Abstract
Coq9 is a polypeptide subunit in a mitochondrial multi-subunit complex, termed the CoQ-synthome, required for biosynthesis of coenzyme Q (ubiquinone or Q). Deletion of COQ9 results in dissociation of the CoQ-synthome, but over-expression of Coq8 putative kinase stabilizes the CoQ-synthome in the coq9 null mutant and leads to the accumulation of two nitrogen containing Q-intermediates, imino-demethoxy-Q6 (IDMQ6) and 3-hexaprenyl-4-aminophenol (4-AP) when para-aminobenzoic acid (pABA) is provided as a ring precursor. To investigate whether Coq9 is responsible for deamination steps in Q biosynthesis, we utilized the yeast coq5-5 point mutant. The yeast coq5-5 point mutant is defective in the C-methyltransferase step of Q biosynthesis, but retains normal steady-state levels of the Coq5 polypeptide. Here we show that when high amounts of 13C6-pABA are provided, the coq5-5 mutant accumulates both 13C6-imino-demethyl-demethoxy-Q6 (13C6-IDDMQ6) and demethyl-demethoxy-Q6 (13C6-DDMQ6). Deletion of COQ9 in the yeast coq5-5 mutant along with Coq8 over-expression and 13C6-pABA labeling leads to the absence of 13C6-DDMQ6, and the nitrogen-containing intermediates 13C6-4-AP and 13C6-IDDMQ6 persist. We describe a coq9 temperature sensitive mutant and show that at the non-permissive temperature, steady state polypeptide levels of Coq9-ts19 increased, while Coq4, Coq5, Coq6, and Coq7 decreased. The coq9-ts19 mutant had decreased Q6 content and increased levels of nitrogen-containing intermediates. These findings identify Coq9 as a multi-functional protein that is required for the function of Coq6 and Coq7 hydroxylases, for removal of the nitrogen substituent from pABA-derived Q-intermediates, and is an essential component of the CoQ synthome.
Keywords: Temperature-sensitive mutant, coenzyme Q, mitochondrial metabolism, Q biosynthetic intermediates, Saccharomyces cerevisiae, mass spectrometry (MS)
Graphical abstract
1. Introduction
Coenzyme Q (ubiquinone or Q)1 is a polyprenylated benzoquinone lipid essential in cellular energy metabolism [1]. Q has a redox active benzoquinone ring connected to a polyisoprenoid side chain, and is anchored to the mitochondrial inner membrane by the polyisoprenyl tail. The polyisoprenyl chain contains six units in Saccharomyces cerevisiae (Q6), eight units in Escherichia coli (Q8) and ten units in humans (Q10) [2]. The reversible reduction and oxidation of the quinone/hydroquinone (Q/QH2) enables its function as an electron and proton carrier in the mitochondrial respiratory chain and as a lipid-soluble antioxidant present in cellular membranes and in lipoproteins [1].
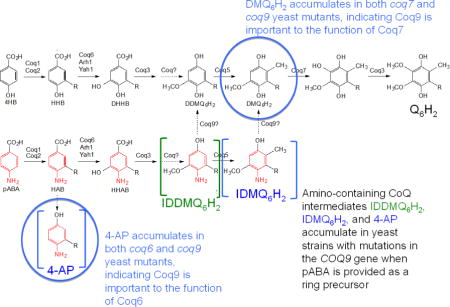
Q biosynthesis in S. cerevisiae requires nine Coq polypeptides (Coq1–Coq9), ferredoxin (Yah1) and ferredoxin reductase (Arh1) [3]. In addition, a Q-binding protein (Coq10) is required for efficient Q biosynthesis and for Q function as an electron carrier in respiratory electron transport [4]. 4-hydroxybenzoic acid (4HB) and para-aminobenzoic acid (pABA) both function as aromatic ring precursors for Q6 biosynthesis in S. cerevisiae [3, 5] (Fig. 1). Coq1 synthesizes the hexaprenyl diphosphate tail, which Coq2 attaches to ring precursors. Coq3 performs two O-methylation steps, Coq5 catalyzes C-methylation, and Coq6 and Coq7 catalyze hydroxylation steps. The proteins responsible for several steps in the Q biosynthesis pathway remain unknown, and the functional roles of the Coq4, Coq8, and Coq9 polypeptides still need further characterization (Fig. 1).
Fig. 1. Biosynthesis of Q in S. cerevisiae from 4HB or pABA.
S. cerevisiae uses either 4-hydoxybenzoic acid (4HB) or para-aminobenzoic acid (pABA) as an aromatic ring precursor for Q6 biosynthesis. Coq1 synthesizes the hexaprenyl-diphosphate tail and Coq2 attaches it to either ring. Coq6 performs the C5-hydoxylation reaction in concert with ferredoxin (Yah1) and ferredoxin reductase (Arh1). Coq3 catalyzes the two O-methylation steps and Coq5 catalyzes the C-methylation step. Coq9 is the putative deaminase that removes the amino groups on imino-demethyl-demethoxy-Q6 (IDDMQ6) or imino-demethoxy-Q6 (IDMQ6). Coq9 is also required for Coq7 to catalyze the penultimate hydroxylation step, and for efficient C5-hydroxylation by Coq6.
