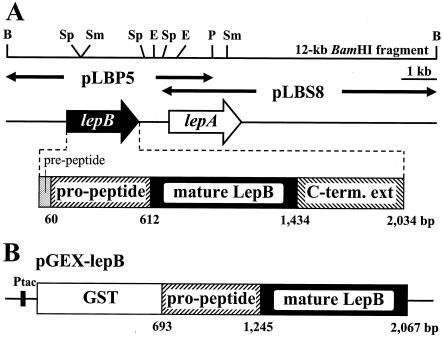
The lepB gene encoding the propeptide and mature-enzyme regions was amplified by LA Taq with GC buffer (Takara Shuzo Co. Ltd., Kyoto, Japan) using two synthetic primers, 5′-GGATCCGCGCCCGCCCTGCGCCCG-3′ (positions 1766 to 1783 in GenBank accession number AB094439) and 5′-GAATTCTCACGGCGCGCCGCCCGAATC-3′ (positions 3139 to 3122) containing BamHI and EcoRI sites (underlined), respectively. The reaction mixture contained 25 μl of 2× GC buffer, 2.5 mM MgCl2, four deoxynucleoside triphosphates (each at 400 μM), 20 ng of pLBP5 as a template (Fig. 1A), 10 pmol of the primers, and 2.5 U of LA Taq DNA polymerase in a final volume of 50 μl. DNA amplification was performed in a temperature cycler (Thermal Cycler Personal; Takara Shuzo), after incubation for 4 min at 94°C, for 30 cycles consisting of a denaturation step for 1 min at 94°C, an annealing step for 1 min at 60°C, and an elongation step for 2 min at 72°C. The amplified 1.4-kb product was inserted into a pGEM T vector (Promega Co., Madison, Wis.), and the nucleotide sequence of the insert DNA was confirmed on both strands. The insert DNA was digested with BamHI and EcoRI and then ligated into an expression vector of GST fusion protein, pGEX-6P-1 (Amersham Pharmacia Biotech AB), and digested with the same restriction enzyme, giving pGEX-lepB (Fig. 1B).
FIG. 1.
Physical maps of the lepB and lepA genes encoding the lysyl endopeptidases from Lysobacter sp. strain IB-9374 (A) and the expression plasmid pGEX-lepB (B). (A) pLBP5 and pLBS8 are the insert DNAs subcloned into pUC118. The arrows indicate the sizes, directions, and locations of the lepB and lepA genes. The lepB gene is depicted below. The restriction site designations are as follows: B, BamHI; E, EcoRI; P, PstI; Sm, SmaI; Sp, SphI. (B) The partial lepB gene containing the propeptide and mature enzyme regions was amplified by PCR, and the DNA fragment was inserted into BamHI and EcoRI sites downstream of the GST gene in pGEX-6P-1. Ptac is the tac promoter.
Expression and purification of recombinant protein.
E. coli JM109 cells carrying pGEX-lepB were grown aerobically on 800 ml of Luria-Bertani broth (19) containing 100 μg of ampicillin/ml at 30°C. When the turbidity of the culture at 600 nm reached ∼0.5, IPTG (isopropyl-1-thio-β-d-galactopyranoside) was added to the culture broth at a concentration of 1 mM, and an additional 6-h cultivation was conducted. The cells were harvested by centrifugation at 8,000 × g for 10 min, washed twice with TE buffer (50 mM Tris-HCl buffer, pH 8.0, containing 2 mM EDTA), resuspended in the same buffer, and disrupted by sonication on ice. After centrifugation at 12,000 × g for 10 min, the resulting precipitate, which contained mostly the GST-fused LepB lacking the C-terminal extension region as inclusion bodies, was washed with TE buffer and dissolved in 50 ml of TE buffer containing 8 M urea and 100 mM 2-mercaptoethanol. After the solution stood for 1 h at room temperature, solid NaCl was added to a final concentration of 0.5 M, and the pH was adjusted to 10.5 with 1 M NaOH. The solution was dialyzed against 500 ml of 20 mM sodium bicarbonate (pH 10.5) containing 4 M urea, and the urea concentration was lowered stepwise from 4 to 2, 1, 0.5, and 0 M every 3 h at room temperature. The dialyzing buffer was then exchanged for 2 liters of 50 mM Tris-HCl (pH 7.2), and dialysis was continued at 4°C for 16 h. The refolded enzyme solution was used as a starting material for the purification of active LepB. The resulting solution was mixed with DEAE-Toyopearl 