Abstract
The marmoset is an important nonhuman primate model for regenerative medicine. For experimental autologous cell therapy based on induced pluripotent (iPS) cells in the marmoset, cells must be able to undergo robust and reliable directed differentiation that will not require customization for each specific iPS cell clone. When marmoset iPS cells were aggregated in a hanging drop format for 3 days, followed by exposure to dual SMAD inhibitors and retinoic acid in monolayer culture for 3 days, we found substantial variability in the response of different iPS cell clones. However, when clones were pretreated with 0.05% – 2% dimethyl sulfoxide (DMSO) for 24 hours, all clones showed a very similar maximal response to the directed differentiation scheme. Peak responses were observed at 0.5% DMSO in two clones and at 1% DMSO in a third clone. When patterns of gene expression were examined by microarray analysis, hierarchical clustering showed very similar responses in all 3 clones when they were pretreated with optimal DMSO concentrations. The change in phenotype following exposure to DMSO and the 6 day hanging drop/monolayer treatment was confirmed by immunocytochemistry. Analysis of DNAcontent in DMSO-exposed cells indicated that it is unlikely that DMSO acts by causing cells to exit from the cell cycle. This approach should be generally valuable in the directed neural differentiation of pluripotent cells for experimental cell therapy.
Keywords: Nonhuman primates, induced pluripotent stem cells, differentiation, dimethyl sulfoxide
Introduction
Nonhuman primates (NHPs) offer many advantages for translational regenerative medicine research because of their relatedness to humans and their similar physiology, particularly with respect to the central nervous system (Qiu et al., 2013). For regenerative medicine, long-term studies of transplanted cell function (>3 years) will be possible in NHPs, but are impossible in rodents. Within NHPs, the common marmoset (Callithrix jacchus), as a small, short-lived, and rapid-breeding NHP species, has some unique advantages for long-term efficacy and safety studies (Abbott et al., 2003; Mansfield, 2003). Marmosets can be housed in a defined environment and have few known comorbidities (Tardif et al., 2011). Several human neurological disorders can be modeled in marmosets (Qiu et al., 2013). The recent publication of the annotated marmoset genome further enhances the attractiveness of this NHP model for biomedical research (Worley et al., 2014).
In order to enable studies on cell therapy in the marmoset, particularly autologous cell transplant experiments, we derived induced pluripotent stem cells (iPS cells) from newborn marmoset skin fibroblasts (Wu et al., 2010; Wu et al., 2012). Subsequently we documented a rapid iterative method for developing neural cell differentiation protocols in marmoset iPS cells (Farnsworth et al., 2013). For autologous cell therapy experiments to be feasible, it must be possible to apply a differentiation protocol to iPS cell clones newly generated from donor animals without the need to customize the protocol for each cell line or for each donor. In an autologous cell transplant experiment, both reprogramming of biopsy-derived cells and the differentiation of the resultant iPS cells to cells ready for transplantation into the donor animal must be accomplished within a period of a few weeks.
In the present experiments, we tested the general applicability of the previously developed neural differentiation protocol in 3 different marmoset iPS cell lines. As expected, the protocol worked efficiently for the cell line on which it was originally developed, but it worked much less well on the other two cell lines. Variability in the responses of different iPS cell clones to differentiation regimens has been repeatedly noted (Chang et al., 2008; Osafune et al., 2008; Hu et al., 2010; Bock et al., 2011). A potential solution to this variability has been proposed, comprising the prior treatment of the iPS cells with DMSO at a concentration of aproximately 1% – 2% (Chetty et al., 2013). In this study, we show that marmoset iPS cell clones that were incubated with 0.5% – 1% DMSO showed greatly enhanced responses to the differentiation protocol. While the changes in gene expression in response to directed differentiation were quite variable among iPS cell clones in the absence of DMSO treatment, they became robust and uniform following exposure to DMSO. Cell cycle analysis demonstrated that the action of DMSO is likely to be more complex than only causing an inhibition of cell replication.
Methods
Marmoset iPS cell clones
Three clonal lines of marmoset iPS cells (B8, 88, 15; Wu et al., 2010) were grown in E8 medium (Chen et al., 2011) supplemented with 10% fetal bovine serum (GlobalStem, Gaithersburg, MD). At the beginning of the differentiation protocols, cells were removed from the dish with Accutase (BioExpress, Kaysville, UT). Cells were then transferred into differentiation medium as described below, or were incubated with DMSO prior to the differentiation treatment.
