Abstract
A genomic library of Bifidobacterium bifidum constructed in Escherichia coli was screened for the ability to hydrolyze the α-(1→2) linkage of 2′-fucosyllactose, and a gene encoding 1,2-α-l-fucosidase (AfcA) was isolated. The afcA gene was found to comprise 1,959 amino acid residues with a predicted molecular mass of 205 kDa and containing a signal peptide and a membrane anchor at the N and C termini, respectively. A domain responsible for fucosidase activity (the Fuc domain; amino acid residues 577 to 1474) was localized by deletion analysis and then purified as a hexahistidine-tagged protein. The recombinant Fuc domain specifically hydrolyzed the terminal α-(1→2)-fucosidic linkages of various oligosaccharides and a sugar chain of a glycoprotein. The stereochemical course of the hydrolysis of 2′-fucosyllactose was determined to be inversion by using 1H nuclear magnetic resonance. The primary structure of the Fuc domain exhibited no similarity to those of any glycoside hydrolases (GHs) but showed high similarity to those of several hypothetical proteins in a database. Thus, it was revealed that the AfcA protein constitutes a novel inverting GH family (GH family 95).
α-l-Fucosyl residues are frequently found at the nonreducing termini of various glycoconjugates, including blood group substances, milk oligosaccharides, gastric and submaxillary mucins, and serum glycoproteins (30, 37). The results of recent studies indicate that such terminal fucosyl residues attached by α-(1→2), α-(1→3), and α-(1→4) linkages play important roles in mammalian cell-to-cell communication mediated through receptor proteins (8). In addition, it has been shown that abnormal fucosylation often occurs in human diseases, including cancer (3). Terminal fucose-mediated biological activity is not limited to higher eukaryotic cells but includes bacterial adhesion to host cells. Campylobacter jejuni is known to bind blood group H(O) antigen [Fucα(1→2)Galβ(1→4)GlcNAc] (31), and Helicobacter pylori has a specific receptor protein, adhesin, that binds Leb antigen {Fucα(1→2)Galβ(1→3)[Fucα(1→4)]GlcNAc} (16), thereby facilitating their infection of host epithelial cells.
α-l-Fucosidase could serve as a powerful tool to elucidate the structure-function relationships of these fucose-containing bioactive glycoconjugates, since it can selectively cleave and remove specific fucosyl residues from the targets without damage, the products of which are then assessed and evaluated as to their biological activities. To date, α-l-fucosidases from various prokaryotic and eukaryotic sources have been purified and characterized (1, 2, 7, 19, 21, 29, 34, 44, 45, 48), and historically, these enzymes have been divided into two groups, one capable of hydrolyzing various types of fucosidic linkages as well as synthetic substrates (EC 3.2.1.51) and the other being active only on α-(1→2) linkages (EC 3.2.1.63). While the genes encoding the former type of fucosidases have been isolated from various sources and are known to constitute glycoside hydrolase (GH) family 29 (4, 9, 10, 23, 28, 42), the genes for the latter type of enzymes have not been cloned yet. With this background, and since the recombinant 1,3-/4-α-l-fucosidase of Streptomyces sp. is now commercially available, the cloning of the 1,2-α-l-fucosidase gene is of great importance not only for the preparation of a pure recombinant enzyme but also for obtaining a better understanding of α-l-fucosidase.
Here we describe the molecular cloning and characterization of 1,2-α-l-fucosidase of Bifidobacterium bifidum. This is the first report describing the isolation of a gene encoding α-(1→2)-specific fucosidase and indicating the existence of a hitherto unknown GH family. Moreover, this report opens the way to elucidating the role of secretory fucosidases of enteric bacteria in the intestinal ecosystem, which could provide important clues for use of bifidobacteria as key commensals that promote a healthy intestinal tract (35).
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study were Bifidobacterium breve 203 (27) and JCM119, B. bifidum ATCC 29251, JCM1254, and JCM7004, Bifidobacterium infantis JCM1222, Bifidobacterium longum 33R and JCM1217, and Escherichia coli DH5α (46).
Media and chemicals.
Luria-Bertani broth was routinely used for the cultivation of E. coli strains (22). Bifidobacteria were grown anaerobically in GAM medium (Nissui, Tokyo, Japan) with AnaeroPack (Mitsubishi Chemical, Tokyo, Japan). Ampicillin and kanamycin were used at final concentrations of 100 and 30 μg/ml, respectively. 2′-Fucosyllactose (Fucα1→2Galβ1→4Glc) was kindly donated by S. Koizumi, Kyowa Hakko Kogyo Co., Ltd. (Tokyo, Japan). 6-Fucosyl-N,N′-diacetylchitobiose [GlcNAcβ1→4(Fucα1→6)GlcNAc] and 3-fucosyllactose [Galβ1→4(Fucα1→3)Glc] were from Sigma. 3-Fucosylgalactose (Fucα1→3Gal), blood group H(II) (Fucα1→2Galβ1→4GlcNAc), blood group A [GalNAcα1→3(Fucα1→2)Gal], and blood group B [Galα1→3(Fucα1→2)Gal] active substances were purchased from Funakoshi (Tokyo, Japan). Milk oligosaccharides, including lacto-N-fucopentaose I (Fucα1→2Galβ1→3GlcNAcβ1→3Galβ1→4Glc), II [Galβ1→3(Fucα1→4)GlcNAcβ1→3Galβ1→4Glc], and V [Galβ1→3GlcNAcβ1→3Galβ1→4(Fucα1→3)Glc] were also from Funakoshi. Cellobiose (Glcβ1→4Glc), N,N′-diacetylchitobiose, kojibiose (Glcα1→2Glc), maltose (Glcα1→4Glc), p-nitrophenyl (pNP)-α-l-fucoside, pNP-β-l-fucoside, and 4-methylumbelliferyl-α-l-fucoside were purchased from Wako Pure Chemical Industries (Tokyo, Japan). Oligosaccharides Galα1→3Galβ1→4Gal and Galα1→4Gal were from Dextra Laboratories Ltd. and ICN, respectively. Pyridylamino (PA)-sugars were purchased from Takara Bio Inc. (Shiga, Japan). The chemicals were all obtained commercially and were not purified further.
Pyridylamination of oligosaccharides and HPLC analysis.