650 M, and the suspension was filtered through a glass filter (26G-3). The filtrate was applied to an AH-Sepharose 4B column (2.5 by 14 cm) previously equilibrated with 50 mM Tris-HCl (pH 8.0), and the enzyme was eluted with a linear gradient of 0 to 1 M NaCl in the same buffer. The active protease fractions were collected, dialyzed against 50 mM Tris-HCl (pH 8.0), and concentrated by ultrafiltration (Amicon YM-10). The concentrated solution was loaded onto a Sephacryl S-200 column (1.5 by 160 cm) equilibrated with 50 mM Tris-HCl (pH 8.0). The active fractions were collected and concentrated to 1.5 ml and stored at −20°C until they were used. Proteins were measured by the Bradford method (2), with bovine serum albumin as a standard. In column chromatography, protein elution patterns were usually measured by absorption at 280 nm. All operations of the purification procedure were done at 4°C. The N-terminal amino acid sequence of the purified enzyme was analyzed with an Applied Biosystems model 494 sequencer.
CD measurements.
Circular dichroism (CD) spectra were measured with a Jasco spectropolarimeter, model J-720W (Japan Spectroscopic Co., Tokyo, Japan), equipped with a thermal incubation system. The far-UV CD spectra of LepB and LepA were measured at a protein concentration of 0.10 mg/ml in 10 mM sodium phosphate (pH 8.0) with a 2-mm-diameter cuvette. The thermal-unfolding profiles of LepB and LepA were monitored by the CD intensity changes at 230 nm at a protein concentration of 0.10 mg/ml in 10 mM sodium phosphate (pH 8.0). The rate of temperature increase was 1°C/min.
Nucleotide sequence accession number.
The nucleotide sequence data for the LepB and LepA genes are available from the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AB094439 and AB045676, respectively.
RESULTS
Cloning of LepB gene and its deduced amino acid sequence.
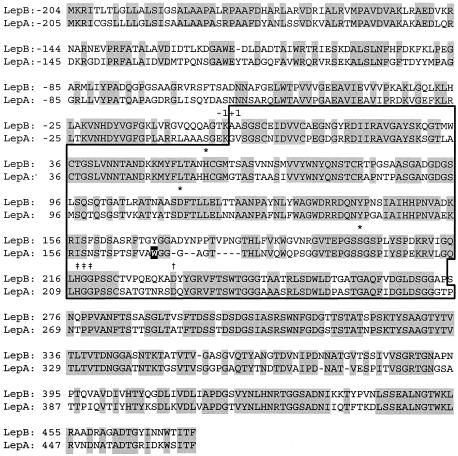
In the previous nucleotide sequence analysis of the cloned lepA gene from Lysobacter sp. strain IB-9374, a gene closely related to the target peptidase gene was detected in a flanking region on the 12-kb BamHI fragment (3). The gene was located immediately upstream from the lepA gene and cloned (Fig. 1A). The newly cloned gene was named the lepB gene, and nucleotide sequence analysis revealed that it consisted of 2,034 nucleotides coding for 678 amino acids. The deduced amino acid sequence was compared with that of LepA, which revealed that the gene product, a LepB precursor, consisted of four distinct peptide fragments, a prepeptide (20 aa), a propeptide (184 aa), a mature LepB (274 aa), and a C-terminal extension (200 aa). The apparent sequence identity between LepB and LepA is 67% for the precursor protein (Fig. 2). More precisely, the sequence identity is highest for the C-terminal extension (75%), followed by the mature-enzyme portion (72%), and it is lowest for the pre- and propeptide portion (54%). In mature LepB, the key amino acids essential for or closely related to the enzyme function of trypsin-type LepA are well conserved: the catalytic-triad amino acids (His57, Asp113, and Ser194; LepA numbers), lysine specificity (Asp225), and substrate binding subsites (His210, Gly211, and Gly212). Amino acid differences exist at various positions; among them, the replacement of Trp169 with tyrosine and the insertion of seven amino acids between positions 172 and 182 were unique and were suspected to be connected to the catalytic function and stability of LepB. By analogy to LepA, proline was temporarily placed at the C terminus of LepB.
