Abstract
The Brahma (BRM) and Brahma-related Gene 1 (BRG1) ATPases are highly conserved homologues that catalyze the chromatin remodeling functions of the multi-subunit human SWI/SNF chromatin remodeling enzymes in a mutually exclusive manner. SWI/SNF enzyme subunits are mutated or missing in man cancer types, but are overexpressed without apparent mutation in other cancers. Here, we report that that both BRG1 and BRM are overexpressed in most primary breast cancers independent of the tumor’s receptor status. Knockdown of either ATPase in a triple negative breast cancer cell line reduced tumor formation in vivo and cell proliferation in vitro. Fewer cells in S phase and an extended cell cycle progression time were observed without any indication of apoptosis, senescence or alterations in migration or attachment properties. Combined knockdown of BRM and BRG1 showed additive effects in the reduction of cell proliferation and time required for completion of cell cycle, suggesting that these enzymes promote cell cycle progression through independent mechanisms. Knockout of BRG1 or BRM using CRISPR/Cas9 technology resulted in loss of viability, consistent with a requirement for both enzymes in triple negative breast cancer cells.
Keywords: SWI/SNF, chromatin remodeling, BRG1, BRM, breast cancer
Introduction
ATP-dependent chromatin remodeling enzymes use the energy generated by ATP hydrolysis to alter histone:DNA contacts and induce alterations in nucleosome structure and/or positioning that can lead to increased or decreased gene expression, replication, repair, and/or recombination (Bartholomew, 2014; Euskirchen et al., 2012; Liu et al., 2011). The enzymes are large complexes consisting of at least 11–15 characterized subunits that include members of multi-gene families and proteins that are expressed in a lineage-restricted manner. Combinatorial assembly can therefore give rise to dozens, and perhaps more, different SWI/SNF complexes that likely have significant to subtle differences in function (Hargreaves and Crabtree, 2011). There are two mutually exclusive enzymatic subunits that provide the DNA-stimulated ATPase activity that drives chromatin remodeling (Wang et al., 1996). BRM and BRG1 share ∼75% identity at the protein level and behave similarly in in vitro chromatin remodeling assays (Phelan et al., 1999). However, they have been reported to preferentially interact with different transcription factors (Kadam and Emerson, 2003) and they show different expression patterns during development and in adult tissues (Reisman et al., 2005). Brg1 deficient mice die early during embryogenesis, underscoring the requirement for Brg1 in mouse development (Bultman et al., 2000) and, indeed, Brg1 is required for nearly every developmental and tissue differentiation event that has been examined (de la Serna et al., 2006; Ho and Crabtree, 2010; Wu, 2012). In contrast, Brm deficient mice developed normally, with strain specific hyperproliferation in some tissues and evidence for increased levels of Brg1, which likely serves as a compensation mechanism for Brm loss (Reyes et al., 1998).
Evidence for widespread connections between SWI/SNF subunits and cancer has been growing in recent years. It has been reported that approximately one out of every five human cancers contains mutations in one or more of the genes encoding SWI/SNF subunits or associated proteins (Kadoch et al., 2013; Shain and Pollack, 2013). There is specificity in the cancer type resulting from mutation of different SWI/SNF subunits (reviewed in (Helming et al., 2014; Hohmann and Vakoc, 2014)), likely reflecting the diversity of complexes in existence and leading to the idea that mutation of different subunits results in different molecular deficiencies that lead to tumor development. Most of these conclusions are based on genome sequencing from patient tumor samples, however, mouse modeling for two of the subunits, Ini1 and Brg1, demonstrate that loss of expression of these proteins induced tumor formation (Bultman et al., 2000; Bultman et al., 2008; Guidi et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000), providing compelling evidence of direct tumor suppressor function.
The large and growing literature on SWI/SNF ATPases in cancer that we will briefly summarize shows that these enzymes have a context-specific function and tumor-specific role in cancer. BRG1 mutations are frequently found in lung tumors (Blanco et al., 2009; Medina et al., 2004; Medina et al., 2008; Reisman et al., 2003; Rodriguez-Nieto et al., 2011) as well as in medullablastomas (Jones et al., 2012; Pugh et al., 2012) and Burkitt’s lymphoma (Love et al., 2012). Silencing of Brm is observed in lung tumors, and mutation or loss of Brg1 and/or Brm correlates with poor prognosis and survival (Fukuoka et al., 2004; Reisman et al., 2003). Epigenetic silencing of BRM was identified by work showing that treatment of BRM deficient cancer cell lines with inhibitors of histone deacetylases (HDACs) reactivated BRM expression (Glaros et al., 2007). Brm loss can promote lung tumorigenesis in response to environmental carcinogens. Prolonged exposure of Brm heterozygous mice to the lung tumor-inducing carcinogen, ethyl carbamate (Tuveson and Jacks, 1999), caused a significant increase in the number of lung adenomas compared to wildtype mice, while Brm null mice had double the number of tumors found in Brm heterozygotes after the same treatment (Glaros et al., 2007). BRM silencing has also been reported in a variety of primary tumor types (Glaros et al., 2007). Moreover, reintroduction of BRG1 or BRM into human cancer cell lines deficient for those subunits slowed cell proliferation (reviewed in (Reisman et al., 2009)), supporting the idea that loss of the ATPase(s) is linked to the highly proliferative nature of these cancer cell lines. The extensive involvement of SWI/SNF ATPases in regulation of cell cycle progression by most of the major classes of tumor suppressors/cell cycle regulators, including Rb, p53, E2F, has provided numerous mechanisms by which BRG1 and/or BRM could contribute to the suppression of unregulated cell growth (Bochar et al., 2000; Dunaief et al., 1994; Hendricks et al., 2004; Hill et al., 2004; Lee et al., 2002; Reisman et al., 2002; Strobeck et al., 2000; Trouche et al., 1997).
However, a positive role for BRG1 in promoting cancer is supported by studies demonstrating that acute myeloid leukemia requires BRG1 for maintenance of leukemia cell proliferation and survival (Buscarlet et al., 2014; Shi et al., 2013). At least part of this requirement for BRG1 was attributed to its chromatin remodeling functions at the c-Myc locus, where it enabled both enhancer binding by hematopoietic transcription factors and enhancer:promoter looping interactions that maintained c-Myc expression (Shi et al., 2013). Another example of a tumor requiring BRG1 was recently reported in small cell lung cancer (SCLC) tumors containing mutations in the Myc associated factor Max (Romero et al., 2014). BRG1 knockdown inhibited cell proliferation and viability in SCLC cells with Max mutations whereas SCLC cells with wildtype Max showed no effect. Clearly, the absence of functional Max in these cells rendered BRG1 function necessary for cell survival. Other data also support a requirement for SWI/SNF ATPases in promoting cancer. At least one of the SWI/SNF ATPases is required for melanoma tumorigenic properties in vitro (Keenen et al., 2010), while HeLa cell proliferation requires BRG1 (Naidu et al., 2009). BRG1 levels are significantly increased in primary and metastatic melanoma, and reduction of BRG1 in melanoma cell lines reduces cell proliferation (Lin et al., 2010; Saladi et al., 2010). Similar results have been reported for colorectal, gastric and prostate cancers (Sentani et al., 2001; Sun et al., 2007; Watanabe et al., 2011).