Coq9 is a polypeptide subunit in the Q biosynthetic complex. Similar to the other Coq polypeptides (with the exception of Coq2, an integral membrane protein), Coq9 is peripherally associated to the inner mitochondrial membrane facing the matrix side [6, 7]. Coq9 co-migrates with Coq3 and Coq4 at high molecular mass and HA tagged Coq9 co-purifies with Coq4, Coq5, Coq6 and Coq7 [6, 7]. Recovery of tagged versions of Coq3, Coq6, or Coq9 from digitonin-extracts of yeast mitochondria results in the recovery of the CoQ-synthome, a multi-subunit Q-biosynthetic complex, containing Coq3–Coq9 polypeptides, Q6, Q6-intermediates, as well as other partner proteins, including the newly identified Coq11 [8]. Deletion of any one of the COQ3–COQ9 genes leads to the decreased steady state of several of the other Coq polypeptides and to the accumulation of two early Q-intermediates, 3-hexaprenyl-4-hydroxybenzoic acid (HHB) and 3-hexaprenyl-4-aminobenzoic acid (HAB) [6, 9]. Sensitive Coq polypeptides were stabilized and late-stage Q-intermediates accumulated in some of the coq3–coq9 null mutants that over-expressed Coq8, a putative kinase [10]. Conserved kinase motifs in Coq8 are essential for the phosphorylation of Coq3, Coq5, and Coq7 [11, 12], and Coq8 over-expression stabilized the Q biosynthetic complex in yeast [7]. These studies suggest that Coq8 over-expression might stabilize the complex by phosphorylation. Recent work identified auto-phosphorylation and ATPase activity in ADCK3, a human ortholog of yeast Coq8 [13, 14].
Several studies suggest that yeast Coq9 is important for formation or stability of the CoQ synthome [7]. Coq8 over-expression suppressed the Q-less phenotype of the coq9 point mutant yeast strain C92 [15]. C92 has a nonsense point mutation in the coq9 gene causing an early stop codon; Coq8 over-expression increased the steady-state level of the Coq9 polypeptide in the C92 mutant [6]. Other work utilizing Coq8 over-expression showed that yeast Coq9 is important for correct function of Coq7 [10]. When Coq8 is over-expressed, intermediates that accumulate in the yeast coq9 null mutant were also found to accumulate in the coq7 null mutant. For example, with Coq8 over-expression, 13C6-DMQ6 accumulates in both yeast coq9 and coq7 null mutants when 13C6-4HB was provided as an aromatic ring precursor [10, 16]. However, when the same strains were provided with 13C6-pABA the yeast coq9 null mutant with Coq8 over-expression accumulated 13C6-imino-demethoxy Q6 (13C6-IDMQ6), while under the same labeling conditions, the yeast coq7 null mutant with Coq8 over-expression still produced 13C6-DMQ6 [10]. This finding suggests that Coq9 is required for Coq7 function, but is also required for deamination of Q-intermediates when pABA is used as a ring precursor. While pABA is utilized to generate Q6 in yeast, it is not a ring precursor for Q biosynthesis in human, mouse, Arabidopsis thaliana or E. coli [17, 18]. Therefore, the important role that Coq9 plays in the deamination of Q-intermediates might be unique to yeast Coq9. Coq9 is also necessary for correct function of Coq6, because in the presence of Coq8 over-expression, both coq6 null and coq9 null mutants accumulate 13C6-4-HP (upon labeling with 13C6-4HB) and 13C6-4-AP (upon labeling with 13C6-pABA) [10]. In this study, we examined the role of yeast Coq9 in mediating the deamination of other nitrogen-containing Q intermediates and employed a temperature sensitive mutant to further clarify its role in stabilizing the CoQ synthome.
2. Materials and Methods
2.1 Yeast strains and growth media
S. cerevisiae strains used in this study are listed in Table 1. Growth media used in this study were prepared as described [19], and included: YPD (2% glucose, 1% yeast extract, 2% peptone), YPEG (1% yeast extract, 2% peptone, 2% ethanol and 3% glycerol), YPGal (1% yeast extract, 2% peptone, 2% galactose, 0.1% dextrose), and YPG (3% glycerol, 1% yeast extract, 2% peptone). Synthetic Dextrose/Minimal medium (SD-complete) consisted of 0.18% yeast nitrogen base without amino acids, 2% dextrose, 0.14% NaH2PO4, 0.5% (NH4)2SO4, and amino acids were added to final concentrations as described [20]. Selective SD/Minimal medium lacking uracil or leucine (SD–Ura, or SD–Leu) were similarly prepared. Agar plate media were prepared as described above and included 2% bacto agar (Fisher).
