Abstract
Bubonic plague is transmitted by fleas whose feeding is blocked by a mass of Yersinia pestis in the digestive tract. Y. pestis and the closely related Y. pseudotuberculosis also block the feeding of Caenorhabditis elegans by forming a biofilm on the nematode head. C. elegans mutants with severe motility defects acquire almost no biofilm, indicating that normal animals accumulate the biofilm matrix as they move through a Yersinia lawn. Using the lectin wheat germ agglutinin as a probe, we show that the matrix on C. elegans contains carbohydrate produced by Yersinia. The carbohydrate is present in bacterial lawns prior to addition of nematodes, indicating that biofilm formation does not involve signaling between the two organisms. Furthermore, biofilm accumulation depends on continuous C. elegans exposure to a lawn of Yersinia bacteria.
Bubonic plague, an acute infection that spreads primarily as a rodent epizootic, killed millions of people in three devastating pandemics (21). Yersinia pestis, the causative agent of plague, is transmitted to rodents and humans by the bites of fleas whose proventriculi are blocked by a dense mass of the bacteria (13). The blockage starves the flea and stimulates it to bite repeatedly in search of blood meals, thus spreading the bacteria to new hosts (4). The Y. pestis hmsHFRS operon (20, 22) is required for flea blockage and disease transmission (14).
Y. pestis also blocks feeding of the laboratory nematode Caenorhabditis elegans (9). This blockage is mediated by a biofilm that forms on the nematode's anterior cuticle, especially the head (9, 15). Like flea blockage, the biofilm requires hmsHFRS, which encodes predicted polysaccharide biosynthetic proteins (9). This suggests that flea blockage by Y. pestis is a biofilm-mediated process and that C. elegans may be an experimentally tractable surrogate for fleas.
Y. pseudotuberculosis, a close relative of Y. pestis, also makes biofilms on nematodes. The two bacterial species are indistinguishable by 16S rRNA sequences (24), and molecular evidence suggests that Y. pestis evolved from Y. pseudotuberculosis only 1,500 to 20,000 years ago (1). Under conditions favorable for nematode observation, Y. pseudotuberculosis biofilm production is more robust than that of Y. pestis (9), and therefore Y. pseudotuberculosis is preferable for many biofilm experiments.
A biofilm is a community of microbes embedded in an organic polymer matrix, usually containing exopolysaccharide, that adheres to a surface that may be biotic or abiotic (6, 11). Some bacterial pathogens use biofilms to adhere directly to human tissues, contributing to human diseases such as endocarditis, osteomyelitis, otitis media, periodontitis, prostatitis, and cystic fibrosis-associated pneumonia (7, 10).
Biofilms are commonly studied on artificial surfaces such as glass and plastic. In the case of C. elegans, the surface is alive and motile. Although the requirement for the predicted polysaccharide biosynthetic operon hmsHFRS strongly suggests that the matrix is of bacterial origin, it has not been ruled out that C. elegans secretes the matrix in response to an hmsHFRS-dependent signal. In the present study, we first show that the lectin wheat germ agglutinin (WGA) is a strong probe for biofilm detection on C. elegans. With this probe we then produce evidence that the matrix on C. elegans is produced by Yersinia and accumulated on the nematode as the animal moves through the bacterial lawn. We further show that biofilm accumulation on C. elegans depends on continuous nematode exposure to a lawn of Yersinia.
MATERIALS AND METHODS
Nematode strains and growth conditions.
C. elegans was grown at 20°C on NGM agar seeded with Escherichia coli OP50 as described previously (25), except that the agar concentration was 2% for both nematode culture and biofilm experiments. The standard wild-type strain N2, strain MT7929 carrying the unc-13(e51) mutation, and strain CB190 carrying the unc-54(e190) mutation were obtained from the Caenorhabditis Genetics Center, University of Minnesota.
Bacterial strains, growth conditions, and biofilm production.
Y. pestis KIM6+ is competent for flea blockage (14) and nematode biofilms (9) but is avirulent in mammals (22). Y. pseudotuberculosis YPIII is a standard laboratory strain (12). For experiments with green fluorescent protein (GFP), strains were transformed with pACYC-GFP (a gift of D. Monack) and grown on medium containing 10 μg of chloramphenicol/ml. For the experiment depicted in Fig. 6, nonfluorescent Yersinia was transformed with the parent plasmid pACYC184 to allow incubation on medium with chloramphenicol.