The roles of BRG1 and BRM in breast cancer are no more definitive. Mice heterozygous for Brg1 developed mammary tumors (Bultman et al., 2000; Bultman et al., 2008), and the combination of Brg1 heterozygosity and complete Brm deficiency did not alter the phenotype (Bultman et al., 2008). These findings led to the seemingly solid conclusion that Brg1 is a breast cancer tumor suppressor. Immunohistochemistry analysis in primary breast cancers revealed about 15% lacked BRM expression (Glaros et al., 2007). Oncogenic Ras had previously been shown to reduce Brm levels as part of Ras-induced fibroblast transformation (Muchardt et al., 1998). Building upon this observation, it has been shown that oncogene-mediated activation of the extracellular signal-regulated kinase (ERK) promotes malignant behavior in mammary epithelial cells through a cascade in which elevated c-Myc levels reduce Brm levels, which induce the C/EBPβ regulator, which then directly up-regulate α5 integrin expression (Damiano et al., 2014). α5 integrin and its ligand fibronectin are upregulated in many tumors, including breast, and are indicators of poor prognosis (Dingemans et al., 2010; Nam et al., 2010; Yao et al., 2007). This work elegantly demonstrates how Brm loss plays a critical role in a key signaling pathway that contributes to the induction of mammary epithelial cell transformation. However, a number of studies are not consistent with BRG1 and BRM being mammary tumor suppressors. Depletion of BRG1 or BRM in immortalized but near diploid, non-tumorigenic MCF10A cells decreases the proliferation rate (Cohet et al., 2010). Conditional knockout of Brg1 in mammary gland does not result in mammary tumors (Serber et al., 2012). Exome sequencing of 507 primary breast cancers failed to identify mutations in BRG1 (Network, 2012). Immunohistochemistry analyses of primary breast tumors showed that nearly all had high expression of BRG1 while high BRG1 expression correlated with poor prognosis (Bai et al., 2013).
We have investigated the roles of the BRG1 and BRM enzymes in breast cancer. We determined that both enzymes are highly expressed in primary breast tumors compared to normal breast tissue. Knockdown of either ATPase in the context of a triple negative breast cancer cell background reduced tumor formation and growth in xenografts and slowed cell proliferation in culture. Combined knockdown of BRG1 and BRM had an additive effect on cell proliferation while CRISPR/Cas9 knockout of BRG1 or BRM impaired cell viability. The results suggest that BRG1 and BRM have at least some non-overlapping roles in promoting breast cancer cell proliferation and recommend both BRG1 and BRM as targets for breast cancer therapy.
MATERIALS AND METHODS
Tumor samples and tissue microarrays
Matched normal and tumor samples were obtained from the University of Massachusetts Medical School Tissue Bank. A tissue array with three-fold redundancy from 63 cases of invasive breast carcinoma, selected from the archives of the Department of Pathology at the Magee-Womens Hospital, University of Pittsburgh Medical Center was described (Domfeh et al., 2008). In addition a commercial breast tumor tissue array was obtained from BioMax (BR1503a; US Biomax Inc.).
Immunohistochemistry
Slides were rehydrated after three 5-min washes with xylene by successive 5-min washes with 100%, 95%, 80% ethanol and finally with distilled water. Antigen retrieval was performed using the ABC kit (Vector Laboratories) following the manufacturer’s protocol. Slides were then treated with 3% hydrogen peroxide for 1 hour to block the endogenous peroxidase activity and incubated overnight with monoclonal rabbit anti-BRG1 antibody (H-88; Santa Cruz Biotechnology; 1:400 dilution) at 4°C. The sections were then incubated for 2 hours with a biotin labeled secondary antibody and then 1 hour with streptavidin-peroxidase. The samples were developed using DAB and nuclei were counterstained with hematoxylin.
Western blots
Tissue samples of 1mg were disrupted with a Brinkman Polytron Homogenizer (Model PT10–35) in 1ml CelLytic MT Cell Lysis Buffer (Sigma-Aldrich) containing Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail (Roche). 30 micrograms of tissue extract or whole cell lysates from 50,000 culture cells were separated on 4–20% gradient SDS-PAGE and transferred to PVDF membrane. Antibodies against phospho-Histone H3-ser10 (D47G5) were obtained from Cell Signaling Technology. BRG1 antibody (H-88; sc107687) was obtained from Santa Cruz Biotechnology. Loading was monitored by Western detection of Histone H4 (EMD Millipore), Lamin B (Santa Cruz; sc377001), GAPDH (Sigma: G9295), or by Coomassie Brilliant Blue staining.
Cell Culture
MDA-MB-231 and MDA-MB-468 cells were maintained in DMEM supplemented with 10% FBS and Penicillin/Streptomycin. Cells inducibly expressing control scrambled shRNA or shRNA against BRG1 or BRM or both were generated as described (Cohet et al., 2010). Gene knockdown was induced by incubation of cells in medium containing 10 ng/mL Doxycycline (Sigma-Aldrich) for 72 hours. siRNA treatment was performed as described (Imbalzano et al., 2013). The BRG1 siRNAs were previously described (Imbalzano et al., 2013). The BRM siRNA pool was obtained from Life Science (SMARCA2 Stealth siRNAs (Set of 3) HSS110000, HSS110001, HSS185952).
Mouse mammary fat pad xenografts
Seven-week-old SCID/NCr mice were given water with or without Doxycycline (500 µg /mL) for 5 days prior to cell injection. One million MDA-MB-231 cells harboring Tet-KRAB (Szulc et al., 2006) (parental cells), scramble sequence shRNA, or shBRG1, were mixed with 50 µL of Matrigel (BD Bioscience) and injected orthotopically into the mammary fat pads. Animals were maintained on water with or without Doxycycline for 5 weeks. Tumors formed in the mammary fat pad were dissected and weighed.
Proliferation Assays
Cell proliferation was monitored by direct cell counting using a Z1 Coulter counter (Beckman Coulter Inc.) or by MTS assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H–tetrazolium) using the CellTiter 96® AQueous One Solution cell proliferation assay (Promega Corporation).
BrdU Incorporation
One hundred thousand non-synchronized cells were seeded on coverslips in a 12-well plate. 5-Bromo-2’-deoxyuridine (BrdU; Roche Diagnostics Corporation/Roche Applied Science) was added to the culture medium to a final concentration of 20 µM and incubated for 20 min at 37°C before immunofluorescence staining.
Time lapse imaging
Cells on coverslips were assembled into observation chambers and filmed by time-lapse microscopy at 37°C for 72 hours. Images were acquired with SimplePCI and HCImage software (Hamamatsu) and exported as AVI movies. Cells imaged at different time points were counted manually and growth curves were plotted in GraphPad Prism.
Clonogenic assay
Two hundred single suspended cells were seeded in each well of 6-well plates and incubated for 7 days. Colonies were fixed with Gluteraldehyde (6.0% v/v), stained with Crystal Violet (0.5% w/v), and counted using a stereomicroscope.
Wound healing assay
A straight scratch was made to a confluent monolayer of cells in 12-well plates using 20 µL filter tips and cell debris was removed by 3 washes with PBS. Pictures were taken at times 0 and 24 hours using an inverted Leica DMI6000.
Adhesion assay
12-well plates were coated with 10 µg/ml Collagen I or Collagen IV and incubated at 4 °C overnight. 105 cells were plated in each well and allowed to adhere at 37 °C for 30 minutes. Adherent cells were fixed with 4% paraformaldehyde, washed 3 times with ice cold PBS and stained with 0.5% Crystal violet.
Senescence assay
Standard Acid β-galalactosidase staining was performed as described (Zhang and Cohen, 2004).
