Abstract
Progesterone (P4), acting through its nuclear receptor (PGR), plays an essential role in ovulation by mediating the expression of genes involved in ovulation and/or luteal formation. To identify ovulatory specific PGR-regulated genes, a preliminary microarray analysis was performed using rat granulosa cells treated with hCG ± RU486 (PGR antagonist). The transcript most highly down-regulated by RU486 was an EST (Expressed Sequence Tag) sequence (gb: BI289578.1) that matches with predicted sequence for Xlr5c-like mRNA. Since nothing is known about Xlr5c-like, we first characterized the expression pattern of Xlr5c-like mRNA in the rat ovary. The level of mRNA for Xlr5c-like is transiently up-regulated in granulosa cells of periovulatory follicles after hCG stimulation in PMSG-primed rat ovaries. The transient induction of Xlr5c-like mRNA was mimicked by hCG treatment in cultured granulosa cells from preovulatory ovaries. We further demonstrated that the LH-activated PKA, MEK, PI3K, and p38 signaling is involved in the increase in Xlr5c-like mRNA. The increase in Xlr5c-like mRNA was abolished by RU486. The inhibitory effect of RU486 was reversed by MPA (synthetic progestin), but not by dexamethasone (synthetic glucocorticoid). Furthermore, mutation of SP1/SP3 and PGR response element sites in the promoter region of Xlr5c-like decreased Xlr5c-like reporter activity. RU486 also inhibited Xlr5c-like reporter activity. ChIP assay verified the binding of PGR and SP3 to the Xlr5c-like promoter in periovulatory granulosa cells. Functionally, siRNA-mediated Xlr5c-like knockdown in granulosa cell cultures resulted in reduced levels of mRNA for Snap25, Cxcr4, and Adamts1. Recombinant Xlr5c-like protein expressed using an adenoviral approach was localized predominantly to the nucleus and to a lesser extent to the cytoplasm of rat granulosa cells. In conclusion, this is the first report showing the spatiotemporally regulated expression of Xlr5c-like mRNA by hCG in rat periovulatory ovaries. P4/PGR mediates the LH-induced increase in Xlr5c-like mRNA. In turn, Xlr5c-like is involved in regulating the expression of specific ovulatory genes such as Snap25, Cxcr4, and Adamts1, possibly acting in the nucleus of periovulatory granulosa cells.
Keywords: LH, progesterone receptor, Xlr5c-like, ovary, granulosa cells
1. INTRODUCTION
The preovulatory LH/FSH surge increases progesterone (P4) production and its nuclear receptor, PGR, expression in periovulatory follicles of the ovary (Chaffin et al, 1999; Espey L.L., 1994; Park & Mayo, 1991; Robker et al, 2000; Slomczynska et al, 2000). This rise in preovulatory progesterone, via binding to its receptor, PGR, has been proved to be essential for successful ovulation and/or luteinization. For instance, the administration of various inhibitors of progesterone biosynthesis or PGR antagonists blocked ovulation and/or luteinization in rats (Brannstrom & Janson, 1989; Pall et al, 2000; Snyder et al, 1984; Tanaka et al, 1992; Uilenbroek et al, 1992), primates (Duffy et al, 1994; Hibbert et al, 1996) and sheep (Murdoch et al, 1986). Studies using Pgr null mice further confirmed the functional significance of preovulatory progesterone/PGR action on ovulation in rodents; follicles develop normally, but fail to ovulate even when given exogenous gonadotropin stimulation (Lydon et al, 1996; Robker et al, 2000).
Upon binding with progesterone, PGR is known to regulate the transcription of a distinct set of genes in various reproductive tissues. In the ovary, considerable efforts have been made to identify downstream targets of PGR to delineate the mechanisms of the ovulatory process. Through the use of PGR null mice and PGR antagonists, over a dozen genes have been identified to be downstream of P4/PGR’s action [reviewed in (Kim et al, 2009a; Robker et al, 2009)]. These PGR-regulated genes encode a diverse array of factors ranging from proteases, secreted peptides, transcription factors, cytokines, and cellular structure proteins, indicating that P4/PGR affects various aspects of intra and extra-cellular events to accomplish ovulation. However, whether these genes are truly the direct transcriptional targets of PGR or indirectly regulated has yet to be determined. Intriguingly, the majority of genes identified as PGR-regulated in periovulatory granulosa cells appear to lack PGR response elements in their promoter regions. Rather, for a few PGR-regulated genes analyzed thus far, their expression was found to be dependent on the binding of Sp1/Sp3 transcription factors to GC-rich elements in their promoter regions (Doyle et al, 2004; Sriraman et al, 2008; Sriraman et al, 2003), thus suggesting that P4/PGR may regulate these genes by enhancing or modulating the activity of Sp1/Sp3 transcription factors.
Our preliminary microarray data using a rat granulosa cell culture model identified an EST (gb: BI289578.1) as the transcript most highly down-regulated by the treatment with PGR antagonist, RU486. Importantly, this transcript matches 100% with the partial cDNA sequence of predicted rat X-linked lymphocyte-regulated 5c-like (Xlr5c-like, also known as synaptonemal complex protein 3-like) gene. Currently, little to nothing is known about Xlr5c-like. The sequence analysis revealed that this gene is highly homologous (86%) to the mouse Xlr5c. Xlr is a multigene family, the prototype of which was found to be expressed in the nucleus of lymphoid cell lineages and suggested to play an important role in lymphocyte differentiation by acting as a transcriptional regulator (Cohen et al, 1985; Escalier et al, 1999; Siegel et al, 1987). Xlr has also been reported to be expressed in the nucleus of oocytes during meiotic prophase I in the mouse ovary (Escalier et al, 1999). However, unlike mouse Xlr genes which are located to the proximal part of the X chromosome, rat Xlr5c-like is localized to chromosome 1. Structurally, the rat Xlr5c-like gene also encodes a protein containing a conserved “Cor1/Xlr” domain that was initially found in SYCP3 (also called COR1), a structural component of the synaptonemal complex (Kolas et al, 2004) and is thought to facilitate the binding of these proteins to chromatin (Ellis et al, 2005).
Our initial computational analysis of the rat Xlr5c-like gene revealed the presence of PGR response elements in the putative promoter region. Together with preliminary microarray data, this information led us to hypothesize that the expression of Xlr5c-like is regulated by hormones, i.e. LH and P4, in a spatiotemporal specific manner in the rat ovary and is a direct transcriptional target of PGR. Therefore, the aims of the present study were to: 1) characterize the expression pattern and localization of Xlr5c-like mRNA in the rat periovulatory ovary, 2) dissect the regulatory mechanisms by which the LH surge increases the transcription of Xlr5c-like, and 3) determine the cellular localization of Xlr5c-like. Lastly, the potential function of Xlr5c-like in periovulatory granulosa cells was assessed using a siRNA approach in vitro.
2. MATERIALS AND METHODS
2.1. Materials
Unless otherwise noted, all chemicals, steroids, and reagents were purchased from Sigma Chemical Co. Molecular biological enzymes, pCRII-TOPO Vector, culture media, Lipofectamine RNAiMAX, siRNAs, and TRIzol reagent were purchased from Invitrogen Life Technologies, Inc. Oligonucleotide primers were purchased from Eurofins MWG operon. Antibodies were purchased from Santa Cruz Biotechnology.
2.2. Animals
All animal experimentations were carried out in accordance with the guidelines and ethics of the University of Kentucky Institutional Animal Care and Use Committee. Sprague Dawley rats were obtained from Harlan, Inc. (Indianapolis, IN) and were maintained in the division of laboratory animal resources, University of Kentucky. Animals were fed with standard rodent diet ad libitum while kept on a 12-h light/12-h dark cycle. Immature female rats (22 or 23 day old) were injected with 10 IU of pregnant mare serum gonadotropin (PMSG) subcutaneously to stimulate follicular development. Forty eight hours later, the rats were injected with 10 IU human chorionic gonadotropin hormone (hCG) subcutaneously to induce ovulation and subsequent formation of corpora lutea. The animals were killed at 0 h (48 h after PMSG), 6 h, 12 h, 24 h or 48 h after hCG administration. In this gonadotropin-induced superovulation model, ovulation occurs within 14–16 h after hCG administration (Jo & Curry, 2006).
2.3. Isolation and culture of rat granulosa cells
Ovaries were collected from immature rats at 48 h after PMSG administration and granulosa cells were isolated as described previously (Jo & Curry, 2006). Granulosa cells were cultured in Opti-MEM media supplemented with 0.05 mg/ml of gentamicin, and 1x ITS-X [Insulin-Transferrin-Selenium-X supplement (Gibco, life technologies)]. The cells were cultured in the absence or presence of various reagents for the time intervals outlined below for each experiment.
2.4. Knockdown of Xlr5c-like expression by siRNA in granulosa cell cultures
Granulosa cells were isolated from ovaries collected at 48 h post-PMSG as described above and the knockdown of Xlr5-like expression was achieved by a siRNA approach as described previously (Park et al, 2010). Briefly, granulosa cells were incubated overnight to acclimatize. The cells were then transfected with siRNA specific for Xlr5c-like or negative control siRNA (Stealth™ RNAi Negative Control Med GC) using Lipofectamine™ RNAiMAX according to the manufacturer’s instruction. Three hours later, the transfected cells were treated with forskolin (FSK, 10 μM) + phorbol 12-myristate 13-acetate (PMA, 20 nM) and further cultured for 12 h. At the end of culture, the cells and conditioned media were collected to isolate total RNA and to measure progesterone, respectively.