FIG. 2.
Amino acid sequence identities between lepB and lepA gene products. The amino acid sequences are numbered from the N-terminal residue of mature enzymes at +1, and the putative mature enzyme regions are boxed. Identical amino acids are shaded. The putative catalytic triad, specificity-determining site, and subsites are indicated by asterisks, dagger, and double daggers, respectively. Trp169 in LepA is shaded black. The dashes in the sequences indicate gaps.
Expression and purification of recombinant LepB.
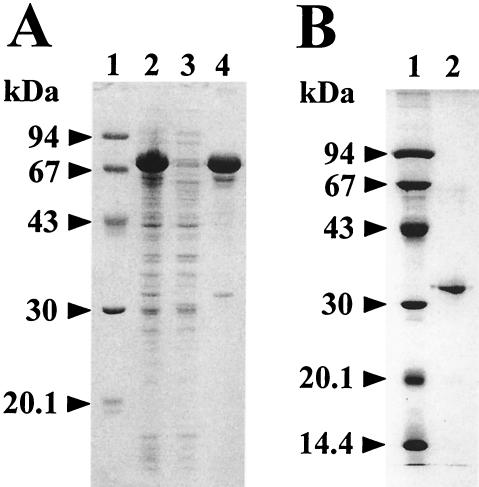
To prepare recombinant LepB, expression of the LepB precursor protein in the periplasm of E. coli cells was attempted under the same conditions used for the API counterpart (16), but it was unsuccessful due to rapid loss of the expression plasmids in host cells upon addition of IPTG. Next, we constructed the expression vector pGEX-lepB, harboring the lepB gene without the coding regions of the prepeptide and the C-terminal extension peptide (starting at Ser275 in Fig. 2), since in the absence of those two regions, the API precursor protein could be expressed to autonomously generate enzyme activity through correct folding (14) (Fig. 1B). E. coli cells carrying pGEX-lepB overproduced 75 kDa of GST-fused LepB, which was collected as inclusion bodies (Fig. 3A). The insoluble proteins were solubilized with 8 M urea and folded to active conformation upon dialyzing to alkaline media. The overall purification of the folded recombinant LepB is summarized in Table 1. Purified LepB migrated as a 32-kDa protein in SDS-12.5% polyacrylamide gel electrophoresis (Fig. 3B). The N-terminal amino acid sequence of the purified protein was determined to be AASGSCEIDVVCAEGNGYRDIIRAVGAYSK, which is totally identical to that predicted for mature LepB (Fig. 2). These results indicate that the expression product of the GST-fused LepB gene cut off the GST and propeptide moiety to produce mature endopeptidase during solubilization and/or the folding process.
FIG. 3.
SDS-polyacrylamide gel electrophoresis (12.5%) analysis of the GST-fused LepB (A) and the purified recombinant LepB (B). (A) Lane 1, standard molecular mass markers; lane 2, 0.1-ml portions at an A600 of 2.5 of E. coli cells that were cultivated at 37°C for 6 h after the addition of 1 mM (final concentration) IPTG; lane 3, soluble fraction of the cells disrupted by sonication; lane 4, insoluble fraction. (B) Lane 1, standard molecular mass markers; lane 2, 6 μg of the purified LepB. The numbers on the left indicate molecular masses.
TABLE 1.
Purification of recombinant LepB
| Step | Protein (mg) | Total activity (U) | Sp act (U/mg of protein) | Purification (n-fold) | Recovery (%) |
|---|---|---|---|---|---|
| Refolded enzyme | 102 | 4.8 | 0.047 | 1 | 100 |
| DEAE-Toyopearl | 26.2 | 4.5 | 0.17 | 4 | 94 |
| AH-Sepharose 4B | 0.81 | 3.0 | 3.7 | 79 | 63 |
| Sephacryl S-200 | 0.54 | 2.3 | 4.3 | 91 | 48 |
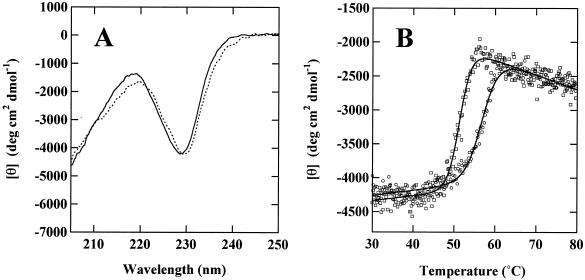
Far-UV CD spectrum of LepB.