Annexin V flow cytometry
200 µL of a cell suspension (2 × 106 cells) were labeled by adding 20 µL of Binding Buffer and 5 µL of Annexin V-Pacific Blue (Invitrogen). Samples were mixed gently and incubated at room temperature in the dark for 15 minutes. 2 µL of DAPI (1 mg/mL) were added to each sample before analysis by flow cytometry. A minimum of 10,000 cells within the gated region was analyzed.
CRISPR Construction
CRISPRs were designed at http://crispr.mit.edu provided by the Zhang laboratory and then cloned into pX330 CRISPR/Cas9 vector (Addgene) following Zhang’s protocol (http://www.genome-engineering.org/crispr/?page_id=23). The target sequences of BRG1 are 187- GGATCCCTACCTTGTGCATC; 189- CTCCTGGCCCGGAAGACATC; 191- CCCGAAGACGGGCCACTGGC. BRM target sequences are 181-CTCCCATCCTATGCCGACGA; 183-CATCGATGGTATACATGACA; 185-GGTATGCGACCACCTCACCC. The control CRISPR sequence is GGCAGAAGGAACACAGGCTC .
Dynamic Cell Proliferation Assay
CRISPR control (GFP) or BRG1 knockout (187, 189, 191) or BRM knockout (181, 183, 185) cells were monitored for proliferation using the xCELLigence RTCA system (ACEA Bioscience, Inc.) following plating on the E-Plates 8 in 30-minute intervals from the time of plating until the end of the experiment (220 hours). Cell Index values for all cell lines were calculated and plotted with the RTCA Software (ACEA Bioscience, Inc.).
Statistical analyses
All quantitative data points represent the mean or representative of three independent experiments +/− standard deviation (SD). Statistical analysis was performed using GraphPad Prism Two-way ANOVA (Graphpad Software Inc.).
RESULTS
BRG1 and BRM are overexpressed in breast tumors
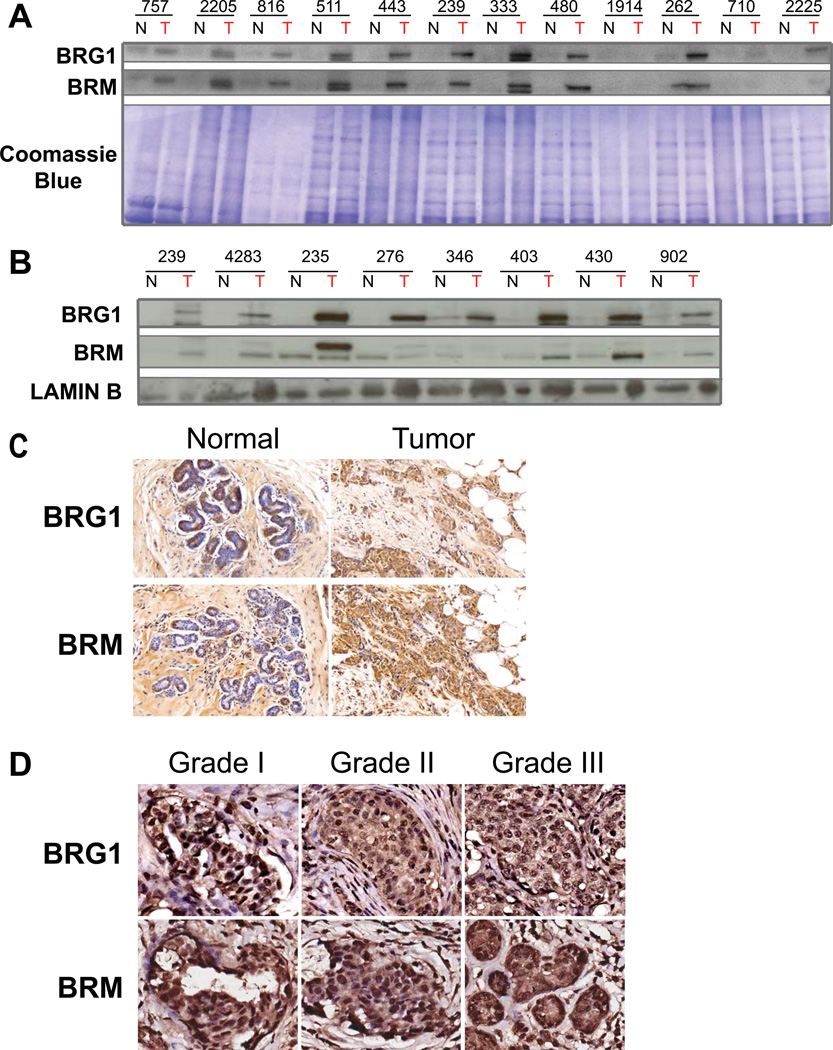
We first measured BRG1 and BRM protein levels in nineteen breast biopsies by Western blotting. Eleven of twelve grade II, ER+/PR+/HER2- tumors showed increased levels of BRG1 relative to adjacent normal tissue, while ten of twelve showed elevated levels of BRM (Fig. 1A; Table S1). Analysis of seven grade III tumors of mixed ER/PR/HER2 status, including three triple negative samples (patients #276, 346, 902), indicated that all showed elevated levels of BRG1 relative to adjacent normal tissue while four showed elevated levels of BRM (Fig. 1B; Table S1). Immunohistochemistry (IHC) of a representative grade II, ER+/PR+/HER2- tumor determined that the invasive breast carcinoma cells had higher BRG1 and BRM levels than adjacent normal tissue and that BRG1 and BRM were observed in myoepithelial cells but not in most ductal epithelial cells (Fig. 1C).
Figure 1. BRG1 and BRM are overexpressed in primary breast tumors.
(A) Western blots showing BRG1 and BRM protein levels in primary breast tumors compared to adjacent normal breast tissues from grade II, ER+, PR+, HER2- patients. N; normal adjacent tissue. T; tumor tissue. Coomassie staining of a duplicate gel was used to demonstrate equal loading between normal and tumor samples from each patient. (B) Western blots showing BRG1 and BRM protein levels in primary breast tumors compared to adjacent normal breast tissues from patients with grade III, mixed ER/PR/HER2 status tumors. N; normal adjacent tissue. T; tumor tissue. Lamin B levels were used as a loading control. (C) Immunohistochemistry of BRG1 and BRM expression in adjacent normal breast tissue (left) and primary tumor (right) sections. This patient was Grade II, ER+, PR+, HER2−. (D) Images of grade I, II, and III tumors stained for BRG1 or BRM extracted from the Biomax 1503a tissue arrays.
Immunohistochemistry was performed on a larger cohort of breast cancer patients derived from two different tissue arrays (Tables S2– S3; see Materials and Methods). Consistent with the previous findings, BRG1 and BRM staining in normal breast tissue adjacent to tumor samples, was limited to myoepithelial and basal cells. In contrast, prominent BRG1 and BRM staining was detected in breast tumor cells compared to normal adjacent tissue. BRG1 was detected in the tumor cells in 176 out of 182 interpretable tumor cores, regardless of subtype, while all 182 interpretable tumor cores had BRM staining. Images of grade I, II, and III tumors stained for BRG1 or BRM extracted from one of the tissue arrays are presented in Fig. 1D. The data show that elevated BRG1 and BRM levels are a common feature of nearly all mammary tumors.