2.5. Analysis of gene expression
Total RNA was isolated from rat ovaries and other rat tissues using a TRIzol reagent and from cultured granulosa cells using a RNeasy mini kit (QIAGEN, Inc.,) according to manufacturer’s instructions. The synthesis of first-strand cDNA was performed by reverse transcription of 500 ng total RNA using superscript III with Oligo(dT)20 primer. Levels of mRNA for rat Xlr5c-like, Cxcr4, Snap25, Adamts1, End2, Pacap, Btg2, Gos2, Mmp10 and S100a3 were measured by real-time PCR. Real-time PCR was performed using SYBR green according to the manufacturer’s protocol (Stratagene, La Jolla, CA). Oligonucleotide primers corresponding to each gene were designed using Primer3 software and listed in Table 1. The specificity for each primer set was confirmed by both running the PCR products on a 2% agarose gel and analyzing the melting (dissociation) curve using the MxPro real-time PCR analysis program after each real-time PCR reaction. All genes were analyzed in duplicate and the amplification efficiency of each transcript primer set was determined by running a standard curve. The relative abundance of the target transcripts was normalized to the endogenous reference gene Ribosomal Protein 32 (L32) and calculated according to the Pfaffl method (Pfaffl, 2001).
Table 1.
List of primers used for Real-time PCR
| Gene name | Accession no | Primer sequence, 5′-3′) |
|---|---|---|
| Xlr5c-like | XM_003748905.1 | ATAGCGGATCATGAAACCAA TAACTCTGTCCTGCACCTT |
| Snap25 | NM_030991.3 | AATTCTGCGGGCTTTGTGTG TTCCCGGGCATCGTTTGTTA |
| Cxcr4 | NM_022205.3 | GCTGAGGAGCATGACAGACA GATGAAGGCCAGGATGAGAA |
| Adamts1 | NM_024400.2 | GCACCTCCGCGGTTCCACAT CGCGACCCGAGTTGCTGGTT |
| Mmp10 | NM_133514.1 | GTCCGAGGAAATGAAGTCCA GTCTCGGGAAGCCTTTATCC |
| Btg2 | NM_017259.1 | TCAAAGCTCCAGGGAACTCC CTAAAACCCACCAGGAATCAGG |
| Endo2 | NM_012549.2 | CTCCCTATGGCCTGGGAAAC CAGCAGTCCACGTCTTGCTA |
| S100a3 | NM_053681 | TGCCATCGTGTGTACCTTCC CACACTCCCGGAACTCACTC |
| Pacap | NM_016989.2 | TTACGATCAGGACGGAAACC TGTCGGCTGGGTAGTAAAGG |
| Gos2 | NM_001009632.1 | TGGCTAAGGAGATGATGA CACACAGTCTCCACTAGA |
2.6. Immunoassay of progesterone
Concentrations of progesterone in spent media were assayed using an Immulite kit on an Immulite 1000 machine (Siemens Healthcare Diagnostics) as described previously (Park et al, 2010). Assay sensitivity was 0.2 ng/ml, and the intra-assay and inter-assay coefficients of variation were 6.3% and 9.1%, respectively.
2.7. In situ localization of rat Xlr5c-like mRNA
Partial cDNA fragments were amplified using primers specific for rat Xlr5c-like (XM_ 003748905, (5′-CAGACTTGAAAGAGGCCAGG-3′, 5′-TTTGCTGACTGCCAATGAAG-3′) and RT reactions of total RNA samples isolated from ovaries at 12 h post-hCG. The amplified PCR fragment was cloned into pCRII-TOPO Vector. Sequences of the cloned DNA were verified commercially (Eurofins Genomics). Plasmids containing partial cDNA for Xlr5c-like were linearized with HpaI and EcoRV to generate sense and antisense riboprobes, respectively. Linearlized plasmids were labeled using a fluorescein RNA labeling kit (Roche Applied Sciences) and T7 and SP6 RNA polymerase, as appropriate. Ovaries collected from immature rats injected with PMSG or PMSG + hCG were sectioned at 10 μm and mounted on Probe On Plus slides (Fisher Scientific). In situ hybridization analysis was carried out as described previously with a slight modification (Jo et al, 2004). Briefly, the ovarian sections hybridized with the probes were incubated with anti-fluorescein antibody (Roche Applied Sciences) at 4°C overnight and the signals for Xlr5c-like mRNA were amplified using a TSA™-plus fluorescein kit (Roche Applied Sciences). The sections were counterstained with propidium iodide for 20 min and fluorescent staining specific for Xlr5c-like mRNA was visualized with an Eclipse E800 Nikon microscope under fluorescent optics.
2.8. Generation of rat Xlr5c promoter-reporter constructs
Rat genomic DNA isolated from rat tail tissues was used to amplify DNA fragments corresponding to promoter regions of the rat Xlr5c-like gene (LOC100366231). The 1879-bp (−1841/+37), 1379-bp (−1841/+37), and 579-bp (−541/+37) fragments of proximal promoter regions of the rat Xlr5c-like gene were amplified using the primers attached with restriction enzyme sites (KpnI and BglII). The cloned fragments were digested with KpnI and BglII enzymes and subcloned into a multiple cloning site of the pGL3 basic vector (Promega Corp). Site-directed deletion mutations of the Xlr5c-like promoter were generated using a QuikChange II site-directed mutagenesis kit (Stratagene). The sequences of the oligonucleotide primers used to generate respective Xlr5c-like promoters and desired deletion mutations in the Xlr5c-like promoter are listed in Table 2. All constructs cloned into the vector were sequenced commercially to verify their authenticity (MWG Biotech, Inc.).
Table 2.
List of primers used for Promoter reporter activity assay
| Reference (Accession no./Primer sets, 5′-3′) |
|---|
| XM_003748905.1 |
| Forward : GGTACCCTCAAGGAGAGGCAAAGGTCTGGG |
| Forward : GGTACCTGATTCACTCAGGGTGGACCAGGA |
| Forward : GGTACCGGAATGTCACCTGACCCTCGACCA |
| Reverse : AGATCTACTTTTCTTCATGGCCTTCTGCT |
2.9. Transient transfection and luciferase reporter assay
Granulosa cells isolated from rat ovaries (48 h post-PMSG) were transfected with respective firefly luciferase reporter plasmids (0.2 μg/well) and Renilla luciferase vector (pRL-TK vector, 0.01 μg/well) using Lipofectamine 2000 (Invitrogen Life Technologies) as described previously (Park et al, 2010). The next day, cells were cultured in the absence or presence of forskolin (FSK), PMA, or FSK+PMA for 8 h, and then harvested to measure firefly and renilla luciferase activities using a dual-luciferase reporter assay system (Promega Corp) and each reaction was monitored for 10 sec by a sequential auto-injection luminometer (Berthold Technologies). Firefly luciferase activities were normalized to Renilla luciferase activities and each experiment was performed in quadruplicate at least 3 times.
2.10. ChIP analysis
ChIP assay was performed for PGR, SP1, and SP3 binding sites in the rat Xlr5c-like promoter region. Briefly, preovulatory granulosa cells were cultured without or with hCG for 8 h. The cells were fixed and then lysed to release nuclei using a ChIP-IT High Sensitivity kit (Active Motif, CA) according to manufacturer’s instruction. The nuclei were sonicated with a Fisher Sonic Dismembrator model 550 to obtain DNA fragments of an average length of approximately 200–1000 bp. The sheared chromatin was immunoprecipitated overnight at 4°C with the antibody (5 μg/reaction) for PGR (sc-538X), SP1 (sc-59X), SP3 (sc-644X), or normal rabbit IgG (sc-2027X). The immunoprecipitated chromatin and 1:10 dilution of input chromatin were analyzed by PCR using the primers designed to amplify fragments spanning PGR, SP1 and SP3 motifs in the Xlr5c-like promoter region (Table 2). After 26 cycles of amplification, PCR products were run on a 2% agarose gel, stained with ethidium bromide, and visualized under UV light.
2.11. Generation of adenovirus expressing Xlr5c-like
The rat Xlr5c-like cDNA was amplified by PCR using total RNA isolated from granulosa cells of rat ovaries (12 h post-hCG). EcoRV and SalI enzyme sites were added to specific primers for Xlr5c-like (forward, 5′- GAT ATC AGC CAT GTC AAG CAA GGA GC -3′, reverse, 5′- GTC GAC AGA TGG TTT CTT CAG AGC AGT CA -3′). The PCR product was cloned into Topo vector and then subcloned into pShuttle-IRES-hrGFP-1 vector (Stratagene). Our cloning strategy removed a stop codon and fused 3 flag tags to the C-terminus of Xlr5c-like to aid the detection of this protein. This vector contains a second open reading frame that directs translation of a hrGFP. This allows the detection of adenovirus infected cells and monitoring of Xlr5c-like expression on a single cell level. The cloned vectors were sequenced commercially to verify their authenticity (MWG Biotech). AdEasyXL Adenovirus vector system was used to generate an adenovirus expressing flag-tagged Xlr5c-like (Ad-Xlr5c-like) according to the manufacturer’s instruction as routinely performed in our laboratory (Park et al, 2013). The adenoviral vectors were linearized by digesting with PacI enzyme and transfected to Ad293 cells where viral particles were further amplified. The viral stocks were prepared by subjecting the cell suspension to 3 rounds of freeze/thaw as described in the manufacturer’s instruction.
2.12. Immunocytochemical detection and Western Blot
Ad-Xlr5c-like and Ad-null viruses were added to preovulatory granulosa cells (48 h post-PMSG) plated onto poly-L- lysin-coated coverslips (Corning) in 24 well culture dishes. Six hours later, the infected cells were washed with fresh media and further cultured in the absence or presence of FSK + PMA for 14 h. To determine the cellular localization of flag tagged Xlr5c-like protein, the infected cells were briefly washed with PBS and fixed in 4% paraformaldehyde + 4% Sucrose in PBS for 15 min. After a PBS rinse, the fixed cells were permeabilized with 0.25% Triton X-100 in PBS and then blocked with 10% BSA before incubating them with the primary FLAG antibody (1 μg/ml, Monoclonal ANTI-FLAG® M2, Sigma). Next day, the cells were incubated with TRITC-conjugate-goat anti-mouse IgG (1:2000 dilution, sigma) and mounted with Fluoroshield™ containing DAPI. To further verify the authenticity of recombinant Xlr5c-like protein, preovulatory granulosa cells plated in 75 mm plates were infected with adenoviruses and cultured as described above. At the end of culture, the cells were briefly washed with ice-cold PBS and lysed in RIPA buffer. The cell lysate was denatured by boiling for 5 min, separated by SDS-PAGE on a 8% polyacrylamide gel and then transferred onto a nitrocellulose membrane. The membrane was incubated with the primary antibody (Monoclonal ANTI-FLAG® M2) in 1% casein solution overnight at 4°C, and then with anti-mouse HRP (1:1000 dilution, Santa Cruz Biotechnology) for 1 hr. Peroxidase activity was visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical Co).
