Abstract
Listeria monocytogenes is a serious food-borne pathogen that can cause invasive disease in humans and other animals and has been the leading cause of food recalls due to microbiological concerns in recent years. In order to test hypotheses regarding L. monocytogenes lineage composition, evolution, ecology, and taxonomy, a robust intraspecific phylogeny was developed based on prfA virulence gene cluster sequences from 113 L. monocytogenes isolates. The results of the multigene phylogenetic analyses confirm that L. monocytogenes comprises at least three evolutionary lineages, demonstrate that lineages most frequently (lineage 1) and least frequently (lineage 3) associated with human listeriosis are sister-groups, and reveal for the first time that the human epidemic associated serotype 4b is prevalent among strains from lineage 1 and lineage 3. In addition, a PCR-based test for lineage identification was developed and used in a survey of food products demonstrating that the low frequency of association between lineage 3 isolates and human listeriosis cases likely reflects rarity of exposure and not reduced virulence for humans as has been previously suggested. However, prevalence data do suggest lineage 3 isolates may be better adapted to the animal production environment than the food-processing environment. Finally, analyses of haplotype diversity indicate that lineage 1 has experienced a purge of genetic variation that was not observed in the other lineages, suggesting that the three L. monocytogenes lineages may represent distinct species within the framework of the cohesion species concept.
Listeria monocytogenes is a ubiquitous gram-positive bacterium that can cause serious invasive disease (listeriosis) in humans and other animals, resulting in severe clinical features, including meningitis, septicemia, and abortion. Contaminated food is believed to be the primary source of human exposure to L. monocytogenes and has been repeatedly linked to sporadic cases and large outbreaks of listeriosis. L. monocytogenes has the highest hospitalization rate (92%) and second-highest case-fatality rate (20%) of any food-borne pathogen and is responsible for more than one-quarter of food-borne disease-related deaths linked to known pathogens (17). The ability of this bacterium to persist in the food-processing environment, its ability to grow at refrigeration temperatures, and its pathogenic potential make L. monocytogenes a unique and significant regulatory problem, which is reflected by the fact that L. monocytogenes contamination has been the leading cause of food recalls due to microbiological concerns in recent years (21, 32).
Two primary evolutionary divisions, or lineages, have been identified within L. monocytogenes on the basis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, ribotyping, and amplified fragment length polymorphism studies (3, 11, 24, 26). However, ribotype and virulence gene polymorphism data were used to describe the existence of a third lineage (25, 33), with some researchers suggesting that this lineage may represent a distinct taxonomic unit requiring recognition as a new species or subspecies (33, 34). Lineage-specific associations with serotypes commonly found in connection with human listeriosis (4b, 1/2b, and 1/2a) and genetic characterization of isolates from human and animal listeriosis cases have led to the suggestion that L. monocytogenes lineages differ in their pathogenic potential and host specificity (13, 33-35). However, disagreement persists about the number and composition of the major phylogenetic divisions within L. monocytogenes (2, 18, 33), the evolutionary history of lineage divergence within L. monocytogenes remains unclear, and perceived differences in virulence or host specificity need to be evaluated with respect to relative frequencies of exposure.
A solid evolutionary framework is essential for understanding the ecology and population dynamics of L. monocytogenes and for evaluating proposals regarding taxonomic revision of this important food-borne pathogen. Therefore, prfA virulence gene cluster (pVGC) sequences from 113 L. monocytogenes isolates, Listeria seeligeri, and Listeria ivanovii were used to develop a robust intraspecific phylogeny for L. monocytogenes. The pVGC is stably integrated in the same chromosomal location in these three Listeria species, and the pVGC of each species contains homologs of six virulence genes: a transcriptional regulator (prfA), two phospholipases (plcA and plcB) and a hemolysin (hly) required for lysis of host phagosomes, a metalloprotease (mpl) involved in extracellular activation of plcB, and a surface protein (actA) responsible for actin-based motility and cell-to-cell spread (31).
The primary objectives of the present study were to (i) determine the number of major phylogenetic divisions within L. monocytogenes, the genetic diversity within each of these lineages, and the distribution of serotypes across lineages; (ii) develop and use an accurate PCR-based approach for lineage identification to evaluate hypotheses of lineage-specific differences in virulence and host specificity with respect to the prevalence of individual lineages in food products; and (iii) combine analyses of phylogeny and historical demography to reconstruct the evolutionary history of lineage divergence within L. monocytogenes and to evaluate the taxonomic status of L. monocytogenes lineages within an appropriate evolutionary framework.
MATERIALS AND METHODS
Isolates and serotype determination.
The L. monocytogenes isolates sequenced in the present study are listed in Table 1. All Listeria isolates were maintained in theAgricultural Research Service Culture Collection (NCAUR, Peoria, Ill.) in liquid nitrogen vapor at −175°C and were cultured at 37°C in brain heart infusion broth or tryptic soy agar containing 0.6% (wt/vol) yeast extract (Difco, Sparks, Md.). Serotype determinations were made by using the 96-well enzyme-linked immunosorbent assay procedure described by Palumbo et al. (22).
TABLE 1.