Targeting BRG1 or BRM reduces colony formation in vitro and tumor growth in vivo
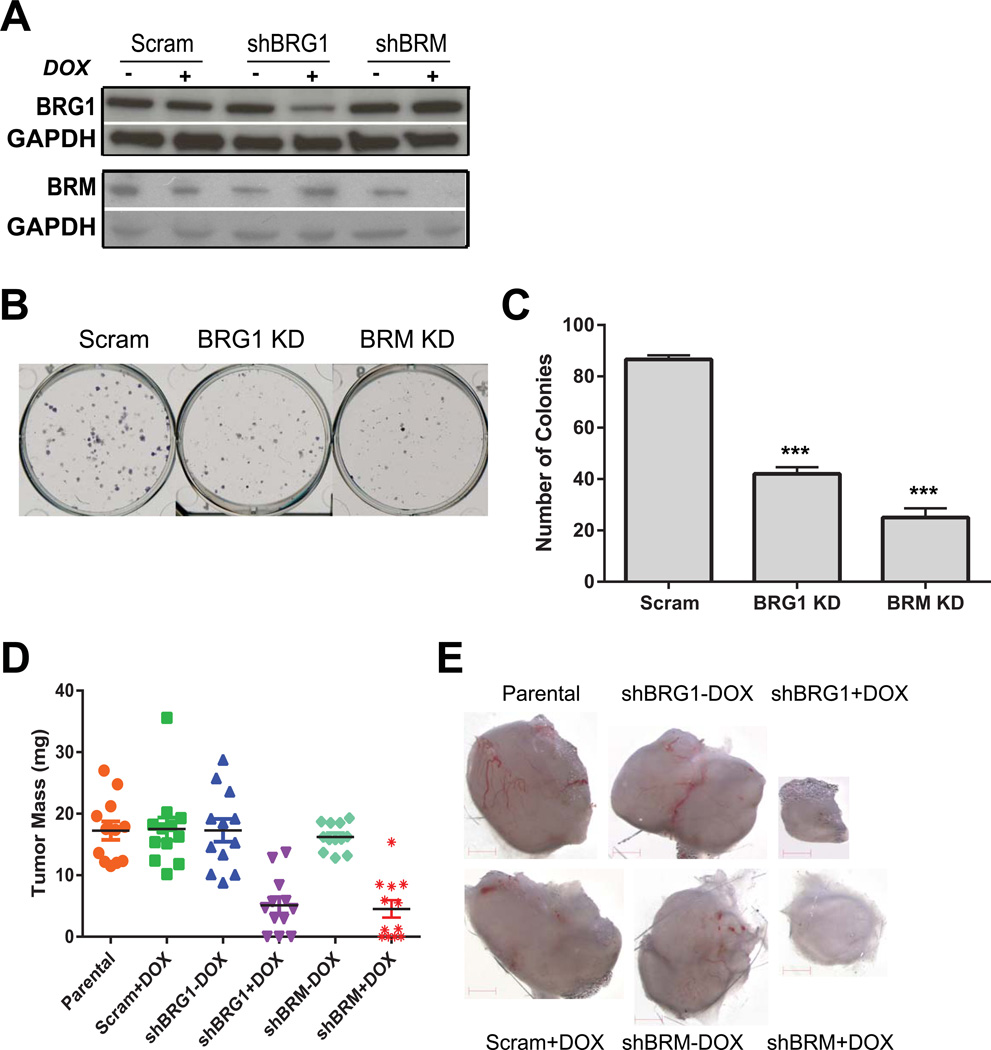
The data from primary tumors suggested that elevated levels of BRG1 and BRM could be related to the transformed state. To further our studies, we utilized the highly metastatic, triple negative breast cancer cell line MDA-MB-231 (Cailleau et al., 1978). These cells express wildtype BRG1 and BRM (COSMIC Cell Lines Project; http://cancer.sanger.ac.uk/cancergenome/projects/cell_lines/ and Broad-Novartis Cancer Cell Line Encyclopedia; http://www.broadinstitute.org/ccle/home). We created MDA-MB-231 cells that inducibly express doxycycline-inducible shRNAs targeting BRG1 or BRM. A scrambled sequence shRNA was used to create control cells. The expression system made use of two lentiviral vectors: one that expressed the doxycycline-inducible tTR-KRAB transcriptional regulator and ds-RED and another that expressed the shRNA and GFP under the control of the tTR-KRAB (Cohet et al., 2010; Szulc et al., 2006). Pools of infected cells were sorted by FACS to generate populations that constitutively expressed ds-RED and expressed GFP in a doxycycline-inducible manner. Western blot analysis confirmed the specificity of knockdown. Cells expressing shBRG1 showed reduced levels of BRG1 with a modest but reproducible increase in BRM (Fig. 2A). Conversely, cells expressing shBRM showed reduced levels of BRM with a modest but reproducible increase in BRG1 (Fig. 2A).
Figure 2. BRG1 or BRM reduction in MDA-MB-231 metastatic breast cancer cells reduced colony formation in culture and tumor growth in vivo.
(A) Western blot indicating BRG1 and BRM levels in MDA-MB-231 cells inducibly expressing a control (Scram) shRNA or shRNA targeting BRG1 or BRM. GAPDH levels were monitored as a loading control. DOX; doxycycline. (B) Colony forming assay demonstrates that BRG1 or BRM knockdown reduces the ability of MDA-MB-231 cells to form colonies when plated at low density. (C) Quantification of colony forming assay. Quantification is the average from 3 independent experiments; error bars, SD. ***P<0.001. (D) BRG1 or BRM knockdown cells formed fewer and smaller tumors after implantation in the mammary fat pad of SCID/NCr mice. Each bar represents averaged results, n=12; error bars, SD. ***P<0.001. (E) Images of representative tumors.
A colonogenic assay was performed to assess the ability of a single cell to grow into a colony in culture when plated at low density. Colony formation by cells knocked down for BRG1 or BRM was decreased by ∼50% compared to the control cell line (Fig. 2B–C). The results indicate a requirement for BRG1 or BRM for MDA-MB-231 cells to form colonies in culture.
To test the possibility that knockdown of BRG1 or BRM might inhibit tumor growth in vivo, the inducible BRG1 or BRM shRNA expressing cells, the control cells expressing an inducible scrambled shRNA, or the parental MDA-MB-231 cells were injected into the mammary fat pads of SCID/Ncr mice given water with or without doxycycline ad libitum. After 35 days, 3 out of 12 mice with BRG1 knockdown xenograft cells and 3 of 11 mice with BRM knockdown xenograft cells failed to form detectable tumors (Fig. 2D). Representative images of tumors are presented in Fig. 2E. The mean tumor mass formed by BRG1 and BRM knockdown cells was 5.1 mg and 4.5 mg, respectively, compared to an average of 17.3 mg when formed by any of the control cells (Fig. 2D). BRG1 or BRM reduction decreased tumor mass by >70% compared to these controls, and ∼25% of mice failed to form tumors after BRG1 or BRM knockdown in xenografts.
Collectively, these data provide evidence that BRG1 and BRM are required for tumor growth in vivo and also suggest that BRG1 or BRM knockdown may delay or attenuate tumor initiation, as evidenced by the colony formation assay. These findings also indicate that targeting BRG1 or BRM expression could be an effective strategy for inhibiting breast tumor cell growth. Therefore, we set out to investigate the mechanism responsible for the observed inhibition in tumor growth properties.