DMSO pretreatment
Following detachment of the cells with Accutase, cells were plated in differentiation medium containing various concentrations (0.05% – 2%) of DMSO (Sigma Aldrich, St. Louis, MO). Differentiation medium comprised DMEM/F12 (Sigma) with 20% KSR (Knockout Serum Replacement; Life Technologies, Grand Island, NY), 0.32 μM dorsomorphin hydrochloride (Tocris, Bristol, UK), 0.32 μM SB431542 (Selleck, Houston, TX), 20 ng/ml FGF2 (Novoprotein, Summit, NJ), 25 μg/ml insulin (Sigma), 1 nM retinoic acid (all-trans retinoic acid, Sigma), and 10 μM ROCK inhibitor Y-27632 (Chemdea, Ridgewood, NJ). Cells were plated in this medium on plates coated with Matrigel (Corning, Tewksbury, MA).
Differentiation protocol
Following 24 hours of incubation in differentiation medium/DMSO, cells were removed from the dish with Accutase and resuspended in differentiation medium without DMSO. Cells were then permitted to form aggregates using 384-well hanging-drop plates (3D Biomatrix, Ann Arbor, MI). Each well of the hanging drop plate received 3000 cells in 30 μl differentiation medium. Plates were placed in a humidified incubator at 37.5 °C for 72 h.
Following this incubation period, the resultant aggregates were collected from the plates. They were then dissociated into single cells by incubation in 1 ml 0.25% trypsin/EDTA (Life Technologies) for 10 min at room temperature. Cells were transferred into DMEM/F12 medium containing 20% KSR to stop the digestion and mechanically dissociated. 100,000 cells were plated per 35 mm Matrigel-coated dish. Cells were plated in DMEM/F12 containing 20% KSR, 2% B27 supplement (Life Technologies), 15 μg/ml transferrin (human, Sigma), 10 μM retinoic acid, 1 μM SB431542, 1 μM dorsomorphin, 3.2 μM SAG (Sonic hedgehog agonist; EMD Chemicals, Billerica, MA) and 10 μM Y-27632. Cells were maintained in the same medium for 72 h with medium changes at 24 h intervals. Following this period, cells were harvested for preparation of RNA.
Quantitative PCR
Total RNA was isolated from cells using RNA Bee (Tel-Test, Friendswood, TX) according to the manufacturer’s instructions. A total of 2 μg of RNA was reverse-transcribed by using superscript II (Life Technologies). Quantitative PCR was conducted using SYBR green detection and an ABI 7900HT system (Applied Biosystems, Foster City, CA). Levels of mRNAs are reported as cycles versus β-actin, using marmoset gene-specific primers. Sequences of marmoset-specific primers are listed in supplemental Table 1. Statistical analyses were performed using Prism software (Graphpad, La Jolla, CA). Responses to differentiation and DMSO pretreatment were analyzed by one-way ANOVA followed by Dunnett’s test of multiple comparisons with a control.
Microarrays
For microarray analysis, pelleted cells were stored in RNAlater (Life Technologies) and then processed to prepare total RNA using RNA Bee, according to the manufacturer’s instructions. Total RNA was quantified using a Nanodrop spectrophotometer. The quality of the RNA was verified by 260/280 ratio and by using a BioRad Experion automated microfluidics system for analysis of RNA. All samples were high-quality RNA preparations. Microarray analysis (probe labeling, hybridization and scanning) was performed using the GeneAtlas Personal Microarray System and marmoset-specific arrays (catalog # 901831; marmoset gene 1.1 ST array strips; 656,668 probes, 21 probes/gene, 33,971 genes).
Data analyses were facilitated using Partek Express Affymetrix Edition software (Partek Inc., St. Louis, MO) and GeneSifter web-based package (PerkinElmer, Waltham, MA). Partek Express uses RMA (Robust Multi-chip Analysis) which includes background correction, quantile normalization, and median polish summarization. The arrays were assessed to be of good quality. For analyses conducted with Partek Express, one-way ANOVA was used to identify significantly expressed genes with a multiple testing correction (Benjamini-Hockberg) for false discovery rate (FDR was 5%). The data were further filtered to omit genes not altered in expression by at least 2-fold or 5-fold. A hierarchical cluster analysis was performed to assess correlations among the samples and genes of interest using Euclidean distance and average linkage statistical methods.
Validation of microarray data by quantitative PCR (qPCR)
The microarray analysis enabled the identification of highly upregulated and highly downregulated genes. 6 of the most upregulated and 6 of the most downregulated genes were selected for further analysis, using cells pretreated with 0.5% or 1% DMSO. qPCR was performed as described above.
Immunocytochemistry
Following the differentiation protocol, cells were detached with Accutase and were plated onto Matrigel-coated glass coverslips. Coverslips were fixed in 4% paraformaldehyde for immunocytochemistry. After washing three times in PBS, the samples were permeabilized with 0.2% Triton X-100 for 15 min. At room temperature, coverslips were rinsed several times in PBS and then incubated in 5% goat serum (Sigma) for 1 hour at room temperature. After the goat serum was aspirated, the samples were incubated with primary antibodies at 4 °C overnight. After rinsing in PBS three times, samples were incubated with fluorochrome-labeled secondary antibodies at room temperature for 1 hour, before rinsing in PBS. Nuclei were stained with 10 μg/ml DAPI (Sigma) for 10 min. Samples were viewed under a Zeiss Axiovert fluorescence microscope. Antibodies used are listed in supplement Table 2.