2.13. Statistical Analysis
All data are presented as means ± SEM. One-way ANOVA was used to test differences in levels of mRNA, luciferase activities of respective Xlr5c-like promoter constructs and progesterone concentration among treatments. If ANOVA revealed significant effects of treatments, the means were compared by Tukey’s test, with p < 0.05 considered significant.
3. RESULTS
3.1. Expression pattern of Xlr5c-like mRNA in rat periovulatory ovaries
In the rOGED database (Jo et al, 2004), the transcript of an EST sequence (gb:BI289578.1) was highly up-regulated after hCG stimulation in granulosa cells and ovaries of PMSG-primed immature rats: the levels began to increase at 6 h and continued to increase at 12 h, whereas the transcript level in residual ovarian tissue (ovary remaining after granulosa cell collection) was undetected at any time points analyzed (Supplemental Fig. 1). Interestingly, the sequence of the EST (512 bp) is a 100% match with the 3′-region of cDNA sequence for predicted rat X-linked lymphocyte-regulated 5c-like (synaptonemal complex protein 3-like) transcript. Because the complete cDNA sequence of rat Xlr5c-like was initially unknown, we began this study by amplifying the complete cDNA of rat Xlr5c-like by PCR using RT samples of total RNA isolated from rat ovaries (12 h post-hCG) and primers designed based on mouse Xlr5c cDNA sequence. Sequence analyses of the amplified cDNA fragment (962 bp) revealed the presence of 5-UTR, open reading frame (708bp), and 3′-UTR (Supplemental Fig. 2). We confirmed that the sequence of this DNA fragment is identical to the complete cDNA sequence of predicted rat Xlr5c-like which became recently available in the GeneBank sequence database (XM_003748905.1).
To investigate whether Xlr5c-like is expressed in the rat ovary and its expression is hormonally regulated in a spatiotemporal manner during the periovulatory period, ovaries of PMSG-primed immature rats were collected before and at defined hours after hCG administration. Real-time PCR data indicated that the level of Xlr5c-like mRNA was transiently increased: the level was highest at 12 h and then declined to near 0 h level at 24 h post-hCG (Fig. 1A). Next, we examined tissue localization of Xlr5c-like mRNA by in situ hybridization analysis. Xlr5c-like mRNA was predominantly localized to the granulosa cell layer of periovulatory follicles in the ovaries obtained at 12 h post-hCG (Fig. 1B-b &d), while little expression of Xlr5c-like mRNA was detected in the ovaries collected at 48 h post-PMSG (0 h-hCG, Fig. 1B-a) and 24 h post-hCG (Fig. 1B-c).
Figure 1.
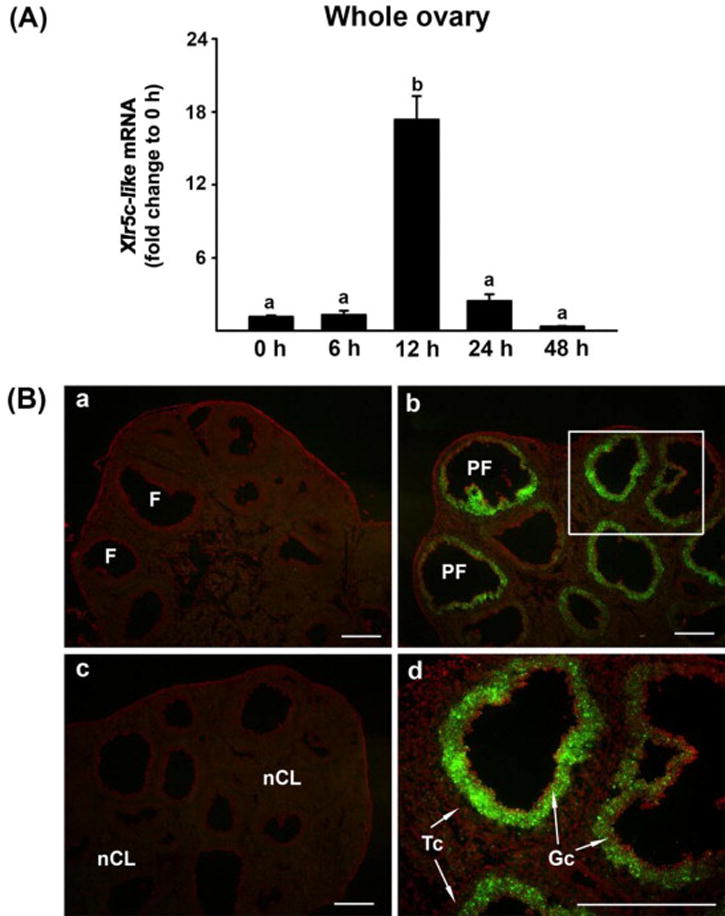
Ovarian expression of Xlr5c-like mRNA during the periovulatory period in the rat. Ovaries were collected at indicated hours (h) after hCG injection from PMSG-primed immature rats (n=4 animals/time point). A) Ovarian levels of Xlr5-like mRNA were measured using Real-time PCR and were normalized to the L32 value in each sample. Bars with no common superscripts are significantly different (p < 0.05). B) Localization of Xlr5c-like mRNA was analyzed using in situ hybridization. Sections of rat ovaries obtained at 0 h (48 h-post PMSG, a), 12 h (b and d) and 24 h post-hCG (c). A box in panel b was magnified in panel d. Scale bar, 500 μm for all the images. F; follicle, PF; periovulatory follicle, nCL; new corpus luteum, Gc; granulosa cell, Tc; theca cell layer
Since nothing is known about Xlr5c-like expression, we also tested whether Xlr5c-like is expressed in other tissues including the heart, spleen, kidney, uterus, liver and skeletal muscle. Xlr5c-like mRNA was detected in heart, kidney, and skeletal muscle, but the levels of mRNA for Xlr5c-like in these tissues were at least 20-fold lower than that in the ovary obtained at 12 h post-hCG (Supplemental Fig. 3).
3.2. Identification of PGR-regulated genes in rat granulosa cells
The action of progesterone through its nuclear receptor PGR is essential for successful ovulatory processes (Duffy et al, 1994; Hibbert et al, 1996; Lydon et al, 1996; Murdoch et al, 1986; Snyder et al, 1984). In an attempt to identify new periovulatory genes that are regulated by P4/PGR, granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were treated with RU486, an antagonist for PGR, in the absence or presence of hCG for 8 h. Total RNA samples were used for preliminary microarray analyses (n=1 /treatment, pooled samples from 3 experiments). Transcripts that are up-regulated at least 2-fold by hCG and down-regulated to 50% by RU486 were selected from the microarray data for further verification. These genes include Snap25, Cxcr4, Adamts1, Pacap, End2, Mmp10, Runx1, Rgc32, Btgs2, Fam110c, Gos2, and S100a3. Our previous reports also showed that the levels of mRNA for Runx1, Rgc32, and Fam110c were up-regulated by hCG treatment, but the increase was inhibited by RU486 in cultured rat granulosa cells (Jo & Curry, 2006; Li et al, 2012; Park et al, 2008). In the present study, real-time PCR confirmed that hCG increased the levels of mRNA for the other 9 genes, while RU486 treatment inhibited the hCG-induced up-regulation of these transcripts in preovulatory rat granulosa cell cultures (Fig. 2A-I).
Figure 2.
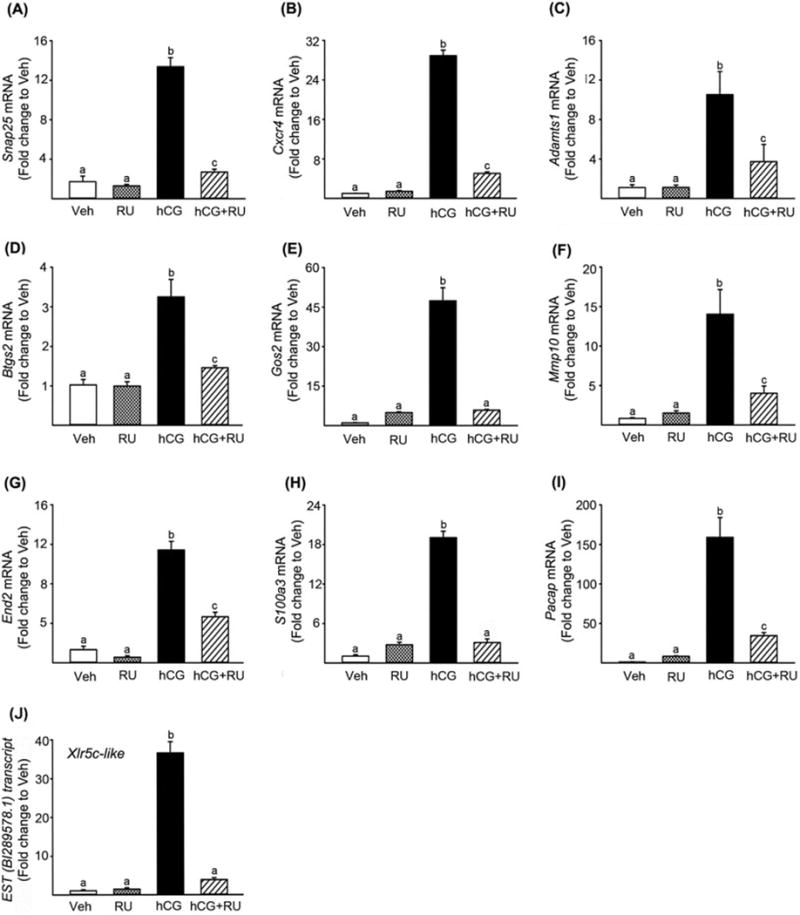
Identification and verification of P4/PGR-downstream genes in rat granulosa cells. Granulosa cells obtained from rat preovulatory ovaries (48 h post-PMSG) were cultured for 8 h in medium alone or with RU486 (PGR antagonist; 1μM) in the absence or presence of hCG (1 IU/ml). The levels of mRNA for Snap25 (A), Cxcr4 (B), Adamts1(C), Btg2 (D), Gos2 (E), Mmp10(F), End2 (G), S100a3 (H), Pacap (I), and Xlr5c-like (J) were measured using real-time PCR and normalized to the L32 value in each sample. Experiments were repeated at least 4 times, each with different granulosa cell samples (mean ± SEM). Bars with no common superscripts are significantly different (p < 0.05).