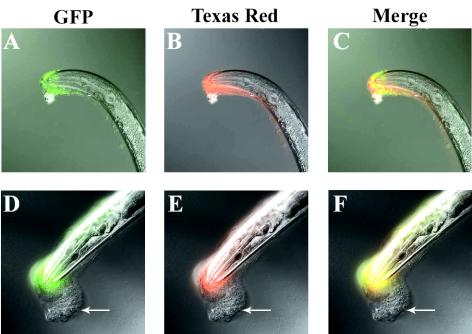
FIG. 6.
New biofilm matrix accumulates on top of old. Columns: GFP, fluorescent bacteria; Texas red, labeled biofilm matrix; Merge, both images. Rows: top, nematodes immediately after 30 min on Y. pseudotuberculosis-GFP incubation and WGA-TR labeling; bottom, nematodes incubated for 30 min on Y. pseudotuberculosis-GFP, labeled with WGA-TR, and then transferred to nonfluorescent Y. pseudotuberculosis for 20 h. Panels: A and D, combined DIC and GFP images; B and E, combined DIC and TR images; C and F, combined DIC, GFP, and Texas red images. Arrows indicate unlabeled new biofilm matrix.
To make lawns for biofilm production, Yersinia strains were cultured with shaking (150 rpm) in Luria-Bertani broth at 26°C for 1 day, and aliquots of 120 μl were pipetted onto NGM agar plates and grown at room temperature for an additional day. Biofilms on adult or fourth-larval-stage C. elegans were produced by placing the animals on Yersinia lawns and incubating them at 20°C.
Lectin labeling of biofilms on C. elegans.
To identify a lectin probe, biofilms were accumulated overnight on about 50 nematodes per plate. The worms were washed off plates with appropriate lectin-binding buffers (Table 1) and then washed twice more in buffer. Centrifugation between washes was at 100 × g for 1 min, which pellets worms but not planktonic bacteria. Nematodes were incubated in 100 μg of each of 12 fluorescein isothiocyanate (FITC)-linked lectins (EY Laboratories, Inc., San Mateo, Calif.)/ml at room temperature for 30 min and then washed twice with buffer to remove unbound lectin. Samples were mounted on a thin layer of 2% (wt/vol) agarose containing 20 mM sodium azide to anaesthetize the nematodes. Differential interference contrast (DIC) and epifluorescence images were captured separately with a monochrome digital camera and then merged and colored with Zeiss AxioVision 3.1 software.
TABLE 1.
Lectins binding Y. pseudotuberculosis biofilm on C. elegans
| FITC-linked lectina | Monosaccharide binding specificityb | Lectin binding bufferc | Lectin binding detected by fluorescence |
|---|---|---|---|
| BPA | GalNAc | 1 | − |
| ConA | α-Man > α-Glu > α-GlcNAc | 2 | − |
| DBA | Terminal α-GalNAc | 1 | − |
| GS-I | α-Gal or α-GalNAc | 3 | − |
| GS-II | Terminal GlcNAc | 3 | − |
| LPA | NANA | 4 | + |
| MPA | α-Gal or α-GalNAc | 1 | − |
| PNA | Terminal β-Gal | 1 | − |
| SBA | GalNAc > Gal | 1 | − |
| UEA-I | Terminal α(1,2)-Fuc | 1 | − |
| WGA | β-GlcNAc > NANA | 1 | ++ |
| Succinylated WGA | β-GlcNAc | 1 | + |
BPA, Bauhinia purpurea agglutinin; ConA, concanavalin A; DBA, Dolichos biflorus agglutinin; GS, Griffonia simplicifolia; MPA, Maclura pomifera agglutinin; PNA, peanut agglutinin; SBA, soybean agglutinin; UEA, Ulex europaeus agglutinin.
Fuc, l-fucose; Gal, d-galactose; GalNAc, N-acetyl-d-galactosamine; Glu, d-glucose; Man, d-mannose; NANA, N-acetyl-d-neuraminic acid (sialic acid). Binding specificities are as reported by the manufacturer (EY Laboratories).