Fluorescence labeling of oligosaccharides with 2-aminopyridine was carried out by the method of Hase et al. (11). The reaction mixture containing PA-oligosaccharides was analyzed by high-pressure liquid chromatography (HPLC) as described previously (18), with excitation and emission wavelengths of 315 and 380 nm, respectively.
Fucosidase assays.
The standard reaction mixture consisted of 100 mM sodium phosphate (pH 6.5), 2 mM substrate, and a protein sample, in a total volume of 100 μl. After incubation at 37°C for an appropriate time, the reaction was stopped by heating for 3 min in a boiling water bath. The amounts of l-fucose liberated from oligosaccharides and glycoproteins were determined by use of fucose dehydrogenase (FDH) from a Pseudomonas strain (14) as described by Cohenford et al. (5). Pseudomonas FDH has been shown to be specific for l-fucose, l-galactose, and, to a lesser extent, d-arabinose (14). One unit of enzyme activity was defined as the amount of enzyme releasing 1 μmol of l-fucose per min from the substrate.
Construction of a B. bifidum genomic library in E. coli and screening for transformants with 1,2-α-l-fucosidase activity.
A genomic library was constructed essentially as described previously (17), using E. coli strain DH5α, with about 800 colonies being obtained. Each transformant was lysed in a small volume of BugBuster reagent (Novagen) and incubated with 2′-fucosyllactose, and then the reaction mixture was analyzed by thin-layer chromatography. The silica gel plate (Silica Gel 60, Merck) was developed with a solvent system of chloroform-methanol-water (3:3:1), dried, and visualized by spraying with orcinol-H2SO4 reagent (13). The strain carrying pSA3 (ColE1 ori rop+ bla+ tet::′afcA) was selected as a fucosidase-positive strain and used for further study.
Genetic techniques.
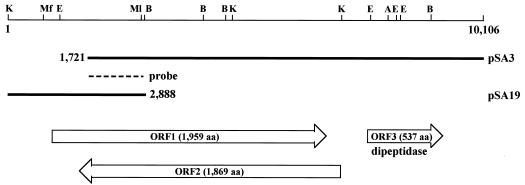
Standard genetic techniques were used (32). DNA sequences were determined for both strands (33) by using a BigDye terminator version 3.0 cycle sequencing ready reaction kit (Applied Biosystems). In order to clone the upstream region of the afcA gene, a genomic library was screened by using the 1.1-kb PstI-BamHI fragment of pSA3 as a specific probe (Fig. 1). Hybridization and subsequent detection were performed essentially as described previously (40).
FIG. 1.
Gene organization of the afcA locus of B. bifidum JCM1254. The DNA sequence (10,106 bp in length) was determined, with the numbering starting at the distal KpnI site. The top line is a restriction map indicating the recognition sites of AflII (A), BamHI (B), EcoRI (E), KpnI (K), MfeI (Mf), and MluI (Ml). The thick lines indicate the lengths of the inserts of the plasmids, and the dashed line indicates the specific probe (bp 1808 to 2888) used for cloning of the upstream region of ORF1 (bp 1 to 2888, pSA19). Neither ORF1 nor ORF2 exhibited similarity in amino acid sequence to proteins with known function in the database. The deduced amino acid sequence of ORF3 showed high similarity to the consensus sequence of Pfam 03577; thus, it probably encodes a dipeptidase. aa, amino acids.
Localization of the catalytic domain.
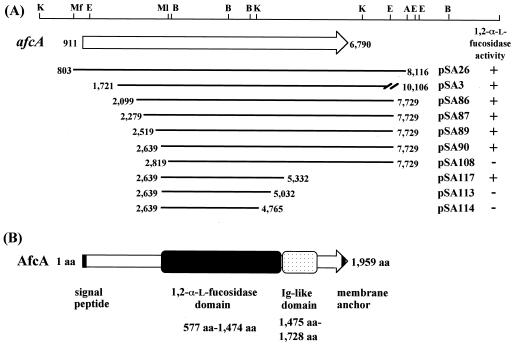
To localize the domain responsible for 1,2-α-l-fucosidase activity, deletion analysis of afcA was carried out. High-fidelity PCR involving KOD polymerase (Toyobo, Tokyo, Japan) was performed, and the amplified fragments were entirely sequenced to ensure that no base change other than those planned had occurred. The N-terminal deletion mutants (Fig. 2) were designed so as to be expressed as fusions with β-galactosidase (N-terminal eight amino acid residues), which was accomplished by inserting the amplified genes into the lacZα gene present on pMW219 (pSC101 ori kan+ lacZα+) (Nippon Gene, Tokyo, Japan). The following primer pairs were used: 5′-CAAGCTTGGTCATCGCCAGTGTCGAGGACGGCG-3′ and 5′-GTTCAGGTCGCGGCGGTATTCGGTG-3′ for pSA86, 5′-CAAGCTTGAACGGTGAGGATAACTACACCATCG-3′ and 5′-GTTCAGGTCGCGGCGGTATTCGGTG-3′ for pSA87, 5′-CAAGCTTGCTGGGCGAGCTCAACAAGTCCGACA-3′ and 5′-GTTCAGGTCGCGGCGGTATTCGGTG-3′ for pSA89, 5′-CAAGCTTGACCGATACCACGAAGACCGCGACGT-3′ and 5′-GTTCAGGTCGCGGCGGTATTCGGTG-3′ for pSA90, and 5′-CAAGCTTGACCGTCTGGGGCGAGGTCAGCCGTGAACGCGTCA-3′ and 5′-TGACGCGTTCACGGCTGACCTCGCCCCAGACGGTCAAGCTTG-3′ for pSA108. The upstream primers contained HindIII sites to allow in-frame fusion with the lacZα gene. The amplified fragments were digested with HindIII and MluI and then ligated with the 8.8-kb HindIII-MluI fragment of pSA23, which contains the same insert as pSA3 on a low-copy-number plasmid, pMW219. After being confirmed by DNA sequencing, the resulting deletion mutants were expressed in E. coli, and then cell extracts (38) were subjected to fucosidase assay. The C-terminal deletion mutants were constructed by introducing stop codons (Fig. 2). To this end, the following primer pairs were used: 5′-CGGTACCGCTGCTTCAAGGGCAACG-3′ and 5′-CGAATTCTTTAGCTCGCCTTCTTCGTGATCGTG-3′ for pSA117, 5′-CGGTACCGCTGCTTCAAGGGCAACG-3′ and 5′-CGAATTCTTTACGTGTAGTTCACGTACTTCTTG-3′ for pSA113, and 5′-AGGTACCGCTAAATCAAGGGCAAGAATTCG-3′ and 5′-CGAATTCTTGCCCTTGATTTAGCGGTACCT-3′ for pSA114. The downstream primers contained EcoRI sites to facilitate plasmid construction. The amplified fragment was digested with KpnI and EcoRI and then ligated with the 6-kb KpnI-EcoRI fragment of pSA90 (Fig. 2) to be sequenced. These C-terminal deletion mutants were similarly examined for fucosidase activity.