Of particular interest, the microarray revealed an EST sequence (gb:BI289578.1) as the transcript most highly down-regulated by RU486. The level of this EST transcript was highly up-regulated by hCG (34-fold higher than that of control treatment) and this increase by hCG was completed abolished by RU486. Since this EST is identical to rat Xlr5c-like, we further confirmed the microarray data by real-time PCR using specific primers for rat Xlr5c-like (Fig. 2J).
3.3. Regulation of Xlr5c-like mRNA in vitro
The in vivo data revealed that hCG administration induces a transient increase in the level of mRNA for Xlr5c-like at 12 h in the rat ovary, close to the time of ovulation. To determine whether the increase in Xlr5c-like mRNA after hCG injection in vivo can be mimicked in vitro and the induction of Xlr5c-like mRNA is mediated by the direct action of LH, granulosa cells isolated from preovulatory rat ovaries (48 h post-PMSG) were cultured in the absence or presence of hCG (1 IU/ml) for 0, 4, 8, 12 or 24 h. The level of mRNA for Xlr5c-like was transiently increased and the level was highest at 8 and 12 h after hCG treatment, comparable to that observed in the rat ovary after hCG administration in vivo (Fig. 3A).
Figure 3.
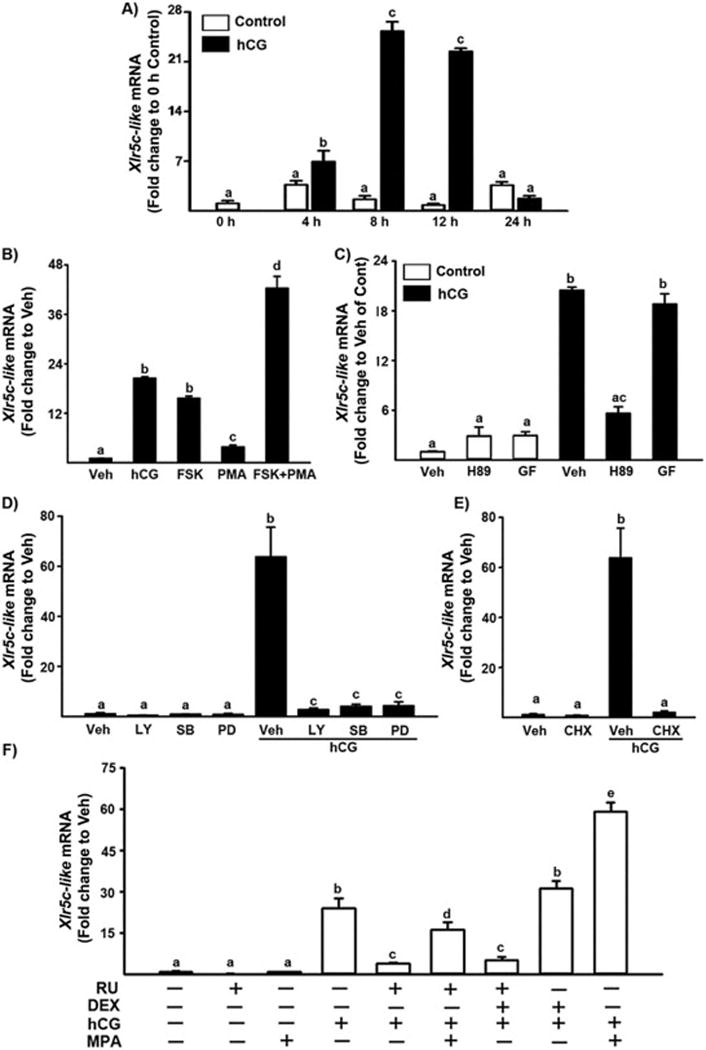
Regulation of Xlr5c-like mRNA in periovulatory granulosa cells in vitro A) Granulosa cells obtained from rat preovulatory ovaries (48 h after PMSG) were cultured in medium alone (Control) or with hCG (1 IU/ml) for 0, 4, 8, 12, 24 h. B) Preovulatory granulosa cells were cultured for 8 h in medium alone (Vehicle, Veh) or with hCG (1 IU/ml), FSK (PKA agonist; 10 μM), PMA (PKC agonist; 20 nM), FSK (10 μM) + PMA (20 nM). C) Preovulatory granulosa cells were cultured for 8 h in medium alone (Veh), or with H89 (PKA inhibitor; 10 μM), GF109203X (PKC inhibitor; 1 μM) in the absence or presence of hCG (1 IU). D) Granulosa cells were cultured for 8 h in medium alone (Veh) or with LY294002 (P13K inhibitor; 25 μM), SB2035850 (p38 inhibitor; 25 μM), PD98059 (MEK inhibitor; 20 μM) in the absence or presence of hCG (1 IU/ml). E) Preovulatory granulosa cells were cultured in medium alone (Veh) or with cyclohexamide (1 μg/ml, CHX) in the presence or absence of hCG (1 IU/ml). F) Granulosa cells were cultured in medium alone (Veh) or with RU486 (PGR antagonist; 1 μM), MPA (synthetic progestin, 10 μM) and DEX (dexamethasone, synthetic glucocorticoid, 10 μM) in the absence or presence of hCG. The level of Xlr5c-like mRNA was measured by real-time PCR (mean ± SEM; n=3 experiments). Bars with no common superscripts in each panel are significantly different (p < 0.05).
The LH surge triggers the ovulatory process in preovulatory follicles. LH binding to its receptor (LHCGR) activates multiple cellular signaling pathways including the protein kinase A (PKA) and protein kinase C (PKC) pathways (Morris & Richards, 1995). Each of the LH-activated signaling pathways regulates the expression of a distinct set of periovulatory genes. To determine which LH-activated signaling pathways are involved in the transcription of Xlr5c-like, granulosa cells isolated from preovulatory ovaries were treated with various activators or inhibitors of cellular signaling molecules in the absence or presence of hCG and cultured for 8 h, the time point of the highest level of Xlr5c-like mRNA. Treatment with forskolin (FSK), an activator of adenylate cyclase or FSK + phorbol 12 myristate 13-acetate (PMA, PKC activator) increased Xlr5c mRNA (Fig. 3B). PMA alone also increased Xlr5c mRNA, but the level was much lower compared to that of hCG. In addition, the stimulatory effect of hCG on Xlr5c like mRNA was inhibited by H89, a specific inhibitor of PKA, but not by GF10920X, a selective PKC inhibitor (Fig. 3C). Furthermore, the stimulatory effect of hCG on Xlr5c-like mRNA was reduced by specific inhibitors of MAPK kinase (MEK, PD98059, 20 μM), p38 kinase (SB2035850, 20 μM), phosphatidylinositol 3-kinase (LY294002, 25 μM) (Fig. 3D). A recent study has shown that hCG treatment induces the activation of MEK and MAPK through the cAMP/PKA-dependent pathway, but also stimulates phosphorylation of p38 kinase in a PKA- and PKC-independent manner in rat granulosa cell cultures (Salvador et al, 2002). Taken together, our data indicated that the increase of Xlr5c-like mRNA in cultured granulosa cells is mediated by the LH-induced activation of various intracellular signaling molecules, including adenylate cyclase, PKA, MEK, p38 kinase and PI3K, suggesting the involvement of multiple signaling pathways in Xlr5c-like mRNA accumulation in periovulatory granulosa cells.
To determine whether the increase in levels of mRNA for Xlr5c-lik by hCG requires de novo protein synthesis, granulosa cells of preovulatory ovaries (48 h-PMSG) were cultured without or with cyclohexamide (CHX), an inhibitor of protein synthesis. As shown in Fig. 3E, the increase in Xlr5c-like mRNA was completely blocked by CHX, indicating that the up-regulation of Xlr5c-like mRNA requires a LH-induced mediator(s) in periovulatory granulosa cells.
Our microarray data suggested that P4/PGR is involved in the increase in Xlr5c-like mRNA. However, RU486 is known to exhibit antagonistic activities for both glucocorticoid receptors and PGR. To determine whether the inhibitory effect of RU486 on Xlr5c-like mRNA is caused by blocking PGR or glucocorticoid receptors, the granulosa cells were also treated with medroxyprogesterone acetate (MPA), synthetic progestin or dexamethasone (DEX), a potent synthetic glucocorticoid to compete with RU486. The treatment with MPA reversed the inhibitory effect of RU486 on hCG-induced Xlr5c-like mRNA expression (Fig. 3F). The addition of MPA further increased hCG-stimulated increase in mRNA for Xlr5c-like. However, DEX had no effect on the basal expression as well as hCG-stimulated increase in Xlr5c-like mRNA. Consistent with the data from in vitro studies, we also found that RU486 treatment reduced the level of Xlr5c-like mRNA in rat periovulatory ovaries obtained at 12 h post-hCG (Supplemental Fig. 5). Taken together, these data indicate that P4/PGR mediates the increase in mRNA for Xlr5c-like in periovulatory rat granulosa cells.
