Abstract
Electron tomography is a powerful method for determining the three-dimensional structures of large macromolecular assemblies, such as cells, organelles, and multiprotein complexes, when crystallographic averaging methods are not applicable. Here we used electron tomographic imaging to determine the molecular architecture of Escherichia coli cells engineered to overproduce the bacterial chemotaxis receptor Tsr. Tomograms constructed from fixed, cryosectioned cells revealed that overproduction of Tsr led to formation of an extended internal membrane network composed of stacks and extended tubular structures. We present an interpretation of the tomogram in terms of the packing arrangement of Tsr using constraints derived from previous X-ray and electron-crystallographic studies of receptor clusters. Our results imply that the interaction between the cytoplasmic ends of Tsr is likely to stabilize the presence of the membrane networks in cells overproducing Tsr. We propose that membrane invaginations that are potentially capable of supporting axial interactions between receptor clusters in apposing membranes could also be present in wild-type E. coli and that such receptor aggregates could play an important role in signal transduction during bacterial chemotaxis.
Over the last three decades, methods for three-dimensional reconstruction of objects (5) imaged with an electron microscope have been used to determine the structures of a variety of biological assemblies by two types of approaches. One approach, which has been used extensively in analyses of large macromolecular assemblies, involves three-dimensional reconstruction of a structure by averaging images recorded from several identical copies oriented randomly relative to the electron beam (11, 31). The other approach, which has been used for reconstruction of objects that cannot be easily averaged, such as whole cells, involves tomographic reconstruction by combining projection images of an object recorded with an electron microscope over a range of tilt angles (4). Electron tomography is therefore a potentially powerful tool for three-dimensional imaging of the spatial arrangement of proteins that make up complex and dynamic assemblies, such as those involved in bacterial chemotaxis.
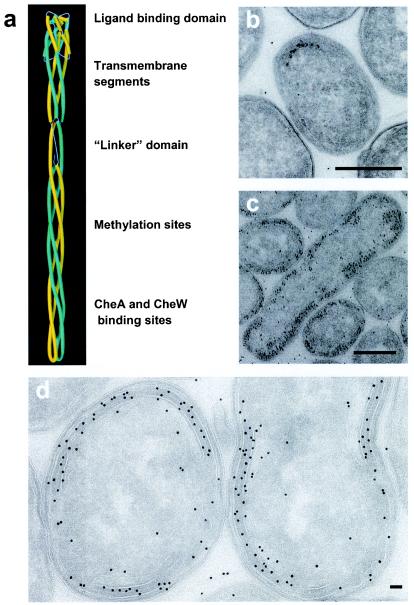
At least 12 proteins act in concert to convert the signal of ligand binding at the periplasmic end of a chemotaxis receptor into rotation of the flagellar motor (6, 27). The principal protein components at the input end include one of the chemotaxis receptors (Tsr, Tar, Trg, Tap, or Aer), and the cytoplasmic signaling proteins CheA and CheW, which are thought to form a noncovalent complex with the chemotaxis receptors. Knowledge of the structure and spatial arrangement of the chemotaxis receptors is therefore fundamental to understanding the structural biology of signaling. X-ray crystallographic studies of the periplasmic fragments of the aspartate receptor fragments have revealed the dimeric organization of the ligand binding domain, in which the ligand binding pocket is located at the dimer interface (20). Structural studies of a cytoplasmic fragment of the related Tsr receptor have shown that it also forms a dimer of extended coiled-coil hairpins (13). The cytoplasmic fragments pack in the three-dimensional crystals used for X-ray crystallographic analysis as trimers of dimers, and this has led to a proposal by Kim et al. (14) that such an arrangement may also be representative of full-length receptors in cell membranes. Figure 1a shows the spatial arrangement of the periplasmic and cytoplasmic domains in the context of an atomic model for the full-length receptor (14) that includes plausible structures for the transmembrane region and the linker region between the membrane domain and the cytoplasmic domain.
FIG. 1.
(a) Structural model for the full-length Tsr dimer based on structures of the periplasmic and cytoplasmic domains, adapted from the work of Kim et al. (14). (b and c) Projection images of thin (70-nm) sections of fixed specimens of either wild-type E. coli (b) or an E. coli Tsr-overexpressing strain (HCB721/pHSe5.tsrQEQE) induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (c). Scale bars = 0.5 μm. (d) Higher magnification of cells overproducing Tsr. Scale bar = 50 nm. The dark spots in the images are from 10-nm gold-conjugated protein A used to locate Tsr.