FIG. 2.
Deletion analysis of the afcA gene for identification of the 1,2-α-l-fucosidase domain (A) and schematic representation of the domain structure of AfcA (B). (A) The top line is a restriction map, as described for Fig. 1. The thick lines indicate the lengths of the inserts of the plasmids. Deletion mutants were designed so as to be expressed as fusions with the eight N-terminal amino acids derived from lacZα on a low-copy-number plasmid, pMW219 (pSA86, -87, -89, -90, -108, -113, -114, and -117). C-terminal deletion mutants were constructed by introducing stop codons at the indicated positions. Each mutant was examined for the ability to liberate l-fucose from 2′-fucosyllactose, with the results shown on the right. (B) The amino acid numbering starts at the probable initiation codon, and the domain responsible for 1,2-α-l-fucosidase activity (amino acids [aa] 577 to 1474) is depicted as a gray box. The region with Ig-like folds (aa 1475 to 1728), which was predicted from the sequence similarity to the consensus sequence (Pfam 02368), is shown by a stippled box. The black bars at the N-terminal and C-terminal ends indicate a signal peptide and a membrane anchor, respectively.
Purification and characterization of 1,2-α-l-fucosidase.
The fucosidase domain of AfcA was purified as a C-terminal hexahistidine-tagged protein. The primer pair 5′-GCATATGGTCATCGCCAGTGTCGAGGACG-3′ (for the upstream end)-5′-GCCCGGGTTTAATGGTGATGGTGATGGTGGCTCGCCTTCTTCGTGATCGTGTAC-3′ (for the downstream end) was used. The upstream and downstream primers contained NdeI and SmaI sites, respectively. After confirmation by sequencing, the fragment was cut off with NdeI and SmaI and then inserted into the NdeI-BamHI (blunt-ended) site of pET3a (Novagen). The resulting plasmid, pSA130, carrying the gene under control of the T7 promoter was introduced into BL21(λDE3) (Novagen). This strain was grown in Luria-Bertani medium at 37°C, and the expression of the domain was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Purification was carried out with a MagneHis system (Promega), followed by size exclusion chromatography (Sephacryl S-300 HR) and Mono Q HR5/5 column chromatography, in which the protein was eluted with a linear gradient of 0 to 1 M NaCl in 10 mM sodium phosphate buffer (pH 6.5). Protein concentrations were determined with the bicinchoninic acid protein assay reagent (Pierce) with bovine serum albumin as a standard. The purity of the protein was judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (20).
Thermostability was evaluated by incubating the protein (0.1 mg/ml) in 10 mM sodium phosphate buffer (pH 6.5) at different temperatures for 30 min prior to assaying. pH stability was assessed by dialyzing the enzyme overnight against buffer (10 mM) of different pHs, followed by standard fucosidase assaying. For determination of the optimal pH, 25 mM citrate-phosphate buffer of different pHs (pH 3 to 8) was used. The reaction was initiated by adding the enzyme dialyzed against 0.2 mM sodium phosphate buffer (pH 6.5), and the mixture was incubated for a short period of time (<1 min) to prevent damage to the enzyme. The Km and Vmax values were determined by double-reciprocal plotting of the data (50 μM to 2 mM 2′-fucosyllactose).
Determination of the stereochemical course of hydrolysis.
The stereochemistry of the hydrolysis was determined by using 1H nuclear magnetic resonance (NMR). All compounds, including the substrate, buffer materials, and enzyme, were dissolved in 99.9% D2O and were repeatedly lyophilized prior to use. 1H NMR spectra were recorded on a JEOL JNM-AL300 spectrometer at 25°C. The reaction mixture, in a total volume of 560 μl, consisted of 10 mM 2′-fucosyllactose and 10 mM sodium phosphate (pH 6.5) in D2O. After recording the reference spectrum (t = 0 min), 60 mU (15 μl) of the purified Fuc domain was added to initiate the reaction. The spectra were recorded at different time intervals (7 min to16 h), with 30 scans. Commercially obtained l-fucose and d-lactose were used as standards.
ESI-MS analysis.
Samples (0.8 to 1 mg) were lyophilized, acetylated in the presence of pyridine and acetic anhydride (1/2, vol/vol; 0.9 ml), and then dissolved in 50% methanol containing 1% acetate to a final concentration of 10 μM. Electrospray ionization-mass spectrometry (ESI-MS) was performed in the positive ion mode with a Mariner mass spectrometer (Applied Biosystems). Neurotensin and angiotensin were used as the internal controls.
Nucleotide sequence accession number.
The DNA sequence of the B. bifidum afcA gene has been deposited in GenBank under accession number AY303700.
RESULTS AND DISCUSSION
Intestinal colonizer strains, such as Bacteroides, Bifidobacteria, and Clostridium strains, are known to produce a variety of exo- and endoglycosidases in secretory forms (1, 21); therefore, they have been good resources for enzymes used in glycotechnology. We sought 1,2-α-l-fucosidase activity in several bifidobacterial strains by using 2′-fucosyllactose as a substrate, and we found that B. infantis JCM1222 and B. bifidum ATCC 29251, JCM1254, and JCM7004 produce l-fucosidases in cell surface-bound and/or extracellular forms, while B. breve 203 and JCM119 and B. longum 33R and JCM1217 do not. A surface-bound l-fucosidase specific for the α-(1→2) linkage was partially purified from B. bifidum JCM1254 cells, but the amount of the enzyme was too small to determine its amino acid sequence. Thus, we used the expression cloning strategy with E. coli strain DH5α, a non-fucosidase-producing bacterium.