3.4. Transcriptional activation of the rat Xlr5c-like gene
To further assess the transcriptional regulation of rat Xlr5c-like gene, we analyzed the putative promoter region and found binding sites for various transcription regulators including PGR and SP1/3 (Supplemental Fig. 4). We generated 3 different Xlr5c-like promoter reporter constructs (−1841/+37, −1341/+37, and −541/+37 bp). These promoter reporter constructs were transfected into preovulatory granulosa cells and stimulated with FSK, PMA, or FSK+PMA for 8 h. FSK and FSK+PMA treatments increased the luciferase activity of all reporter constructs compared with that of control cultures, whereas PMA had no stimulatory effects on any of the Xlr5c-like promoter constructs. Importantly, the full-length Xlr5c-like promoter construct (−1841/+37bp) showed significantly higher transcriptional activities than those of the truncated promoter reporter constructs (−1341/+37 bp and −541/+37 bp). The distal region of the full-length promoter construct contains specific binding sites for SP1, SP3, and PGR (Fig. 4A). To determine the significance of consensus SP1, SP3, and PGR binding sites within the −1841/+37bp region of the Xlr5c-like promoter, deletion mutations for the SP3 binding site (−1593/−1585 bp, mutant A), overlapping binding sites for SP1/SP3 and PGR (−1573/−1560 bp, mutant B, see Supplemental Fig. 4), and double mutation of these two regions (mutant AB) were generated. Mutated promoter constructs were transfected into cultured preovulatory granulosa cells in the presence of FSK+PMA. The results indicated that mutation of these binding sites decreased the FSK+PMA-induced luciferase activity (Fig. 4B). The double mutation further reduced the luciferase activity. Together, these data indicated that the binding sites for SP1/SP3 and PGR are involved in transcriptional activation of the rat Xlr5c-like promoter.
Figure 4.
Regulation of transcriptional activities of the Xlr5c-like promoter. Preovulatory granulosa cells were isolated from gonadotropin-primed immature rats (48 h-post-PMSG). A) The cells were transfected with empty luciferase reporter vector (LUC), 1879-bp (−1841/+37), 1379-bp (−1841/+37), and 579-bp (−541/+37) Xlr5c-like luciferase reporter constructs, treated with FSK+PMA and further cultured for 8 h. B) The cells were transiently transfected with wild type, mutant-A, mutant-B, and mutant-AB Xlr5c-like luciferase reporter constructs, and stimulated with FSK+PMA for 8 h. C) ChIP detection of PGR binding to the rat Xlr5c-like promoter region in periovulatory granulosa cells. Chromatin samples prepared from 0 h or 12 h after hCG stimulation were immunoprecipated with antibodies for SP1, SP3, PGR. The immunoprecipitated chromatins were analyzed by PCR using the primers (arrows) designed to amplify fragments spanning SP1, SP3 and PGR binding motifs in the Xlr5c-like promoter. The experiment was repeated at least 3 times with different granulosa cell samples. D) Preovulatory granulosa cells transfected with the wild type 1379-bp (−1841/+37) Xlr5c-like luciferase reporter construct were treated without or with RU486 (PGR antagonist; 1 μM) and then stimulated with FSK+PMA for 12 h. Firefly luciferase activity was normalized to Renilla luciferase activity in each sample. Experiments for A, B, and D were repeated 4 times, each with different granulosa cell sample (mean ± SEM). Bars with no common superscripts in each panel are significantly different (p < 0.05).
Next, to determine whether PGR, SP1, and/or SP3 specifically bind to these candidate sites in the Xlr5c-like promoter, we performed ChIP assays on chromatin samples extracted from granulosa cells cultured with hCG for 12 h. PCR data indicate that immunoprecipitation with antibodies for PGR and SP3, but not SP1 enriches chromatin fragments containing these binding sites in the promoter region compared to that with normal rabbit IgG (Fig. 4C), indicating PGR and, with a lesser prevalence, SP3 binds to the specific region in the rat Xlr5c-like promoter.
Lastly, to determine whether P4/PGR is involved in transcriptional activation of the rat Xlr5c-like promoter, granulosa cells transfected with the full-length Xlr5c-like promoter construct (−1841/+37bp) were treated with RU486 in the absence or presence of FSK+PMA. As expected, the treatment with FSK+PMA increased the transcriptional activity of the Xlr5c-like promoter reporter construct, but the stimulatory effect was significantly reduced by RU486 (Fig. 4D).
3.5. Identification of genes regulated by Xlr5c-like in luteinizing granulosa cells
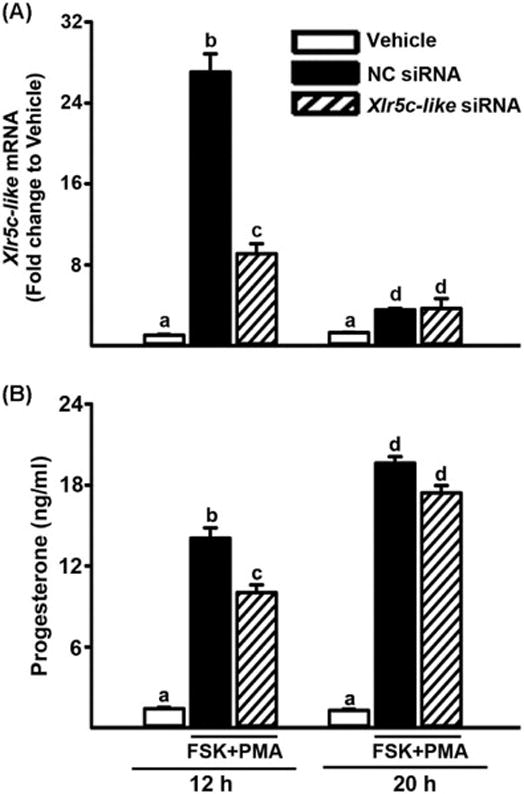
To begin to decipher the potential function of Xlr5c-like, the level of mRNA for Xlr5c-like was reduced using a siRNA approach. Granulosa cells were transfected with siRNA specific for Xlr5c-like mRNA and then stimulated with FSK+PMA for 12 h or 20 h. The levels of Xlr5c-like mRNA were reduced in Xlr5c-like siRNA-treated cells compared to that in negative control (NC) siRNA-treated cells (Fig. 5A). First, the impact of Xlr5c-like silencing on luteinizing granulosa cells was assessed by measuring the levels of progesterone, a hallmark of luteinization. As expected, FSK+PMA treatment steadily increased progesterone accumulation. But, the levels of progesterone were lower in Xlr5c-like siRNA-treated cells compared to those in NC siRNA-treated cells at 12 h, although the difference was not detected at 20 h. These data suggested that Xlr5c-like expression may be associated with initial progesterone production (Fig. 5B).
Figure 5.
Knockdown of Xlr5c-like expression by Xlr5c-like siRNA and its effect on Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were transfected with negative control siRNA (NC siRNA) or Xlr5c-like siRNA, and stimulated with FSK+PMA for 12 h or 20 h. A) The level of mRNA for Xlr5c-like was measured by real-time PCR and normalized to the L32 value in each sample. B) Concentration of progesterone was measured in the spent media of granulosa cells transfected with siRNAs and cultured with FSK+PMA for 12 h or 20 h. Experiments were repeated 4 times, each with different granulosa cell sample (mean ± SEM). Bars with no common superscripts are significantly different (p< 0.05).
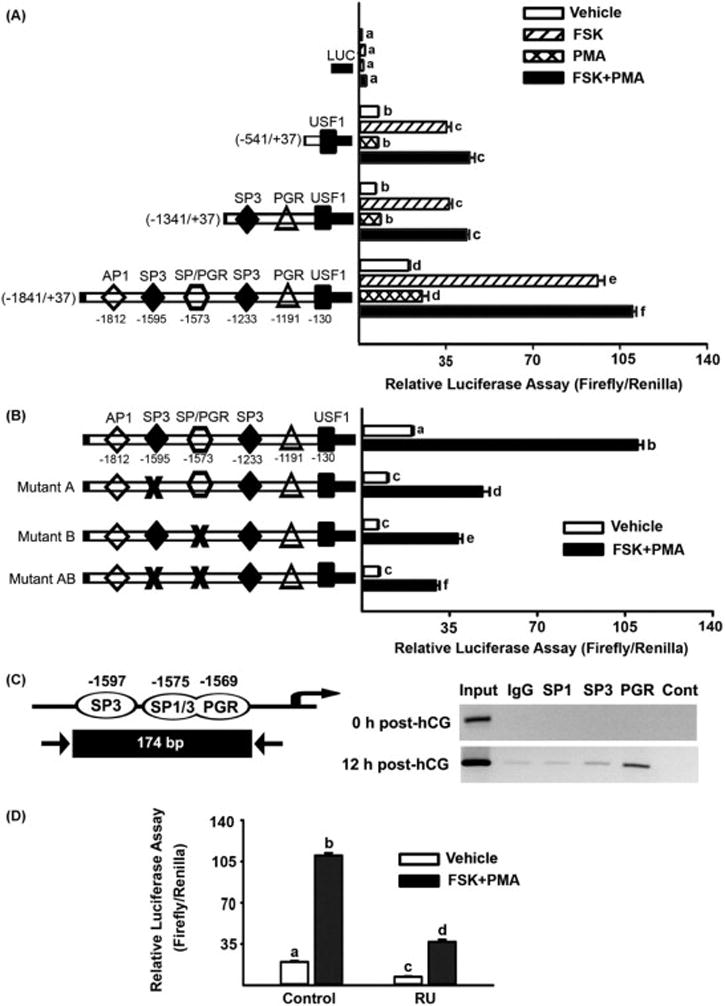
The Xlr5c-like gene showed high sequence and structure homology to mouse Xlr family genes that are found to be expressed in the nuclei (Ellis et al, 2005), suggesting that rat Xlr5c-like may be a nuclear protein. Thus, we explored the possibility that Xlr5c-like affects gene expression in luteinizing granulosa cells. Since the present study demonstrated that Xlr5c-like expression is under control of PGR’s action, we tested whether the expression of known PGR-regulated genes was affected in Xlr5c-like siRNA-treated cells. Interestingly, the level of mRNA for Snap25, Cxcr4, and Adamts1 was decreased in the cells treated with Xlr5c-like siRNA compared to NC siRNA. In contrast, the level of Gos2, MMP10, and Btg2 mRNA was increased by Xlr5c-like silencing, while Pacap, End2, and S1003a expression was not changed. These data suggest the selective regulation/impact of Xlr5c-like on the expression of LH-induced and PGR-downstream genes (Fig. 6).