Ultrastructural studies of bacterial cells in combination with immunoelectron microscopy have revealed that chemotaxis receptors, as well as CheA and CheW, are predominantly localized to the poles of the cells (18, 25). Although biochemical studies have implicated the involvement of extended interactions among receptor subunits in the membrane during signaling (1) and many computational models for clustering have been proposed (19, 24), there is only limited direct structural evidence that such interactions occur under physiological conditions. Approaches such as electron tomography provide tools which can be used to begin to unravel the molecular arrangement of these protein components in the cell because of the potential for integrating structural information from X-ray crystallography with the lower-resolution data obtained from electron microscopic imaging. As a first step in this direction, we obtained three-dimensional images of the assemblies and networks formed in cells by overexpression of the full-length chemotaxis receptor Tsr. Projection images recorded from negatively stained membrane extracts obtained from cells have shown that Tsr is organized in receptor arrays that have the appearance of either zipper-like or micelle-like entities (32). The novel feature of these assemblies was the evidence that there are direct interactions between Tsr molecules both in the plane of the membrane and in the axial direction involving the cytoplasmic domains. In this study, we extended this analysis into the third dimension with a description of the overall architecture of cells engineered to overproduce Tsr. The three-dimensional structural analyses demonstrated that the receptor arrays form an extended membrane network that extends into the cytoplasm, resulting in the formation of extended sheets and tubules, which are converted into the zippered and rounded assemblies seen in the isolated membrane extracts. We also describe similar analyses performed with wild-type cells to evaluate the presence of local invaginations in the cytoplasmic membrane.
MATERIALS AND METHODS
Fixation and embedding of cells.
Escherichia coli strain RP437 was used as the control for wild-type experiments. Plasmids pHSe5.tsrQQQQ and pHSe5.tsrQEQE were used to produce Tsr in HCB721 cells, which do not express the chemotaxis-related proteins Tar, Tsr, Trg, Tap, CheA, CheW, CheR, and CheB, as described previously (32). Harvested cells were fixed at room temperature for 2.5 h in a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in the presence of 60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 50 mM HEPES (pH 6.9), 4 mM MgCl2, and 20 mM EGTA. The fixed cells were collected by centrifugation and resuspended in prewarmed 0.1 M phosphate buffer containing 12% gelatin. After the cell-containing gelatin pellets were solidified on ice, they were cut into 1-mm cubes and incubated with a solution containing 2.3 M sucrose and 0.1 M sodium phosphate buffer (pH 7.4). Cubes of gelatin were frozen on the surfaces of aluminum pins by plunging them into liquid nitrogen and were sectioned with a cryoultramicrotome at −120°C. Labeling with anti-Tsr antibody (which specifically reacts with the conserved signaling domain) and protein A-gold and subsequent embedding in methyl cellulose with uranyl acetate with reagents were carried out as described previously (22).
Electron microscopy.
The projection images shown in Fig. 1 were recorded with a Gatan 2K charge-coupled device camera mounted on a Tecnai 12 electron microscope (FEI Corporation, Hillsboro, Oreg.) equipped with an LaB6 filament operating at 120 kV. For tomography, a series of images were recorded at room temperature with the aid of a Gatan 2K charge-coupled device (magnification, ∼×47,500) by tilting the specimen from −70° to 70° in increments of 0.5° in a Tecnai F30 microscope equipped with a field emission gun tip operating at 300 kV. Images were recorded at underfocus values that were between 2 and 3 μm along the tilt axis. A back-projection algorithm, as implemented in the IMOD reconstruction package (15), was used to convert the information present in the series of tilted projection images into three-dimensional density maps.
Preparation of Tween 80-extracted membranes.
Membrane preparations (16) isolated on sucrose gradients were typically incubated with Tween 80 at a protein/Tween 80 molar ratio of 0.004 for about 4 h as described previously (32).
Segmentation and rendering.
The tomogram was segmented in the environment of the program Amira (TGS Inc., San Diego, Calif.) by marking all regions in the volume where the bilayer (white lines in the slice) could be visualized clearly in three dimensions. An isosurface was created by tracing the path of the bilayer in each slice of the tomogram. Structural models of the two types of receptor assemblies shown in Fig. 3d and e were docked onto the isosurface by using the program 3dsmax (DISCREET, Montreal, Quebec, Canada). The coordinates for the receptor dimer were the coordinates in the model described by Kim et al. (14) and kindly provided by Sung-Hou Kim. Starting from the model of the dimer, a variety of plausible arrangements for higher-order arrangements, such as the trimer of dimers shown in Fig. 3d and e, were then generated by using the electron microscopic images as a guide. One set of the plausible arrangements is shown in Fig. 3.
FIG. 3.