Cloning of the gene encoding 1,2-α-l-fucosidase (afcA) from B. bifidum.
As described in Materials and Methods, a genomic library of B. bifidum JCM1254 constructed in E. coli was screened for the ability to hydrolyze the α-(1→2) linkage of 2′-fucosyllactose, with one recombinant, designated SA3, being selected. The ability of the cell extract of SA3 to liberate l-fucose was confirmed by FDH assay and HPLC analysis. When PA-2′-fucosyllactose was used for the reaction and then analyzed by HPLC, the peak of PA-2′-fucosyllactose decreased and a new peak corresponding to PA-lactose appeared (data not shown), whereas no peak change was observed when the control strain (DH5α/pBR322) (39) was used. Time- and dose-dependent liberation of l-fucose from PA-2′-fucosyllactose was also observed in the FDH assay (data not shown). In this reaction, a total of 21.0 nmol of PA-2′-fucosyllactose was consumed and 22.4 and 21.2 nmol of l-fucose and PA-lactose, respectively, were produced, demonstrating the stoichiometric scheme of the reaction. These results suggested that the 1,2-α-l-fucosidase gene was successfully cloned and expressed in E. coli.
Sequence analysis.
Sequence analysis of plasmid pSA3 revealed that the cloned gene contains two large truncated open reading frames (ORFs) (designated ORF1 and ORF2) and one intact ORF (ORF3) that exhibits high identity (>90%) to the consensus sequence of the peptidase family U34 (Pfam 03577) (Fig. 1). The flanking DNA segment (pSA19) containing the N-terminal part of ORF1 (the C-terminal part of ORF2) was isolated from the B. bifidum genomic library by use of a standard hybridization method, and the intact forms of ORF1 and ORF2 appeared as shown in Fig. 1. The sense strands of ORF1 and ORF2 overlapped to a large extent in reverse, but this is not so surprising because such cases are sometimes found in the recently determined genome sequence of B. longum NCC2705 (35). In order to determine which ORF actually encodes 1,2-α-l-fucosidase, each ORF (the MfeI-AflII fragment) was placed under the control of the lac promoter and its expression was induced by the addition of IPTG. While no increase in 1,2-α-l-fucosidase activity was observed when ORF2 was induced (from 1.0 to 1.1 mU/mg), the activity was significantly elevated when ORF1 was induced (from 1.3 to 20 mU/mg), indicating that ORF1 encodes 1,2-α-l-fucosidase (hereafter referred to as AfcA). The codon usage of the afcA gene was quite similar to that of other genes of B. bifidum in the database (Codon Usage Database at Kazusa DNA Research Institute [http://www.kazusa.or.jp/codon/]), and Southern hybridization analysis with the 2.3-kb KpnI fragment as a specific probe revealed that the afcA gene exists as a single copy on the genome of B. bifidum JCM1254 (data not shown).
Analysis of the primary structure by the use of the SignalP (http://www.cbs.dtu.dk/services/SignalP-2.0) (26) and PSORT (http://psort.ims.u-tokyo.ac.jp) (24) programs revealed the presence of a signal peptide and a membrane anchor at the N and C termini, respectively. Cell surface anchoring motifs such as LPXTG and GW repeats (25) were absent in the amino acid sequence. When B. bifidum cells grown in 500 ml of GAM medium were subjected to subcellular fractionation (38), 6.2 mU of α-l-fucosidase activity was detected in the cell wall and membrane fractions, 2.2 mU was detected in the culture fluid fraction, and 0.32 mU was detected in the cytoplasmic fraction. These results were considered to well reflect its sequential features. A possible ribosome-binding site was located 6 bp from a probable initiation codon, and a promoter-like sequence was also found in the upstream region as deduced with the Genetyx-Mac 10.1 software. The AfcA protein consists of 1,959 amino acid residues with a calculated molecular mass of 205 kDa.
Domain structure.
The primary structure of AfcA did not exhibit any similarity to those of known glycosidase families; therefore, we attempted to localize a catalytic domain that is essential for the hydrolysis of 2′-fucosyllactose. The fact that an E. coli strain with pSA3 (carrying an artificial tet′-′afcA translational fusion) showed 1,2-α-l-fucosidase activity indicated that the catalytic domain was located within this insert (downstream of base 1721 [Fig. 2A]). The N-terminal and C-terminal deletion mutants were constructed and expressed as described in Materials and Methods, and then their activities were assessed. Consequently, as shown in Fig. 2, the N-terminal 576 amino acid residues (bases 911 to 2638) and the C-terminal 485 amino acid residues (bases 5333 to 6790) were found to be removable without loss of fucosidase activity. Thus, it was revealed that the region consisting of amino acid residues 577 to 1474 (Fig. 2B) constitutes the catalytic domain. A detailed description of this domain (the Fuc domain) with respect to enzymatic properties and sequence similarity with other proteins is described below.
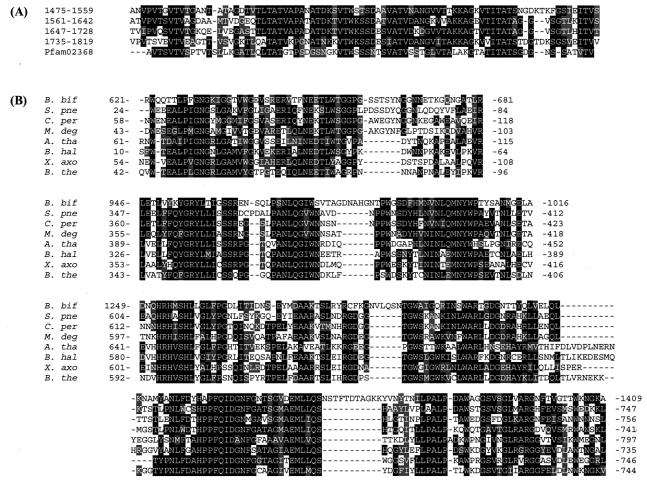
The region of amino acid residues 1475 to 1728 contained four repetitive sequences with immunoglobulin (Ig)-like folds, the so-called bacterial Ig-like domain B (Pfam 02368) (Fig. 3A). Although the function of this domain is not clear, it is highly likely that this domain, 254 amino acids in length, at least acts to display the fucosidase domain of AfcA so that it protrudes from the cell surface, thereby enabling B. bifidum cells to gain access to and degrade the fucosyl residues present on glycoconjugates of enterocytes.