BRG1 and BRM promote breast cancer cell proliferation
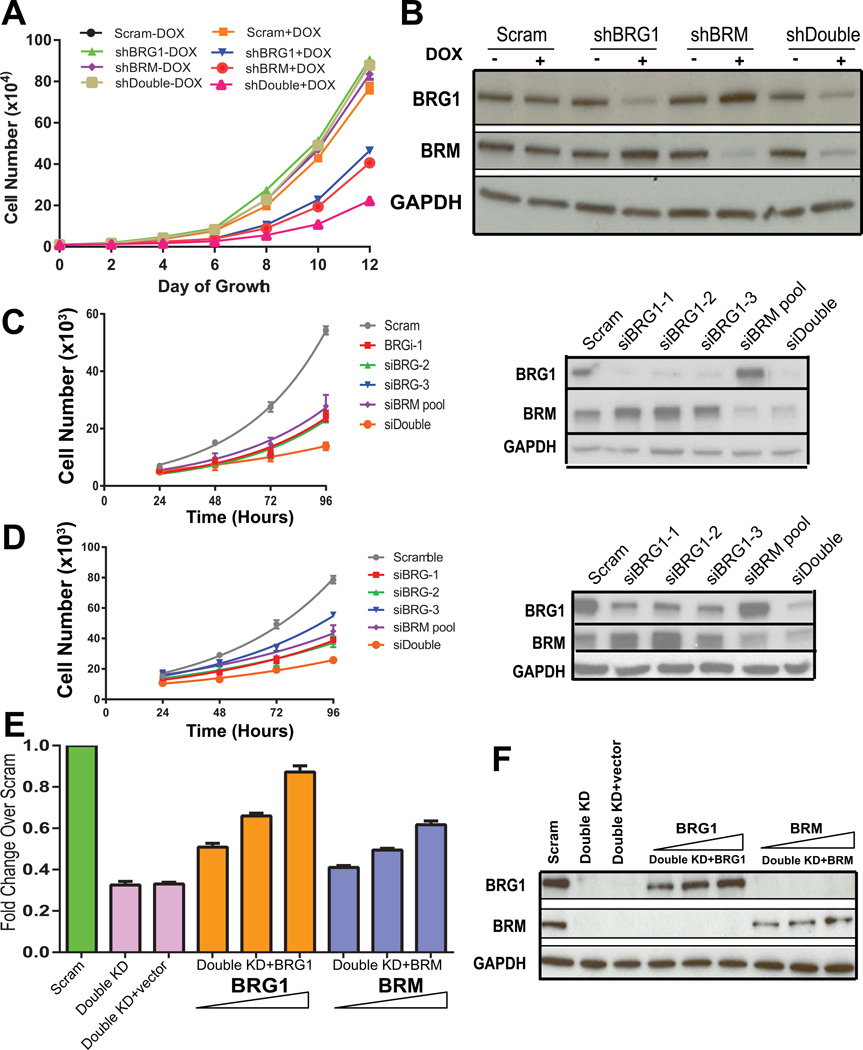
The high frequency of elevated BRG1 and BRM in breast tumors and the inhibition of colony formation and xenograft formation when BRG1 or BRM was knocked down suggested that the BRG1 and BRM ATPases might promote breast cancer cell proliferation. Each of the existing, MDA-MB-231 cell populations that inducibly express shRNAs against BRG1, BRM or a control sequence were tested for proliferative abilities in culture in the presence an absence of doxcycline. In addition, another cell line that inducibly expresses both shRNAs against BRG1 and BRM was created and tested in parallel to gain insight into whether the effects of BRG1 and BRM were redundant or independent.
All cells grown in the absence of doxycycline showed similar proliferation kinetics, as did the scramble control cells grown in the presence of doxycycline (Fig. 3A). BRG1 and BRM knockdown cells showed reduced rates of proliferation, and the double knockdown cells showed a further decrease that appeared additive in nature (Fig. 3A). Western blot analysis confirmed the knockdown of BRG1, BRM, or both (Fig. 3B).
Figure 3. Knockdown of BRG1 and/or BRM reduces triple negative breast cancer cell proliferation.
(A) Cell number was monitored over time to examine cell proliferation of each of the cell lines in the induced and uninduced state. DOX; doxycycline. (B) Corresponding western blot measuring the level of BRG1 and/or BRM knockdown. GAPDH is shown as a loading control. Data represent the average of three independent experiments; error bars, SD. (C) MDA-MB-231 cells were treated with one of three distinct siRNAs targeting BRG1 (Imbalzano et al., 2013) or with a pool of siRNAs targeting BRM or with both siRNA pools. Cell proliferation was monitored, and western blots confirmed knockdown of the target protein(s). (D) MDA-MB-468 cells were treated and experimentally evaluated exactly as in (C). (E) Increasing levels of BRG1 or BRM cDNAs were introduced to double knockdown MDA-MB-231 cells and proliferation rate was monitored. Results are presented as fold-change relative to the proliferation rate of the scram siRNA treated cells. (F) A representative western blot measuring BRG1 and BRM levels from the experiments described in (E).
We performed additional experiments to demonstrate the specificity of knockdown and the generality of the findings. We treated both MDA-MB-231 cells and another triple negative breast cancer cell line, MDA-MB-468, with one of three siRNAs targeting distinct regions of the BRG1 transcript. Each siRNA reduced BRG1 levels and caused a significant inhibition of cell proliferation relative to a scrambled siRNA control (Fig. 3C–D). A pool of siRNAs targeting BRM reduced BRM levels and similarly reduced the proliferation rate of both the MDA-MB-231 and the MDA-MB-468 cells (Fig. 3C–D). Combining the BRM siRNA pool with the BRG1 siRNA pool reduced the protein levels of both BRM and BRG1 and further reduced the proliferation of both triple negative cell lines, seemingly in an additive manner (Fig. 3C–D), which is consistent with the results presented in Fig. 3A. Western blot analysis confirmed the knockdown of BRG1, BRM, or both (Fig 3C–D). These data demonstrate a requirement for BRG1 and BRM in promoting breast cancer cell proliferation in two triple negative breast cancer cell lines. Furthermore, the evidence suggests that the effects of BRG1 and BRM on cell proliferation are mediated via mechanisms that are at least partially independent.
To address the specificity of the involvement of BRG1 and BRM in mediating cell proliferation, we re-introduced BRG1 or BRM cDNAs into the double knockdown cells. Re-introduction of BRG1 or BRM gave a dose-dependent rescue of proliferation rate (Fig. 3E). Re-introduction of BRG1 gave nearly complete rescue, while re-introduction of BRM gave a partial rescue (Fig. 3E). Western blot analysis provided evidence of the re-expression of both proteins (Fig. 3F).