Figure 4A shows the far-UV CD spectra of LepB and LepA. The spectra are the same for the two proteins, indicating that LepB possesses a secondary structure quite similar to that of LepA. To clarify folding similarity and dissimilarity, thermal-unfolding profiles were compared by monitoring the ellipticity changes of LepB and LepA at 230 nm (Fig. 4B). Ellipticity in the far-UV region was unchanged at 30 to ∼45°C and decreased steeply at 50 to 60°C. The cooperative unfolding curves recorded for LepB and LepA indicate that the two proteins retain their own stable tertiary structures up 50°C, followed by thermal transition at different midpoint temperatures of thermal unfolding (Tm). The apparent Tm values of LepB and LepA were 51.5 and 57.2°C, respectively, indicating that LepB is structurally less stable than LepA.
FIG. 4.
CD spectra and thermal-unfolding profiles of LepB and LepA. (A) Far-UV CD spectra of LepB (dotted line) and LepA (solid line) were measured at 20°C. (B) Thermal-unfolding profiles of LepB (squares) and LepA (circles) were monitored by changes in the CD intensity at 230 nm. The solid lines show a conventional two-state equation of the thermal-unfolding process.
Enzyme properties.
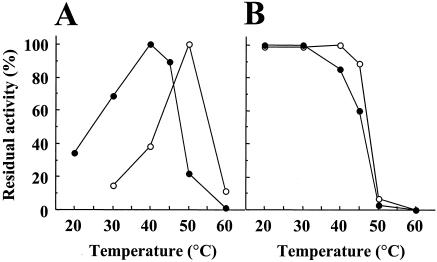
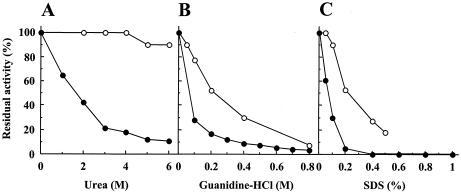
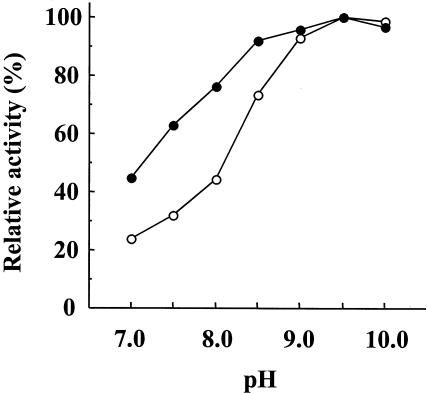
Purified recombinant LepB was most active at pH 8.5 to 10.0 and stable at pH 4.0 to 11.0 at 4°C for 24 h. The amidolytic activity was lost with 10 mM diisopropyl fluorophosphate or 0.1 mM N-tosyl-l-lysine chloromethyl ketone. LepB was most active at 40°C and moderately stable up to 30°C (Fig. 5). LepB lost 82, 72, and 70% of its amidolytic activity after incubation with 4 M urea, 0.1 M guanidine-HCl, and 0.1% SDS, respectively, at 30°C for 20 min (Fig. 6). LepB hydrolyzed Bz (or Ac)-Lys-pNA moderately and, under the same conditions, Lys-pNA was hydrolyzed slowly, yielding pNA only at 6%. All other acylated or unacylated pNA-type substrates tested, including Arg, Ala, His, Phe, Tyr, and Leu in place of Lys, were insensitive to hydrolysis (data not shown). The lysyl peptide bond in glucagon at pH 9.2 was exclusively hydrolyzed at a lower rate than that with LepA (data not shown). The pH dependences and catalytic potencies of LepB and LepA in peptidase function were compared using Boc-Val-Leu-Lys-MCA, a most efficient tripeptide substrate for API (18, 23, 24). The pH-activity experiment showed that the pH range of 50 to 100% activity was 7.0 to 10.0 for LepB and was broader than pH 8.0 to 10.0 for LepA (Fig. 7). The kcat/Km ratio and Km of LepB were one-sixth and 8.5-fold for LepA, respectively, although the kcats were of the same order (Table 2). However, a similar difference between the kcat/Km ratios of these two peptidases was not observed for small substrates, such as Bz-Lys-pNA, Ac-Lys-pNA, and Lys-pNA (Table 2), as reported for API and API[W169Y] (23). Accordingly, these results suggest that the binding subsite of LepB is composed of three subsites, S1 to S3 (the notation is used as described by Schechter and Berger [22]), toward the N terminus from the scissile bond, as in the case of API (LepA) (18).