FIG. 3.
Comparison of repetitive sequences in the Ig-like domain of AfcA with the consensus sequence of Pfam 02368 (A) and alignment of the amino acid sequences of the Fuc domain of AfcA and homologous proteins in the database (B). (A) The four repetitive sequences constituting the bacterial Ig-like domain were aligned with the consensus sequence of bacterial Ig-like domain group 2. White letters on a black background indicate identical residues in more than three sequences, and conservative amino acid changes are shown in gray. (B) The primary structure of the Fuc domain was aligned with those of homologues by using the Clustal W program (43), and highly homologous regions are shown. White letters on a black background indicate identical residues in more than four sequences, and conservative amino acid changes are shown in gray. B. bif, AfcA of B. bifidum JCM1254; S. pne, S. pneumoniae R6 (GenBank accession number NP_359091) (15); C. per, C. perfringens strain 13 (GenBank accession number NP_562791) (36); M. deg, M. degradans 2-40 (GenBank accession number ZP_00064866); A. tha, A. thaliana (GenBank accession number AY125494); B. hal, B. halodurans (GenBank accession number NP_241708) (41); X. axo, X. axonopodis pv. Campestris strain ATCC 33913 (GenBank accession number NP_637123) (6); B. the, B. thetaiotaomicron VPI 5482 (GenBank accession number NP_809923) (47).
For the N-terminal domain (amino acid residues 1 to 576), neither sequence similarity to other ORFs nor a functional motif was found in the sequence.
Characterization of the Fuc domain.
We tried to overexpress and purify the AfcA protein; however, probably due to the extremely long length of the protein, an intact form of the protein was not obtained. Therefore, the catalytic domain (Fuc domain) was expressed and purified as described in Materials and Methods and then used for characterization.
The ability of the Fuc domain to hydrolyze the α-(1→2)-fucosidic linkage of 2′-fucosyllactose was further confirmed by ESI-MS and NMR analyses. When the substrate was incubated in the presence of the purified enzyme and subjected to ESI-MS analysis, two major peaks appeared at m/z 355.0936 and 701.1820, which correspond to the sodium adducts of acetylated fucose [M + Na]+ (355.1005) and of acetylated lactose [M + Na]+ (701.1905), respectively, whereas only one peak appeared at m/z 931.2735 in the case of the substrate alone (the sodium adduct of acetylated 2′-fucosyllactose, 931.2695). The NMR spectrum of the hydrolysate was exactly the same as that of the solution containing the commercially available l-fucose and d-lactose; the peaks of the anomeric regions are shown in Fig. 4. Thus, the protein with the novel amino acid sequence (the Fuc domain) was found to indeed be 1,2-α-l-fucosidase.
FIG. 4.
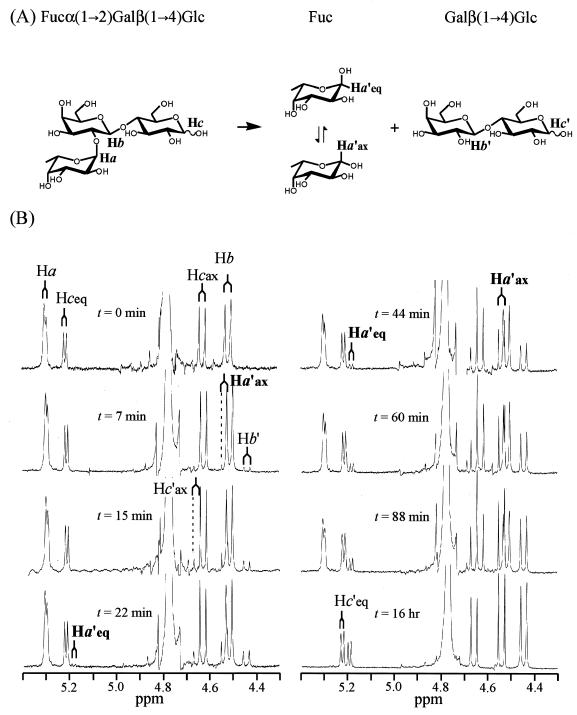
1H NMR spectra showing the stereochemical course of the hydrolysis of 2′-fucosyllactose by 1,2-α-l-fucosidase. (A) Reaction scheme and assignment of the anomeric protons. (B) The reaction was monitored by 1H NMR at different times, and the signals for the region of anomeric protons (4.3 to 5.4 ppm) are shown. The resonance signal of the axial anomeric proton of fucose (Ha′ax) (δ 4.54, J = 8.1 Hz) (β-l-fucose) appeared first (t = 7 min), followed by the appearance of the signal of the equatorial anomeric proton (Ha′eq) (δ 5.19, J = 3.9 Hz) (α-l-fucose) (t = 22 min) as a consequence of mutarotation. See the text for a detailed explanation of the chemical shifts.