L. monocytogenes isolates used in analyses of intraspecific phylogeny and serotype evolution
| NRRL no.a | Equivalent no. | Sourceb | Origin | Serotype | Lineage | NNRL no.a | Equivalent no. | Sourceb | Origin | Serotype | Lineage | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 33001 | RM2205 | WRRC | Human | 4b | 1 | |||||||
| 33002 | RM2212 | WRRC | Food | 1/2a | 2 | |||||||
| 33004 | RM2215 | WRRC | Food | 4b | 1 | |||||||
| 33005 | RM2216 | WRRC | Food | 1/2b | 1 | |||||||
| 33007 | RM2218 | WRRC | Food | 4b | 1 | |||||||
| 33008 | RM2387 | WRRC | Food | 4b | 1 | |||||||
| 33009 | RM2388 | WRRC | Food | 1/2a | 2 | |||||||
| 33010 | G3990 | CFSAN | NA | 4e or 4b | 1 | |||||||
| 33011 | G3982 | CFSAN | Human | 4e or 4d | 1 | |||||||
| 33012 | H7550 | CFSAN | Human | 4e or 4b | 1 | |||||||
| 33013 | Scott A | CFSAN | Human | 4b | 1 | |||||||
| 33014 | 12443 | CFSAN | Animal | 1/2a | 2 | |||||||
| 33015 | 12375 | CFSAN | Animal | 4b | 1 | |||||||
| 33022 | DSM20600 | DSMZ | Animal | 1/2a | 2 | |||||||
| 33027 | OB001075 | FSIS | Food | 1/2a | 2 | |||||||
| 33028 | OB001102 | FSIS | Food | 1/2b | 1 | |||||||
| 33029 | OB001124 | FSIS | Food | 1/2c | 2 | |||||||
| 33030 | OB001171 | FSIS | Food | 1/2b | 1 | |||||||
| 33031 | OB001183 | FSIS | Food | 1/2a | 2 | |||||||
| 33032 | OB001186 | FSIS | Food | 1/2b | 1 | |||||||
| 33033 | OB001206 | FSIS | Food | 1/2b | 1 | |||||||
| 33034 | OB001241 | FSIS | Food | 1/2a | 2 | |||||||
| 33035 | OB001270 | FSIS | Food | 1/2a | 2 | |||||||
| 33036 | OB001325 | FSIS | Food | 1/2b or 3b | 1 | |||||||
| 33037 | OB001350 | FSIS | Food | 1/2b | 1 | |||||||
| 33038 | OB001385 | FSIS | Food | 1/2b | 1 | |||||||
| 33039 | OB001410 | FSIS | Food | 1/2c | 2 | |||||||
| 33040 | OB001411 | FSIS | Food | 1/2a | 2 | |||||||
| 33041 | OB001412 | FSIS | Food | 1/2a | 2 | |||||||
| 33042 | OB000208F | FSIS | Food | 1/2b | 1 | |||||||
| 33043 | OB000217B | FSIS | Food | 1/2a | 2 | |||||||
| 33044 | OB000220(IA) | FSIS | Food | 1/2a | 2 | |||||||
| 33045 | OB000223C | FSIS | Food | 1/2b or 3b | 1 | |||||||
| 33046 | OB000255J | FSIS | Food | 1/2b | 1 | |||||||
| 33047 | 2202 | NADC | Human | 4b | 1 | |||||||
| 33049 | 2395 | NADC | Human | 4b | 1 | |||||||
| 33056 | 2220 | NADC | Human | 4b | 1 | |||||||
| 33064 | 2064 | NADC | Animal | 1/2a | 2 | |||||||
| 33068 | 8058 | NADC | Animal | 1/2b | 1 | |||||||
| 33069 | 2070 | NADC | Food | 1/2a | 2 | |||||||
| 33073 | 3883 | NADC | Animal | 1/2b | 1 | |||||||
| 33074 | 8054 | NADC | Animal | 1/2b | 1 | |||||||
| 33077 | 7035 | NADC | Animal | 4b | 3 | |||||||
| 33078 | 7680 | NADC | Animal | 4b | 1 | |||||||
| 33080 | 7679 | NADC | Animal | 1/2b | 1 | |||||||
| 33083 | 2632 | NADC | Food | 4b | 1 | |||||||
| 33090 | 7675 | NADC | Animal | 1/2b | 1 | |||||||
| 33092 | 7678 | NADC | Animal | 4b | 3 | |||||||
| 33094 | 3889 | NADC | Animal | 4b | 1 | |||||||
| 33095 | 7037 | NADC | Animal | 4b | 1 | |||||||
| 33098 | 2427 | NADC | Food | 4b | 1 | |||||||
| 33100 | 2612 | NADC | Animal | 1/2a | 2 | |||||||
| 33105 | 7676 | NADC | Animal | 4b | 3 | |||||||
| 33106 | 2420 | NADC | Food | 1/2a | 2 | |||||||
| 33114 | 2613 | NADC | Animal | 1/2b | 1 | |||||||
| 33115 | 3890 | NADC | Animal | 4c | 3 | |||||||
| 33116 | 2847 | NADC | NA | 4d | 1 | |||||||
| 33120 | 2848 | NADC | NA | 4b | 1 | |||||||
| 33123 | 2110 | NADC | Environmental | 1/2b | 1 | |||||||
| 33124 | 2111 | NADC | Food | 1/2b or 3b | 1 | |||||||
| 33125 | 3869 | NADC | Animal | 4b | 1 | |||||||
| 33126 | 7034 | NADC | Animal | 1/2b | 1 | |||||||
| 33127 | 2063 | NADC | Animal | 1/2a | 2 | |||||||
| 33128 | 2153 | NADC | Food | 1/2a | 2 | |||||||
| 33130 | 2071 | NADC | Food | 1/2b | 1 | |||||||
| 33140 | 2617 | NADC | Animal | 4b | 1 | |||||||
| 33141 | 2218 | NADC | Human | 4b | 1 | |||||||
| 33143 | 2149 | NADC | Human | 4b | 1 | |||||||
| 33144 | 2112 | NADC | Food | 4b | 1 | |||||||
| 33145 | 2401 | NADC | Human | 4b | 1 | |||||||
| 33148 | 5713 | NADC | Environmental | 1/2b | 1 | |||||||
| 33152 | 2072 | NADC | Food | 1/2a | 2 | |||||||
| 33154 | 2364 | NADC | Food | 1/2b | 1 | |||||||
| 33157 | 2355 | NADC | Environmental | 4b | 1 | |||||||
| 33160 | 3682 | NADC | Food | 1/2b | 1 | |||||||
| 33164 | 5712 | NADC | Food | 1/2b | 1 | |||||||
| 33166 | 2196 | NADC | Human | 4b | 1 | |||||||
| 33167 | 2362 | NADC | Environmental | 1/2a | 2 | |||||||
| 33169 | SE 106 | CFSAN | NA | 1/2a | 2 | |||||||
| 33171 | H 6900 | CFSAN | Human | 1/2a | 2 | |||||||
| 33176 | 20240-954 | LDDC | Animal | 1/2b | 1 | |||||||
| 33177 | 28838-95 | LDDC | Animal | 4c | 3 | |||||||
| 33178 | 32736-96 | LDDC | Animal | 1/2b | 1 | |||||||
| 33179 | 25734-97 | LDDC | Animal | 4b | 1 | |||||||
| 33180 | 41966-97 | LDDC | Animal | 1/2a | 2 | |||||||
| 33181 | 1709-98 | LDDC | Animal | 4b | 3 | |||||||
| 33182 | 7259-98 | LDDC | Animal | 4c | 3 | |||||||
| 33183 | 20842-98 | LDDC | Animal | 4b | 3 | |||||||
| 33184 | 11466-01 | LDDC | Animal | 4c | 3 | |||||||
| 33185 | 12459-01 | LDDC | Animal | 4b | 3 | |||||||
| 33186 | 20674-01 | LDDC | Animal | 1/2b | 1 | |||||||
| 33187 | 22409-01 | LDDC | Animal | 4b | 3 | |||||||
| 33188 | 23594-01 | LDDC | Animal | 4c | 3 | |||||||
| 33189 | 32285-01 | LDDC | Animal | 1/2a | 2 | |||||||
| 33190 | 36087-01 | LDDC | Animal | 4b | 3 | |||||||
| 33191 | 50301-01 | LDDC | Animal | 4b | 3 | |||||||
| 33215 | LMB0027 | ADRU | Food | 1/2ac | 2 | |||||||
| 33216 | LMB0033 | ADRU | Food | 1/2ac | 2 | |||||||
| 33218 | LMB0338 | ADRU | Environmental | 1/2bc | 1 | |||||||
| 33219 | LMB0340 | ADRU | Environmental | 1/2ac | 2 | |||||||
| 33220 | LMB0345 | ADRU | Human | 1/2bc | 1 | |||||||
| 33221 | LMB0347 | ADRU | Human | 4bc | 1 | |||||||
| 33223 | LMB0366 | ADRU | Human | 1/2cc | 2 | |||||||
| 33225 | LMB0455 | ADRU | NA | 3ac | 2 | |||||||
| 33226 | LMB0456 | ADRU | NA | 3cc | 2 | |||||||
| 33227 | LMB0459 | ADRU | NA | 4cc | 3 | |||||||
| 33229 | LMB0487 | ADRU | Human | 4cc | 3 | |||||||
| 33230 | LMB0291 | ADRU | Food | 4bc | 3 | |||||||
| 33231 | MFS 108 | ERRC | Food | 4cd | 3 | |||||||
| 33232 | MFS 53 | ERRC | Food | 4bd | 1 | |||||||
| 33233 | MFS 96 | ERRC | Food | 4bd | 1 | |||||||
| 33234 | MFS 110 | ERRC | Food | 1/2ad | 2 |
NRRL, U.S. Department of Agriculture, Agricultural Research Service Culture Collection, Peoria, Ill.