FIG. 5.
Effects of temperature on the amidolytic activities of LepB and LepA. (A) Optimum temperature. The amidolytic activities of LepB (solid circles) and LepA (open circles) were measured at various temperatures, as indicated. (B) Heat stability. After 1.1 μg of LepB or LepA was incubated at various temperatures, as indicated, for 30 min, the residual amidolytic activities against Bz-Lys-pNA were measured.
FIG. 6.
Effects of various concentrations of urea (A), guanidine-HCl (B), and SDS (C) on the amidolytic activity of LepB. After 1.1 μg of the purified LepB (solid circles) or LepA (open circles) was incubated at 30°C for 20 min in the presence of various concentrations of urea, guanidine-HCl, or SDS, as indicated, the amidolytic activities against Bz-Lys-pNA were measured in the presence of the same denaturant at a given concentration.
FIG. 7.
Relative pH-activity profiles of LepB and LepA. The amidolytic activities of LepB and LepA against Boc-Val-Leu-Lys-MCA were measured at various pHs of 20 mM Tris-HCl. The relative activities of LepB and LepA are indicated by solid and open circles, respectively.
TABLE 2.
Kinetic parameters of LepB and LepA
| Substrate | LepB
|
LepA
|
||||
|---|---|---|---|---|---|---|
| Km (μM) | kcata (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcata (s−1) | kcat/Km (μM−1 s−1) | |
| Boc-Val-Leu- Lys-MCAb | 11 | 168 | 15 | 1.3 | 123 | 95 |
| Bz-Lys-pNAc | 67 | 3.0 | 0.045 | 65 | 5.7 | 0.088 |
| Ac-Lys-pNAc | 42 | 14 | 0.33 | 18 | 4.5 | 0.25 |
| Lys-pNAc | 100 | 0.21 | 0.0021 | 77 | 0.18 | 0.0023 |
Parameters were determined by Lineweaver-Burk plots, and the molecular masses of 28,916.2 and 27,735.9 Da calculated from the deduced amino acid sequences were used for the kcat values of LepB and LepA, respectively.
Reaction mixtures containing 0.5 to 30 μM Boc-Val-Leu-Lys-MCA and 20 mM Tris-HCl (pH 9.0) were incubated at 30°C for 5 min, and the reaction was initiated by the addition of LepB or LepA to a final concentration of 1 pM in a total volume of 3 ml. Increased fluorescence of liberated 7-amino-4-methylcoumarin was monitored at 440 nm upon excitation at 380 nm for 4 min.
Reaction mixtures containing 40 to 250 μM Bz-Lys-pNA, 20 to 200 μM Ac-Lys-pNA, or 35 to 250 μM Lys-pNA and 180 mM Tris-HCl (pH 9.2) were incubated at 30°C for 5 min, and the reaction was initiated by the addition of LepB or LepA to a final concentration of 16.6 nM for Bz-Lys-pNA, 4.1 nM for Ac-Lys-pNA, and 170 nM for Lys-pNA in a total volume of 0.75 ml. After 10 min of incubation, the reaction was terminated by adding 0.25 ml of 45% (vol/vol) acetic acid, and the absorbance at 405 nm was measured.
Modeling of LepB based on the three-dimensional structure of LepA (API).