Cell cycle analysis
For cell cycle analysis cells were fixed in 70% ethanol and were stained with propidium iodide by incubation in a solution comprising 0.1% Triton X-100, 20 mg/ml propidium iodide and 0.2 mg/l RNAse A (catalog #4087, Cell Signaling Technology, Danvers, MA). Cell cycle analysis was performed using a Becton Dickinson FACSCalibur (UTHSCSA Flow Cytometry Core).
Results
We investigated how well different marmoset iPS cell clones responded to a previously developed directed differentiation protocol (Farnsworth et al., 2013). In this protocol, the cells are formed into aggregates in hanging-drop plates for 3 days and then exposed to differentiation factors (dual SMAD inhibitors plus retinoic acid) for 3 days in monolayer culture. Responses were measured as increase in expression of a set of 7 genes that were previously used to develop the protocol (Fig. 1).
Figure 1.
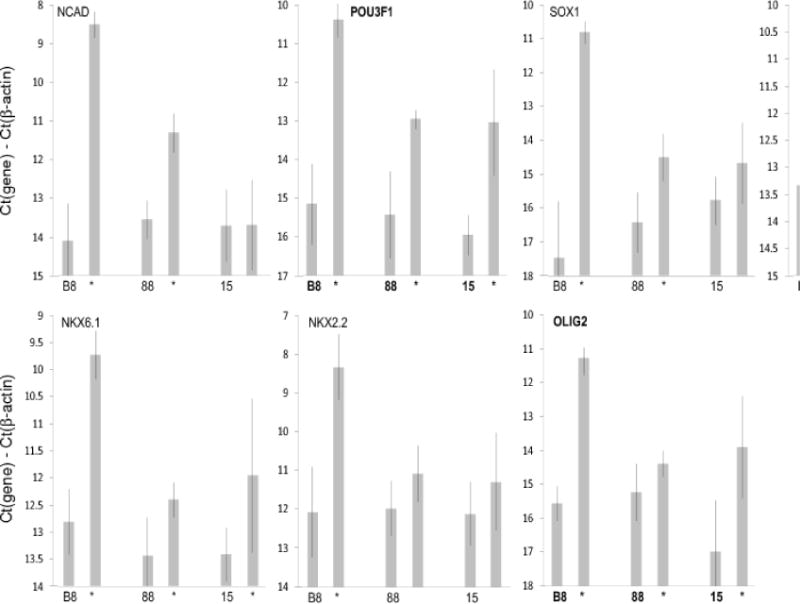
Cell line-specific variation in response to a neural differentiation protocol. Three marmoset iPS cell lines (B8, 88, 15) were subjected to a 6-day differentiation protocol involving 3 days of aggregate formation using a hanging-drop format followed by 3 days of exposure to dual SMAD inhibitors and retinoic acid in monolayer culture (see Materials and Methods). Levels of various neuroectodermal gene mRNAs before and after differentiation were assessed by qPCR. mRNA levels are plotted as Ct(gene)-Ct(β-actin) (log2 scale). For each gene and cell line, the first value is the level in the undifferentiated iPS cells and the second is the value following the differentiation protocol. Values ±S.D. (n = 3); statistically significant changes (p<0.05) indicated by asterisks.
The clone that we was used in iterative process of improving differentiation (B8; Farnsworth et al., 2013) responded as expected, with increases of up to 100-fold in the tested genes. However, two other clones (88, 15) that were previously validated as iPS cells via teratoma formation (Wu et al., 2010), responded much less well (up to about 8-fold, but also failing to respond at all in some cases). Moreover these clones showed considerable variability in response in repeated experiments.
For experimental autologous cell therapy to feasible, unselected iPS cell clones must respond reliably and robustly to differentiation protocols. A previously reported set of experiments using DMSO pretreatment was shown to improve the differentiation potential of a wide variety of pluripotent stem cell lines (Chetty et al., 2013). We therefore investigated whether 24 hours of incubation in 0.05% – 2% DMSO would enable refractory clones to respond to differentiation factors. During the 24 h incubation in DMSO cells showed some evidence of toxicity at the higher concentrations but the pretreatment did not affect the hanging-drop aggregation and there were no noticeable differences in cell morphology at the end of the 6 day differentiation protocol (Fig. 2).
Figure 2.