WRRC, U.S. Department of Agriculture, Western Regional Research Center, Albany, Calif.; CFSAN, U.S. Food and Drug Administration, Center for Food Safety and Applied Nutrition, Washington, D.C.; NADC, U.S. Department of Agriculture, National Animal Disease Center, Ames, Iowa; LDDC, Livestock Disease Diagnostic Center, University of Kentucky, Lexington, Ky.; ADRU, U.S. Department of Agriculture, Animal Disease Research Unit, Pullman, Wash.; ERRC, U.S. Department of Agriculture, Eastern Regional Research Center, Wyndmoor, Pa.
Serotype data reported by M. Borucki and D. R. Call, unpublished data.
Serotype information provided with strain histories.
DNA sequencing.
DNA isolation was performed as described by Fliss et al. (9). Primers were designed to amplify and sequence overlapping segments of the pVGC from 112 L. monocytogenes isolates (Table 1), L. seeligeri isolate NRRL 33019 (LMG11386; Belgian Coordinated Collections of Microorganisms, Ghent, Belgium), L. ivanovii subsp. ivanovii isolate NRRL 33017 (LMG11388; Belgian Coordinated Collections of Microorganisms), and L. ivanovii subsp. londoniensis isolate NRRL 33021 (DSM12491; Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany). Amplifications were performed with Platinum Taq DNA Polymerase High-Fidelity (Invitrogen Life Technologies, Carlsbad, Calif.), and amplification products were purified by using Montage PCR cleanup filter plates (Millipore, Billerica, Mass.). Sequencing reactions were performed by using ABI BigDye version 3.0 sequencing chemistry (Applied Biosystems, Foster City, Calif.). Reaction products were purified via ethanol precipitation and run on an ABI3100 or an ABI3730 genetic analyzer (Applied Biosystems). Primer sequences and PCR protocols are presented in the supplemental material.
Phylogenetic analyses and genetic distance estimation.
DNA sequences were edited and aligned by using Sequencher (version 4.1.2; Gene Codes, Ann Arbor, Mich.). In addition to the sequences generated in the present study, the pVGC sequence from L. monocytogenes strain EGD-e (GenBank accession no. AL591824) was included in the phylogenetic analyses. Prior to phylogenetic analyses, ambiguously aligned characters and nonunique pVGC haplotypes identified by using Collapse (version 1.1 [http://inbio.byu.edu/Faculty/kac/crandall_lab/programs.htm]) were removed from the data set.
Phylogenetic reconstructions were performed under both distance and maximum-parsimony frameworks. Distance analyses were performed by using the neighbor-joining algorithm and the Kimura two-parameter model of molecular evolution (15) as implemented in MEGA version 2.1 (http://www.megasoftware.net). Maximum-parsimony analyses were conducted by using the tree-bisection and reconnection method of branch swapping and the heuristic search algorithm of PAUP* version 4.0b (Sinauer Associates, Sunderland, Mass.). Relative support for individual nodes was assessed by nonparametric bootstrapping (8, 23) with 1,000 pseudoreplications of the data. For the combined pVGC data, bootstrap analyses were performed under both maximum-parsimony and distance frameworks. However, due to computational constraints, bootstrap analyses for the individual pVGC genes were performed only with the neighbor-joining algorithm. Genetic distance estimates were obtained as described for phylogenetic analyses with MEGA version 2.1, with standard errors estimated by using the bootstrap method and 1,000 pseudoreplications of the data. The significance of differences in genetic distance estimates was assessed by using one-tailed t tests and infinite degrees of freedom.
Development of an ASO-PCR multiplex for lineage identification.
Three sets of primers were designed from pVGC sequences for the specific identification of isolates from each of the three L. monocytogenes lineage groups via an allele-specific oligonucleotide PCR (ASO-PCR) multiplex (Table 2). Amplifications were performed in 10-μl volumes with 0.5 μM concentrations of each primer, 2 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.5 U of AmpliTaq Polymerase (Applied Biosystems), and 100 ng of genomic DNA. Amplifications consisted of 25 cycles of 15 s at 94°C, 10 s at 56°C, and 10 s at 72°C. Amplification products were resolved on 1.5% (wt/vol) agarose gels, and scored relative to a 100-bp DNA size ladder (Invitrogen Life Technologies, Carlsbad, Calif.).
TABLE 2.
ASO-PCR primer sequences and predicted product sizes.
| Lineage | Primer | Primer sequencesa (5′-3′) | PCR product size (bp) |
|---|---|---|---|
| 1 | actA1-f | AATAACAACAGTGAACAAAGC | 373 |
| actA1-r | TATCACGTACCCATTTACC | ||
| 2 | plcB2-f | TTGTGATGAATACTTACAAAC | 564 |
| plcB2-r | TTTGCTACCATGTCTTCC | ||
| 3 | actA3-f | CGGCGAACCATACAACAT | 277 |
| plcB3-r | TGTGGTAATTTGCTGTCG |
Underlined nucleotides are specific to the L. monocytogenes lineage listed in the first column.
Nucleotide sequence accession numbers.
DNA sequences have been deposited in the GenBank database under accession numbers AY510072 to AY510074 and AY512391 to AY512502.
RESULTS
Intraspecific phylogeny and L. monocytogenes lineage evolution.
The sequenced region consists of 8,750 aligned nucleotides, includes the entire pVGC with the exception of the last 12 bp of the prfA gene, and corresponds to nucleotides 203652 to 212294 in the complete genome sequence for L. monocytogenes strain EGD-e (GenBank accession no. AL591824). After ambiguously aligned characters were excluded, 61 unique pVGC haplotypes were identified among the sequenced L. monocytogenes isolates.