Figure 6.
Effects of Xlr5c-like silencing on the expression of ovulatory genes in cultured granulosa cells. Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were transfected with negative control siRNA (NC siRNA) or Xlr5c-like siRNA, and stimulated with FSK+PMA for 12 h. The level of mRNA for Snap25 (A), Cxcr4 (B), Adamts1(C), Btg2 (D), Gos2 (E), Mmp10 (F), End2 (G), S100a3 (H), and Pacap (I) was measured by real-time PCR and normalized to the L32 value in each sample. Experiments were repeated at least 4 times, each with different granulosa cell sample (mean ± SEM). Bars with no common superscripts are significantly different (p< 0.05).
3.6. The expression of Xlr5c-like protein in rat granulosa cells
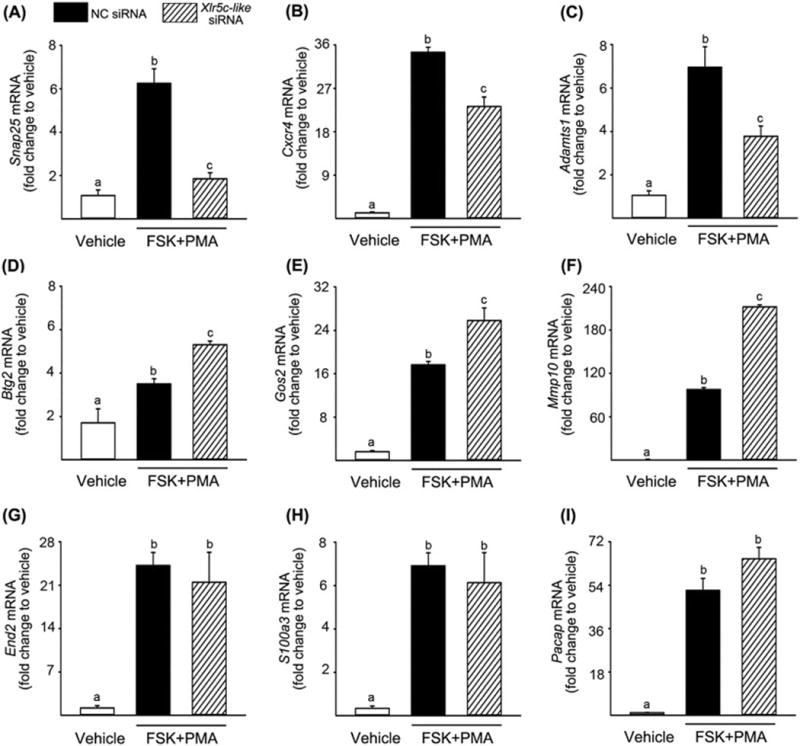
To determine the subcellular localization of Xlr5c-like, preovulatory granulosa cells were infected with an adenovirus expressing flag-tagged Xlr5c-like (Ad-Xlr5c-like) and stimulated with FSK+PMA. The cells were immunostained with a monoclonal anti-flag antibody. As expected, the cells infected with the adenovirus (GFP expressing cells) were staining positively for flag-tagged Xlr5c-like (Fig. 7A-b&c). Flag-tagged Xlr5c-like was predominantly localized to the nucleus of infected cells. Flag-tagged Xlr5c-like was also detected in the cytoplasm, albeit with lower signal than in the nucleus. The rat Xlr5c-like cDNA can be translated into 235 amino acids. This putative encoded protein has a predicted molecular mass of 26.9 kDa. Western blot analysis using the anti-flag antibody detected a single band corresponding to ~30 kDa in cultured rat granulosa cells infected with Ad-Xlr5c-like (Fig. 7B). The difference in size can be accounted for by the 3 flag tags (~3 kDa) fused to Xlr5c-like (26.9 kDa).
Figure 7.
The expression of flag-tagged rat Xlr5c-like in granulosa cells. Granulosa cells isolated from rat preovulatory ovaries (48 h post-PMSG) were infected with an adenovirus containing null vector (Ad-ϕ) or expressing flag-tagged Xlr5c-like (Ad-Xlr5c-like), and stimulated with FSK+PMA for 14 h. A) Flag-tagged Xlr5c-like protein was immunostained with monoclonal anti-flag antibody (M2), followed by TRITC-conjugate-goat anti-mouse IgG (red staining, b & e). The cells infected with Ad-Xlr5c-like also express hrGFP (green staining, c). Note that hrGFP freely defuses to the nucleus, staining for both the nucleus and cytoplasm. Nuclei were stained by DAPI (blue staining, a & d). Panel f represents the merged image of d and e. Scale bar, 20 μm for all the images. B) Cell lysates from adenovirus-infected cells were subjected to Western blots using anti-flag antibody (M2). β-Actin was used as a loading control.
4. DISCUSSION
It is well established that the preovulatory gonadotropin (LH/FSH) surge initiates the ovulatory process. Yet, the coordinated actions of a variety of local hormones produced in periovulatory follicles are required to bring about the complex process of follicular rupture and the release of the oocytes [reviewed in (Richards et al, 2002)]. One such local regulator is progesterone. In response to the LH/FSH surge, preovulatory follicles increase progesterone production/secretion (Smith et al, 1975) and its nuclear receptor, PGR, expression in mural granulosa cells (Park & Mayo, 1991). A long list of experimental evidence has proven that the action of progesterone through PGR is a critical step that links extracellular signals of the gonadotropin surge and the final steps of ovulation [reviewed in (Robker et al, 2009)]. Therefore, it is essential to identify progesterone/PGR-downstream genes and events to understand the cellular and molecular mechanisms underlying successful ovulation. Upon binding of progesterone, PGR undergoes a conformational change which promotes DNA binding, allowing the transcriptional activation or repression on its target genes (Allan et al, 1992). Thus, it is believed that follicular progesterone binds to PGR in periovulatory granulosa cells and regulates the expression of ovulatory genes. In the present study, we have determined that Xlr5c-like is a direct transcriptional target of progesterone/PGR and plays a role in modulating the expression of specific ovulatory genes in rat periovulatory granulosa cells (Fig. 8).
Figure 8.
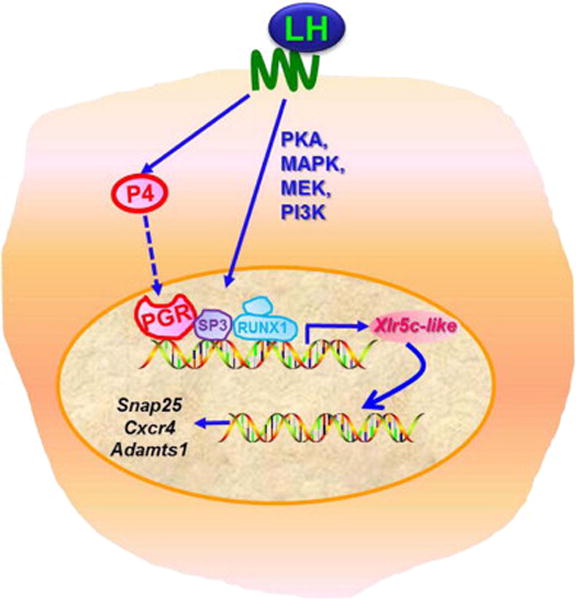
A hypothetical model of regulatory pathways involved in Xlr5c-like expression and potential function of Xlr5c-like in granulosa cells of rat periovulatory follicles. hCG increases transcription of the Xlr5c-like gene by activating signaling molecules including PKA, MAPK, MEK and PI3K. At the promoter level, PGR, RUNX1 and SP3 are involved in transcriptional activation of the Xlr5c-like gene. In turn, Xlr5c-like affects the expression of specific ovulatory genes such as Snap25, Cxcr4, and Adamts1.
The present study is also the first report of Xlr5c-like expression in various tissues of rats including the ovary, heart, kidney, and skeletal muscle (Supplemental Fig. 3). More importantly, in the ovary we found that Xlr5c-like mRNA increases exclusively and transiently in the granulosa cell compartment immediately prior to ovulation. After ovulation, the expression of Xlr5c-like mRNA was barely detected in the newly forming corpus luteum. Using a culture model we further demonstrated that the transient induction of Xlr5c-like mRNA after hCG injection in vivo can be mimicked by stimulating preovulatory granulosa cells with hCG in vitro. These data indicated that Xlr5c-like expression resulted from the direct action of LH/hCG on preovulatory granulosa cells, and subsequently the activation of LH/hCG-induced signaling. Indeed, the experiments with inhibitors of various signaling molecules revealed the involvement of LH/hCG-activated multiple intracellular signaling pathways in the induction of Xlr5c-like mRNA including PKA, MEK, p38 kinase and PI3Kinase. In addition, we found that Xlr5c-like expression requires the de novo synthesis of protein(s) induced by LH/hCG in periovulatory granulosa cells, indicating the involvement of regulatory mediator(s). Amongst the candidate regulatory protein(s) is PGR. PGR expression transiently increases in periovulatory granulosa cells in response to LH/hCG stimulation and its peak expression is only a few hours prior to that of Xlr5c-like (Ko et al, 1999). Moreover, the promoter region of rat Xl5c-like contains progesterone response elements (PRE)(Supplemental Fig. 4), suggesting a possible causal relationship between PGR and Xlr5c-like expression. The present study validated this notion by showing that 1) PGR antagonist (RU486) treatment completely abolished the hCG-induced increase in the level of Xlr5c-like transcript in granulosa cell cultures, 2) the PGR binding site in the Xlr5c-like promoter is involved in the agonist-induced transcriptional activity of this gene, and 3) PGR directly binds to the promoter region of the Xlr5c-like gene that contains a PRE.