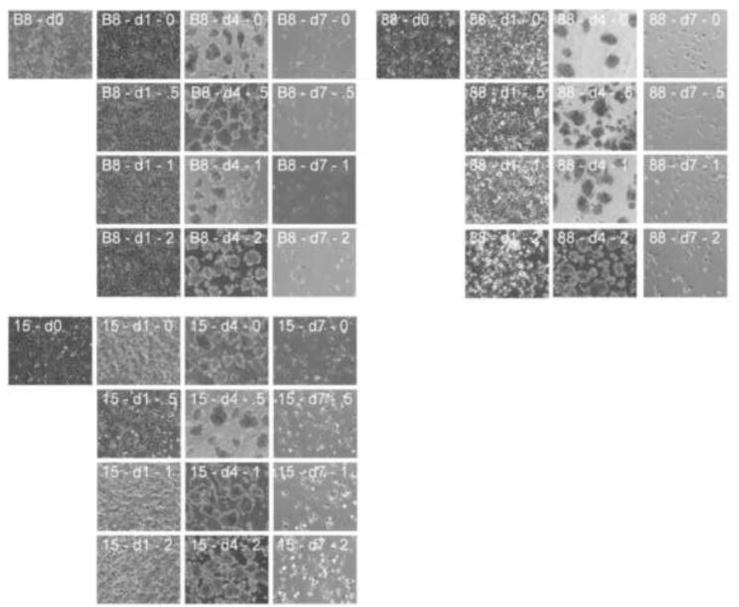
DMSO pretreatment of marmoset iPS cells and differentiation protocol. Cells were exposed to 0.5%, 1% or 2% DMSO (1 day) prior to hanging-drop aggregate formation (4 days) and exposure to differentiation factors in monolayer culture (7 days). Representative images of marmoset iPS cell clones, B8, 88, and 15.
The DMSO pretreatment had striking effects on the response of the 3 iPS cells clones to the differentiation protocol (Fig. 3). All clones showed increased levels of the 7 mRNAs that were originally used to develop the differentiation protocol. Clones 88 and 15, that were refractory to the protocol in the absence of DMSO, responded robustly following DMSO pretreatment. For example, NCAD mRNA increased by about 4-fold in clone 88 and showed almost no effect in clone 15; following DMSO, NCAD increase 45-fold in clone 88 and 30-fold in clone 15. The same pattern was seen in all genes. Clone B8, which responded well in the absence of DMSO, also showed increases in several mRNAs. For example, SOX10 increased 8-fold in the absence of DMSO and 80-fold following DMSO pretreatment. The optimal DMSO concentration differed among clones; 1% treatment resulted in the largest increases in mRNA levels in clone B8, and 0.5% gave the largest increases in clones 88 and 15. The highest concentration used, 2%, did not improve responses in clones 88 and 15. We therefore selected 1% DMSO pretreatment in clone B8 for further experiments and 0.5% for clones 88 and 15.
Figure 3.
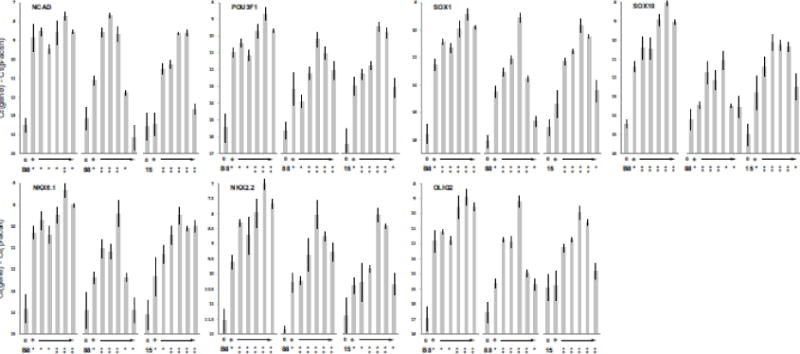
DMSO pretreatment improves response of marmoset iPS cells to the differentiation protocol. Neuroectodermal gene mRNAs were measured following hanging-drop aggregate formation and exposure to differentiation factors as described in Fig. 1, with the addition of 24 hours DMSO pretreatment (details in Materials and Methods). Levels of mRNAs are plotted as Ct(gene)-Ct(β-actin) (log2 scale). For each gene and cell line, values indicated as “0” are from the undifferentiated iPS cells; “+” indicates values following differentiation, without DMSO; the arrow indicates the values obtained following differentiation with the addition of 24 hours pretreatment with a range of DMSO concentrations (0.05%, 0.1%, 0.5%, 1% or 2% DMSO). Values ±S.D. (n = 3); statistically significant changes (p<0.05) versus the undifferentiated cells indicated by asterisks in the upper row; statistically significant changes (p<0.05) for DMSO treatment versus the differentiated cells without DMSO pretreatment indicated by asterisks in the lower row.