Table 1.
Genotype and Source of Yeast Strains
| Strain | Genotype | Source |
|---|---|---|
| W3031B | MAT α ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303Δcoq4 | MAT a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq4::TRP1 | [41] |
| W303Δcoq7 | MAT a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq7::LEU2 | [42] |
| W303Δcoq9 | MAT a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq9::URA3 | [15] |
| BY4741Δcoq9 | MAT a coq9Δ::kanMX4 his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | [43]b |
| CH316-6B | MAT α coq5-5 trp1-1 ura3-1 | [27] |
| CH316-6BΔcoq9 | MAT α coq5-5 trp1-1 ura3-1 coq9::kanMX4 | This study |
Dr. Rodney Rothstein, Department of Human Genetics, Columbia University
European S. cerevisiae Archive for Functional Analysis (EUROSCARF), available on-line
2.2 Disruption of COQ9 in W3031B and CH316-6B (coq5-5 point mutant) yeast strains
A PCR product containing the KanMX4 gene was amplified with the genomic DNA isolated from BY4741Δcoq9 (used as template) and with primers that annealed to 100 bp upstream and downstream of the COQ9 ORF. The sequences of the primers utilized were: 5′-TTTGGGCCTACATAAGGTACTTC-3′ and 5′-CGCACAGACCAATAAATCTGCC-3′. The PCR product was then transformed into the yeast W3031B to create W303Δcoq9K. Transformants that grew on YPD + 200 μg/ml G418 (Geneticin) were selected. Proteins were extracted from these transformants as described [21] and separated by SDS-PAGE with a 10% polyacrylamide gel. Proteins were transferred to an Immobilon-P transfer membrane (Millipore) and analyzed by immunoblotting as described [7]. The primary antibody against Coq9 was used at a 1:1000 dilution and the secondary antibody, goat anti-rabbit IgG H&L chain specific peroxidase conjugate (Calbiochem), at a 1:10,000 dilution. The absence of Coq9 polypeptide confirmed that COQ9 was replaced with KanMX4. The resulting null mutant is W303Δcoq9K. To generate the double mutant, coq5 point mutant with coq9 knocked out, PCR product was transformed into the yeast coq5-5 point mutant strain (CH316-6B) and transformants were screened and verified as described above. The resulting double mutant is CH316-6BΔcoq9.
2.3 Construction of plasmids
Plasmids used in this study are listed in Table 2. Over-expression of Coq8 made use of the p4HN4 plasmid (mcCOQ8), which contains the COQ8 gene in pRS426, a multi-copy yeast shuttle vector [22]. To construct the plasmid pRS315COQ9 (COQ9), the genomic DNA of W3031B was isolated using the Wizard Genomic DNA purification kit (Promega). The COQ9 gene was then amplified with Taq polymerase and primers XhoI400upCoq9F (5′-CTCGAGCCGGGTTCAGAGGTAAAAGG-3′ −400 to −380 of COQ9 with XhoI restriction site at the 5′ end) and BamHI240downCoq9R (5′-GGATCCGGGACAAGCAGGAAGAACTA-3′ +220 to +240 with BamHI restriction site at the 5′ end). PCR products were inserted into the TOPO vector using the TOPO TA Cloning kit (Invitrogen) resulting in a plasmid named TOPOCOQ9. pRS315 and TOPOCOQ9 were digested with the restriction digestion enzymes XhoI and BamHI (New England BioLabs) and separated by gel electrophoresis. DNA fragments that contained the digested pRS315 or COQ9 were purified from agarose gel using the Purelink quick DNA gel extraction kit (Invitrogen) and then ligated with T4 DNA Ligase (New England BioLabs) resulting in pRS315COQ9. The correct nucleotide sequence of the COQ9 ORF in pRS315COQ9 was verified (Laragen, Los Angeles). pRS315COQ9 was shown to rescue growth of W303ΔCOQ9 on medium containing a non-fermentable carbon source, YPG.
Table 2.