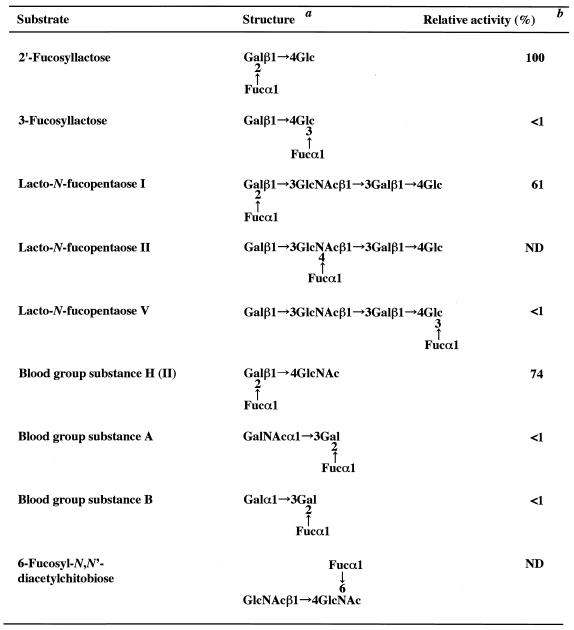
Next, the substrate specificity was examined by using naturally occurring substrates. As shown in Table 1, of the milk oligosaccharides examined, the most readily hydrolyzed substrates were 2′-fucosyllactose and lacto-N-fucopentaose I, both of which contain l-fucose bound to galactose through the α-(1→2) linkage at the nonreducing termini. The enzyme showed a very limited activity for α-(1→3)-linked l-fucosyl residues of 3-fucosyllactose and lacto-N-fucopentaose V and apparently had no action at all on the α-(1→4) linkage of lacto-N-fucopentaose II. The enzyme did not hydrolyze 3-fucosylgalactose [Fucα(1→3)Gal] or 4-fucosyl-N-acetylglucosamine [Fucα(1→4)GlcNAc] (data not shown). Blood group H(II) active substance [Fucα(1→2)Galβ(1→4)GlcNAc] was found to be a good substrate; however, the enzyme showed very limited action on blood group A and B active substances, which have α-(1→3)-GalNAc and α-(1→3)-Gal residues, respectively, in addition to α-(1→2)-Fuc residues, at the nonreducing termini. The α-(1→6) linkage of 6-fucosyl-N,N′-diacetylchitobiose was not cleaved by this enzyme. In addition to fucose-containing oligosaccharides, a high-molecular-weight glycoprotein with terminal α-(1→2)-linked l-fucosyl residues, e.g., porcine gastric mucin (H), was also found to be a substrate of the Fuc domain (data not shown).
TABLE 1.
Substrate specificity of the Fuc domain of AfcA from B. fifidum JCM1254
Gal, d-galactose; Glc, d-glucose; Fuc, l-fucose; GlcNAc, N-acetyl-d-glucosamine; GalNAc, N-acetyl-d-galactosamine.
The reaction mixture, consisting of 100 mM sodium phosphate (pH 6.5), 2 mM substrate, and 50 mU of the purified Fuc domain in a total volume of 100 μl, was incubated for 1 min to 12 h at 37°C. The value obtained with 2′-fucosyllactose was taken as 100%. ND, not detectable.
The initial velocity of the hydrolysis of 2′-fucosyllactose (1.5 mM) was not affected by the addition of monosaccharides (N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, dl-arabinose, dl-fucose, dl-galactose, d-glucose, d-mannose, l-rhamnose, and dl-xylose) at a final concentrations of 10 mM, but it was significantly decreased in the presence of the disaccharide Fucα(1→2)Gal; HPLC and PA-2′-fucosyllactose were used for analysis because some of the monosaccharides are known to act as substrates of FDH. The enzyme are inactive against various N-linked-type oligosaccharides [the terminal linkages examined were Manα(1→2)Man, N-acetylneuraminic acid α(2→3)Gal, Manα(1→3)Man, Manα(1→6)Man, and GlcNAcβ(1→2)Man]; terminal linkages of Forssman pentasaccharide [GalNAcα(1→3)GalNAc], globotriose [Galα(1→4)Gal], asialo GM1 tetrasaccharide [Galβ(1→3)GalNAc], and globo-N-tetraose [GalNAcβ(1→3)Gal], and the linkage of Galα(1→3)Gal (all of these were analyzed by HPLC with PA-labeled sugar chains). The Fuc domain did not liberate fucose from any of the artificial substrates examined (pNP-α-l-fucoside, pNP-β-l-fucoside, and 4-methylumbelliferyl-α-l-fucoside) and did not hydrolyze PA-labeled disaccharides Glcβ(1→4)Glc, GlcNAcβ(1→4)GlcNAc, Glcα(1→2)Glc, and Glcα(1→4)Glc.
Judging from these results, it was concluded that the Fuc domain is highly specific for the terminally linked α-(1→2)-fucosyl residue. The catalytic domain showed maximum activity at pH 5, suggesting the involvement of an acidic residue(s) in the catalysis, and was stable below 35°C for 30 min and at pH 6.5 to 7.5 for 12 h. The Km and Vmax values for 2′-fucosyllactose were determined to be 0.53 mM and 1.6 μmol/min/mg, respectively.
Stereochemistry of hydrolysis of 2′-fucosyllactose by AfcA.
In order to determine the stereochemical course of the reaction catalyzed by AfcA, the hydrolysis of 2′-fucosyllactose was monitored by 1H NMR. Figure 4 shows the resonance signals of the anomeric protons of the substrate 2′-fucosyllactose (Ha, Hb, and Hc) and released products fucose and lactose (Ha′, Hb′, and Hc′) at different reaction times. A reference spectrum (t = 0 min) was recorded before the enzyme was added to the reaction mixture. The doublets centered at δ 4.63 (J = 8.1 Hz) and at δ 5.22 (J = 3.6 Hz) are from the axial and equatorial protons (Hcax and Hceq), respectively, of the glucose residue of the substrate. The peak of Hbax of the galactose residue is observed at δ 4.52 (J = 7.8 Hz), and the resonance of Haeq of the fucose moiety is at δ 5.31 (J = 2.4 Hz). The large signal around δ 4.8 is from HDO.
After the addition of the enzyme, the Ha′ax doublet of liberated fucose (β-l-fucose) appeared at δ 4.54 (J = 8.1 Hz) (t = 7 and 15 min), and simultaneously, the signals for Hb′ (δ 4.45, J = 7.8 Hz, axial) and Hc′ (δ 4.68, J = 8.1 Hz, axial) of lactose appeared. The Hc′eq doublet of lactose overlapped with the Hceq doublet of 2′-fucosyllactose (δ 5.22, J = 3.6 Hz). At 22 min, a small doublet (δ 5.19, J = 3.9 Hz) corresponding to Ha′eq of the liberated fucose (α-l-Fuc) appeared as a consequence of mutarotation of the initial product β-l-fucose at an α/β ratio of 9/91. As the reaction proceeded (t = 44, 60, and 88 min), the resonance signals derived from the liberated products were increased while those from the substrate were decreased, and the α/β anomer ratio of the liberated l-fucose gradually changed (α/β ratios of 19/81, 21/79, and 23/77). The spectrum at 16 h indicates completion of the hydrolysis as well as equilibration of the mutarotation of the liberated Fuc (α/β ratio of 28/72). These data clearly show that the hydrolysis catalyzed by AfuA proceeds with inversion of the anomeric configuration.