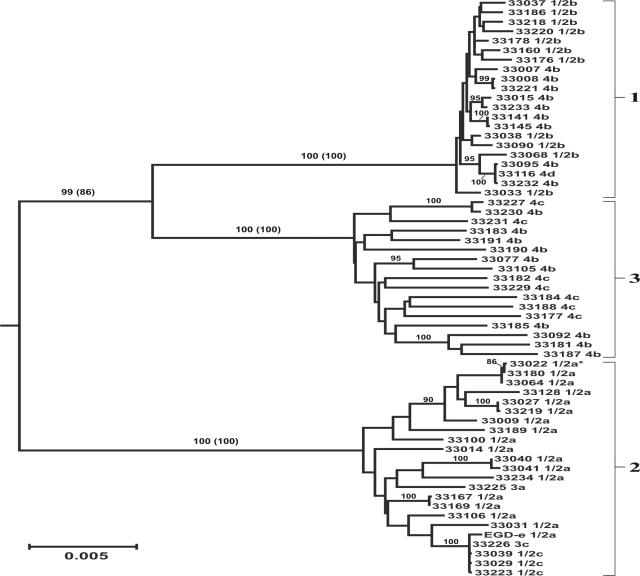
Phylogenetic analyses of the combined pVGC data resolved three distinct L. monocytogenes lineages, with each of the lineages recovered as monophyletic groups in 100% of bootstrap replicates from both neighbor-joining and maximum-parsimony analyses (Fig. 1). Lineage designations were assigned according to the convention of Rasmussen et al. (25), by including partial hly sequences from this previous study into phylogenetic analyses of the pVGC haplotypes reported here (not shown). Based on these phylogenetic reconstructions 21, 23, and 17 unique haplotypes were identified within the pVGC data for L. monocytogenes lineages 1, 2, and 3, respectively (Fig. 1). Neighbor-joining and maximum-parsimony analyses both resolved L. monocytogenes lineages 1 and 3 as sister groups that formed a larger monophyletic group referred to here as the L1/L3 clade (Fig. 1). These lineage relationships were supported by 99 and 86% of bootstrap replicates from neighbor-joining and maximum parsimony analyses, respectively. Topological constraints that forced lineages 2 and 3 to form a single monophyletic group required nine additional steps in maximum-parsimony analyses, and constraints that forced lineages 1 and 2 into a sister-group relationship provided the worst fit to the observed data with respect to lineage relationships, requiring 15 additional steps in maximum-parsimony analyses.
FIG. 1.
Neighbor-joining phylogram inferred from analysis of the combined pVGC sequence data. Strains are identified by their NRRL numbers and serotype designations. Lineages are demarcated with numbered brackets, and the L. monocytogenes type strain is marked with an asterisk. The tree was rooted with L. ivanovii (NRRL 33017 and NRRL 33021) and L. seeligeri (NRRL 33019) sequences (not shown). The frequency (percent) with which a given branch was recovered in 1,000 neighbor-joining bootstrap replications is shown above branches recovered in more than 70% of bootstrap replicates, with bootstrap values from maximum-parsimony analysis given in parentheses.
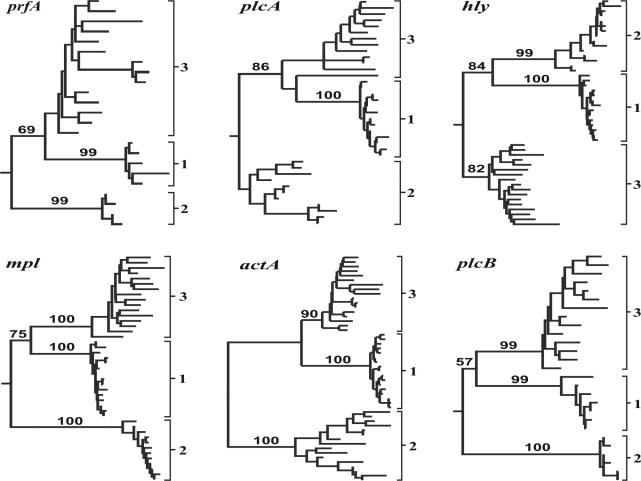
With the exception of a single lineage 3 isolate (NRRL 33227), which was recovered as the nearest relative of a monophyletic lineage 1 in the plcA neighbor-joining tree, the three L. monocytogenes lineages identified in analyses of the combined data also were resolved as monophyletic clades within individual gene trees derived from each of the six genes within the pVGC (Fig. 2). Clustering of NRRL 33227 with lineage 1 was not supported by bootstrap analyses, and examination of individual character differences revealed that no character states were uniquely shared between NRRL 33227 and lineage 1 isolates. In addition, all three lineages were recovered as monophyletic groups in maximum-parsimony analyses, suggesting that the paraphyletic distribution of lineage 3 isolates in the plcA neighbor-joining tree is an artifact resulting from shared-ancestral character states and relatively long terminal branches within lineage 3.
FIG. 2.
Neighbor-joining phylogram inferred from analysis of individual pVGC genes, with lineages demarcated by numbered brackets. With the exception of the actA gene tree, which was midpoint rooted, individual gene trees were rooted with L. ivanovii (NRRL 33017 and NRRL 33021) and L. seeligeri (NRRL 33019) sequences (data not shown). The frequency (percent) with which a given branch was recovered in 1,000 neighbor-joining bootstrap replications is shown above branches recovered in >50% of the bootstrap replicates.
Neighbor-joining and maximum-parsimony trees derived from each of the pVGC genes except hly were congruent with the results of the combined data analyses in supporting a monophyletic L1/L3 clade exclusive of lineage 2 (Fig. 2). However, due to the inability to adequately assess positional homology between L. monocytogenes and the outgroup species L. seeligeri and L. ivanovii, the actA gene tree was rooted by the midpoint method (along the longest branch in the phylogeny). Although midpoint rooting indicated that lineages 1 and 3 are more closely related to each other than either is to lineage 2, bootstrap support for lineage relationships in the actA gene tree could not be assessed without the ability to root the tree with an outgroup sequence. In contrast to the results of the combined data analyses and gene trees recovered from the other pVGC genes, lineages 1 and 2 formed a clade exclusive of lineage 3 in neighbor-joining and maximum parsimony trees from hly. These lineage relationships were supported by 84% of neighbor-joining bootstrap replicates and likely reflect recombination between the ancestors of extant lineage 1 and lineage 2 haplotypes.