Plasmid constructs used in this study
| Plasmid | Relevant genes | Copy number | Source |
|---|---|---|---|
| pRS315 | Yeast shuttle vector | Low copy | [44] |
| pRS426 | Yeast shuttle vector | Multicopy | [43, 44] |
| p4HN4 (mcCOQ8) | Yeast ABC1/COQ8 | Multicopy | [45] |
| pRS315COQ9 (COQ9) | Yeast COQ9 | Low copy | This work |
| TS19 | Yeast COQ9 | Low copy | This work |
| E55G | Yeast COQ9 | Low copy | This work |
| R107G | Yeast COQ9 | Low copy | This work |
| Q256L | Yeast COQ9 | Low copy | This work |
| a-12g | Yeast COQ9 | Low copy | This work |
| a-93g | Yeast COQ9 | Low copy | This work |
| E55GR107G | Yeast COQ9 | Low copy | This work |
| E55GQ256L | Yeast COQ9 | Low copy | This work |
| R107GQ256L | Yeast COQ9 | Low copy | This work |
| R107GQ256L | Yeast COQ9 | Low copy | This work |
| E55GR107GQ256L | Yeast COQ9 | Low copy | This work |
| R107GQ256La-93g | Yeast COQ9 | Low copy | This work |
| R107GQ256La-12g | Yeast COQ9 | Low copy | This work |
| E55GR107Ga-12g | Yeast COQ9 | Low copy | This work |
2.4 Construction of coq9 temperature-sensitive mutants using polymerase chain reaction (PCR) mutagenesis
Temperature-sensitive coq9 yeast strains were generated using error-prone PCR, followed by in vivo homologous recombination [23]. COQ9 with 400 bp 5′- and 240 bp 3′-flanking regions was cloned into pRS315, resulting in the plasmid pRS315COQ9. COQ9 was amplified using PCR with primers designed 180 bp upstream (5′-ACTGGAAAGCGGGCAGTGA-3′) and 240 downstream (5′-CAAGTGTAGCGGTCACGCTG-3′) of the multiple cloning region in the presence of 0.2 mM dCTP, 0.2 mM dTTP, 0.2 mM dGTP, 0.05 mM dATP, 0.8 mM MgCl2, and 0.6–0.8 mM MnCl2. The amplified fragments were purified and co-transformed with linearized pRS315 into the yeast null mutant W303Δcoq9. Leu+ transformants were then selected and screened for growth at 25°C and 37°C on YPEG plates. One of the plasmids generated (TS19) using the method outlined here was used in this study.
2.5 Site-Directed Mutagenesis of S. cerevisiae COQ9
Mutagenesis of the wild-type yeast COQ9 was carried out with either the QuikChange or QuikChange Lightning site-directed mutagenesis kit from Agilent following the manufacturer’s protocol. The primers used to generate single mutant plasmids are listed in Table 4: for E55G mutant, forward primer E55Gf and reverse primer E55Gr; for R107G mutant, forward primer R107Gf and reverse primer R107Gr; for Q256L mutant, forward primer Q256Lf and reverse primer Q256Lr; for a-12g mutant, forward primer a-12gf and reverse primer a-12gr; for a-93g mutant, forward primer a-93gf and reverse primer a-93gr. Single mutant plasmids were transformed into E. coli as described in the manufacturer’s protocol (Agilent) and then were purified from 3 ml cultures. The identities of the mutations were verified by DNA sequencing (Laragen). To generate secondary point mutations, single mutant plasmids were used as templates: E55GR107G mutant was generated using E55G as template and the R107Gf and R107Gr as primers; E55GQ256L mutant was generated using E55G as template and the Q256Lf and Q256Lr as primers; R107GQ256L mutant was generated using R107G as template and the Q256Lf and Q256Lr as primers. To generate tertiary point mutations, double mutant plasmids were used as templates: E55GR107GQ256L mutant was generated using R107GQ256L as template and the E55Gf and E55Gr as primers; R107GQ256La-12g mutant was generated using R107GQ256L as template and the a-12gf and a-12gr as primers; R107GQ256La-93g mutant was generated using R107GQ256L as template and the a-93gf and a-93gr as primers; E55GR107Ga-12g mutant was generated using E55GR107G as template and the a-12gf and a-12gr as primers. The identities of these mutations were verified by DNA sequencing (Laragen).
Table 4.