Quite recently, α-l-fucosidase of GH family 29 was revealed to be a retaining enzyme by means of chemical rescue of an inactive mutant by azide (4) and also by trapping a covalently attached fucosyl-enzyme intermediate (42). Thus, in this regard too, B. bifidum 1,2-α-l-fucosidase is different from other fucosidases of GH family 29.
Structural similarity of the Fuc domain to hypothetical proteins in the database.
The primary structure of the Fuc domain was subjected to a BLAST search, but no significant similarity with proteins with defined functions was found (12). Scrutiny of the AfcA sequence in reference to conserved motifs of GH family 29 did not lead to the finding of any indications of the family; instead, a very distant relationship to GH families 15 and 65 was observed in a PSI-BLAST search (Bernard Henrissat, personal communication).
In a BLAST search, high scores (score, >307 bits; expected value, <5e-82 in BLAST) were obtained with several hypothetical proteins, the amino acid sequences of which are compared and the highly homologous regions are shown in Fig. 3B. Of particular interest is the finding that the Fuc domain is homologous to CPE1875 in the genome sequence of Clostridium perfringens strain 13 (36), because it has been shown that C. perfringens produces a high-molecular-mass (ca. 200 kDa) 1,2-α-l-fucosidase in culture medium (1). The deduced amino acid sequence of CPE1875 has a typical signal sequence at the N terminus (26), and its molecular mass is estimated to be 165 kDa. Thus, CPE1875 might encode 1,2-α-l-fucosidase.
Other homologous proteins were found from the genome sequences of Streptococcus pneumoniae R6 (15), Bacteroides thetaiotaomicron VPI-5482 (47), Microbulbifer degradans 2-40, Bacillus halodurans (41), Xanthomonas campestris pv. Campestris ATCC 33913 (6), and Arabidopsis thaliana. Among these, B. thetaiotaomicron is also known to secrete α-l-fucosidase (47). Although it remains to be elucidated whether these homologues indeed show 1,2-α-l-fucosidase activity, the finding of a new conservative sequence indicates the existence of a novel GH family (GH family 95) (Bernard Henrissat, personal communication).
In conclusion, the B. bifidum afcA gene encoding 1,2-α-l-fucosidase has been cloned, and its DNA sequence was determined. The AfcA protein, consisting of 1,959 amino acid residues, can be divided into three domains: the N-terminal domain with unknown function, the catalytic domain, and the C-terminal bacterial Ig-like domain. The purified catalytic domain specifically hydrolyzed the terminal α-(1→2)-fucosidic linkages of oligosaccharides and glycoproteins through the inverting mechanism. The primary structure of the catalytic domain exhibited similarity to those of ORFs of unknown function. These results revealed the existence of a novel glycosidase family.
Although B. bifidum cells cannot ferment l-fucose, occurrence of α-l-fucosidase does benefit the cells, because while B. bifidum cells were capable of degrading and fermenting 2′-fucosyllactose, none of the bifidobacterial strains that do not secrete fucosidase could degrade 2′-fucosyllactose without the aid of exogenously added fucosidase (purified Fuc domain). These results suggest that, in preference to non-fucosidase-producing bacteria, B. bifidum cells are able to degrade several types of substrates present in the intestinal epithelium and mucosa, and they imply the biological importance of a secretory fucosidase of a gut microbe for the intestinal ecosystem.
Acknowledgments
We are very grateful to Bernard Henrissat for valuable comments on the AfcA sequence and to S. Koizumi for providing 2′-fucosyllactose.
This work was partly supported by a Grant-in-Aid for Scientific Research (B), no. 14360055, from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; by a Grant-in-Aid for Scientific Research from Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (JST); and by a Grant-in-Aid for Food Science Research from the Food Science Institute Foundation.
REFERENCES
- 1.Aminoff, D., and K. Furukawa. 1970. Enzymes that destroy blood group specificity. I. Purification and properties of α-l-fucosidase from Clostridium perfringens. J. Biol. Chem. 245:1659-1669. [PubMed] [Google Scholar]
- 2.Bahl, O. P. 1970. Glycosidases of Aspergillus niger. II. Purification and general properties of 1,2-α-l-fucosidase. J. Biol. Chem. 245:299-304. [PubMed] [Google Scholar]
- 3.Chambers, W., S. Thompson, A. W. Skillen, C. O. Record, and G. A. Turner. 1993. Abnormally fucosylated haptoglobin as a marker for alcoholic liver disease but not excessive alcohol consumption or non-alcoholic liver disease. Clin. Chim. Acta 219:177-182. [DOI] [PubMed] [Google Scholar]
- 4.Cobucci-Ponzano, B., A. Trincone, A. Giordano, M. Rossi, and M. Moracci. 2003. Identification of the catalytic nucleophile of the family 29 α-l-fucosidase from Sulfolobus solfataricus via chemical rescue of an inactive mutant. Biochemistry 42:9525-9531. [DOI] [PubMed] [Google Scholar]
- 5.Cohenford, M. A., A. Abraham, J. Abraham, and J. A. Dain. 1989. Colorimetric assay for free and bound l-fucose. Anal. Biochem. 177:172-177. [DOI] [PubMed] [Google Scholar]
- 6.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, Jr., L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. A. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, Jr., E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. N. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, Jr., A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 7.Eneyskaya, E. V., A. A. Kulminskaya, N. Kalkkinen, N. E. Nifantiev, N. P. Arbatskii, A. I. Saenko, O. V. Chepurnaya, A. V. Arutyunyan, K. A. Shabalin, and K. N. Neustroev. 2001. An α-l-fucosidase from Thermus sp. with unusually broad specificity. Glycoconj. J. 18:827-834. [DOI] [PubMed] [Google Scholar]