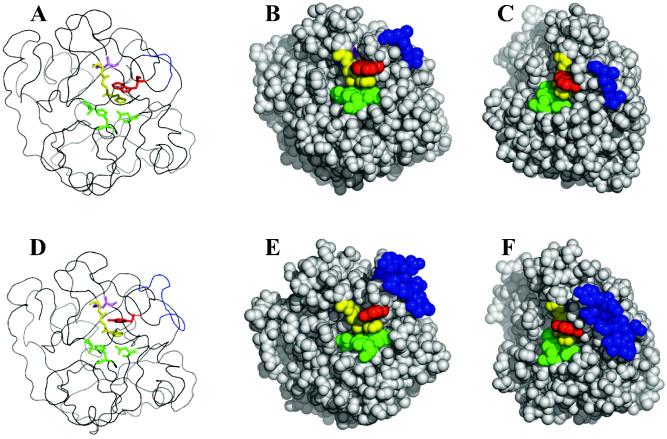
The tertiary structure of LepB was modeled by the program MODELLER 6 (8) based on coordinates of LepA (API) deposited in the Protein Data Bank (code 1ARB) (Fig. 8). In this model, the main chain of LepB constitutes essentially the same tertiary fold as in LepA, except for the region of seven amino acids inserted in the middle of the peptide chain. It is interesting that the far-UV CD spectra of LepB and LepA (Fig. 4A) are not typical spectra characteristic of α-helix or β-sheet structures with peaks at 222 or 218 nm but are trypsin-type spectra with a characteristic peak at 230 nm, which has been observed for serine proteases (21). The above-mentioned extra peptide segment can form a folded structure outside the molecule with no significant perturbation of the surroundings other than those making contact with atoms newly integrated into the extra folding domain. This local folded domain is near the active site but has no direct contact with it. Key active-site amino acids detectable upon sequence alignment are seen mostly at the same loci as the counterparts of LepA and are possibly able to play pivotal roles in the peptidase function of LepB as well. Accessible surface areas were calculated for individual amino acids based on the LepB model, which revealed apparently quite similar physical properties of surface atoms for LepA and LepB (Fig. 8). In particular, the key residues in LepB—catalytic triad, subsite, Tyr169, and Asp232—are structurally identical to those in LepA. Exceptions are seen in the sites of the highly exposed (>80%) Gln31, Asp91, Arg165, Tyr174, Asn192, and Gln227 in LepB.
FIG. 8.
Modeling structure of LepB based on the tertiary structure of LepA. Shown are models for LepA (A to C) and LepB (D to F). The green and yellow residues represent catalytic triad and subsite residues, respectively. The red residues represent Trp169 (LepA) or Tyr169 (LepB). The pink residues represent Asp225 (LepA) or Asp232 (LepB). The blue residues represent the gap sequences (LepA, Gly172-Thr175; LepB, Ala172-Gly182).
DISCUSSION
Detection of a new gene, coding for another lysine-specific serine protease, upstream of the LepA gene was unexpected, since this less stable peptidase isoform had been undetectable at the protein level in the broth of Lysobacter sp. strain IB-9374. In the case of API-producing A. lyticus M497-1, two additional less active lysyl endopeptidases (APII and APIII) had been detectable at the protein level (11), but a search for genes coding for either of these two endopeptidases had not been made for genomic DNA of the Achromobacter strain. The extracellular lysyl endopeptidase-producing Achromobacter and Lysobacter strains are able to lyse bacteria by collaboration with α- and β-lytic protease (1, 7). Whether the lepB gene is a relic of a gene encoding highly active LepA during its evolution or a product to actually serve as active peptidase synthesized in weakly alkaline medium to effect lysis of target cells remains unclear.
The putative amino acid sequence of mature LepB, with lower enzymatic activity and reduced structural stability compared to LepA, shows 72% identity with that of the latter endopeptidase. All essential amino acids are well conserved, except Trp169, which serves to generate the high catalytic potency and wide pH optimum of LepA (23, 24). In the primary structure, the extra peptide segment present only in LepB is aligned to the region that the peptide segment of Gly173-Ala174-Gly175-Thr176 occupies to compose an exposed loop connecting the β8 strand to the β9 strand in LepA. The proline-rich 11-aa peptide segment, including 4 aa originally present in LepA, is folded and protrudes toward the solvent from the protein surface. This likely location of the inserted peptide segment in the modeled structure is suggestive of a possibility that it is an internal candidate for the locus into which an appropriate peptide fragment can be incorporated. It is interesting that the cape of this protruding peninsula-like structural domain holds a mostly exposed phenol ring of Tyr174 that is absent in LepA. At present, it is not known whether this exposed aromatic ring is related to the function and/or stability of LepB.