These experiments suggested that DMSO pretreatment was capable of normalizing the responses of refractory clones. Maximal levels of mRNAs in the 3 clones were similar following incubation with DMSO, while they were quite different without DMSO pretreatment. In order to assess whether the similarity of response following DMSO extended to a larger number of genes, we performed a microarray analysis on the 3 iPS cell clones following optimal DMSO pretreatment and differentiation. We found that the robustness of mRNA increases was a general response extending to several thousand genes (Fig. 4). Cluster analysis showed that the three differentiated samples closely resembled each other and were distinct from the three undifferentiated iPS cell clones.
Figure 4.
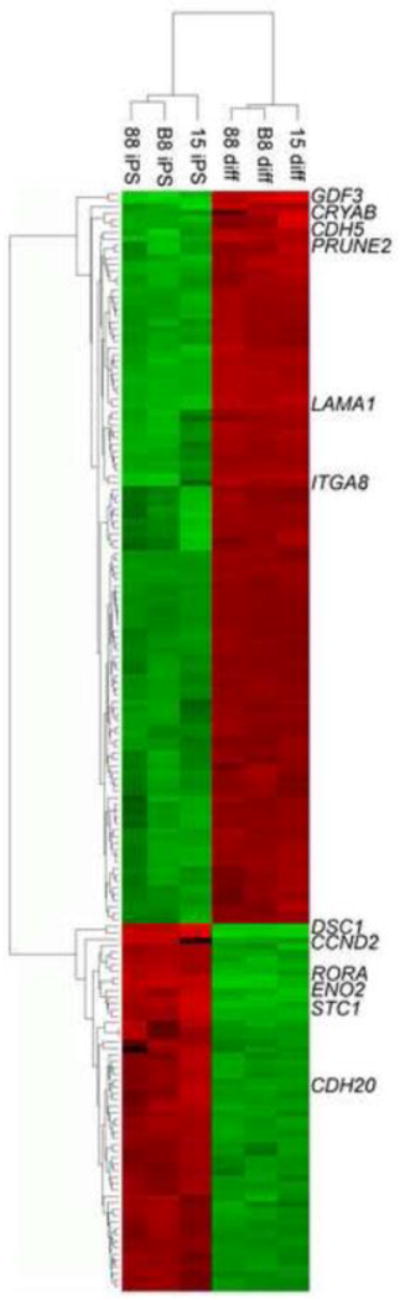
mRNA microarray analysis of DMSO/differentiation protocol in 3 marmoset iPS cell lines; heat maps and selected upregulated and downregulated genes (see Results). Red indicates mRNAs elevated following DMSO/differentiation and green indicates downregulated mRNAs.
In order to verify that the effect of DMSO would apply to genes not previously selected as part of the differentiation protocol, we selected 6 upregulated and 6 downregulated genes from the microarray results to assess the effect of DMSO. qPCR verified that DMSO pretreatment increased the responses of the 6 upregulated genes in the differentiation protocol (Fig. 5). Some selected genes (LAMA1, CRYAB, ITGA8) showed very little response in the absence of DMSO, but strong responses following DMSO pretreatment. For some other mRNAs weak responses became more robust following DMSO. For the 6 downregulated genes, the patterns were more complex, but in all cases DMSO pretreatment caused the mRNA levels following differentiation to be very simiar, while they were quite dissimilar in the absence of DMSO.
Figure 5.
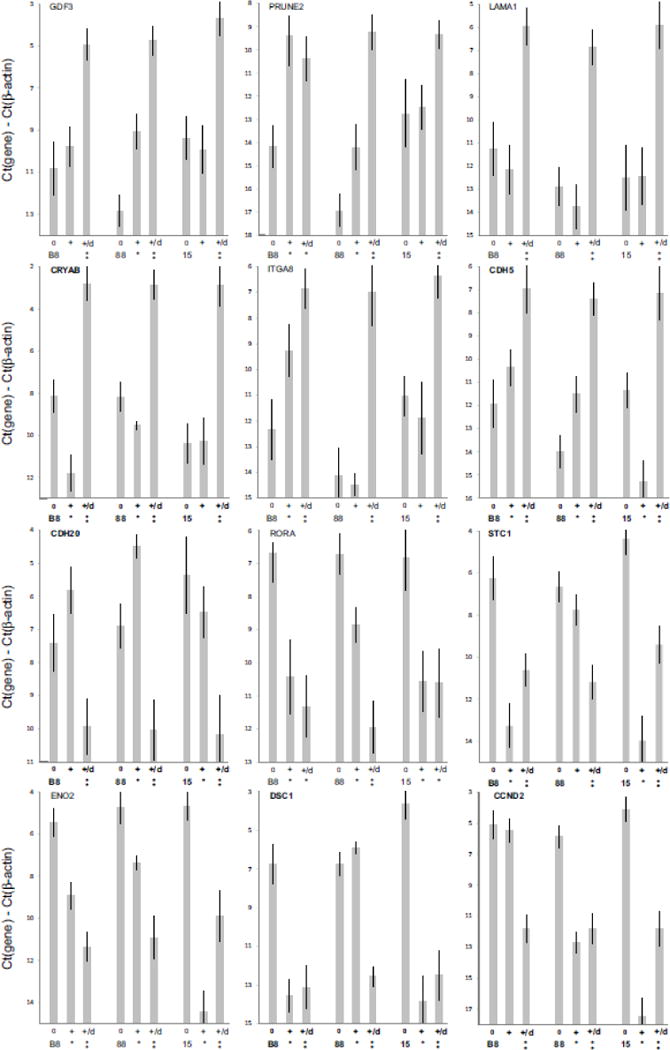
qPCR validation of microarray analysis. Upregulated and downregulated mRNAs were selected from the microarray experiment (Fig. 4) mRNAs were measured following the 6 day differentiation protocol described in Fig. 1, with the addition of 24 hours DMSO pretreatment (1% DMSO for clone B8, 0.5% DMSO for clones 88 and 15). Levels of mRNAs are plotted as Ct(gene)-Ct(β-actin) (log2 scale). For each gene and cell line, values indicated as “0” are from the undifferentiated iPS cells; “+” indicates values following differentiation, without DMSO; “+/d” indicates the values obtained following differentiation with the addition of 24 hours pretreatment DMSO. Values ±S.D. (n = 3); statistically significant changes (p<0.05) versus the undifferentiated cells indicated by asterisks in the upper row; statistically significant changes (p<0.05) for DMSO treatment versus the differentiated cells without DMSO pretreatment indicated by asterisks in the lower row.