So far, a dozen genes have been identified to be downstream of P4/PGR-regulated pathways [reviewed in (Robker et al, 2009)], but none were verified to be a direct transcriptional target of PGR in periovulatory granulosa cells. Therefore, our data from ChIP analysis and promoter reporter activity assay are the first experimental evidence suggesting that PGR is directly involved in the transcriptional induction of Xlr5c-like in periovulatory granulosa cells of the rat ovary. In addition, the sequence analysis of the rat Xlr5c-like promoter region revealed the presence of binding sites for various transcriptional regulators including RUNX and C/EBP (Supplemental Fig. 4), suggesting that these transcriptional regulators may be involved in transcriptional activation of the Xlr5c-like gene. For instance, Runx1 expression increases in periovulatory granulosa cells of the rat, mouse and human after the LH surge or hCG administration to mimic the LH surge (Jo & Curry, 2006; Park et al, 2010; Shimada et al, 2006). We have previously reported that RUNX1 is involved in the up-regulation of key periovulatory genes in rat granulosa cells (Jo & Curry, 2006; Liu et al, 2009; Liu et al, 2010). Moreover, our preliminary experiment showed that the knockdown of Runx1 expression by Runx1 siRNA decreased the level of Xlr5c-like mRNA in granulosa cell cultures, suggesting that RUNX1 is involved in Xlr5c-like expression (Supplemental Fig. 6). Together, these data indicate that the up-regulation of Xlr5c-like expression resulted from the activation of multiple signaling pathways by LH/hCG and, at the transcriptional level, the action of critical mediators of the ovulatory process, including PGR and RUNX1.
Regarding its function, nothing is known about Xlr5c-like. Nonetheless, mouse Xlr, the sequence of which exhibits a high degree of homology (> 73%) to the rat, was found to be expressed in the nucleus of lymphoid cells and suggested to play an important role in lymphocyte differentiation by acting as a transcriptional regulator (Cohen et al, 1985; Escalier et al, 1999; Siegel et al, 1987). More recently, the second member of Xlr family, SLX2 (SYCP3-like X-linked 2) was identified and found to be expressed in the nuclei of meiotic germ cells in the mouse testis and ovary (Shi et al, 2013; Zhuang et al, 2014). Like all mouse Xlr proteins, rat Xlr5c-like contains a conserved domain (referred as “Cor1 /Xlr”) that is believed to promote nuclear localization and DNA binding (Ellis et al, 2005). Of note, computational analysis of Xlr5c-like protein mapped a stretch of amino acid residues in the N-terminal region (positions 9 to 19) as a putative nuclear localization signal (Supplemental Fig. 2). Experimentally, we also showed that flag-tagged Xlr5c-like expressed in cultured rat granulosa cells is predominantly localized to the nuclear compartment. Although it remains to be confirmed whether Xlr5c-like is indeed expressed and localized to the nucleus, the present data suggests that Xlr5c-like may function in the nucleus of periovulatory granulosa cells.
Based on the present finding that Xlr5c-like is only expressed in periovulatory granulose cells at the time of ovulation and is localized to the nuclear compartment, it is conceivable that Xlr5c-like may be involved in the ovulatory process by regulating periovulatory gene expression. As a first step toward determining the function of Xlr5c-like, the expression of Xlr5c-like was knocked down by a siRNA approach and its effect on ovulatory characteristics was assessed in granulosa cell cultures. Distinct characteristics of periovulatory granulosa cells during the ovulatory period include the increase in progesterone production and ovulatory gene expression. In our granulosa cell cultures, the knockdown of Xlr5c-like expression had only a minor impact on progesterone accumulation. Next, to identify ovulatory genes whose expression may be affected by Xlr5c-like, we began with the list of genes identified as PGR-regulated genes in the present study. Our rationale was that the expression of these genes was confirmed to be highly up-regulated by hCG at the time when Xlr5c-like expression was highest in our granulosa cell cultures. Notably, we found that the expression of Snap25, Cxcr4, and Adamts1 was reduced by Xlr5c-like siRNA, indicating that Xlr5c-like expression is involved in the up-regulation of Snap25, Cxcr4, and Adamts1 expression in periovulatory granulosa cells. The question still remains as to how Xlr5c-like expression affects these gene expressions, thus warranting further investigation. It is also noteworthy that this is the first report showing the transient induction of Snap25, Cxcr4, and Adamts1 expression by hCG in rat granulosa cell cultures (Supplemental Fig. 7), although other laboratories have previously reported a similar expression pattern of these genes in mouse granulosa cells in vivo and/or in vitro (Hernandez-Gonzalez et al, 2006; Kim et al, 2009b; Robker et al, 2000; Shimada et al, 2007). More importantly, a previous study has demonstrated that the periovulatory expression of Adamts1, a multifunctional protease capable of cleaving extracellular matrix proteoglycan, is involved in the remodeling of the ovulating follicle wall and COC matrix and is crucial for successful ovulation (Brown et al, 2010). Snap25 is a component of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein which is required for intracellular membrane fusion events during exocytosis of vesicles. Shimada et al. (Shimada et al, 2007) have recently demonstrated that Snap25 expression is hormonally regulated in mouse granulosa cells of periovulatory follicles during the ovulatory period and it promotes cytokine/chemokine exocytosis. As for the Cxcr4, a chemokine receptor specific for CXCL12 (also called SDF1), Kryczek et al. (Kryczek et al, 2005) have reported the expression of CXCL12 and Cxcr4 in human periovulatory granulosa cells. Moreover, they also demonstrated that CXCL12 facilitates T lymphocyte recruitment and increases granulosa cell survival through Cxcr4 (Kryczek et al, 2005). Together, these studies indicated Adamts1, Snap25, and Cxcr4 contribute distinct, yet intertwined aspects of the ovulatory process such as tissue remodeling, secretory process, and chemokine response. Therefore, the effect of Xlr5c-like on the expression of these genes indicates that Xlr5c-like may contribute to diverse aspects of the ovulatory process by regulating the expression of ovulatory genes.
Supplementary Material
Highlights.
hCG increases Xlr5c-like mRNA in granulosa cells of rat periovulatory follicles.
The increase in Xlr5c-like mRNA is mediated by P4/PGR.
The knockdown of Xlr5c-like mRNA affects the expression of ovulatory genes.
Recombinant Xlr5c-like is localized dominantly to the nucleus of granulosa cells.
Acknowledgments
We thank Drs. Thomas E. Curry, Linah Al-Alem and Ms. Katherine Rosewell and Kourtney Trudgen for critical reading for the manuscript. We also thank Ms. Georgia Bryant for technical help with steroid assays. This work was supported by National Institutes of Health Grant R01HD061617 and PO1HD71875.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have nothing to disclose
References
- Allan GF, Tsai SY, Tsai MJ, O’Malley BW. Ligand-dependent conformational changes in the progesterone receptor are necessary for events that follow DNA binding. Proc Natl Acad Sci USA. 1992;89:11750–11754. doi: 10.1073/pnas.89.24.11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannstrom M, Janson PO. Progesterone is a mediator in the ovulatory process of the in vitro-perfused rat ovary. Biol Reprod. 1989;40:1170–1178. doi: 10.1095/biolreprod40.6.1170. [DOI] [PubMed] [Google Scholar]
- Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, Russell DL. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod. 2010;83:549–557. doi: 10.1095/biolreprod.110.084434. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Stouffer RL, Duffy DM. Gonadotropin and steroid regulation of steroid receptor and aryl hydrocarbon receptor messenger ribonucleic acid in macaque granulosa cells during the periovulatory interval. Endocrinology. 1999;140:4753–4760. doi: 10.1210/endo.140.10.7056. [DOI] [PubMed] [Google Scholar]
- Cohen DI, Steinberg AD, Paul WE, Davis MM. Expression of an X-linked gene family (XLR) in late-stage B cells and its alteration by the xid mutation. Nature. 1985;314:372–374. doi: 10.1038/314372a0. [DOI] [PubMed] [Google Scholar]
- Doyle KM, Russell DL, Sriraman V, Richards JS. Coordinate transcription of the ADAMTS-1 gene by luteinizing hormone and progesterone receptor. Mol Endocrinol. 2004;18:2463–2478. doi: 10.1210/me.2003-0380. [DOI] [PubMed] [Google Scholar]
- Duffy DM, Hess DL, Stouffer RL. Acute administration of a 3 beta-hydroxysteroid dehydrogenase inhibitor to rhesus monkeys at the midluteal phase of the menstrual cycle: evidence for possible autocrine regulation of the primate corpus luteum by progesterone. J Clin Endocrinol Metab. 1994;79:1587–1594. doi: 10.1210/jcem.79.6.7989460. [DOI] [PubMed] [Google Scholar]
- Ellis PJ, Clemente EJ, Ball P, Toure A, Ferguson L, Turner JM, Loveland KL, Affara NA, Burgoyne PS. Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Human molecular genetics. 2005;14:2705–2715. doi: 10.1093/hmg/ddi304. [DOI] [PubMed] [Google Scholar]
- Escalier D, Allenet B, Badrichani A, Garchon HJ. High level expression of the Xlr nuclear protein in immature thymocytes and colocalization with the matrix-associated region-binding SATB1 protein. Journal of immunology. 1999;162:292–298. [PubMed] [Google Scholar]