Primer sequences (site-directed mutagenesis of S. cerevisiae COQ9)
| Primer name | Sequence |
|---|---|
| E55Gf | 5′-AGAGAAACCGTGCCCG GGAACAAAC-3′ |
| E55Gr | 5′-GTTTGTTCCCGGGCACGGTTTCTCT-3′ |
| R107Gf | 5′-GGGTTGATTCCTTCAGTTAAACGATACCCTTTATCTACC-3′ |
| R107Gr | 5′-GGTAGATAAAGGGTATCGTTTAACTGAAGGAATCAACCC-3′ |
| Q256Lf | 5′-CCCCTAACTAATAGAGATTTGATTAAATTTA CCGTAGACA-3′ |
| Q256Lr | 5′-TGTCTACGGTAAATTTAATCAAATCTC TATTAGTTAGGGG-3′ |
| a-12gf | 5′-GAGATAACAGAGTCTTTACCGCATTATAAATC-3′ |
| a-12gr | 5′-GATTTATAATGCGGTAAAGACTCTGTTATCTC-3′ |
| a-93gf | 5′-GCAATAACAATAGTAAGAAACGATAATACGGGG-3′ |
| a-93gr | 5′-CCCCGTATTATCGTTTCTTACTATTGTTATTGC-3′ |
2.6 Lipid extraction and detection of Q6-intermediates by HPLC and tandem mass spectrometry
The designated strains of coq5-5 yeast mutants were labeled with 13C6-pABA followed by lipid analysis. Labeling media were prepared with 50 μg/ml 13C6-pABA dissolved in ethanol. The final concentration of ethanol in the medium was 0.2%. Yeast mutants without plasmids or harboring p4HN4 (mcCOQ8), were grown in 100 ml of SD complete or SD−Ura, respectively. To label cells, yeast cultures were diluted to 0.5 A600nm/ml in 100 ml of fresh SD complete or SD-URA with 13C6 pABA and labeled for 6 hours. The final cell density was 2–3 A600nm/ml. For lipid extraction cells were collected by centrifugation and 145 pmol Q4 was added to each cell pellet to serve as an internal standard. Lipid extracts were analyzed by RP-HPLC-MS/MS [10]. Briefly, a phenyl-hexyl column (Luna 5u, 100 × 4.60 mm, 5-μm, Phenomenex) was used for liquid chromatography. The mobile phase includes Solvent A (methanol/isopropanol, 95:5, with 2.5 mM ammonium formate) and Solvent B (isopropanol, 2.5 mM ammonium formate). From 0 to 6 min, Solvent B was increased linearly from 0 to 5% and the flow rate was increased from 600 to 800 μl/min. At 7 min, the flow rate and mobile phase were changed back to 100% Solvent A and a flow rate of 600 μl/min. The 4000 QTRAP linear MS/MS spectrometer from Applied Biosystems (Foster City, CA) was used for multiple reaction monitoring mode (MRM) analysis. Data were processed with Analyst version 1.4.2 software (Applied Biosystems).
To quantify Q6 content and determine de novo synthesis of Q6-intermediates in temperature-sensitive yeast mutants at permissive (25 °C) and non-permissive (37 °C) temperatures, yeast cells were labeled with 13C6-pABA followed by lipid analysis as described above. Labeling media were prepared with 10 μg/ml 13C6-pABA dissolved in ethanol (ethanol was 0.2% final concentration). Cells were collected (a total of 30 A600nm) as pellets after 5 hours of labeling. Q4 was added (145 pmol) to each cell pellet as an internal standard. The exact amounts (total pmol) of 12C-Q6 and 13C6-Q6 were calculated by normalizing the peak areas of 12C6-Q6 (sum of oxidized and reduced) and 13C6-Q6 (sum of oxidized and reduced) by the peak areas of Q4 (sum of oxidized and reduced); the pmol amounts were then determined from the Q6 standard curve. After the pmol of 12C-Q6 and 13C6-Q6 was calculated, they were further normalized by the wet weight of yeast pellets. Chemical standards for Q-intermediates 4-AP, IDMQ6 and DMQ6 are not available. To quantify these intermediates, the peak areas for each were normalized by the recovery of Q4 (sum of oxidized and reduced peaks). Finally, calculated values were further normalized by the wet weight of yeast pellets.
2.7 Mitochondrial isolation and immunoblot analyses with temperature-sensitive mutants
To study the protein levels in temperature-sensitive mutants, mitochondria were isolated from yeast cells and analysed by immunoblot. Yeast cultures were grown to 3–4 A600nm in YPGal medium at different temperatures (W3031B and W303Δcoq9:TS19 were grown at 25°C and 37°C for 18.5 hours; W303ΔCOQ9, W303ΔCOQ7, and W303ΔCOQ4 were grown at 30°C overnight). Crude mitochondria were isolated from a total volume of 1 L of culture as described [24]. Next, crude mitochondria were further purified with an OptiPrep discontinuous iodixanol gradient and then solubilized with digitonin as described in [7]. Solubilized mitochondria (15 μg based on total protein measured by the bicinchoninic acid assay from Thermo) were separated by SDS-PAGE with 10% polyacrylamide gels. Proteins were transferred to Immobilon-P transfer membranes (Millipore) and analyzed by immunoblotting as described [7]. The source and use of primary antibodies is described in Table 3. The secondary antibody used was goat anti-rabbit IgG H&L chain specific peroxidase conjugate (Calbiochem), 1:10,000.
Table 3.