As a result of these studies we modified our differentiation protocol by adding a 24 h incubation with 0.5% – 1% DMSO prior to 3 days of hanging drop aggregate formation and 3 days of incubation in monolayer culture with differentiation factors. To verify that this protocol was useful, we performed immunocytochemistry on the differentiated cells. Fig. 6 shows that there was robust expression of NCAD, SOX10, NKX6.1 and NKX2.2 as well as the general neural marker βIII tubulin, whereas there was minimal expression of these proteins in the undifferentiated iPS cells. We therefore concluded that adding a DMSO pretreatment to the differentiation protocol would be of general value in experiments that require rapid and robust differentiation.
Figure 6.
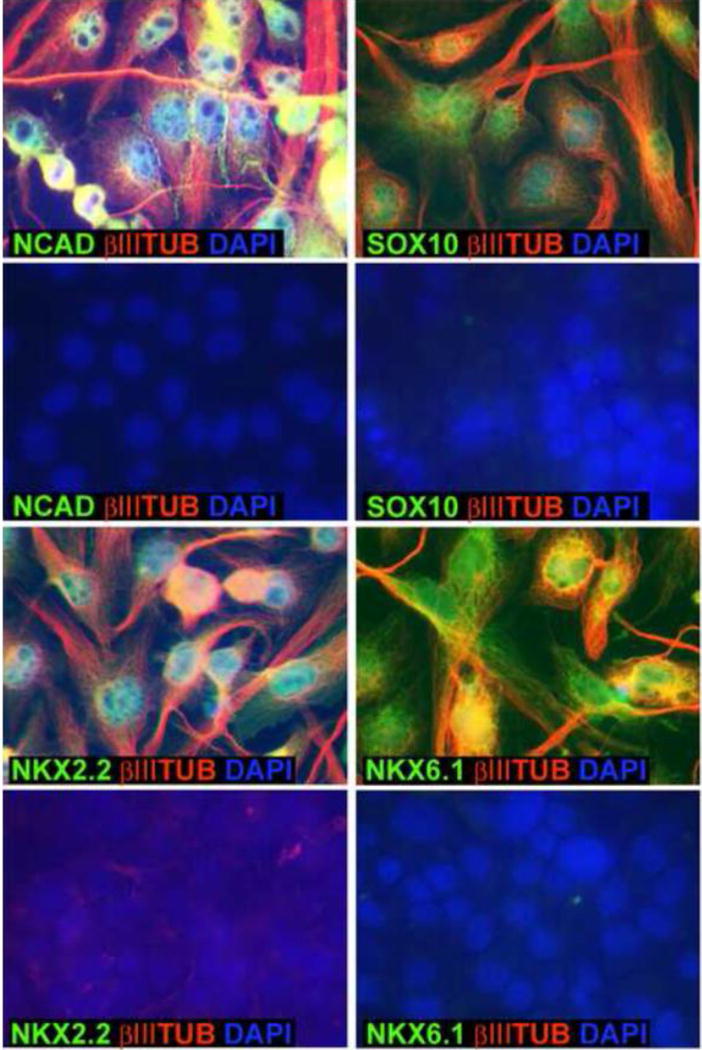
Immunocytochemical confirmation of expression of some DMSO-responsive genes. Marmoset iPS clone B8 cells were incubated for 24 hours in 1% DMSO and then subjected to the 6 day differentiation protocol described in Fig. 1. Cells were plated on glass coverslips for immunocytochemistry with antibodies against NCAD, Sox10, Nkx2.2 and Nkx6.1 (green fluorescence; see Materials and Methods). In each case the cells were also stained with an antibody against βIII tubulin (red) and were costained with DAPI (blue). For each of 4 the genes the upper images are of the cells following differentiation and the lower images are of the undifferentiated iPS cells.