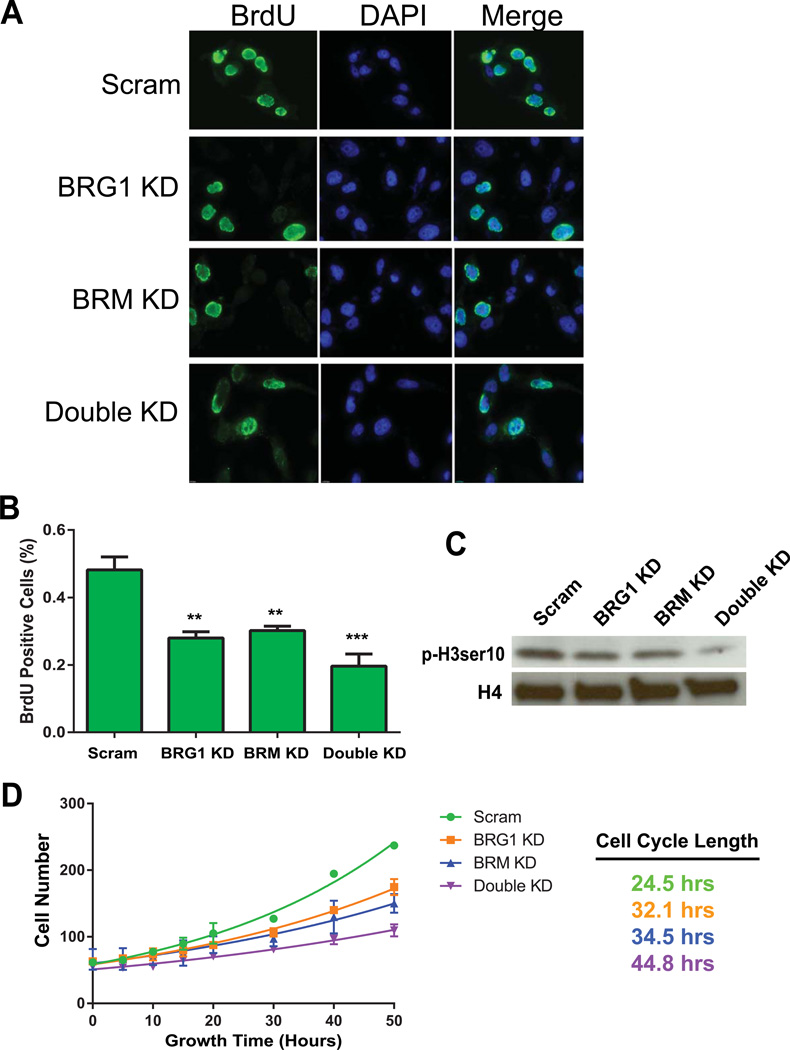
After knockdown of BRG1, BRM, or both, the number of cells in S phase as measured by BrdU incorporation was reduced compared to control cells (Fig. 4A–B). The decrease observed by BRG1 or BRM knockdown was similar. There was a further reduction in the percentage of BrdU positive cells when both BRG1 and BRM were knocked down, though the decrease did not prove to be statistically significant (Fig. 4B). We also examined levels of the mitotic marker, serine-10 phosphorylated Histone 3 (p-H3ser10) by western blot analysis. Cells expressing reduced levels of BRG1 or BRM had reduced levels of p-H3ser10 (Fig. 4C). Consistent with previous results, knockdown of both BRG1 and BRM caused a further reduction in p-H3ser10 levels (Fig. 4C). The reduction in the number of S and M phase cells suggested that cells expressing lower levels of BRG1 and/or BRM might have an overall decrease in the ability to traverse the cell cycle. When single cells were monitored by time-lapse video, the average cell cycle progression time was increased from 24.5 hours for the control cells to 32.1 and 34.5 hours for BRG1 and BRM knock down cells, respectively (Fig. 4D). Double knockdown cells were further reduced in cell cycle length, taking 44.8 hours to complete the cell cycle (Fig. 4D). Collectively, the results indicate that BRG1 and BRM are required for cell cycle progression in these breast cancer cells and that reinforces the idea that the contributions of BRG1 and BRM to cell proliferation are at least partially independent.
Figure 4. BRG1 and/or BRM knockdown results in a longer cell cycle.
(A) Knockdown of BRG1 resulted in fewer cells in S-phase as measured by BrdU incorporation. Representative images are presented. (B) Quantified data are the average of three independent experiments; three fields of at least 100 nuclei were counted per experiment. error bars, SD. **P<0.01. (C) The mitotic marker histone H3 phospho-serine 10 was reduced in BRG1 and/or BRM knockdown cells as measured by Western blotting. (D) Proliferation curves of control and BRG1, BRM, and double knockdown cells as determined by time-lapse videography.
The effects of knockdown of BRG1, BRM, or both on cell proliferation and cell cycle progression were not due to apoptosis as measured by Annexin V staining (Fig. 5A), nor did we observe permanent withdrawal from the cell cycle as measured by senescence-associated-β-galactosidase staining (Fig. 5B). Thus, knockdown of BRG1 and/or BRM decreased breast cancer cell proliferation mainly by decreasing the rate of cell cycle progression. Furthermore, knockdown of BRG1 and/or BRM caused no changes in cell migration as measured by a wound healing assay (Fig. 5C) or in adhesion to different collagen substrates (Fig. 5D).
Figure 5. BRG1 and/or BRM knockdown in MDA-MB-231 cells did not alter apoptosis, senescence, migration, or attachment to different collagen substrates.
(A) BRG1, BRM, or double knockdown did not cause apoptosis as judged by Annexin V staining. (B) BRG1, BRM, or double knockdown did not induce senescence as measured by senescence associated-β-galactosidase staining. (C) BRG1, BRM, or double knockdown did not change the ability of cells to fill the gap scratched into a monolayer of cells in a standard wound healing assay. (D) BRG1, BRM, or double knockdown did not affect attachment to collagen I- or collagen IV- coated plates.
CRISPR/Cas9 mediated knockout of BRG1 or BRM results in breast cancer cell death
The data resented thus far correlate BRG1 and BRM expression with breast tumorigenesis and indicate a role for BRG1 and BRM in promoting breast cancer cell proliferation, both in culture and in xenograft experiments. The observations raise the question of whether BRG1 and/or BRM are required for breast cancer cell viability. Prior evidence suggests that mouse embryonic fibroblasts can survive and proliferate in the absence of Brg1 (Bultman et al., 2000) and a number of cancer cell lines lack BRG1, BRM or both (Decristofaro et al., 2001; Fukuoka et al., 2004; Reisman et al., 2003; Reisman et al., 2002).
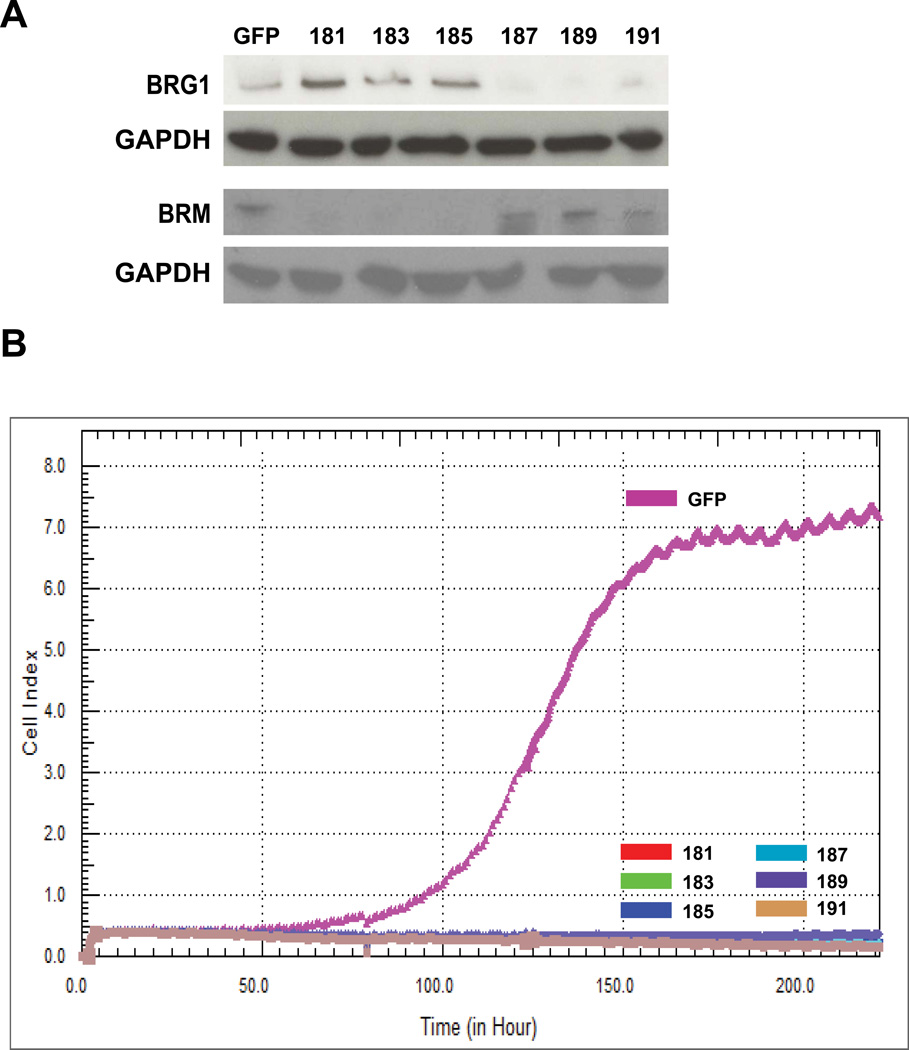
Viral vectors harboring 3 different guide RNA sequences targeting BRG1 or BRM were used to achieve BRG1 or BRM knockout in MDA-MB-231 cells. Significant cell death was observed during creation of the knockout cell lines. Western blotting was performed to confirm that majority of BRG1 or BRM proteins were depleted (Figure 6A). Dynamic cell proliferation was monitored in 30-minute intervals over 220 hours using xCELLigence RTCA system. In both cases, BRG1 or BRM knockout cells did not proliferate (Figure 6B), which suggested a severe low viability. After long-term passage, cell growth was resumed in the knockout cells. When examined by western blot, BRG1 or BRM expression was restored in those cells, presumably due to the expansion of a small population of cells that escaped knockout (data not shown). Overall, complete loss of BRG1 or BRM impaired cell viability.