In this context, the role of another exposed aromatic amino acid, Tyr169, not far from Tyr174, should be considered. If the extra peptide segment present only in LepB has no direct connection to the difference between the catalytic functions of LepB and the LepA peptidase, the presence of Tyr169 of LepB in place of Trp169 of LepA is likely to be a major factor causing the difference between their catalytic functions. This possibility is supported by the fact that the kcat/Km ratio of LepB is the same as that of the LepA mutant containing Tyr169 (23-25). The Tyr169 in the LepB model is also superimposable on Tyr169 in API[W169Y], modeled based on API in the same manner. It has been shown that, in API, the indole ring of Trp169 stacks on the imidazole ring of His210, the S1 subsite for substrate binding, and is responsible for the creation of its distinct enzymatic properties, such as 1 order of magnitude-higher peptidase activity at pH 8.5 to 10.5 (9, 23, 24). If Tyr169 and His217 in LepB are disposed in the same manner as Tyr169 and His210 in API[W169Y], the difference in kcat and Km detected between the API mutant and LepB would be attributable to the difference in the tertiary structures derived from amino acid substitutions and a 7-aa insertion. To further study this point, site-directed mutagenesis experiments and the analysis of the three-dimensional structure of LepB are in progress in our laboratory.
REFERENCES
- 1.Ahmed, K., S. Chohnan, H. Ohashi, T. Hirata, T. Masaki, and F. Sakiyama. 2003. Purification, bacteriolytic activity, and specificity of β-lytic protease from Lysobacter sp. IB-9374. J. Biosci. Bioeng. 95:27-34. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1980. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chohnan, S., J. Nonaka, K. Teramoto, K. Taniguchi, Y. Kameda, H. Tamura, Y. Kurusu, S. Norioka, T. Masaki, and F. Sakiyama. 2002. Lysobacter strain with high lysyl endopeptidase production. FEMS Microbiol. Lett. 213:13-20. [DOI] [PubMed] [Google Scholar]
- 4.Elliott, B. W., and C. Cohen. 1986. Isolation and characterization of a lysine-specific protease from Pseudomonas aeruginosa. J. Biol. Chem. 261:11259-11265. [PubMed] [Google Scholar]
- 5.Jekel, P. A., W. J. Weijer, and J. J. Beintema. 1983. Use of endoproteinase Lys-C from Lysobacter enzymogenes in protein sequence analysis. Anal. Biochem. 134:347-354. [DOI] [PubMed] [Google Scholar]
- 6.Kawata, Y., F. Sakiyama, and H. Tamaoki. 1988. Amino-acid sequence of ribonuclease T2 from Aspergillus oryzae. Eur. J. Biochem. 176:683-697. [DOI] [PubMed] [Google Scholar]
- 7.Li, S. L., S. Norioka, and F. Sakiyama. 1990. Molecular cloning and nucleotide sequence of the β-lytic protease gene from Achromobacter lyticus. J. Bacteriol. 172:6506-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 9.Masaki, T., T. Fujihashi, K. Nakamura, and M. Soejima. 1981. Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. II. specificity and inhibition studies of Achromobacter protease I. Biochim. Biophys. Acta 660:51-55. [DOI] [PubMed] [Google Scholar]
- 10.Masaki, T., T. Fujihashi, and M. Soejima. 1984. Effect of various inhibitors on the activity of Achromobacter protease I. Nippon Nogeikagaku Kaishi 58:865-870. (In Japanese.) [Google Scholar]
- 11.Masaki, T., K. Nakamura, M. Isono, and M. Soejima. 1978. A new proteolytic enzyme from Achromobacter lyticus M497-1. Agric. Biol. Chem. 42:1443-1445. [Google Scholar]