Branching patterns observed in the combined pVGC phylogeny (Fig. 1) suggest that the sampled lineage 1 haplotypes shared a single common ancestor more recently than haplotypes in the other two L. monocytogenes lineages. In order to test this hypothesis, the average genetic distance between haplotypes within each of the three lineages was determined. The average genetic distance between sampled haplotypes was significantly (P < 0.001) less for lineage 1 (0.29% ± 0.04%) than for either lineage 2 (0.73% ± 0.05%) or lineage 3 (1.17% ± 0.08%). Although this could result from biased sampling of highly related lineage 1 isolates or from differences in population substructure, these explanations are unlikely as the maximum genetic distance between lineage 2 (1.29%) or lineage 3 haplotypes (1.58%) was >2.5-fold the maximum genetic distance between lineage 1 haplotypes (0.49%). In addition, the average genetic distance between lineage 1 isolates after exclusion of the lower quartile of values (0.33% ± 0.05%) also was significantly (P < 0.001) less than the average values for the other two lineages.
Serotype distributions.
Unambiguous serotype determinations were made for 93 of the 96 L. monocytogenes isolates tested, with three isolates ambiguously typed as 1/2b or 3b (Table 1). However, due to conflicts with strain history data and previously reported problems distinguishing among serotypes 4b, 4e, and 4d (22), four serotype 4e isolates were retested. Upon retesting, one isolate was confirmed as serotype 4e, two isolates were identified as serotype 4b (in agreement with strain histories), and a fourth isolate was identified as serotype 4d, confirming the previously reported difficulties in distinguishing among the 4b, 4e, and 4d serotypes. In addition to the serotype data collected here, L. monocytogenes strain EGD-e has been reported as serotype 1/2a (10), and serotype information was previously published (1) or provided with strain histories for 16 isolates from which pVGC sequence data were collected (Table 1). Serotypes were almost exclusively associated with one of the three L. monocytogenes lineages. Serotypes 4b, 1/2b, 4e and 4d were identified among lineage 1 isolates. Serotypes 1/2a, 1/2c, 3a and 3c were identified among lineage 2 isolates. However, in addition to the 4a and 4c serotypes identified among lineage 3 isolates, 10 (59%) of the 17 unique pVGC haplotypes identified within lineage 3 were from serotype 4b isolates (Fig. 1).
Lineage identification by using an ASO-PCR multiplex.
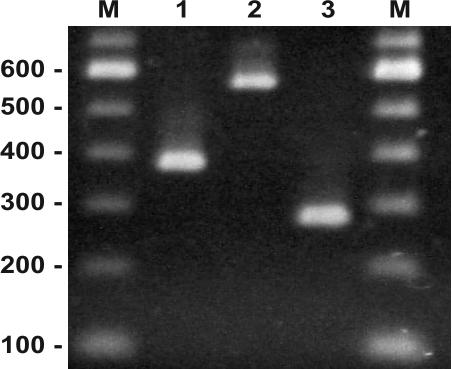
An ASO-PCR multiplex was used to determine the lineage of individual L. monocytogenes isolates (Fig. 3). The accuracy of this test was evaluated by comparing the ASO-PCR multiplex results with lineage identifications based on pVGC sequence data for the 112 L. monocytogenes isolates used in the phylogenetic analyses. The ASO-PCR multiplex produced a single amplicon of the correct size for each of these isolates. In addition, no target amplicons were produced with isolates from any of the other Listeria species when the test was applied to four L. innocua, four L. ivanovii, two L. grayi, two L. welshimeri, and one L. seeligeri strain. In order to evaluate the utility of the test with a panel of isolates for which lineage identity was unknown and to estimate the frequency of the three L. monocytogenes lineages in food products, the ASO-PCR multiplex was also applied to 99 L. monocytogenes isolates from food products surveyed by the U.S. Department of Agriculture's Food Safety and Inspection Service (FSIS) (Table 3). A single target amplicon was produced for each of the 99 food isolates, with lineages 1 (47%) and 2 (51%) present at nearly equal frequencies and lineage 3 (2%) nearly absent.
FIG. 3.
ASO-PCR multiplex amplification results for representative L. monocytogenes strains from lineage 1, NRRL 33176 (lane 1); lineage 2, NRRL 33180 (lane 2); and lineage 3, NRRL 33185 (lane 3). Amplification products were scored relative to a 100-bp DNA size ladder (lane M).
TABLE 3.
ASO-PCR lineage identification for L. monocytogenes food isolates
| NRRL no.a | FSIS equivalent no. | Origin | Lineage | NRRL no.a | FSIS equivalent no. | Origin | Lineage | |
|---|---|---|---|---|---|---|---|---|
| 33235 | OB1441 | Beef and pork franks | 2 | |||||
| 33236 | OB1520 | Beef and pork weiners | 2 | |||||
| 33237 | OB1547 | Beef and pork franks | 1 | |||||
| 33238 | OB1548 | Beef jerky | 2 | |||||
| 33239 | OB1549 | Beef and pork franks | 1 | |||||
| 33240 | OB1550 | Beef and pork franks | 1 | |||||
| 33241 | OB1566 | Cooked apple sausage | 2 | |||||
| 33242 | OB1597 | Roast beef | 1 | |||||
| 33243 | OB1608 | Cooked beef | 2 | |||||
| 33246 | OB1648 | White chicken salad | 2 | |||||
| 33247 | OB1649 | Roast beef | 2 | |||||
| 33248 | OB1650 | Barbeque chicken | 1 | |||||
| 33250 | OB1720 | Boneless smoked ham-steak | 1 | |||||
| 33252 | OB1777 | Embotido | 1 | |||||
| 33253 | OB1778 | Cooked ham | 2 | |||||
| 33254 | OB1779 | Roast beef | 1 | |||||
| 33255 | OB1780 | Chinese sausage | 2 | |||||
| 33256 | OB1781 | Chinese sausage | 2 | |||||
| 33257 | OB10002 | Roast beef | 2 | |||||
| 33258 | OB10003 | Smoked boneless ham | 1 | |||||
| 33259 | OB10008 | Cooked chicken meat strips | 2 | |||||
| 33260 | OB10016 | Beef sausage links | 2 | |||||
| 33261 | OB10017 | Beef jerky | 3 | |||||
| 33262 | OB10022 | Boneless cooked country ham | 1 | |||||
| 33264 | OB10065 | Sliced cooked beef | 2 | |||||
| 33265 | OB10068 | Boneless cooked country ham | 1 | |||||
| 33276 | OB10106 | Mechanically separated chicken | 2 | |||||
| 33281 | OB10112 | Dried sausage | 2 | |||||
| 33282 | OB10113 | Duck breast | 2 | |||||
| 33283 | OB10114 | Chicken base | 2 | |||||
| 33284 | OB10115 | Boneless cooked country ham | 1 | |||||