Buffer 1 = 0.01 M phosphate and 0.15 M NaCl (pH 7.3). Buffer 2 = 0.05 M Tris and 0.15 M NaCl (pH 7.0). Buffer 3 = 0.01 M phosphate, 0.15 M NaCl, and 0.5 mM CaCl2 (pH 7.3). Buffer 4 = 0.05 M Tris, 0.15 M NaCl, and 0.01 M CaCl2 (pH 8.0).
After selection of WGA as a probe, the same protocol was used except that nematodes were incubated on Yersinia lawns for various periods depending on the experiment, and 20 μg of FITC-linked WGA (WGA-FITC) or Texas red-linked WGA (WGA-TR) (EY Laboratories)/ml was used for detection. The specificity of WGA binding was demonstrated by preincubation of WGA-FITC with increasing concentrations of N-acetyl-d-glucosamine (GlcNAc) for 1 h, or with 1 M d-glucose as a control, followed by incubation of the lectin with nematodes as described above.
Labeling of biofilm carbohydrate in bacterial lawns.
A total of 2 ml of buffer 1 (Table 1) was pipetted onto a Y. pseudotuberculosis lawn, and the bacteria were scraped into the liquid with a sterile glass rod. After suspension by gentle pipetting, the cells were washed twice in the same buffer, with centrifugation at 16,100 × g for 5 min between washes. The pellet was resuspended in 1.5 ml of buffer containing WGA-FITC at 20 μg/ml, followed by incubation at room temperature for 30 min. The bacteria were washed twice with the buffer to remove unbound WGA-FITC and resuspended in 100 μl of buffer. To observe WGA-FITC-reactive material in the lawns, 5 μl of the treated sample was transferred to a thin layer of 2% (wt/vol) agarose on a glass slide and examined by epifluorescence microscopy. E. coli OP50 was treated identically as a negative control. To show transfer of WGA-FITC-reactive material to nematodes, 100 μl of lectin-treated sample per plate was pipetted onto the center of fresh NGM agar and dried for 1 h at 37°C, at which temperature the bacteria do not produce new matrix material. About 50 adult nematodes were placed on each newly formed lawn and incubated for 1, 3, or 20 h and then examined by epifluorescence microscopy. As one negative control, nematodes were incubated with WGA-FITC and placed on untreated Y. pseudotuberculosis lawns. For a second negative control, yersiniae were incubated with the lectin GS-II, which binds neither biofilms nor nematode cuticles.
Removal of biofilm matrix material from lawns by washing.
Y. pseudotuberculosis was scraped from agar, washed, and resuspended in 1.5 ml of buffer 1 as described above. The pH was then adjusted to 10.6 by the addition of 1 M NaOH (∼4 μl/ml). The suspension was incubated at room temperature for 20 min with vortexing for 5 s at 5-min intervals. The pH was then adjusted back to 7.3 by the addition of 1 M HCl (∼4 μl/ml), and the bacteria were immediately washed twice (with centrifugation at 16,100 × g for 5 min) in pH 7.3 buffer. The pellet was resuspended in 100 μl of buffer, pipetted onto fresh NGM agar, and incubated for 1 h at 37°C to make a new, dry lawn. About 50 adult C. elegans organisms were transferred to the lawn, followed by incubation at 20°C. Nematodes were assayed for biofilm with WGA-FITC at 1, 3, and 20 h. Unwashed lawns and lawns washed in buffer 1 without pH adjustments were used as controls.
Transfer of biofilm-positive C. elegans to E. coli lawns.
About 100 adult C. elegans organisms were placed on each lawn of Y. pseudotuberculosis expressing GFP, followed by incubation at 20°C. After 5 min, 15 min, or 2 h, the nematodes were suspended in sterile distilled water and then washed twice with water (with centrifugation at 100 × g, 1 min) to remove planktonic bacteria. A sample of these worms was examined immediately by epifluorescence microscopy. The remainder were transferred to E. coli OP50 lawns and incubated at 20°C for 20 h and then washed and examined.
Simultaneous labeling of bacteria and biofilm carbohydrate.