Description and Source of Antibodies
| Antibody | Working Dilution | Source |
|---|---|---|
| Atp2 | 1:4000 | Carla M. Koehlera |
| Coq4 | 1:250 | [46] |
| Coq5 | 1:5000 | [27] |
| Coq6 | 1:250 | [47] |
| Coq7 | 1:1000 | [48] |
| Coq9 | 1:1000 | [6] |
Dr. Carla M. Koehler, Department of Chemistry and Biochemistry, UCLA
2.8 RNA extraction and Northern blot analyses with temperature-sensitive mutants
To determine the COQ mRNA levels in temperature-sensitive mutants, samples of yeast total RNA were analysed by Northern blot. Yeast cells were grown to 0.5 A600nm in YPGal; W3031B and W303Δcoq9:TS19 were grown at 25 °C and 37 °C for 18.5 hours; W303ΔCOQ9, W303ΔCOQ7, and W303ΔCOQ4 were grown at 30 °C overnight. Aliquots (25 ml) of each culture were harvested by centrifugation at 1000 × g for 5 min at 4°C, and cell pellets were washed with water and frozen in liquid nitrogen. RNA was extracted as described in [25] with some modifications. Briefly, 500 μl of RNA-SDS buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM EDTA, pH 8.0, 2% SDS), 400 μl of acid‐washed glass beads (Sigma), and 500 μl RNA Phenol-Chloroform (Fisher) were added. The tubes were vortexed for 1 min and incubated for 6 min at 65 °C. An aliquot (450 μl) of the aqueous top layer was added to 450 μl fresh RNA Phenol-Chloroform for the second extraction. RNA was precipitated with 1 ml ethanol and 40 μl 3 M sodium acetate (pH 5.2). The RNA pellets were then washed with 450 μl 70% ethanol (v/v) and resuspended in distilled water.
Samples of RNA (5 μg) were denatured at 55 °C for one hour with 5 volumes of glyoxal buffer. (Glyoxal buffer contains 60.9% DMSO (v/v) (SIGMA), 20.3% deionized glyoxal (v/v) (Fluka), 4.87% glycerol (v/v), 0.04 mg/ml ethidium bromide, and 12.2% 10× BPTE (v/v)). 10× BPTE contains 100 mM PIPES (SIGMA), 300 mM BIS-TRIS (SIGMA) and 10 mM EDTA. Samples of denatured RNA were separated by 1.2% agarose–1× BPTE gels and transferred to Hybond N+ nylon membranes (GE Healthcare) in 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) as described [26]. Blots were crossed-linked and hybridized with Church hybridization buffer containing probes as described [25].
Probes were generated as described [25], with some modifications. Briefly, PCR products were generated with genomic DNA extracted from W3031B and the primers annealed to the ORF of COQ4, COQ5, COQ6, COQ7 or COQ9 as listed in Table 5. Next, 4 μl of PCR products (100–200 ng/μl) were used as a template and mixed with 2.1 μl of distilled H2O, 4 μl of the mixture of rATP, rCTP, and Rgtp (2.5 mM each), 2.4 μl of 100 μM rUTP, 2 μl of 100 mM DTT, 1.25 μl of [α-32P] UTP (6,000 Ci/mmol; 40 μCi/μl; Perkin Elmer), 4 μl of 5× Promega transcription buffer, and 0.25 μl of T3 RNA polymerase (Promega). The in vitro transcription reaction was incubated at 37 °C for 1 h. To generate the SCR1 probe, 0.2 μl of 5′-GTCTAGCCGCGAGGAAGG-3′ oligo (100 μM) was mixed with 1 μl of T4 polynucleotide kinase (PNK), 1 μl of 10x PNK buffer, 3 μl of [γ−32P] ATP (3,000 Ci/mmol; 10 μCi/μl; Perkin Elmer), and 4.8 μl of distilled H2O. The reaction was incubated at 37 °C for 30 min.