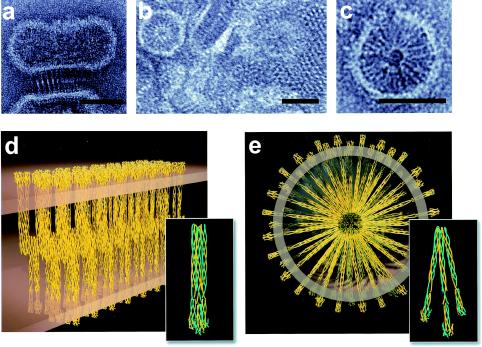
(a to c) Negatively stained electron micrographs obtained from a Tween 80 extract of cells expressing Tsr. The extract preserved the overall appearance of the membrane morphologies in the tomogram, and the images show in more detail the organization of the zippered structures in cross section (a) and top view (b), as well as rounded vesicular structures (c). Scale bars = 50 nm. (d) Molecular model for packing in the zippered regions based on the atomic model for the structure of Tsr shown in Fig. 1a and the electron crystallographic analysis of the crystalline regions (32). The packing in the crystalline areas of the membrane (b) roughly corresponds to a lattice with the following constants: a, ∼75 Å; b, ∼75 Å; and γ, ∼60°. The cross-sectional area of each unit in the crystal is ∼5,000 Å2, which can accommodate at most three Tsr dimers, based on the presence of four helices per Tsr monomer at the wider, periplasmic end of the receptor (20) and the known values of about 180 Å2 that characterize the cross-sectional areas of helices in well-packed two-dimensional crystalline arrays (12). The array shown is one of many possible arrangements of the three dimers and is presented primarily to provide an indication of the density of receptor packing in the membrane. The white regions at the center of the zippered structure in panel a are interpreted as high-density regions where the cytoplasmic ends of the Tsr trimer are presumed to be interdigitated. The depth of the interdigitation is consistent with the proportional width of this region in the micrograph in panel a. The average internal spacing of the zippered structures in tomographic slices such as the slice shown in Fig. 2c is 31.5 + 2.5 nm (averaged over 26 separate measurements). This compares well with the value for the same internal spacing in these negatively stained specimens (31.3 ± 0.6 nm [averaged over 10 measurements], as reported by Weis et al. [32]). (e) Molecular model for packing in the vesicular regions (sectional view). The outer white ring of the circular structure in panel c is interpreted as the surface of the membrane region enclosing the volume, and the less clear inner ring is interpreted as a high-density region where the cytoplasmic ends of Tsr come together.
RESULTS
We carried out a three-dimensional structural analysis of E. coli cells engineered to overexpress the 60-kDa membrane protein Tsr (16) in order to explore whether the novel assemblies observed in membrane preparations isolated from disrupted cells (32) could also be detected in intact cells and imaged. Immunolabeling of fixed cryosections of wild-type E. coli (Fig. 1b) showed that Tsr was predominantly located at the polar region(s) of the cells, which is consistent with previously reported data (18). In contrast, in cells overproducing Tsr, the receptor was located throughout the periphery and in some interior regions of the cell (Fig. 1c). In all areas where the immunolabel appeared to be localized to the interior of the cell, there was evidence of membranous material, implying that the high level of Tsr resulted in extension of the membrane so that it was no longer confined to the periphery of the cell. A closer examination of a region of the cell displaying invaginations (Fig. 1d) confirmed that the immunolabel, which was directed against the C-terminal region of Tsr, was localized primarily to the interior portions of the zippered regions.
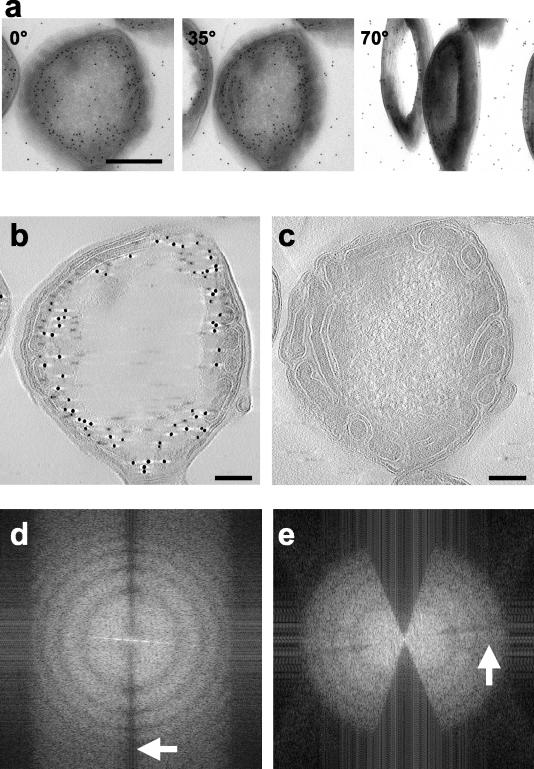
In order to investigate the three-dimensional structure of the cells overproducing Tsr, we recorded a series of projection images by tilting the specimen relative to the electron beam, typically over a range of ±70° (Fig. 2a). The images were then combined by using weighted back-projection methods to reconstruct the three-dimensional structure. Representative slices from an electron tomogram constructed from images such as those shown in Fig. 2a are shown in Fig. 2b and c. These slices provide a glimpse of the extensive internal organization of the membranes and the improved clarity relative to the projection images. The slice in Fig. 2b is close to the upper edge of the section on the grid and shows extensive labeling of Tsr, as expected. The slice in Fig. 2c is from an interior region of the section, and the absence of immunolabeling reflects the lack of penetration of the anti-Tsr antibody and/or the protein A-gold conjugates into the section. An MPEG movie of the tomogram is available at http://hrem.nci.nih.gov/Lefman_et_al_J.Bact_2004/E.coli_tomogram.