For the experiment shown in Fig. 6, C. elegans was incubated for 30 min on lawns of Y. pseudotuberculosis expressing GFP, and the accumulated biofilm carbohydrate was labeled with WGA-TR. A sample of the nematodes was examined immediately by microscopy and photographed. The remaining nematodes were transferred to lawns of Y. pseudotuberculosis that did not express GFP and examined after 20 h. The plates contained 10 μg of chloramphenicol/ml to maintain pACYC-GFP in the original bacteria.
RESULTS
WGA is a probe for biofilm detection on C. elegans.
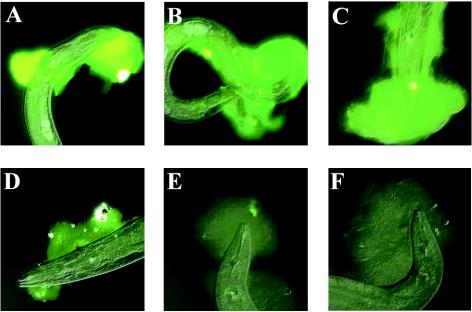
Lectins are proteins that specifically bind carbohydrates. We screened 12 FITC-linked lectins with different specificities (Table 1) for binding of Yersinia biofilms on C. elegans. Positive results were obtained for Limulus polyphemus agglutinin (LPA), WGA, and succinylated WGA (Table 1). The fluorescence intensity was strongest for WGA, and it was therefore chosen for subsequent experiments. The specificity of WGA binding was confirmed by preincubating the lectin with GlcNAc, which competed for lectin binding in a concentration-dependent manner (Fig. 1). Preincubation with 1 M d-glucose had no inhibitory effect (data not shown).
FIG. 1.
GlcNAc inhibits WGA binding of biofilm on C. elegans. WGA was preincubated with no GlcNAc (A) and with GlcNAc concentrations of 0.1 M (B), 0.2 M (C), 0.4 M (D), 0.8 M (E), and 1.0 M (F). Exposures were identical for all pictures.
Less biofilm forms on C. elegans motility mutants.
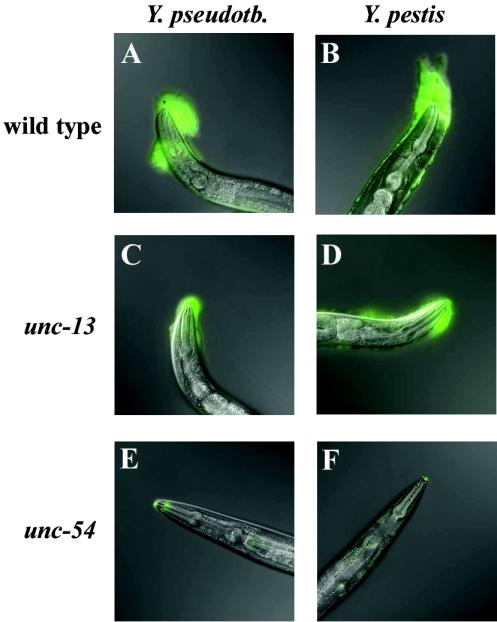
Wild-type C. elegans move almost constantly on bacterial lawns. In contrast, unc-13 mutants move sporadically and slowly because of a defect in neurotransmitter release (17). An even stronger phenotype is seen in unc-54 mutants, which are essentially paralyzed due to a myosin defect in locomotion muscles (23). Incubation on Y. pestis or Y. pseudotuberculosis lawns for 4 h resulted in large biofilms on wild-type animals, whereas much smaller biofilms were observed on unc-13 mutants (Fig. 2A to D). No biofilm was visible by light microscopy alone on unc-54 mutants, but with WGA-FITC a trace of biofilm was detected on the mouths (Fig. 2E and F). Thus, the size of the biofilm correlated with the motility of the worms. These results suggest that the matrix accumulates on C. elegans as a result of their motion through the bacterial lawn.
FIG. 2.
Less biofilm forms on C. elegans motility mutants. Nematodes of the indicated genotype were placed on Yersinia lawns for 4 h and then treated with WGA-FITC to detect biofilms and photographed.
Biofilm carbohydrate is in bacterial lawns in the absence of C. elegans.