| 33285 | OB10118 | Smoked boneless turkey breast | 2 | |||||
| 33286 | OB10119 | Cooked sausage | 2 | |||||
| 33287 | OB10120 | Cooked roast beef brisket | 1 | |||||
| 33288 | OB10123 | Sliced cooked beef | 2 | |||||
| 33289 | OB10142 | Sweet sopressata | 1 | |||||
| 33290 | OB10145 | Quesadilla with beef | 2 | |||||
| 33291 | OB10146 | Portuguese sausage with egg wrap | 1 | |||||
| 33292 | OB10147 | Smoked boneless turkey breast | 2 | |||||
| 33293 | OB10149 | Cooked pork meat | 1 | |||||
| 33294 | OB10151 | Cooked charbroil beef patty | 1 | |||||
| 33295 | OB10153 | Chorizo | 2 | |||||
| 33296 | OB10154 | Boneless cooked country ham | 1 | |||||
| 33297 | OB10158 | Dried sausage | 2 | |||||
| 33298 | OB10167 | Cooked sausage | 2 | |||||
| 33299 | OB10169 | Cooked sausage | 2 | |||||
| 33304 | OB10205 | Cooked pork sausage | 1 | |||||
| 33305 | OB10206 | Chicken bacon | 1 | |||||
| 33306 | OB10219 | Chicken chow mein | 1 | |||||
| 33307 | OB10334 | Chicken breast tenders | 2 | |||||
| 33308 | OB10335 | Seasoned chicken or beef | 1 | |||||
| 33309 | OB10341 | Wieners | 1 | |||||
| 33310 | OB10347 | Ham | 2 | |||||
| 33311 | OB10348 | Quesadilla with chicken | 2 | |||||
| 33312 | OB10349 | Cooked ham | 1 | |||||
| 33313 | OB10350 | Smoked ham | 1 | |||||
| 33315 | OB10388 | Semiboneless ham | 1 | |||||
| 33316 | OB10390 | Roast beef | 2 | |||||
| 33317 | OB10391 | Deli turkey | 2 | |||||
| 33318 | OB10392 | Deli turkey cheese | 2 | |||||
| 33319 | OB10393 | Beef franks | 2 | |||||
| 33320 | OB20002 | Franks | 1 | |||||
| 33321 | OB20004 | Roast duckling | 2 | |||||
| 33322 | OB20009 | Pork barbeque | 1 | |||||
| 33323 | OB20012 | Pork barbeque | 1 | |||||
| 33324 | OB20017 | Pork spring rolls | 2 | |||||
| 33325 | OB20061 | Barbeque sauce w/pork | 1 | |||||
| 33326 | OB20062 | Hungarian paprika salami | 2 | |||||
| 33327 | OB20065 | Smoked turkey drumsticks | 1 | |||||
| 33329 | OB20091 | Corn beef brisket | 1 | |||||
| 33330 | OB20097 | Liquid unpast whole egg | 3 | |||||
| 33331 | OB20114 | Chicken burrito | 1 | |||||
| 33332 | OB10004 | Beef or pork smoked sausage | 2 | |||||
| 33334 | OB10216 | Pork | 1 | |||||
| 33335 | OB020094 | Smoked pork chops | 2 | |||||
| 33336 | OB020122 | Boneless pork chops | 2 | |||||
| 33337 | OB020132 | Chicken burrito | 1 | |||||
| 33338 | OB020428 | Pork links | 2 | |||||
| 33339 | OB020429 | Spicy cashew chicken egg roll | 2 | |||||
| 33340 | OB020552 | Kayseri soujouk | 2 | |||||
| 33341 | OB020632 | Ham bologna | 1 | |||||
| 33342 | OB020663B | Turkey pastrami | 2 | |||||
| 33343 | OB020709 | Pork hash dumpling | 1 | |||||
| 33344 | OB020735 | Polish sausage | 2 | |||||
| 33345 | OB020760 | Buffet style ham | 1 | |||||
| 33346 | OB030003 | Boneless deli ham | 1 | |||||
| 33347 | OB030094 | Sliced beef in barbeque sauce | 1 | |||||
| 33348 | OB030115 | Sweet bologna | 2 | |||||
| 33349 | OB030116 | Smoked pork chops | 2 | |||||
| 33350 | OB030145 | Cooked hot Italian sausage | 2 | |||||
| 33351 | OB030159 | Chicken in chipotle sauce burrito | 1 | |||||
| 33352 | OB030205 | Sliced roast beef | 2 | |||||
| 33353 | OB030305 | Boneless ham | 1 | |||||
| 33354 | OB030306 | Cooked beef brisket | 2 | |||||
| 33355 | OB030469 | Sliced sausage for pizza | 1 | |||||
| 33356 | OB030631 | Cooked pork pattie | 1 | |||||
| 33357 | OB030758 | Cajun chicken salad | 1 | |||||
| 33358 | OB030759 | Cooked sweet Italian sausage | 1 | |||||
| 33359 | OB030774 | Pizza pocket | 1 |
NRRL, U.S. Department of Agriculture, Agricultural Research Service Culture Collection, Peoria, Ill.
DISCUSSION
L. monocytogenes lineage composition.
The results of the multigene phylogenetic analyses presented here clearly demonstrate that L. monocytogenes comprises at least three primary evolutionary divisions (Fig. 1 and 2), corresponding to lineages proposed by Rasmussen et al. (25) and Wiedmann et al. (33). In contrast, Mereghetti et al. (18) concluded on the basis of ribotyping and random amplification of polymorphic DNA (RAPD) data that L. monocytogenes is composed of only two lineages, with lineage 3 interpreted as a branch of the lineage 1 group. Similarly, only two primary divisions were recognized by Borucki et al. (2) based on microarray analyses. However, the results presented here demonstrate that the average genetic distance between pVGC haplotypes was significantly (P < 0.001) greater for lineage 3 than for lineage 1 and that lineage 1 haplotypes share a common ancestor more recently than haplotypes from the other two lineages. These results demonstrate that lineage 3 cannot be considered a branch of the lineage 1 group and that there are at least three primary evolutionary divisions within L. monocytogenes.
Salcedo et al. (28) have suggested that the three primary divisions of L. monocytogenes are evident only from analyses of specific genes associated with virulence, and that housekeeping genes or random genetic markers are unable to distinguish more than two lineages. However, they did not include lineage 3 isolates in their analyses. In addition, all three lineages were monophyletic in analyses of mixed genome microarray data (2) and data from ribotyping and RAPD typing (18). Recognition of only two major divisions by the authors of these studies was likely due to an underestimation of diversity within lineage 3, since both included only two lineage 3 isolates, representing only one of the three known serotypes from this lineage, and no more than 2 of the 17 unique lineage 3 haplotypes reported here (Fig. 1). Therefore, the identification of three primary divisions within L. monocytogenes is not restricted to analyses of virulence associated genes but may have been hampered in some studies by inadequate sampling of variation within lineage 3.