- Espey LL, H L. The physiology of Reproduction. 2. 1994. Ovulation; pp. 725–780. [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hibbert ML, Stouffer RL, Wolf DP, Zelinski-Wooten MB. Midcycle administration of a progesterone synthesis inhibitor prevents ovulation in primates. Proc Natl Acad Sci USA. 1996;93:1897–1901. doi: 10.1073/pnas.93.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Curry TE., Jr Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol. 2006;20:2156–2172. doi: 10.1210/me.2005-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr, Ko C. Development and application of a rat ovarian gene expression database (rOGED) Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Jo M, Komar CM, Fortune JE. Gonadotropin surge induces two separate increases in messenger RNA for progesterone receptor in bovine preovulatory follicles. Biol Reprod. 2002;67:1981–1988. doi: 10.1095/biolreprod.102.004366. [DOI] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Control of ovulation in mice by progesterone receptor-regulated gene networks. Molecular human reproduction. 2009a;15:821–828. doi: 10.1093/molehr/gap082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Signaling by hypoxia-inducible factors is critical for ovulation in mice. Endocrinology. 2009b;150:3392–3400. doi: 10.1210/en.2008-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C, In YH, Park-Sarge OK. Role of progesterone receptor activation in pituitary adenylate cyclase activating polypeptide gene expression in rat ovary. Endocrinology. 1999;140:5185–5194. doi: 10.1210/endo.140.11.7149. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Yuan L, Hoog C, Heng HH, Marcon E, Moens PB. Male mouse meiotic chromosome cores deficient in structural proteins SYCP3 and SYCP2 align by homology but fail to synapse and have possible impaired specificity of chromatin loop attachment. Cytogenetic and genome research. 2004;105:182–188. doi: 10.1159/000078188. [DOI] [PubMed] [Google Scholar]
- Kryczek I, Frydman N, Gaudin F, Krzysiek R, Fanchin R, Emilie D, Chouaib S, Zou W, Machelon V. The chemokine SDF-1/CXCL12 contributes to T lymphocyte recruitment in human pre-ovulatory follicles and coordinates with lymphocytes to increase granulosa cell survival and embryo quality. American journal of reproductive immunology. 2005;54:270–283. doi: 10.1111/j.1600-0897.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- Li F, Jang H, Puttabyatappa M, Jo M, Curry TE., Jr Ovarian FAM110C (family with sequence similarity 110C): induction during the periovulatory period and regulation of granulosa cell cycle kinetics in rats. BiolReprod. 2012;86:185. doi: 10.1095/biolreprod.112.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park ES, Jo M. Runt-related transcription factor 1 regulates luteinized hormone-induced prostaglandin-endoperoxide synthase 2 expression in rat periovulatory granulosa cells. Endocrinology. 2009;150:3291–3300. doi: 10.1210/en.2008-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Park ES, Jo M. Periovulatory Expression of Hyaluronan and Proteoglycan Link Protein 1 (Hapln1) in the Rat Ovary: Hormonal Regulation and Potential Function. Mol Endocrinol. 2010;24(6):1203–1217. doi: 10.1210/me.2009-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Conneely OM, O’Malley BW. Reproductive phenotpes of the progesterone receptor null mutant mouse. J Steroid Biochem Mol Biol. 1996;56:67–77. doi: 10.1016/0960-0760(95)00254-5. [DOI] [PubMed] [Google Scholar]
- Morris JK, Richards JS. Luteinizing hormone induces prostaglandin endoperoxide synthase-2 and luteinization in vitro by A-kinase and C-kinase pathways. Endocrinology. 1995;136:1549–1558. doi: 10.1210/endo.136.4.7895665. [DOI] [PubMed] [Google Scholar]
- Murdoch WJ, Peterson TA, Van Kirk EA, Vincent DL, Inskeep EK. Interactive roles of progesterone, prostaglandins, and collagenase in the ovulatory mechanism of the ewe. Biol Reprod. 1986;35:1187–1194. doi: 10.1095/biolreprod35.5.1187. [DOI] [PubMed] [Google Scholar]
- Pall M, Mikuni M, Mitsube K, Brannstrom M. Time-dependent ovulation inhibition of a selective progesterone-receptor antagonist (Org 31710) and effects on ovulatory mediators in the in vitro perfused rat ovary. Biol Reprod. 2000;63:1642–1647. doi: 10.1095/biolreprod63.6.1642. [DOI] [PubMed] [Google Scholar]
- Park ES, Choi S, Muse KN, Curry TE, Jr, Jo M. Response Gene to Complement 32 Expression Is Induced by the Luteinizing Hormone (LH) Surge and Regulated by LH-Induced Mediators in the Rodent Ovary. Endocrinology. 2008;149:3025–3036. doi: 10.1210/en.2007-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park ES, Lind A-K, Dahm-Kahler P, Brannstrom M, Carletti MZ, Christenson LK, Curry TE, Jr, Jo M. RUNX2 transcription factor regulates gene expression in luteinizing granulosa cells of rat ovaries. Mol Endocrinol. 2010;24(4):846–858. doi: 10.1210/me.2009-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Jang H, Curry TE, Sakamoto A, Jo M. Prostate androgen-regulated mucin-like protein 1: a novel regulator of progesterone metabolism. Molecular Endocrinol. 2013;27:1871–1886. doi: 10.1210/me.2013-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OK, Mayo KE. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol. 1991;5:967–978. doi: 10.1210/mend-5-7-967. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Ochsner S, Hsieh M, Doyle KH, Falender AE, Lo YK, Sharma SC. Novel signaling pathways that control ovarian follicular development, ovulation, and luteinization. Recent Prog Horm Res. 2002;57:195–220. doi: 10.1210/rp.57.1.195. [DOI] [PubMed] [Google Scholar]
- Robker RL, Akison LK, Russell DL. Control of oocyte release by progesterone receptor-regulated gene expression. Nucl Recept Signal. 2009;7:e012. doi: 10.1621/nrs.07012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker RL, Russell DL, Espey LL, Lydon JP, O’Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci USA. 2000;97:4689–4694. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador LM, Maizels E, Hales DB, Miyamoto E, Yamamoto H, Hunzicker-Dunn M. Acute signaling by the LH receptor is independent of protein kinase C activation. Endocrinology. 2002;143:2986–2994. doi: 10.1210/endo.143.8.8976. [DOI] [PubMed] [Google Scholar]
- Shi YQ, Zhuang XJ, Xu B, Hua J, Liao SY, Shi Q, Cooke HJ, Han C. SYCP3-like X-linked 2 is expressed in meiotic germ cells and interacts with synaptonemal complex central element protein 2 and histone acetyltransferase TIP60. Gene. 2013;527:352–359. doi: 10.1016/j.gene.2013.06.033. [DOI] [PubMed] [Google Scholar]
- Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol Endocrinol. 2006;20:1352–1365. doi: 10.1210/me.2005-0504. [DOI] [PubMed] [Google Scholar]
- Shimada M, Yanai Y, Okazaki T, Yamashita Y, Sriraman V, Wilson MC, Richards JS. Synaptosomal associated protein 25 (Snap25) gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol Endocrinol. 2007;21(10):2487–502. doi: 10.1210/me.2007-0042. [DOI] [PubMed] [Google Scholar]
- Siegel JN, Turner CA, Klinman DM, Wilkinson M, Steinberg AD, MacLeod CL, Paul WE, Davis MM, Cohen DI. Sequence analysis and expression of an X-linked, lymphocyte-regulated gene family (XLR) The Journal of experimental medicine. 1987;166:1702–1715. doi: 10.1084/jem.166.6.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomczynska M, Krok M, Pierscinski A. Localization of the progesterone receptor in the porcine ovary. Acta histochemica. 2000;102:183–191. doi: 10.1078/S0065-1281(04)70027-6. [DOI] [PubMed] [Google Scholar]
- Smith MS, Freeman ME, Neill JD. The control of progesterone secretion during the estrous cycle and early pseudopregnancy in the rat: prolactin, gonadotropin and steroid levels associated with rescue of the corpus luteum of pseudopregnancy. Endocrinology. 1975;96:219–226. doi: 10.1210/endo-96-1-219. [DOI] [PubMed] [Google Scholar]
- Snyder BW, Beecham GD, Schane HP. Inhibition of ovulation in rats with epostane, an inhibitor of 3 beta-hydroxysteroid dehydrogenase. Proc Soc Exp Biol Med. 1984;176:238–242. doi: 10.3181/00379727-176-41865. [DOI] [PubMed] [Google Scholar]
- Sriraman V, Eichenlaub-Ritter U, Bartsch JW, Rittger A, Mulders SM, Richards JS. Regulated expression of ADAM8 (a disintegrin and metalloprotease domain 8) in the mouse ovary: evidence for a regulatory role of luteinizing hormone, progesterone receptor, and epidermal growth factor-like growth factors. Biol Reprod. 2008;78:1038–1048. doi: 10.1095/biolreprod.107.066340. [DOI] [PubMed] [Google Scholar]
- Sriraman V, Sharma SC, Richards JS. Transactivation of the progesterone receptor gene in granulosa cells: evidence that Sp1/Sp3 binding sites in the proximal promoter play a key role in luteinizing hormone inducibility. Mol Endocronol. 2003;17:436–449. doi: 10.1210/me.2002-0252. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Espey LL, Stacy S, Okamura H. Epostane and indomethacin actions on ovarian kallikrein and plasminogen activator activities during ovulation in the gonadotropin-primed immature rat. Biol Reprod. 1992;46:665–670. doi: 10.1095/biolreprod46.4.665. [DOI] [PubMed] [Google Scholar]
- Uilenbroek JT, Sanchez-Criado JE, Karels B. Decreased luteinizing hormone-stimulated progesterone secretion by preovulatory follicles isolated from cyclic rats treated with the progesterone antagonist RU486. Biol Reprod. 1992;47:368–373. doi: 10.1095/biolreprod47.3.368. [DOI] [PubMed] [Google Scholar]
- Zhuang XJ, Shi YQ, Xu B, Chen L, Tang WH, Huang J, Lian Y, Liu P, Qiao J. SLX2 interacting with BLOS2 is differentially expressed during mouse oocyte meiotic maturation. Cell cycle. 2014;13(14):2231–2237. doi: 10.4161/cc.29265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