As noted above, the use of DMSO pretreatment in this modified protocol was based on prior data that suggested that this may be of general use in the differentiation of pluripotent cells (Chetty et al., 2013). In the prior work, it was suggested that DMSO might act by causing the cells to exit from the cell cycle. We tested whether marmoset iPScells are caused to exit from the cell cycle by exposure to DMSO (Fig. 7). Flow cytometric analysis of propidium iodide-stained cells showed no effect of DMSO on cell cycle distribution when used at 0.5%, 1% and 2%.
Figure 7.
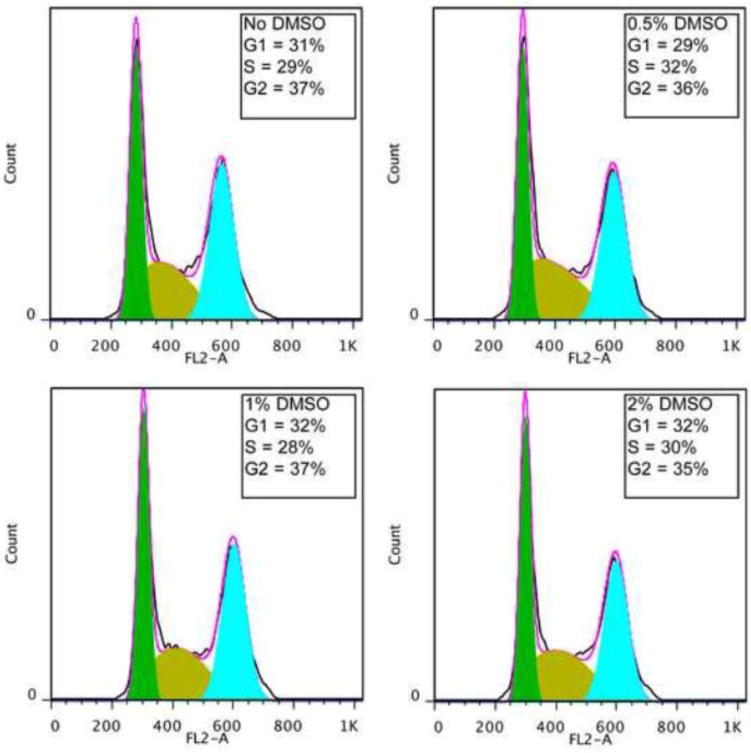
Effect of DMSO on cell cycle in marmoset iPS cells. Cells of clone B8 were exposed to the indicated concentrations of DMSO in differentiation medium, as described in Methods, for 24 hours. Following this incubation cells were fixed and stained with propidium iodide for cell cycle analysis. The percentages of cells under each condition in G1, S and G2 are indicated.
Discussion
In experimental autologous cell therapy in NHPs, rapid and reliable differentiation is essential for the accomplishment of the aims of the procedure in a feasible length of time. Given that biopsies of tissues from a living animal can be used to prepare iPS cells via validated reprogramming methods (Mishra et al., 2014), it is then critical to be able to perform directed differentiation of the cells using protocols that can be applied to any iPS cell clone that is generated during reprogramming. Autologous cell therapy would be much more difficult if optimization of differentiation protocols were required to be performed for each iPS cell clone. Nevertheless, the reported variability in response of pluripotent cells to directed differentiation (Chang et al., 2008; Osafune et al., 2008; Hu et al., 2010; Bock et al., 2011) forms an obstacle that must be addressed.
Here, we confirmed the variability in responsiveness of pluripotent cells, using marmoset iPS cell clones subjected to a directed neural differentiation protocol. A clone that was used to develop the protocol responded well in terms of expected changes in gene expression, while 2 others responded variably and less well. However, following pretreatment with DMSO for 24 hours, all clones responded with large changes in gene expression, and the clone used for the protocol development also showed increased expression of most genes. The uniformity of the changes in gene expression was confirmed using a microarray analysis of the 3 clones following DMSO pretreatment and directed differentiation. A further analysis of 6 genes that were highly upregulated and 6 genes that were highly downregulated showed that DMSO pretreatment caused a high degree of uniformity of response in these 12 genes among the 3 iPS cell clones. Using the pretreatment with DMSO as an added step in an improved protocol, we showed robust expression of several proteins by immunocytochemistry.