Figure 6. Loss of BRG1 or BRM impaired cell viability.
(A) Western blots demonstrate the efficacy of CRISPR/Cas9 mediated knockout of BRG1 or BRM in MDA-MB-231 cells. Cell lines were derived using each of three different guide RNAs for BRG1 (187, 189, 191) or three different guide RNAs for BRM (181, 183, 185). GAPDH was monitored as a loading control. (B) Dynamic cell proliferation measured by xCELLigence RTCA system. 10,000 cells of each line were plated on E-Plate 8-well plates. Cell proliferation was monitored in 30-minute intervals over 220 hours. Cell Index values for all cell lines were calculated and plotted with the RTCA Software.
DISCUSSION
Carcinogenesis and malignant progression have proven to be very complicated multi-stage processes. As a natural response to complexity, cancer biologists developed simple inductive models and hypotheses to organize the vast amount of data available and to guide experimental design. One of the most powerful of these has been the Oncogene Model (Huebner and Todaro, 1969). In this view, the inappropriate expression of a gene or a mutated form of a gene transformed cells to a malignant phenotype. It was originally proposed that such genes in the human genome were derived from RNA viruses (Huebner and Todaro, 1969), known since 1911 to cause some tumors (Rous, 1911). However, evidence emerged that proto-oncogenes in mammalian genomes were the precursors to viral forms (Bishop, 1981; Weinberg, 1983). It is the proto-oncogenes, present in every cell, that are mutated or inappropriately expressed in human cancers.
A later and complimentary model, the Tumor Suppressor Model (Klein, 1987) was more consistent with experimental cell fusions between tumor and normal cells where the hybrid cell, almost invariably, had non-tumor phenotypes (Harris et al., 1969; Klinger, 1982; Sager, 1986; Wiener et al., 1974). Many tumor suppressor genes have been identified (Lee and Muller, 2010; Sherr, 2004) including some, like p53, that were identified first as oncogenes and then as tumor suppressors, a difference correlating with p53 mutations (Levine and Oren, 2009).
Clearly, both models of cancer development are over-simplified. Cancer is a disease of complex systems interactions (Du and Elemento, 2014; Gentles and Gallahan, 2011). That this is true of breast cancer is evidenced by the Cancer Genome Atlas cataloging of 30,626 somatic mutations and 3,662 significant mRNA expression level changes in 510 human breast tumors (Network, 2012). Epigenetics, and the ability of epigenetic mechanisms to organize complex programs of gene expression, offers a promising approach to experimentally addressing the complexity of malignant progression.
In our results, most primary breast tumors had elevated levels of BRG1 and BRM compared to normal breast tissue. Knockdown of BRG1 or BRM inhibited MDA-MB-231 xenograft growth and the proliferation of multiple breast cancer cell lines. Our BRG1 results are consistent with a prior report that almost half of the primary breast tumors have elevated BRG1 expression and that high BRG1 expression in patient samples correlates with poor survival (Bai et al., 2013). We also confirmed that BRG1 knockdown inhibited proliferation in MDA-MB-231 cells (Bai et al., 2013) and extended this result to xenografts. We further showed that breast tumor cells have a similar requirement for BRM, the alternative ATPase subunit of SWI/SNF, for normal proliferation.
Our results are not consistent with a tumor suppressor model for BRG1 or for BRM in breast cancer. The correlation of higher levels in breast tumors fits, but it does not prove, an oncogene model for SWI/SNF chromatin remodeling enzymes in breast cancer. Proof would require the demonstration that forced over-expression of BRG1 or BRM drive cells to transformation. This may prove to be an unrealistic experiment to perform, requiring simultaneous forced over-expression of many subunits of the 2 megadalton SWI/SNF multi-protein complex (Kwon et al., 1994). In any case, the oncogene model may be too simple to apply to an epigenetic factor capable of regulating complex programs in gene expression. The emergent properties of those programs in some cell and microenvironment contexts may be tumor suppression, while in other contexts, like breast cancer, it may be tumor progression.
The observation that BRG1 and BRM are necessary for breast tumor cell proliferation may recommend them as targets for cancer therapy. As epigenetic regulators of large programs in gene expression, we propose that BRG1 and BRM may be required for phenotypic changes in cells (de la Serna et al., 2006; Ho and Crabtree, 2010). Cancer is a progression of many state changes, with coordinate changes in the expression of sets of genes. For example, one programmed change of genes may drive cells to greater proliferation while another may drive them to a cancer metabolic phenotype (Cairns et al., 2011). Disruption of whole programs of transcriptional change by targeting epigenetic regulators like BRG1 and BRM may be useful in slowing the progression of states that comprise cancer.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Wang, B. Bautista, S. LeBlanc, and J. Feng for technical assistance, C. Baron for assistance with figures, and A. R. Barutcu, I. L. de la Serna, H. Zhang, M. C. Barton, L. M. Shaw, S. N. Jones, Y-J Hu, and T. Padilla-Benavides for suggestions and comments on the manuscript. This work was supported by NIH grants P01 CA82834 (GSS, JBL, JLS, AJV, JAN, ANI), R01 EB014869 (JAN), and R01 GM30758 (GSS). A.N.I. is a member of the UMMS DERC, which was supported by NIDDK grant 5P30 DK32520.