FIG. 2.
(a) Selected projection images (recorded at tilt angles of 0°, 35°, and 70°) from a 250-nm-thick section obtained from cells overproducing Tsr. (b and c) Slices from an electron tomogram constructed by using a series of tilted images from −70° to 70° at tilt intervals of 0.5°. The slice in panel b is from a region closer to the surface of the section, while the slice in panel c is from an interior region that is not accessible to the protein A-gold particles. Bars, 0.17 μm (b and c). (d) Computed Fourier transform from a region in the x-y slice of the tomogram, showing Thon rings at a resolution of 15 Å−1 in the x-y plane (indicated by the arrow). The region of the slice used to calculate the transform contains a flagellar filament whose helical structure is responsible for producing the elongated spots in the transform in the horizontal direction. (e) Transform in the x-z plane from a similar-size region, showing the expected missing wedge due to the absence of data between tilt angles of 70° and 90° and Thon rings at a resolution of ∼22 Å−1 (indicated by the arrow). The edge of the transform in panels d and e is at 13 Å−1.
A measure of the quality of both the collected data and the reconstruction procedure was obtained from analysis of a small region extracted from the central region of the tomogram used to obtain the images shown in Fig. 2. The Fourier transforms (Fig. 2d and e) computed from individual horizontal (x-y) and vertical (x-z) slices in the tomogram showed that there were well-defined Thon rings (30). The first zero crossing of the contrast transfer function was at ∼40 Å−1, and the spacing of the rings matched the values expected for the defocus values used (∼2.6-μm underfocus along the tilt axis). The overall contrast in the slice arises primarily from the presence of the uranyl acetate used for staining. However, the intrinsic resolution limit in a given slice of the tomogram relevant for detection of molecular structures is expected to be limited by the damage that occurred during specimen preparation and by the radiation-induced damage that occurs upon illumination of the specimen. For these reasons, the tomograms were not corrected for the effects of the contrast transfer function.
Inspection of the tomographic slice in Fig. 2c suggests that the internal volume is laced with an extensive network of twisted sheet-like features that have the appearance of stacked layers in certain regions and rounded features that appear to be vesicular in cross section in other regions. From their location and continuity with the cytoplasmic membrane, it is clear that the white lines defining the boundaries of the stacks and the rounded structures represent the bilayer membrane. The formation of these structures is a consequence of invaginations of the surface of the cytoplasmic membrane in response to the high level of expression of Tsr. The uniformity of the spacing between the membrane layers (there was <5% variation across the length of an individual stretch) in the stacks is consistent with the idea that the membrane network is stabilized by specific interactions between protein components in the bilayer membrane. Since these extended stacks and rounded structures were not observed in either wild-type E. coli or control cells not expressing Tsr, the simplest explanation is that they are formed in response to the high concentration of Tsr in the membrane and are stabilized by interactions between Tsr molecules located in adjacent bilayer membranes.
Further structural insights into the molecular organization of the membrane stacks were developed from analyses of membrane extracts obtained from lysed cells (32). Tsr represents about 90% of the polypeptide present in these membrane extracts (16). Micrographs (Fig. 3a to c) obtained from negatively stained membrane specimens treated with small amounts of the detergent Tween 80 (at molar ratios of 1:250) indicated that there was a rich variety of connected membranous morphologies (see the legend to Fig. 3 for details of the dimensions), with the principal components being represented by (i) zipper-like features (Fig. 3a) with dimensions matching those observed for the cross sections of the sheet-like regions detected in the tomogram, (ii) small crystalline arrays (Fig. 3b) with pseudohexagonal ordering, and (iii) rounded structures (Fig. 3c) which had a micellar appearance in the projection images and had a radius of curvature similar to that observed for the rounded structures in the tomogram. Previous electron crystallographic analyses (32) have suggested that the packing of the small crystalline patches in the plane is consistent with an arrangement in which the repeating unit has three Tsr dimers with a packing density of approximately 200 Å2/helix. The overall appearance of these structures is thus similar to the appearance of structures seen in cells, although the detergent treatment may have resulted in a slight enhancement of the packing density of protein in the membranes.