- 12.Masaki, T., T. Takiya, S. Tsunasawa, S. Kuwahara, F. Sakiyama, and M. Soejima. 1994. Hydrolysis of S-2-aminoethylcysteinyl peptide bond by Achromobacter protease I. Biosci. Biotechnol. Biochem. 58:215-216. [DOI] [PubMed] [Google Scholar]
- 13.Masaki, T., M. Tanabe, K. Nakamura, and M. Soejima. 1981. Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. I. Purification and some enzymatic properties. Biochim. Biophys. Acta 660:44-50. [DOI] [PubMed] [Google Scholar]
- 14.Norioka, S., S. Ohta, T. Ohara, S. L. Li, and F. Sakiyama. 1994. Identification of three catalytic triad constituents and Asp-225 essential for function of lysine-specific serine protease, Achromobacter lyticus protease I. J. Biol. Chem. 269:17025-17029. [PubMed] [Google Scholar]
- 15.Oda, Y., Y. Kitayama, H. Yamaguchi, Y. Matsuura, Y. Katsube, T. Masaki, T. Tanaka, S. Matsuura, S. Norioka, and F. Sakiyama. 1996. Crystallization and preliminary X-ray diffraction analysis of two lysinal derivatives of Achromobacter lyticus protease I. Acta Crystallogr. D 52:1027-1029. [DOI] [PubMed] [Google Scholar]
- 16.Ohara, T., K. Makino, H. Shinagawa, A. Nakata, S. Norioka, and F. Sakiyama. 1989. Cloning, nucleotide sequence, and expression of Achromobacter lyticus protease I gene. J. Biol. Chem. 264:20625-20631. [PubMed] [Google Scholar]
- 17.Sakiyama, F., and T. Masaki. 1994. Lysyl endopeptidase of Achromobacter lyticus. Methods Enzymol. 244:126-137. [DOI] [PubMed] [Google Scholar]
- 18.Sakiyama, F., M. Suzuki, K. Yamamoto, S. Aimoto, S. Norioka, T. Masaki, and M. Soejima. 1990. Mapping of substrate binding site of a lysine-specific serine protease, Achromobacter protease I. J. Protein Chem. 9:297-298. [Google Scholar]
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schechter, N. M., G. Y. Eng, T. Selwood, and D. R. McCaslin. 1995. Structural changes associated with the spontaneous inactivation of the serine proteinase human tryptase. Biochemistry 34:10628-10638. [DOI] [PubMed] [Google Scholar]
- 22.Schechter, I., and A. Berger. 1967. On the size of the active site in protease. I. Papain. Biochem. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 23.Shiraki, K., S. Norioka, S. L. Li, and F. Sakiyama. 2002. Contribution of an imidazole-indole stack to high catalytic potency of a lysine-specific serine protease, Achromobacter protease I. J. Biochem. 131:213-218. [DOI] [PubMed] [Google Scholar]
- 24.Shiraki, K., S. Norioka, S. Li, K. Yokota, and F. Sakiyama. 2002. Electrostatic role of aromatic ring stacking in the pH-sensitive modulation of a chymotrypsin-type serine protease, Achromobacter protease I. Eur. J. Biochem. 269:4152-4158. [DOI] [PubMed] [Google Scholar]
- 25.Shiraki, K., and F. Sakiyama. 2002. Histidine 210 mutant of a trypsin-type Achromobacter protease I shows broad optimum pH range. J. Biosci. Bioeng. 93:331-333. [DOI] [PubMed] [Google Scholar]
- 26.Tsunasawa, S., T. Masaki, M. Hirose, M. Soejima, and F. Sakiyama. 1989. The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. J. Biol. Chem. 264:3832-3839. [PubMed] [Google Scholar]
- 27.Tsunasawa, S., A. Sugihara, T. Masaki, F. Sakiyama, Y. Takeda, T. Miwatani, and K. Narita. 1987. Amino acid sequence of thermostable direct hemolysin produced by Vibrio parahaemolyticus. J. Biochem. 101:111-121. [DOI] [PubMed] [Google Scholar]
- 28.Wachsmuth, E. D., I. Fritze, and G. Pfleiderer. 1966. An aminopeptidase occurring in pig kidney. I. An improved method of preparation. Physical and enzymatic properties. Biochemistry 5:169-174. [DOI] [PubMed] [Google Scholar]