The requirement for nematode motility, as well as rapid matrix accumulation after the animals are placed on a lawn, suggests that the material is present in Yersinia lawns prior to addition of the worms. If this is the case, it should be possible to label the biofilm carbohydrate with lectin, independent of the presence of C. elegans. We removed bacteria and associated material from the plates, incubated them with WGA-FITC, and examined the samples by epifluorescence microscopy. Fluorescence was observed in both Y. pestis and Y. pseudotuberculosis lawns but not in lawns of E. coli strain OP50, the standard C. elegans food (data not shown). To demonstrate that this labeled material is transferred to nematode cuticles, we treated bacterial lawns with WGA-FITC and replated the material on agar. When C. elegans was added, FITC-positive biofilms formed on the animals (Fig. 3C, F, and I). In the reciprocal experiment, C. elegans organisms themselves were incubated with WGA-FITC and then placed on untreated Y. pseudotuberculosis lawns; no label appeared in the resulting biofilms (Fig. 3A, D and G). The labeling of biofilms was specific for WGA, since lectin GSII-FITC did not label the biofilms (Fig. 3B, E and H). The biofilm matrix on the nematodes was entirely FITC-positive at 1 h after the transfer (Fig. 3C), but after longer incubations the distal portion was unlabeled (Fig. 3F and I). The presence of this unlabeled material, evidently produced as the bacteria grew after the labeling reaction, indicates that WGA-FITC does not interfere with accumulation of additional matrix on the nematodes.
FIG. 3.
C. elegans accumulates prelabeled biofilm carbohydrate. “Labeled worm” indicates nematodes that were treated with WGA-FITC and then transferred to untreated Y. pseudotuberculosis lawns and incubated for the indicated times. “Labeled biofilm” indicates Y. pseudotuberculosis lawns that were labeled with the indicated lectin conjugated to FITC and then replated on agar; untreated nematodes were added and incubated for the indicated times.
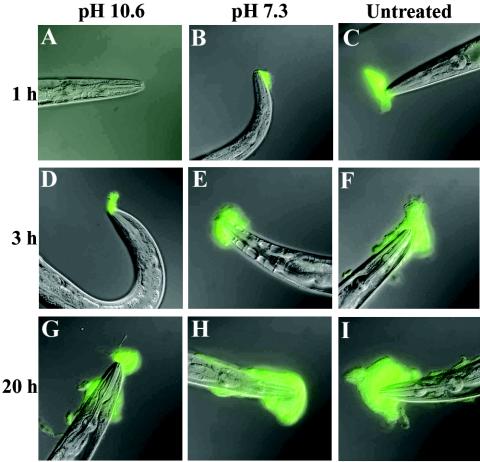
Biofilm matrix material is removed by washing the bacterial lawn.
If biofilm material is of bacterial origin, it should be possible to prevent biofilm formation by removing the material from the lawn before nematodes are added. In preliminary experiments, we found that biofilms can be removed from C. elegans itself by a wash with buffer 1 adjusted to pH 10.6 but not with the same buffer at pH 7.3 (data not shown). A bacterial lawn was washed with the high-pH buffer and replated on agar, after which nematodes were added. No biofilms formed after 1 h (Fig. 4A), a period sufficient to accumulate easily detected biofilms when control bacteria were either washed with pH 7.3 buffer (Fig. 4B) or left untreated (Fig. 4C). This indicated that the high-pH treatment removed the matrix material from the lawn. The treatment did not affect bacterial viability, as assayed by serial dilution and colony counting (data not shown). Consistent with continued bacterial viability, biofilms formed on nematodes with incubations of 3 and 20 h on the washed lawns (Fig. 4D and G), presumably because the bacteria synthesized new matrix.
FIG. 4.
High-pH wash removes biofilm matrix material. In the left column, nematodes were incubated for the indicated times on Y. pseudotuberculosis lawns that had been washed at pH 10.6. In the middle column, nematodes were incubated on lawns washed at pH 7.3. In the right column, nematodes were incubated on unwashed lawns.
Biofilm development stops after removal of nematodes from Y. pseudotuberculosis.