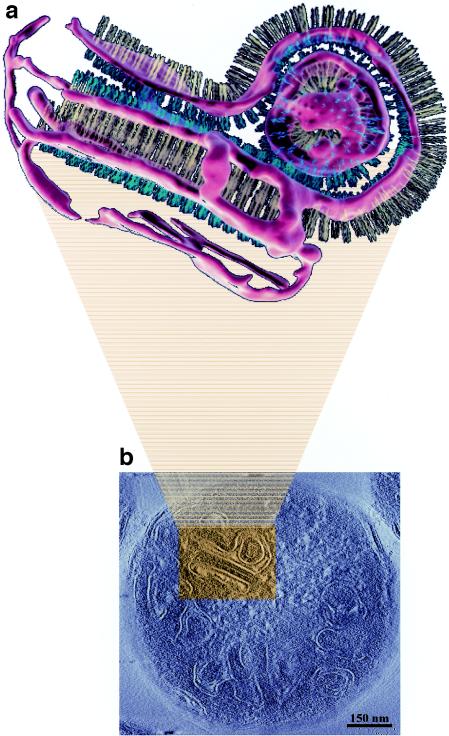
The constraints for receptor packing in the isolated membrane preparations suggest how the interactions between Tsr cytoplasmic domains from two adjacent bilayers could lead to the formation of a pair of stacked membranes whose appearance (Fig. 3d) could correspond to that of a sheet (membrane stack) in a tomogram (Fig. 2c) and a zipper (Fig. 3a) or quasicrystalline array (Fig. 3b) in a projection view. Similarly, it is possible to derive a plausible molecular model for the rounded structures (Fig. 3e) in which the lengths of the cytoplasmic domain of Tsr are well-matched with the experimentally measured radii in the electron micrographs (Fig. 3c) and with packing densities of the cytoplasmic ends of Tsr in its interior comparable to the densities observed in the zipper-like structures. Construction of plausible models for these assemblies allowed placement of the assemblies into the tomographic volume, thereby providing a powerful tool for interpreting the large amount of information contained in the tomogram at the molecular level (Fig. 4).
FIG. 4.
(a) Segmented representation of a small region from a cellular tomogram. (b) Slice of the tomogram. The region indicated by the box (width, 230 nm) corresponds to a slice in the region that was segmented. Aside from the interaction between the cytoplasmic domains (yellow) in apposing bilayers, a complementary interaction between the periplasmic domains (blue) also appears to be likely. The uniformity in spacing between the rounded vesicular region and the invaginated membrane surrounded by it suggests that the entire membrane network is stabilized by interactions at both ends of the receptor. The segmented region is composed of two sets of stacked membranes. The lower membrane sheet is wrapped around a tubular invagination (see panel 4 in Fig. 5 for a schematic representation of the geometry). The segmentation was carried out to be faithful to the density in the tomogram and is therefore noisy, as seen, for example, in the patchy appearance of the rounded end of the tubular invagination.
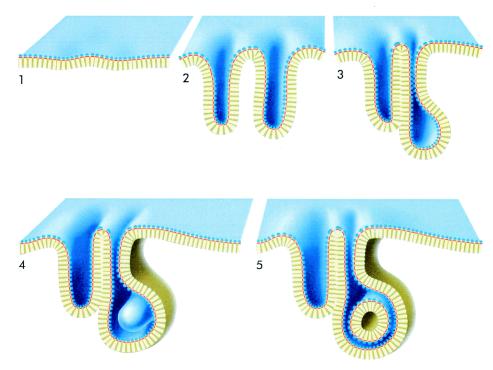
What is the mechanism underlying the formation of these structures? In wild-type E. coli, the surface area covered by the cytoplasmic membrane is adequate to accommodate normal levels of expressed membrane proteins and lipids. In the cells overproducing Tsr, as the Tsr levels began to exceed the amount of membrane proteins normally present in the cell, the membrane invaginates in order to accommodate the excess protein (Fig. 5, panels 1 and 2). Evidence for these intermediate stages is clearly present in other regions of the tomographic slice shown in Fig. 2c. The interaction between the invaginated membranes can thus lead to the formation of two kinds of stacking interactions (Fig. 5, panels 3 and 4) depending on whether they involve the periplasmic side (narrower spacing) or the cytoplasmic side (wider spacing). The interactions at the periplasmic domain may help accommodate the unique packing interaction between complementary curved surfaces (i.e., between the rounded ends at the ends of two separately invaginated regions). As shown in Fig. 5, panel 5, these interdigitated surfaces can sometimes suggest the presence of free-standing vesicular regions in a given slice. However, all such regions inspected by tomography were found to represent fortuitous cross sections of tubular invaginations arising from a different plane of the membrane.
FIG. 5.