- 8.Erbe, D. V., S. R. Watson, L. G. Presta, B. A. Wolitzky, C. Foxall, B. K. Brandley, and L. A. Lasky. 1993. P- and E-selectin use common sites for carbohydrate ligand recognition and cell adhesion. J. Cell Biol. 120:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, K. J., and N. N. Aronson, Jr. 1989. Isolation and sequence analysis of a cDNA encoding rat liver α-l-fucosidase. Biochem. J. 264:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushima, H., J. R. de Wet, and J. S. O'Brien. 1985. Molecular cloning of a cDNA for human α-l-fucosidase. Proc. Natl. Acad. Sci. USA 82:1262-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hase, S., T. Ikenaka, and Y. Matsushima. 1978. Structure analysis of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem. Biophys. Res. Commun. 85:257-263. [DOI] [PubMed] [Google Scholar]
- 12.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes, E. W., and J. S. O'Brien. 1979. Separation of glycoprotein-derived oligosaccharides by thin-layer chromatography. Anal. Biochem. 93:167-170. [PubMed] [Google Scholar]
- 14.Horiuchi, T., T. Suzuki, M. Hiruma, and N. Saito. 1999. Purification and characterization of l-fucose (l-galactose) dehydrogenase from Pseudomonas sp. no. 1143. Agric. Biol. Chem. 53:1493-1501. [Google Scholar]
- 15.Hoskins, J. A., W. Alborn, Jr., J. Arnold, L. Blaszczak, S. Burgett, B. S. DeHoff, S. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. Kraft, R. LaGace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. McAhren, M. McHenney, K. McLeaster, C. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilver, D., A. Arnqvist, J. Ogren, I.-M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 17.Katayama, T., H. Suzuki, T. Koyanagi, and H. Kumagai. 2000. Cloning and random mutagenesis of the Erwinia herbicola tyrR gene for high-level expression of tyrosine phenol-lyase. Appl. Environ. Microbiol. 66:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, T., K. Fujita, M. Takeuchi, K. Kobayashi, S. Natsuka, K. Ikura, H. Kumagai, and K. Yamamoto. 2002. Identification of an endo-β-N-acetylglucosaminidase gene in Caenorhabditis elegans and its expression in Escherichia coli. Glycobiology 12:581-587. [DOI] [PubMed] [Google Scholar]
- 19.Kurimura, Y., Y. Tsuji, K. Yamamoto, H. Kumagai, and T. Tochikura. 1995. Efficient production and purification of extracellular 1,2-α-l-fucosidase of Bacillus sp. K40T. Biosci. Biotechnol. Biochem. 59:585-594. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Larson, G., P. Falk, and L. C. Hoskins. 1988. Degradation of human intestinal glycosphingolipids by extracellular glycosidases from mucin-degrading bacteria of the human fecal flora. J. Biol. Chem. 263:10790-10798. [PubMed] [Google Scholar]
- 22.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Muller-Taubenberger, A., M. Westphal, A. Noegel, and G. Gerisch. 1989. A developmentally regulated gene product from Dictyostelium discoideum shows high homology to human α-l-fucosidase. FEBS Lett. 246:185-192. [DOI] [PubMed] [Google Scholar]
- 24.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-35. [DOI] [PubMed] [Google Scholar]
- 25.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 27.Nunoura, N., K. Ohdan, K. Tanaka, H. Tamaki, T. Yano, M. Inui, H. Yukawa, K. Yamamoto, and H. Kumagai. 1996. Cloning and nucleotide sequence of the β-d-glucosidase gene from Bifidobacterium breve clb, and expression of β-d-glucosidase activity in Escherichia coli. Biosci. Biotechnol. Biochem. 60:2011-2018. [DOI] [PubMed] [Google Scholar]
- 28.Occhiodoro, T., and D. S. Anson. 1996. Isolation of the canine α-l-fucosidase cDNA and definition of the fucosidosis mutation in English Springer spaniels. Genome 7:271-274. [DOI] [PubMed] [Google Scholar]
- 29.Ogata-Arakawa, M., T. Muramatsu, and A. Kobata. 1977. α-l-Fucosidase from almond emulsin: characterization of the two enzymes with different specificities. Arch. Biochem. Biophys. 181:353-358. [DOI] [PubMed] [Google Scholar]
- 30.Podolsky, D. K. 1985. Oligosaccharide structures of human colonic mucin. J. Biol. Chem. 260:8262-8271. [PubMed] [Google Scholar]
- 31.Ruiz-Palacios, G. M., L. E. Cervantes, P. Ramos, B. Chavez-Munguia, and D. S. Newburg. 2003. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278:14112-14420. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano, M., K. Hayakawa, and I. Kato. 1992. Purification and characterization of α-l-fucosidase from Streptomyces species. J. Biol. Chem. 267:1522-1527. [PubMed] [Google Scholar]
- 35.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, J.-F., M.-Q. Weng, S.-M. Wu, and Q.-C. Xia. 2002. Analysis of neutral saccharides in human milk derivatized with 2-aminoacridone by capillary electrophoresis with laser-induced fluorescence detection. Anal. Biochem. 304:126-129. [DOI] [PubMed] [Google Scholar]
- 38.Sprott, G. D., S. F. Koval, and C. A. Schnaitman. 1994. Cell fractionation, p. 72-103. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 39.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki, H., T. Katayama, K. Yamamoto, and H. Kumagai. 1995. Transcriptional regulation of tyrosine phenol-lyase gene of Erwinia herbicola AJ2985. Biosci. Biotech. Biochem. 59:2339-2441. [DOI] [PubMed] [Google Scholar]
- 41.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, Y. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarling, C. A., S. He, G. Sulzenbacher, C. Bignon, Y. Bourne, B. Henrissat, and S. G. Withers. 2003. Identification of the catalytic nucleophile of the family 29 α-l-fucosidase from Thermotoga maritima through trapping of a covalent glycosyl-enzyme intermediate and mutagenesis. J. Biol. Chem. 278:47394-47399. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuji, Y., K. Yamamoto, T. Tochikura, T. Seno, Y. Ohkubo, and H. Yamaguchi. 1990. Purification and characterization of α-l-fucosidase from Bacillus circulans grown on porcine gastric mucin. J. Biochem. 107:324-330. [DOI] [PubMed] [Google Scholar]
- 45.Wong-Madden, S. T., and D. Landry. 1995. Purification and characterization of novel glycosidases from the genus Xanthomonas. Glycobiology 5:19-28. [DOI] [PubMed] [Google Scholar]
- 46.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 48.Yano, T., K. Yamamoto, H. Kumagai, T. Tochikura, T. Yokoyama, T. Seno, and H. Yamaguchi. 1985. Purification and characterization of a novel α-l-fucosidase from Fusarium oxysporum grown on sludge. Agric. Biol. Chem. 49:3179-3187. [Google Scholar]