Biofilms on C. elegans increase in size when the animals remain on a Y. pseudotuberculosis lawn (Fig. 4C, F, and I). One possible explanation is that, as the nematodes move, they continue to pick up matrix material present in the lawn. An alternative is that the later material is synthesized by the bacteria that are initially trapped in the matrix. To distinguish between these possibilities, we generated biofilms on C. elegans by using Y. pseudotuberculosis expressing GFP. After 5 min, 15 min, or 2 h of accumulation, the worms were washed to remove planktonic yersiniae and transferred to lawns of E. coli, which does not form biofilms on C. elegans. When examined 20 h later, the size of the biofilm in each case was the same as when the animal was removed from Yersinia (compare Fig. 5A to D, B to E, and C to F), indicating that the Yersinia trapped in the initial matrix are not sufficient for continued biofilm production. E. coli does not inhibit biofilm production because biofilms are formed on C. elegans on mixed lawns of yersiniae and E. coli (data not shown).
FIG. 5.
Biofilm development stops after removal of nematodes from Yersinia lawns. Nematodes kept on a Y. pseudotuberculosis-GFP lawn for 5 min (A and D), 15 min (B and E), or 2 h (C and F). The top row shows nematodes photographed immediately after removal from Y. pseudotuberculosis. In the bottom row, nematodes were photographed after 20 h of incubation on E. coli lawns.
Yersinia is static within the biofilm matrix on C. elegans.
To further explore the dynamics of biofilm development, we exposed C. elegans to Y. pseudotuberculosis expressing GFP for 30 min and then labeled the resulting biofilms with WGA-TR. This allowed us to mark the boundary of the original biofilm matrix (red fluorescence) while separately tracking the bacteria that were originally embedded in the biofilm (green fluorescence). Immediately after WGA-TR labeling, we transferred the animals to Y. pseudotuberculosis that did not express GFP and examined them after 20 h. If the bacteria in the original biofilm were motile and/or dividing rapidly, some GFP signal would be expected distal to the boundary of the original matrix. No such signal was observed (Fig. 6). Instead, red and green signals substantially overlapped at 20 h, just as they did immediately after WGA-TR labeling (Fig. 6C and F). The absence of GFP-expressing bacteria in the new biofilm matrix (Fig. 6, arrows) indicates that Y. pseudotuberculosis is largely static within the original biofilm matrix. This is further evidence suggesting that the matrix is picked up by motile C. elegans and that continued biofilm accumulation does not require significant activity by Yersinia embedded in the early matrix.
DISCUSSION
We describe here an experimental system in which a biofilm adheres to a living, movable surface: the cuticle of C. elegans. Previous studies have shown that these biofilms were produced by four different strains of Y. pestis (9, 15) and by 24% of Y. pseudotuberculosis strains examined (15). We have not observed biofilm production by the third pathogenic member of the genus, Y. enterocolitica (C. Darby, unpublished data). Biofilm production on C. elegans by other genera apparently is not common. The model nematode has been exposed experimentally to at least 18 gram-negative and 8 gram-positive pathogens (2), many of which are known to produce biofilms under some conditions. Only the gram-negative insect pathogen Xenorhabdus nematophila, which, like Yersinia sp., belongs to the Enterobacteriaceae, produced a biofilm-like structure on C. elegans (8). Neither mucoid Pseudomonas aeruginosa, which produces copious amounts of extracellular polysaccharide, nor uropathogenic E. coli, which makes a biofilm-like structure in some experimental infections (3), produced a nematode biofilm (Darby, unpublished).
In most laboratory models, biofilms form on abiotic, stationary surfaces such as glass and plastic, so that the source of the matrix is obvious. In the C. elegans system, the surface is alive, so that it was not immediately apparent whether the matrix is produced by the bacteria or the nematode. One possibility was that the matrix is produced in whole or in part by the nematode in response to some bacterial signal. This did not seem likely for three reasons. First, biofilms are a well-known bacterial phenomenon (6). Second, the biofilm on C. elegans requires the Yersinia operon hmsHFRS, which encodes predicted polysaccharide biosynthetic proteins (9). Third, C. elegans has been studied intensively in dozens of laboratories and has been subjected to extensive mutagenesis; a biofilm-like secretion would be easily observed but has not yet been reported for any mutant.