Schematic illustration of the formation of membrane networks such as the network shown in Fig. 3. (Panel 1) Cytoplasmic membrane of wild-type cell or overproducing cell when the levels of Tsr expression are not very high. (Panel 2) Invaginations of the cytoplasmic membrane at different regions to accommodate the increased levels of Tsr by increasing the ratio of membrane surface to cytoplasmic volume. (Panel 3) Formation of two kinds of interactions between proximal invaginated membranes. The interactions between the cytoplasmic domains (yellow) in adjacent membranes lead to a zipper-like structure with wider spacing (about 270 Å), while the interactions between the periplasmic domains lead to the zipper-like structure with narrower spacing (about 150 Å). Note that the diagram is drawn so that it indicates that not all regions of the membrane are part of the zippered structures. (Panel 4) The rounded regions at the ends of the invaginations are occasionally large enough to accommodate an invagination from a different region of the membrane, as shown by the rounded tubule beginning to emerge from the backside. (Panel 5) The secondary invagination can become large enough that its growth is limited by the boundaries of other surrounding membranes. A cross-sectional view through such a region would have the appearance of a free-standing vesicular region. This diagram corresponds to the tomogram shown in Fig. 4b (rotated clockwise about 70°).
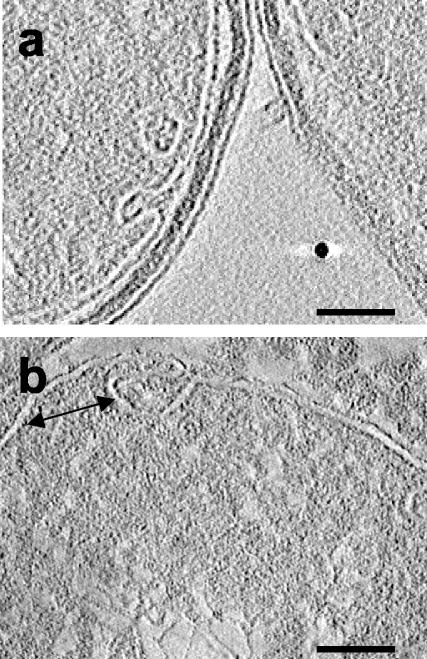
Are the modes of interaction which we observed in cells overproducing Tsr relevant to chemotaxis receptors in wild-type cells? While the resolution of electron tomograms is not adequate at present to detect individual Tsr dimers in wild-type cells, we reasoned that a prerequisite for the formation of axially interacting receptor clusters would be local curvature variations in the cytoplasmic membrane which might be detectable in tomograms. Tomograms recorded from wild-type cells (RP437) indeed showed the presence of small local invaginations, as indicated in two representative slices shown in Fig. 6. These invaginations were about five times more abundant in the polar regions of the cell, and their overall frequency of occurrence was low; fewer than 10 such invaginations were typically observed in tomograms of 1,000-Å-thick slices of wild-type cells. This is not surprising since the level of expression of Tsr in the overproducing strain is ∼2 orders of magnitude higher than the combined levels of expression of all chemotaxis receptors in wild-type E. coli. The dimensions of some of the invaginations (Fig. 6a) appear to be too small to accommodate the zippered assemblies, such as those shown in Fig. 4, or the receptor-CheA-CheW complexes observed in vitro by mixing purified components (10). However, other invaginations (Fig. 6b) resulted in membrane geometries that were, in principle, long enough to accommodate such a longitudinal receptor-transducer assembly. Invaginations such as those seen in wild-type cells were also observed in control cells (HCB721) lacking chemotaxis receptors, indicating that chemoreceptors are not required in the formation of these invaginations. At present, it is not possible to establish whether some of the invaginations in wild-type cells contain axially interacting clusters of chemoreceptors. We also cannot exclude the possibility that the invaginations in the wild-type cells were an artifact induced by chemical fixation. Nevertheless, we think that it is conceivable that the initial formation of the different shapes could be an intrinsic property of the cytoplasmic membrane that is preferential to polar regions of the such cells. Once they are formed, such invaginations could provide a structural context for axial interaction signaling between receptors which may already be clustered (1) in the plane of the membrane. Structural analyses of cells fixed by rapid freezing instead of chemical methods, as well as resolution improvements in tomography, are likely to provide a greater understanding of the structure and arrangement of chemoreceptor assemblies in bacterial cells.
FIG. 6.
Representative slices from tomograms of sections obtained from wild-type cells processed in the same way that the strain overproducing Tsr was processed. Invaginations in the cytoplasmic membrane are evident in both tomograms. The invagination in panel a appears to be too small to accommodate an axially interacting cluster of chemotaxis receptors, while the invagination in panel b has the potential to accommodate such an interdigitated receptor cluster at the locations indicated by the arrow (width, ∼50 nm). Scale bars = 50 nm.