Table 5.
Primer sequences (Riboprobe generation)
| Primer name | Sequence |
|---|---|
| COQ4F | 5′-ACAGCTACTTTGCCAGTGAAATGCC-3′ |
| COQ4T3R | 5′-AATTAACCCTCACTAAAGGGAAGTCGTGGCTCGTTTCTGTGAGTTGT-3′ |
| COQ5F | 5′-TGTTGATTTCTTCACGGATCGTTCG-3′ |
| COQ5T3R | 5′-AATTAACCCTCACTAAAGGGAGCCAGCAGATTTGAATCCTGCCTTC-3′ |
| COQ6F | 5′-CAGGATTGTCAGTGTTACGCCTAGATC-3′ |
| COQ6T3R | 5′-AATTAACCCTCACTAAAGGGAGGGCAACTCTATCAGTGCAATAACGA-3′ |
| COQ7F | 5′-GCAGAGGCTTTTCCGTCTTATCATCT-3′ |
| COQ7T3R | 5′-AATTAACCCTCACTAAAGGGAGCCATATACGAATCATGCTTGATAGCGG-3′ |
| COQ9F | 5′-ATGCTTTGTCGCAATACTGCCAGAACG-3′ |
| COQ9T3R | 5′-AATTAACCCTCACTAAAGGGATACTCACCCAAACGCATGACCCTA-3′ |
3. Results
3.1 The deletion of COQ9 in coq5-5 point mutant yeast leads to the accumulation of unique nitrogen-containing Q-intermediates
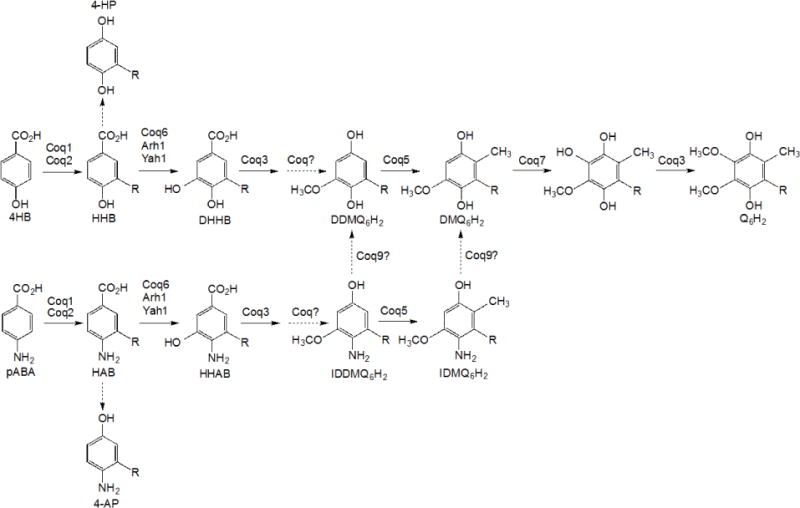
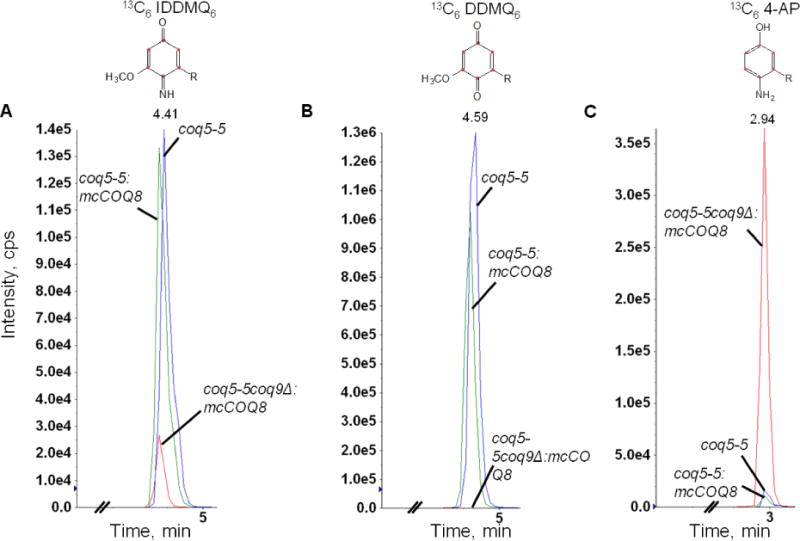
In addition to Coq6 and Coq7 function, Coq9 also appears to be necessary to convert IDMQ6 to DMQ6 (Fig. 1). To investigate whether Coq9 may act to remove the amino/imino group from other Q-intermediates, we utilized the CH316-6B yeast strain that harbors the coq5-5 point mutation. This mutant lacks C-methyltransferase activity but retains steady state levels of Coq5 and other Coq polypeptides [27] and accumulates DDMQ6 as a late-stage Q-intermediate [28]. To examine the effect of COQ9 on the DDMQ6 intermediate, we deleted the COQ9 gene in CH316-6B yeast to generate a double mutant strain (coq5-5 Δcoq9), and overexpressed Coq8 to stabilize the CoQ-synthome. We labeled the coq5-5, coq5-5:mcCOQ8, and coq5-5 Δcoq9:mcCOQ8 yeast strains with 13C6-pABA for 6 hours and used HPLC with tandem mass spectrometry to detect Q6 intermediates in the yeast lipid extracts. We found that both coq5-5 and coq5-5:mcCOQ8 strains accumulated predominant amounts of 13C6-DDMQ6 as well as readily detectable levels of 13C6-IDDMQ6 (blue and green traces in Fig. 2A and 2B). However, in the double mutant coq5-5 Δcoq9:mcCOQ8, 13C6-DDMQ6 disappeared (Fig. 2B), while intermediates containing the nitrogen group, 13C6-IDDMQ and 13C6-4-AP, accumulated (red traces in Fig. 2A and 2C). 13C6-DDMQ6 was identified by its retention time (4.59 min), precursor-to-product ion transition (553.4/159.0), and fragmentation spectrum [28]. 13C6-IDDMQ6 was identified by its retention time (4.41 min), precursor-to-product ion transition (552.4/158.0), and fragmentation spectrum (Fig. 3). 13C6-4-AP was identified by its retention time (2.94 min), precursor-to-product ion transition (524.4/128.0), and fragmentation spectrum [7]. The results suggest that Coq9 is essential for converting the amino or imino group to a hydroxyl group in Q intermediates derived from pABA.
Fig. 2. The deletion of COQ9 in a yeast coq5-5 point mutant leads to the accumulation of 3-hexaprenyl-4-aminophenol (4-AP) and the disappearance of demethyl-demethoxy-Q6 (DDMQ6), but imino-demethyl-demethoxy-Q6 (IDDMQ6) is still present.