We have now addressed the origin of the matrix experimentally and confirmed that it is largely if not exclusively a bacterial product. Indirect evidence for bacterial origin is the requirement for nematode motility, as revealed by experiments with uncoordinated (Unc) mutants. A large reduction of biofilm occurred when unc-13 or unc-54 mutants, which have strong motility defects, were substituted for wild-type animals (Fig. 2). unc-13 functions intracellularly in neurotransmitter release (17), whereas unc-54 encodes a major myosin expressed in C. elegans body wall muscles (23). It is not apparent how defects in interior tissues of unc-13 and unc-54 mutants could directly affect binding of a biofilm matrix to the nematode exterior. The most plausible explanation for our observations is that the defective animals accumulate little biofilm simply because they cannot move well. This interpretation is supported by the correlation between motility and biofilm formation. Wild-type worms accumulated the most biofilm, moderately defective unc-13 animals acquired substantially less, and severely defective unc-54 mutants picked up almost none (Fig. 2). A wild-type nematode moving through a Yersinia lawn is like a car moving through wet snow, accumulating sticky materials in front of it as it goes. The mutants are like cars that remain parked.
Lectin-binding experiments provide direct evidence that the biofilm matrix is produced by Yersinia. First, WGA-reactive material was detected in Yersinia lawns in the absence of nematodes. Next, lawns were fluorescently labeled with WGA-FITC and nematodes added; the label appeared in the biofilms that then accumulated (Fig. 3). Finally, it was possible to prevent biofilm formation by washing the bacterial lawn in a high- pH buffer prior to the addition of nematodes (Fig. 4). Together, these experiments show that the biofilm matrix material is largely produced by Yersinia and that the material is present extracellularly before addition of the nematodes. Although we cannot rule out the possibility that a small component of the matrix is produced by C. elegans, we have no evidence for such a contribution.
Early in biofilm development, Yersinia cells are trapped in the matrix that adheres to the worm. If there were signaling between the two organisms, it would be expected that the proximity of these initial bacteria would facilitate the process. However, biofilms stopped developing when C. elegans were removed from Yersinia lawns (Fig. 5), despite the continuous presence of matrix-embedded Yersinia. This shows that continued biofilm accumulation on C. elegans is not due to activity of the bacteria embedded in the early matrix, and it suggests that there is no important signaling between the two organisms. Other experiments indicate that the biofilm expands because the nematode continuously moves through a bacterial lawn and picks up additional matrix material (Fig. 4 and 6). Thus, it appears that biofilm formation is not a response by Yersinia to the presence of nematodes and is not initiated by direct bacterial adhesion to the animals. Rather, Yersinia biofilm accumulation on C. elegans is a two-stage process. Initially, matrix material attaches directly to the nematode cuticle as the animal moves through the bacterial lawn. Later, the matrix becomes progressively thicker as additional material accumulates on top of the initial layer (Fig. 5 and 6).
In the majority of laboratory models, biofilms are made on abiotic, stationary surfaces, typically glass or plastic. Biofilm development under such conditions is a dynamic bacterial process that is generally divided into three phases: initial attachment to the surface; migration on the surface and formation of microcolonies; and differentiation into mature, exopolysaccharide-encased biofilms (7, 10, 11). The Yersinia-C. elegans system that we describe here is of a different sort. The nematode surface brings itself to the bacteria and bacterial secretions, and continuous motion of the surface is responsible for the increasing size of the matrix. Yersinia does not appear to sense the nematode surface or interact with it, inasmuch as the matrix material has already been secreted into the lawn prior to nematode addition. Furthermore, the bacterial cells trapped in the matrix contribute minimally, if at all, to biofilm expansion (Fig. 5).
Yersinia-C. elegans biofilms are a potential model for aspects of Y. pestis colonization of its vector, the flea. The two phenomena overlap molecularly: the hmsHFRS operon, which includes predicted polysaccharide-biosynthetic genes, is required for both flea transmission of plague (14) and biofilm formation on C. elegans (9). There is a striking functional similarity as well, since in both cases Yersinia prevents invertebrate feeding by physically blocking the digestive tract. The exterior location of the biofilm on C. elegans is advantageous experimentally, because the material is readily observable and easily accessible for biochemical studies. Although nematode motility is required in our experiments, this is because the bacteria and their secretions remain external to the animal. This should not be a limitation in modeling flea infection, because in fleas Y. pestis organisms are trapped in the gut and therefore would secrete carbohydrate directly into the digestive lumen. Moreover, even if motion is required to firmly attach the matrix to flea tissues, this could be supplied by the peristalsis of the insect's feeding and digestive muscles.