DISCUSSION
It is instructive to identify specific features revealed by the tomographic analysis that are not evident in projection images recorded from isolated membrane assemblies. One of the goals of the tomographic studies was to understand the nature of membrane continuity in the cells overproducing Tsr. Two-dimensional images do not allow determination of whether the striations and rounded regions observed in images such as those in Fig. 3a to c result from free or connected entities in the cell. Our tomographic analysis revealed that in all cases examined, the invaginations showed evidence of physical continuity with the cytoplasmic membrane. An illustration of the insight gained by this approach is shown in Fig. 5, panel 5, which suggests how a series of sectional views alone could lead to an incorrect interpretation. Another new insight from the tomography analysis is the identification of a well-preserved shorter spacing corresponding to the face-to-face interactions between the periplasmic sides of the membrane (Fig. 4). This interaction, which is presumably weaker than the interdigitating interaction on the cytoplasmic side, was an unexpected finding and explains how a three-dimensional network of membranes could be stabilized in the cell by combining the cytoplasmic and periplasmic pairs of interactions. Finally, it is important to note that there is significant improvement in resolution in a tomographic slice compared to the resolution in a projection image. For example, the image in Fig. 2c, which corresponds to a 1.4-nm-thick computational slice, shows more structural detail than one would expect from a conventional thin section that is 30 to 50 nm thick.
The accumulation of membrane-rich structures as a result of overproduction of membrane proteins has been documented previously for both prokaryotic cells (3) and eukaryotic cells (2). There are also examples of membrane deposits that have been found naturally, such as the deposits found in certain cyanobacteria (23). Nevertheless, the observation of these extended structures provides an interesting view of the effects of overexpression of membrane proteins in a bacterial cell. It is very likely that high levels of expression of a membrane protein may be required before invaginations form, but it is equally possible that high levels of expression can be sustained in this case because of the stabilization of the membrane by the interactions which we describe here. The receptor arrangements reflect the intrinsic tendency of Tsr to associate, and extension of these studies to analysis of cells overproducing Tsr, CheA, and CheW is likely to provide further insights into the organization of receptor-transducer assemblies in chemotaxis.
The presence of the invaginations in wild-type cells could provide a new dimension to considerations of the molecular mechanism of receptor signaling. The propensity of Tsr to interact axially suggests that the interactions might stabilize the formation of complexes of CheA and CheW with the receptor in which these signaling proteins bind at the junction of the two cytoplasmic tail regions. Such an arrangement has indeed been experimentally observed in mixtures prepared from purified CheA, CheW, and a Tar receptor in which the transmembrane region was replaced by a stretch of mostly nonpolar residues (10). What is especially striking is that the combined length (∼30 nm) of the apposing cytoplasmic portions in the pair of axially interacting receptors in this ternary complex is essentially the same as the length observed in the zippered Tsr assemblies reported here (Fig. 3) which did not contain either CheA or CheW. It is therefore reasonable to suppose that the type of interaction observed here in the cells overproducing Tsr in the absence of CheA and CheW may also be relevant to signaling in wild-type bacteria. In cells that lack expression of chemotaxis receptors, CheA and CheW do not localize to the poles, while in cells with normal levels of chemotaxis receptors but without CheA and CheW the extent of polar localization of receptors is considerably reduced (17). The idea that invaginations near the polar regions may provide a mechanism for the selective clustering of an assembly of chemoreceptors, CheA, and CheW is fully consistent with these findings.
The zippered and rounded assemblies that we describe here have some parallels to structural features observed in eukaryotic cells. Weak homotypic interactions between cytoplasmic domains of membrane proteins expressed in the endoplasmic reticulum are known to induce the formation of membrane stacks known as crystalloid (33) or organized smooth (26) endoplasmic reticulum. All eukaryotic cells also have molecular machinery that is capable of triggering the conversion of flat regions of membrane into rounded structures that can be used for transporting soluble and membrane-bound components. At least four eukaryotic proteins have been identified that can induce changes in membrane curvature: dynamin (28), amphiphysin (29), endophilin (7), and epsin (8). Furthermore, the recruitment of clathrin to membranes via epsin (8) or Ap180 (9) can trigger bud formation, an event that precedes vesicle formation. The novel aspect of our work is the discovery that when suitable protein-protein interactions are present, phenomena strongly resembling curvature induction and tubulation (21) may be mimicked even in a simple prokaryotic model system. The rounded regions in the cell appear to be poised for release as small vesicles, but they stay attached in the absence of a mechanism such as detergent addition (Fig. 2c) or an energy source that triggers their release. We expect that the methods used here to reveal the organization of the bacterial membrane networks will be directly applicable to imaging similar structures in eukaryotic cells and that the strategy of combining tomographic, electron, and X-ray crystallographic approaches can be a tool that is generally applicable for constructing plausible three-dimensional models for protein networks in cells at molecular resolution.
Acknowledgments
We thank Anas Chalah for assistance with preparation of membranes and Stanton Lee for generous assistance with recording tomograms.