REFERENCES
- Bai J, Mei P, Zhang C, Chen F, Li C, Pan Z, Liu H, Zheng J. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One. 2013;8(3):e59772. doi: 10.1371/journal.pone.0059772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JM. Enemies within: the genesis of retrovirus oncogenes. Cell. 1981;23(1):5–6. doi: 10.1016/0092-8674(81)90263-4. [DOI] [PubMed] [Google Scholar]
- Blanco R, Iwakawa R, Tang M, Kohno T, Angulo B, Pio R, Montuenga LM, Minna JD, Yokota J, Sanchez-Cespedes M. A gene-alteration profile of human lung cancer cell lines. Human mutation. 2009;30(8):1199–1206. doi: 10.1002/humu.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102(2):257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27(4):460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B, Crabtree GR, Sauvageau G, Thibault P, Lessard JA. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123(11):1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro. 1978;14(11):911–915. doi: 10.1007/BF02616120. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cohet N, Stewart KM, Mudhasani R, Asirvatham AJ, Mallappa C, Imbalzano KM, Weaver VM, Imbalzano AN, Nickerson JA. SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells. J Cell Physiol. 2010;223(3):667–678. doi: 10.1002/jcp.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano L, Stewart KM, Cohet N, Mouw JK, Lakins JN, Debnath J, Reisman D, Nickerson JA, Imbalzano AN, Weaver VM. Oncogenic targeting of BRM drives malignancy through C/EBPbeta-dependent induction of alpha5 integrin. Oncogene. 2014;33(19):2441–2453. doi: 10.1038/onc.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186(1):136–145. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Dingemans AM, van den Boogaart V, Vosse BA, van Suylen RJ, Griffioen AW, Thijssen VL. Integrin expression profiling identifies integrin alpha5 and beta1 as prognostic factors in early stage non-small cell lung cancer. Mol Cancer. 2010;9:152. doi: 10.1186/1476-4598-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domfeh AB, Carley AL, Striebel JM, Karabakhtsian RG, Florea AV, McManus K, Beriwal S, Bhargava R. WT1 immunoreactivity in breast carcinoma: selective expression in pure and mixed mucinous subtypes. Mod Pathol. 2008;21(10):1217–1223. doi: 10.1038/modpathol.2008.69. [DOI] [PubMed] [Google Scholar]
- Du W, Elemento O. Cancer systems biology: embracing complexity to develop better anticancer therapeutic strategies. Oncogene. 2014 doi: 10.1038/onc.2014.291. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79(1):119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem. 2012;287(37):30897–30905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, Hewitt S, Travis WD, Jen J. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10(13):4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Gallahan D. Systems biology: confronting the complexity of cancer. Cancer Res. 2011;71(18):5961–5964. doi: 10.1158/0008-5472.CAN-11-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26(49):7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21(10):3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223(5204):363–368. doi: 10.1038/223363a0. [DOI] [PubMed] [Google Scholar]
- Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26(3):309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24(1):362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, de la Serna IL, Veal TM, Imbalzano AN. BRCA1 interacts with dominant negative SWI/SNF enzymes without affecting homologous recombination or radiation-induced gene activation of p21 or Mdm2. J Cell Biochem. 2004;91(5):987–998. doi: 10.1002/jcb.20003. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30(8):356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Todaro GJ. Oncogenes of RNA tumor viruses as determinants of cancer. Proc Natl Acad Sci U S A. 1969;64(3):1087–1094. doi: 10.1073/pnas.64.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One. 2013;8(2):e55628. doi: 10.1371/journal.pone.0055628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2010;29(1):81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G. The approaching era of the tumor suppressor genes. Science. 1987;238(4833):1539–1545. doi: 10.1126/science.3317834. [DOI] [PubMed] [Google Scholar]
- Klinger HP. Suppression of tumorigenicity. Cytogenet Cell Genet. 1982;32(1–4):68–84. doi: 10.1159/000131688. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1(6):500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277(25):22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harb Perspect Biol. 2010;2(10):a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wong RP, Martinka M, Li G. BRG1 expression is increased in human cutaneous melanoma. Br J Dermatol. 2010;163(3):502–510. doi: 10.1111/j.1365-2133.2010.09851.x. [DOI] [PubMed] [Google Scholar]
- Liu N, Balliano A, Hayes JJ. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chembiochem : a European journal of chemical biology. 2011;12(2):196–204. doi: 10.1002/cbic.201000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers CR, Naresh KN, Evens AM, Chadburn A, Gordon LI, Czader MB, Gill JI, Hsi ED, Greenough A, Moffitt AB, McKinney M, Banerjee A, Grubor V, Levy S, Dunson DB, Dave SS. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Carretero J, Fraga MF, Esteller M, Sidransky D, Sanchez-Cespedes M. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer. 2004;41(2):170–177. doi: 10.1002/gcc.20068. [DOI] [PubMed] [Google Scholar]
- Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Human mutation. 2008;29(5):617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J. 1998;17(1):223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009;28(27):2492–2501. doi: 10.1038/onc.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JM, Onodera Y, Bissell MJ, Park CC. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Res. 2010;70(13):5238–5248. doi: 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3(2):247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2005;13(1):66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63(3):560–566. [PubMed] [Google Scholar]
- Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21(8):1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97(25):13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Nieto S, Canada A, Pros E, Pinto AI, Torres-Lanzas J, Lopez-Rios F, Sanchez-Verde L, Pisano DG, Sanchez-Cespedes M. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Human mutation. 2011;32(2):E1999–E2017. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- Romero OA, Torres-Diz M, Pros E, Savola S, Gomez A, Moran S, Saez C, Iwakawa R, Villanueva A, Montuenga LM, Kohno T, Yokota J, Sanchez-Cespedes M. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2014;4(3):292–303. doi: 10.1158/2159-8290.CD-13-0799. [DOI] [PubMed] [Google Scholar]
- Rous P. A Sarcoma of the Fowl Transmissible by an Agent Separable from the Tumor Cells. J Exp Med. 1911;13(4):397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R. Genetic suppression of tumor formation: a new frontier in cancer research. Cancer Res. 1986;46(4 Pt 1):1573–1580. [PubMed] [Google Scholar]
- Saladi SV, Keenen B, Marathe HG, Qi H, Chin KV, de la Serna IL. Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Mol Cancer. 2010;9:280. doi: 10.1186/1476-4598-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentani K, Oue N, Kondo H, Kuraoka K, Motoshita J, Ito R, Yokozaki H, Yasui W. Increased expression but not genetic alteration of BRG1, a component of the SWI/SNF complex, is associated with the advanced stage of human gastric carcinomas. Pathobiology. 2001;69(6):315–320. doi: 10.1159/000064638. [DOI] [PubMed] [Google Scholar]
- Serber DW, Rogala A, Makarem M, Rosson GB, Simin K, Godfrey V, Van Dyke T, Eaves CJ, Bultman SJ. The BRG1 chromatin remodeler protects against ovarian cysts, uterine tumors, and mammary tumors in a lineage-specific manner. PLoS One. 2012;7(2):e31346. doi: 10.1371/journal.pone.0031346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8(1):e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, Campbell AE, Kawaoka S, Shareef S, Zhu Z, Kendall J, Muhar M, Haslinger C, Yu M, Roeder RG, Wigler MH, Blobel GA, Zuber J, Spector DL, Young RA, Vakoc CR. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, Knudsen ES. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000;97(14):7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, Li B. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67(2):203–213. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3(2):109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A. 1997;94(21):11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18(38):5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15(19):5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Semba S, Yokozaki H. Regulation of PTEN expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. Br J Cancer. 2011;104(1):146–154. doi: 10.1038/sj.bjc.6606018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. Alteration of the genomes of tumor cells. Cancer. 1983;51(11):1971–1975. doi: 10.1002/1097-0142(19830601)51:11<1971::aid-cncr2820511102>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Wiener F, Klein G, Harris H. The analysis of malignancy by cell fusion. V. Further evidence of the ability of normal diploid cells to suppress malignancy. J Cell Sci. 1974;15(1):177–183. doi: 10.1242/jcs.15.1.177. [DOI] [PubMed] [Google Scholar]
- Wu JI. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]
- Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67(2):659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18(24):3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.