In its effect on enhancing differentiation of a large variety of human pluripotent stem cells, the effect of DMSO was thought to be mediated by causing an exit of the pluripotent cells from the cell cycle and greater effectiveness of differentiation factors in G1 (Chetty et al., 2013). However, in our experiments we observed clear effects of DMSO at very low concentrations, which are unlikely to have an effect on cell growth rate, and a direct test of higher concentrations of DMSO showed no effect of this agent on cell cycle distribution. It is unlikely that in this case DMSO acts by creating a G1 phase in cells that normally lack it. It is likely that the effects of DMSO are more complex. DMSO has long been established as a potent agent for the differentiation of pluripotent and multipotent stem cells of various kinds (Preisler and Giladi, 1975; Gusella et al., 1976; McBurney et al., 1982; Santos et al., 2003; Iwatani et al., 2006) and in these cases it has not been possible to reproduce its actions using other factors that cause exit from the cell cycle.
In conclusion, DMSO pretreatment in conjunction with an established neural differentiation protocol will be valuable, if not essential, for autologous cell therapy experiments in the marmoset.
Supplementary Material
Highlights.
Nonhuman primate models are critical for developing autologous cell therapy.
Marmoset iPS cell clones responded variably to neural differentiation factors.
Prior treatment of the clones with 0.5% or 1% DMSO normalized their responses.
Inclusion of a DMSO treatment step may be essential for practical autologous cell therapy.
Acknowledgments
This work was supported by VA grant I01BX001454, NIH grants R21AG033286 and R03AG045481, and by grants from the Owens Medical Research Foundation and the Ted Nash Long Life Foundation. Miao Li acknowledges the support of the China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ. Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med. 2003;53:339–50. [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–52. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Nelson AM, Fields PA, Hesson JL, Ulyanova T, Cao H, Nakamoto B, Ware CB, Papayannopoulou T. Diverse hematopoietic potentials of five human embryonic stem cell lines. Exp Cell Res. 2008;314:2930–40. doi: 10.1016/j.yexcr.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S, Pagliuca FW, Honore C, Kweudjeu A, Rezania A, Melton DA. A simple tool to improve pluripotent stem cell differentiation. Nat Methods. 2013;10:553–6. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth SL, Qiu Z, Mishra A, Hornsby PJ. Directed neural differentiation of induced pluripotent stem cells from non-human primates. Exp Biol Med. 2013;238:276–284. doi: 10.1177/1535370213482442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella J, Geller R, Clarke B, Weeks V, Housman D. Commitment to erythroid differentiation by friend erythroleukemia cells: a stochastic analysis. Cell. 1976;9:221–9. doi: 10.1016/0092-8674(76)90113-6. [DOI] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani M, Ikegami K, Kremenska Y, Hattori N, Tanaka S, Yagi S, Shiota K. Dimethyl sulfoxide has an impact on epigenetic profile in mouse embryoid body. Stem Cells. 2006;24:2549–56. doi: 10.1634/stemcells.2005-0427. [DOI] [PubMed] [Google Scholar]
- Mansfield K. Marmoset models commonly used in biomedical research. Comp Med. 2003;53:383–92. [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–7. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Mishra A, Qiu Z, Farnsworth SL, Hemmi JJ, Li M, Pickering AV, Hornsby PJ. Induced pluripotent stem cells from nonhuman primates. Methods in Molecular Biology. 2014 doi: 10.1007/7651_2014_159. (in press) Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–5. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Preisler HD, Giladi M. Differentiation of erythroleukemic cells in vitro: irreversible induction by dimethyl sulfoxide (DMSO) J Cell Physiol. 1975;85:537–46. doi: 10.1002/jcp.1040850305. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Farnsworth SL, Mishra A, Hornsby PJ. Patient-specific induced pluripotent stem cells in neurological disease modeling: The importance of nonhuman primate models. Stem Cells Cloning. 2013;6:19–29. doi: 10.2147/SCCAA.S34798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos NC, Figueira-Coelho J, Martins-Silva J, Saldanha C. Multidisciplinary utilization of dimethyl sulfoxide: pharmacological, cellular, and molecular aspects. Biochem Pharmacol. 2003;65:1035–41. doi: 10.1016/s0006-2952(03)00002-9. [DOI] [PubMed] [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, Ziegler TE. The marmoset as a model of aging and age-related diseases. ILAR J. 2011;52:54–65. doi: 10.1093/ilar.52.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Warren WC, Rogers J, Locke D, Muzny DM, Mardis ER, Weinstock GM, Tardif SD, Wilson RK. The common marmoset genome provides insight into primate biology and evolution. Nat Genet. 2014;46:850–7. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Mishra A, Tardif SD, Hornsby PJ. Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res. 2010;4:180–188. doi: 10.1016/j.scr.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Mishra A, Qiu Z, Farnsworth S, Tardif SD, Hornsby PJ. Nonhuman primate induced pluripotent stem cells in regenerative medicine. Stem Cells Int. 2012;2012:767195. doi: 10.1155/2012/767195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