Direct correlations between the three L. monocytogenes lineages and the most common serotypes have previously been reported, with lineage 1 containing serotypes 4b, 1/2b, 3b, and 3c; lineage 2 containing serotypes 1/2a, 1/2c, and 3a; and lineage 3 containing serotypes 4a and 4c (20). Such correlations are of interest because serotypes represent the traditional common language of L. monocytogenes subtyping, and because strains with serotypes 4b, 1/2b, and 1/2a are responsible for the vast majority of human listeriosis cases (7, 29). Similar correlations were observed in the present study, particularly with respect to serotypes 1/2a, 1/2c, and 1/2b. However, comparisons between serotype and lineage for 106 L. monocytogenes isolates for which both were unambiguously determined (Table 1 and Fig. 1) revealed for the first time that serotype 4b, which is responsible for the majority of human listeriosis cases (16) and virtually all major outbreaks of listeriosis in humans (13), is prevalent (59% of unique haplotypes) among strains from lineage 3, which is rarely associated with human listeriosis (13). These results demonstrate that serotype 4b isolates do not represent a distinct evolutionary group within L. monocytogenes and that serotype 4b cannot be used as a proxy for lineage identification.
Lineage identification, host specificity, and virulence differences.
L. monocytogenes lineage-specific variation identified during the analyses of pVGC sequences was used to develop an ASO-PCR multiplex test for the specific identification of evolutionary lineage for individual L. monocytogenes isolates. This test proved to be 100% sensitive and specific in accurately assessing the lineage for 112 L. monocytogenes isolates for which lineage identity had been confirmed phylogenetically. Previously, Jinneman and Hill (14) developed a PCR-based assay for L. monocytogenes lineage identification based on sequences from the hly gene but reported that one of the lineage 3 isolates produced target amplicons indicative of both lineage 2 and lineage 3. Analyses of the hly sequences reported here indicated that by using the Jinneman and Hill test, multiple target amplicons would be produced with nine of the 17 unique lineage 3 haplotypes and that only the amplicon specific to lineage 2 isolates would be produced for one of the lineage 3 haplotypes. These predictions were confirmed by performing the Jinneman and Hill test on the isolates in Table 1. In addition to problems with specificity caused by undersampling variation in lineage 3, this test also requires at least three separate PCRs. Similar problems exist with a lineage identification test developed by Moorehead et al. (19) that is based on only 23 isolates and also requires at least three separate PCRs.
Accurate, inexpensive, and high-throughput methods for L. monocytogenes lineage identification have the potential to inform studies of the population genetics, ecology, and epidemiology of this important food-borne pathogen and can also aid in understanding the biological and regulatory significance of the evolutionary lineages that have been identified within this species. For instance, the fact that lineage 3 isolates are rarely associated with human listeriosis but are common among animal isolates led Wiedmann et al. (33), and later Jeffers et al. (13), to suggest that lineage 3 isolates show a host specificity for nonprimate mammals and limited virulence in humans. However, application of the ASO-PCR multiplex to 99 L. monocytogenes isolates surveyed by FSIS provided the first direct estimate of the prevalence of individual lineages from a broad array of food products, and indicated that lineage 3 accounts for only 2% of L. monocytogenes isolates from food (Table 3). If we assume that contaminated food is the primary cause of listeriosis in humans, the frequency of lineage 3 isolates among human sporadic cases (1%) reported by Jeffers et al. (13) is entirely consistent with the relative frequency of lineage 3 isolates in food products (Table 3). Therefore, the low frequency of association between lineage 3 isolates and human listeriosis cases likely reflects rarity of exposure and not reduced virulence for humans or specificity for nonhuman hosts. In addition, the prevalence of lineage 3 among animal isolates (37% of the animal isolates in Table 1) and the near absence among food isolates suggests that lineage 3 may be better adapted to the animal production environment than the food processing environment. Systematic comparisons of lineage-specific fitness in different environments are needed to fully evaluate this hypothesis. However, De Jesús and Whiting (6) have found that strains from lineage 3 are less likely to survive thermal inactivation than strains from the other two lineages of L. monocytogenes, indicating that lineages 1 and 2 may be better adapted to the food-processing environment than are lineage 3 isolates.
Comparison of the relative frequencies of the three L. monocytogenes lineages in food products (Table 3) and human listeriosis cases (13) suggests that lineage 1 is overrepresented and lineage 2 is underrepresented among isolates from human listeriosis cases. However, it is unclear if this reflects enhanced virulence for humans or unique ecological adaptations such as enhanced psychrotolerance and growth at refrigeration temperatures. Prevalence studies alone are insufficient to clearly demonstrate lineage-specific differences in virulence or ecological adaptations. However, the availability of complete genome sequences for L. monocytogenes lineage 1 (http://www.tigr.org) and lineage 2 (10) isolates will facilitate functional genomic studies and additional analyses of genomic variation within and between lineages that will complement comparative evaluations of virulence and comprehensive surveys of lineage prevalence in different environments. A combination of such studies will be required to fully test hypotheses regarding lineage-specific differences in virulence, host range, or ecology and to understand the genetic and evolutionary basis of such differences.
Lineage relationships and taxonomy.
Previous analyses of relationships within L. monocytogenes based on shotgun DNA microarray data suggested that the single lineage 3 isolate examined in that study was distinct from a more derived group consisting of lineages 1 and 2 (36). However, these data were highly homoplasious (homoplasy index = 0.6491) in that they contained a high proportion of character state similarities that were not due to inheritance from a common ancestor, with over half of the polymorphisms distributed among polyphyletic groups (36). As noted by Zhang et al. (36), several comparative studies of Listeria genomes suggest a bias toward cell surface-related differences in genome content (4, 10, 12), indicating that different combinations of genes encoding cell surface characteristics may be favored by selection (36). This suggests that polymorphism data from such genome content studies may be inherently less reliable for use in phylogenetic reconstruction because these studies can be biased toward nonessential genes that may be lost independently in multiple evolutionary lineages or classes of genes that may be frequently involved in lateral gene transfer events due to selection.
Accurate reconstruction of the evolutionary relationships between the three L. monocytogenes lineages is essential to understanding the evolution of virulence traits and ecological adaptations within this species and is also critical in evaluating proposals to reassess the taxonomic rank of individual lineages. Both neighbor-joining and maximum-parsimony analyses of the combined pVGC data strongly support a sister-group relationship between lineages 1 and 3 (Fig. 1). The L1/L3 clade was also recovered in five of the six gene trees constructed from individual pVGC genes (Fig. 3). Lineages 1 and 2 were most closely related in the hly gene tree, which appears to reflect historical recombination between ancestors of present-day lineage 1 and lineage 2 haplotypes. However, despite the discordant hly gene tree, a sister-group relationship between lineages 1 and 2 provided the worst fit to the combined pVGC data, and the combined analyses of pVGC sequences strongly support the conclusion that lineages 1 and 3 share a common ancestor exclusive of lineage 2 (Fig. 1). This conclusion is congruent with the midpoint-rooted phylogenetic tree derived from combined analyses of ribotyping and RAPD typing data (18) and phylogenetic analyses based on mixed genome microarray data (2). In addition, the single serotype 4a isolate included in the multilocus enzyme electrophoresis study conducted by Piffaretti et al. (24) clustered with lineage 1 isolates. Although lineage association was not directly determined for this isolate, the 4a serotype appears to be specific to lineage 3 (Table 1) (20) and the other serotypes that have been identified within lineage 3 were either absent (serotype 4c) from the Piffaretti et al. (24) study or clustered with the lineage 1 group (serotype 4b), a finding consistent with the conclusion that lineages 1 and 3 are sister-groups.