The Yersinia-C. elegans system may prove useful as a general model for biofilm formation on a living surface. Because of the great versatility of C. elegans as an experimental animal, the nematode surface can be experimentally altered, allowing investigation of the host factors involved in biofilm attachment. Characterization of these factors could produce insights into the means by which bacterial pathogens adhere to host tissues in biofilm-mediated infectious disease.
As a practical matter, our results demonstrate the utility of lectins as in situ probes for biofilm detection on nematodes. Because WGA does not bind the wild-type C. elegans cuticle (16), its use allows clear delineation between the biofilm matrix and the nematode itself (e.g., Fig. 3). Moreover, WGA detection is sensitive: traces of biofilms that were not seen on unc-54 mutants by bright-field microscopy were observed with WGA-FITC (Fig. 2E and F). WGA has affinity for β-GlcNAc and, to a lesser extent, N-acetyl-d-neuraminic acid (sialic acid) (5). LPA and succinylated WGA, which also bind the Yersinia biofilm, specifically bind sialic acid (19) and β-GlcNAc (18), respectively. This suggests that the Yersinia biofilm on C. elegans contains both β-GlcNAc and sialic acid. Purification of the biofilm carbohydrate for compositional analysis is in progress.
Acknowledgments
This study was supported by a Frederick G. Cottrell postdoctoral enhancement award to L.T. and by institutional development funds of the University of Alabama at Birmingham. Nematode strains were supplied by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alegado, R. A., M. C. Campbell, W. C. Chen, S. S. Slutz, and M. W. Tan. 2003. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell. Microbiol. 5:435-444. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 4.Bacot, A. W., and C. J. Martin. 1914. Observations on the mechanism of the transmission of plague by fleas. J. Hyg. 13:423-439. [PMC free article] [PubMed] [Google Scholar]
- 5.Bhavanandan, V. P., and A. W. Katlic. 1979. The interaction of wheat germ agglutinin with sialoglycoproteins. J. Biol. Chem. 254:4000-4008. [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Couillault, C., and J. J. Ewbank. 2002. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect. Immun. 70:4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 10.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne, W. M., Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemski, P., J. R. Lazere, T. Casey, and J. A. Wohlhieter. 1980. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect. Immun. 28:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinnebusch, B. J. 1997. Bubonic plague: a molecular genetic case history of the emergence of an infectious disease. J. Mol. Med. 75:645-652. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 15.Joshua, G. W. P., A. V. Karlyshev, M. P. Smith, K. E. Isherwood, R. W. Titball, and B. W. Wren. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221-3229. [DOI] [PubMed] [Google Scholar]
- 16.Link, C. D., M. A. Silverman, M. Breen, K. E. Watt, and S. A. Dames. 1992. Characterization of Caenorhabditis elegans lectin-binding mutants. Genetics 131:867-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruyama, H., T. L. Rakow, and I. N. Maruyama. 2001. Synaptic exocytosis and nervous system development impaired in Caenorhabditis elegans unc-13 mutants. Neuroscience 104:287-297. [DOI] [PubMed] [Google Scholar]
- 18.Monsigny, M., A. C. Roche, C. Sene, R. Maget-Dana, and F. Delmotte. 1980. Sugar-lectin interactions: how does wheat-germ agglutinin bind sialoglycoconjugates? Eur. J. Biochem. 104:147-153. [DOI] [PubMed] [Google Scholar]
- 19.Muresan, V., V. Iwanij, Z. D. Smith, and J. D. Jamieson. 1982. Purification and use of limulin: a sialic acid-specific lectin. J. Histochem. Cytochem. 30:938-946. [DOI] [PubMed] [Google Scholar]
- 20.Pendrak, M. L., and R. D. Perry. 1991. Characterization of a hemin-storage locus of Yersinia pestis. Biol. Met. 4:41-47. [DOI] [PubMed] [Google Scholar]
- 21.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachat, F. H., H. E. Harris, and H. F. Epstein. 1977. Two homogeneous myosins in body-wall muscle of Caenorhabditis elegans. Cell 10:721-728. [DOI] [PubMed] [Google Scholar]
- 24.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labeled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood, W. B. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Plainview, N.Y.