This work was supported by the intramural program at the National Cancer Institute.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. G., L. Orci, M. S. Brown, L. M. Garcia-Segura, and J. L. Goldstein. 1983. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J. Cell Sci. 63:1-20. [DOI] [PubMed] [Google Scholar]
- 3.Arechaga, I., B. Miroux, S. Karrasch, R. Huijbregts, B. de Kruijff, M. J. Runswick, and J. E. Walker. 2000. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(0) ATP synthase. FEBS Lett. 482:215-219. [DOI] [PubMed] [Google Scholar]
- 4.Baumeister, W., R. Grimm, and J. Walz. 1999. Electron tomography of molecules and cells. Trends Cell Biol. 9:81-85. [DOI] [PubMed] [Google Scholar]
- 5.DeRosier, D. J., and A. Klug. 1968. Reconstruction of three-dimensional structures from electron micrographs. Nature 217:130-134. [DOI] [PubMed] [Google Scholar]
- 6.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford, M. G., I. G. Mills, B. J. Peter, Y. Vallis, G. J. Praefcke, P. R. Evans, and H. T. McMahon. 2002. Curvature of clathrin-coated pits driven by epsin. Nature 419:361-366. [DOI] [PubMed] [Google Scholar]
- 9.Ford, M. G., I. G. Mills, B. J. Peter, Y. Vallis, G. J. Praefcke, P. R. Evans, and H. T. McMahon. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291:1051-1055. [DOI] [PubMed]
- 10.Francis, N. R., M. N. Levit, T. R. Shaikh, L. A. Melanson, J. B. Stock, and D. J. DeRosier. 2002. Subunit organization in a soluble complex of Tar, CheW, and CheA by electron microscopy. J. Biol. Chem. 277:36755-36759. [DOI] [PubMed] [Google Scholar]
- 11.Frank, J. 1996. Three-dimensional electron microscopy of macromolecular assemblies. Academic Press, New York, N.Y.
- 12.Heymann, J., R. Sarker, T. Hirai, D. Shi, J. L. S. Milne, P. C. Maloney, and S. Subramaniam. 2001. Projection structure and molecular architecture of OxlT, a membrane transporter. EMBO J. 20:4408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 14.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 16.Li, G., and R. M. Weis. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357-365. [DOI] [PubMed] [Google Scholar]
- 17.Lybarger, S. R., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 19.Mello, B. A., and Y. Tu. 2003. Quantitative modeling of sensitivity in bacterial chemotaxis: the role of coupling among different chemoreceptor species. Proc. Natl. Acad. Sci. USA 100:8223-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milburn, M. V., G. G. Privé, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, J. D. E. Koshland, and S. H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342-1347. [DOI] [PubMed] [Google Scholar]
- 21.Nossal, R., and J. Zimmerberg. 2002. Endocytosis: curvature to the ENTH degree. Curr. Biol. 12:R770-R772. [DOI] [PubMed] [Google Scholar]
- 22.Peters, P. J., and W. Hunziker. 2001. Subcellular localization of Rab17 by cryo-immunogold electron microscopy in epithelial cells grown on polycarbonate filters. Methods Enzymol. 329:210-225. [DOI] [PubMed] [Google Scholar]
- 23.Porta, D., R. Rippka, and M. Hernandez-Marine. 2000. Unusual ultrastructural features in three strains of Cyanothece (cyanobacteria). Arch. Microbiol. 173:154-163. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, T. S., S. V. Aksenov, and D. Bray. 2003. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J. Mol. Biol. 329:291-309. [DOI] [PubMed] [Google Scholar]
- 25.Skidmore, J. M., D. D. Ellefson, B. P. McNamara, M. M. Couto, A. J. Wolfe, and J. R. Maddock. 2000. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J. Bacteriol. 182:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2002. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock, J. B., M. N. Levit, and P. M. Wolanin. 2002. Information processing in bacterial chemotaxis. Sci. STKE 2002:PE25. [DOI] [PubMed]
- 28.Sweitzer, S. M., and J. E. Hinshaw. 1998. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93:1021-1029. [DOI] [PubMed] [Google Scholar]
- 29.Takei, K., V. I. Slepnev, V. Haucke, and P. De Camilli. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1:33-39. [DOI] [PubMed] [Google Scholar]
- 30.Thon, F. 1966. On the defocusing dependence of phase contrast in electron microscopical images. Z. Naturforsch. 21a:476-478. [Google Scholar]
- 31.van Heel, M., B. Gowen, R. Matadeen, E. V. Orlova, R. Finn, T. Pape, D. Cohen, H. Stark, R. Schmidt, M. Schatz, and A. Patwardhan. 2000. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33:307-369. [DOI] [PubMed] [Google Scholar]
- 32.Weis, R. M., T. Hirai, A. Chalah, M. Kessel, P. J. Peters, and S. Subramaniam. 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 185:3636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto, A., R. Masaki, and Y. Tashiro. 1996. Formation of crystalloid endoplasmic reticulum in COS cells upon overexpression of microsomal aldehyde dehydrogenase by cDNA transfection. J. Cell Sci. 109:1727-1738. [DOI] [PubMed] [Google Scholar]