Wiedmann et al. (33, 34) have suggested that lineage 3 represents a distinct taxonomic unit separate from lineages 1 and 2 and that lineage 3 should be recognized as a new species or subspecies because the small number of lineage 3 isolates examined had a distinctive ribotype fragment, a unique 16S rRNA sequence, and 70 to 76% DNA-DNA homology with the L. monocytogenes type strain from lineage 2 (27) and were predominantly serotype 4a or 4c. However, these differences were not evaluated relative to the phylogenetic history of lineage divergence within L. monocytogenes. The results of the phylogenetic analyses presented here strongly support a monophyletic L1/L3 clade exclusive of lineage 2 (Fig. 1). Therefore, recognizing lineage 3 as a new species or subspecies without equivalent recognition for lineage 1 would make L. monocytogenes paraphyletic, which is inconsistent with the modern systematic principles that taxonomy should reflect evolutionary history and taxonomic groups should comprise individuals that uniquely share a most recent common ancestor. In addition, the use of genetic or phenotypic features to circumscribe new species in the absence of an evolutionary framework for interpreting species boundaries is arbitrary and is not supported by population genetic or evolutionary theory.
The cohesion species concept proposed by Templeton (30) provides an evolutionary framework for understanding species as groups of organisms whose divergence is constrained by microevolutionary forces that maintain species as genetically and phenotypically cohesive groups. For species such as L. monocytogenes, which has a largely clonal population structure (4, 24, 25), evolutionary theory indicates that genetic drift and natural selection are the primary forces influencing species cohesion (5, 30). The results presented here, indicating a more recent coalescence for lineage 1 haplotypes than haplotypes from the other two lineages (Fig. 1), are interesting because they suggest that lineage 1 was exposed to a purge of genetic variation not observed in the other two lineages. This indicates a limitation on the extent to which these lineages are bound together by natural selection and suggests that they may represent distinct species within the framework of the cohesion species concept. Evidence from the survey of lineage prevalence in food products (Table 3) and from previous studies (6, 13), suggesting lineage-specific differences in ecological niche adaptations further support this interpretation. Given the phylogenetic relationships supported by the combined pVGC data and the fact that the L. monocytogenes type strain belongs to lineage 2 (Fig. 1), species recognition for lineage 1 would require reclassification of lineage 1 and lineage 3 isolates into two new species. However, the results of the present study should be viewed as hypothesis-generating with respect to taxonomic revision of L. monocytogenes, which will require a greater understanding of the ecology and demographic exchangeability of L. monocytogenes lineages and evaluations of the demographic history of these lineages based on genetic variation sampled from additional regions of the L. monocytogenes chromosome.
Supplementary Material
Acknowledgments
We are indebted to Douglas Abbott, James Donahue, Robert Duvall, Richard Raybourne, Jeff Call, Danielle Janssens, Reiner Kroppenstedt, and Irene Wesley for generously supplying isolates used in this study. We also thank Thomas Usgaard and Amy Morgan for excellent technical assistance and Joseph Bielawski, Alejandro Rooney, Kerry O'Donnell, and Cletus Kurtzman for helpful discussions and criticisms of the manuscript.
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Borucki, M., and D. R. Call. 2003. Listeria monocytogenes serotype identification using PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 3.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 6.De Jesús, A. J., and R. C. Whiting. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611-1617. [DOI] [PubMed] [Google Scholar]
- 7.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Fliss, I., E. Emond, R. E. Simard, and S. Pandian. 1991. A rapid and efficient method of lysis of Listeria and other gram-positive bacteria using mutanolysin. BioTechniques 11:453-457. [PubMed] [Google Scholar]
- 10.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charcit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueño, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 11.Graves, L. M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, J. K. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 14.Jinneman, K. C., and W. E. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 16.McLauchlin, J. 1990. Distribution of serovars of Listeria monocytogenes isolated from different categories of patients with listeriosis. Eur. J. Clin. Microbiol. Infect. Dis. 9:210-213. [DOI] [PubMed] [Google Scholar]
- 17.Mead, P., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mereghetti, L., P. Lanotte, V. Savoye-Marczuk, N. Marquet-Van Der Mee, A. Audrier, and R. Quentin. 2002. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 68:2849-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moorehead, S. M., G. A. Dykes, and R. T. Cursons. 2003. An SNP-based PCR assay to differentiate between Listeria monocytogenes lineages derived from phylogenetic analysis of the sigB gene. J. Microbiol. Methods 55:425-432. [DOI] [PubMed] [Google Scholar]
- 20.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505-515. [PubMed] [Google Scholar]
- 22.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penny, D., and M. D. Hendy. 1985. Testing methods of evolutionary tree construction. Cladistics 1:266-272. [DOI] [PubMed] [Google Scholar]
- 24.Piffaretti, J.-C., H. Kressebuch, M. Aeschenbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein, and listeriolysin O genes. Microbiol. 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 26.Ripabelli, G., J. McLauchlin, and E. J. Threlfall. 2000. Amplified fragment length polymorphism (AFLP) analysis of Listeria monocytogenes. Syst. Appl. Microbiol. 1:132-136. [DOI] [PubMed] [Google Scholar]
- 27.Rocourt, J., F. Grimont, P. A. D. Grimont, and H. P. R. Seeliger. 1982. DNA realtedness among serovars of Listeria monocytogenes sensu lato. Curr. Microbiol. 7:383-388. [Google Scholar]
- 28.Salcedo, C., L. Arreaza, B. Alcalá, L. de la Fuente, and J. A. Vázquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Templeton, A. R. 1989. The meaning of species and speciation: a genetic perspective, p. 3-27. In D. Otte and J. E. Endler (ed.), Speciation and its consequences. Sinauer Associates, Inc., Sunderland, Mass.
- 31.Vázquez-Boland, J. A., G. Domínguez-Bernal, B. González-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 32.Wallace, F. M., J. E. Call, A. C. S. Porto, G. J. Cocoma, J. B. Luchansky, et al. 2003. Recovery rate of Listeria monocytogenes from commercially prepared frankfurters during extended refrigerated storage. J. Food Protect. 66:584-591. [DOI] [PubMed] [Google Scholar]
- 33.Wiedmann, M., J. L. Bruce, C. Keating, A. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in their pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 35.Wiedmann, M. 2003. An integrated science-based approach to dairy food safety: Listeria monocytogenes as a model system. J. Dairy Sci. 86:1865-1875. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.