Yeast coq5-5 point mutants with COQ8 over-expressed (coq5-5:mcCOQ8), without COQ8 over-expressed (coq5-5), or with the COQ9 gene deleted and COQ8 over-expressed (coq5-5 coq9Δ:mcCOQ8), were cultured in SD complete or SD−Ura with 50 μg/ml 13C6-pABA and 2 μl ethanol/ml medium at 0.5 A600nm/ml and collected after 6 hours. Q4 (145.4 pmol) was added prior to extraction to serve as an internal standard. Lipid extracts prepared from the cell pellets were analyzed by RP-HPLC-MS/MS. Multiple reaction monitoring (MRM) detected precursor-to-product ion transitions 552.4/158.0 (13C6 –IDDMQ6), 553.4/159.0 (13C6-DDMQ6), and 524.4/128.0 (13C6-4-AP). Just the oxidized forms of IDDMQ6 and DDMQ6 were detected, while only the reduced form of 4-AP was present. 13C6-DDMQ6 accumulates in the coq5-5 and coq5-5:mcCOQ8 yeast mutants (B), and 13C6-IDDMQ6 is readily detected (A). 13C6-IDDMQ6 and 13C6-4-AP accumulate in coq5-5 coq9Δ:mcCOQ8 (A and C), but 13C6-DDMQ6 is not detected (B). In all panels, the blue traces designate the Q-intermediate signals in coq5-5 and green traces designate the Q-intermediate signals in coq5-5:mcCOQ8, and the red traces indicate the Q-intermediate signals in coq5-5 coq9Δ:mcCOQ8.
Fig. 3. Identification of de novo imino-demethyl-demethoxy-Q6 (13C6-IDDMQ6).
Yeast coq5-5 coq9Δ:mcCOQ8 was cultured and labeled with 50 μg/ml 13C6 pABA as described in Figure 2. Total lipids were extracted and analyzed by RP-HPLC-MS/MS. The fragmentation spectra are shown for 13C6-IDDMQ6 [M+H]+ precursor ion (13C6 12C31H56NO2+; monoisotopic mass 552.4), the 13C6-IDDMQ6 tropylium ion [M]+ (13C6 12C2H10NO2+; 158.1), and the 13C6-IDDMQ6 chromenylium ion [M]+(13C6 12C5H14NO2+; 198.1).
3.2 Characterization of a coq9 temperature-sensitive mutant (coq9-ts19)
To better understand the role of Coq9 in Q biosynthesis, we generated conditional coq9 mutants. Using error-prone PCR and in vivo recombination, we mutagenized the cloned COQ9 gene. One clone, TS19, was selected for further analysis (Fig. 4). At the permissive temperature of 25 °C, the yeast coq9 null mutant harboring TS19 grew as well as wild type, while at the non-permissive temperature of 37 °C, it grew poorly when compared to either the wild type at 37 °C, or to growth at the permissive temperature, 25 °C.
Fig. 4. Characterization of the mutations responsible for temperature sensitivity of the Coq9-TS19 polypeptide.
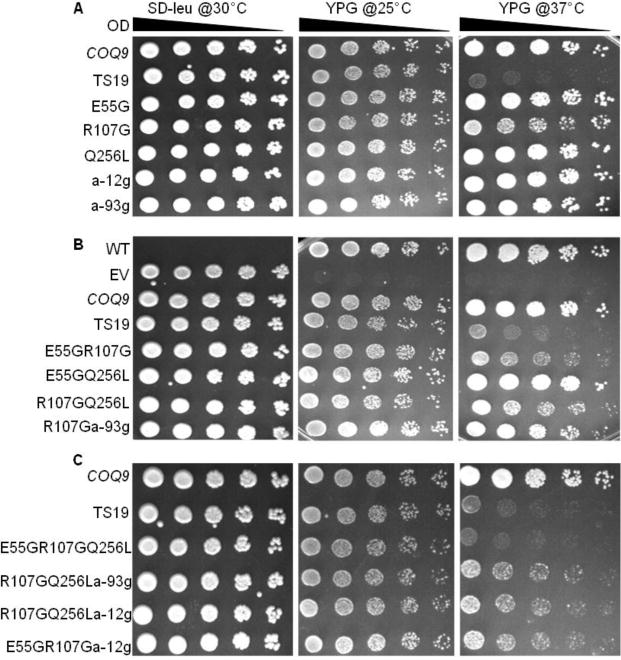
The yeast coq9 null mutant (W303ΔCOQ9) was transformed with plasmids containing the designated coq9 mutations. pRS315 empty vector (EV), and yeast wild-type COQ9 in pRS315 (COQ9), were included as controls. Yeast wild-type W3031B and the transformants were cultured over-night at 30 °C in YPD or SD−Leu media, respectively. Cell cultures were diluted to 0.2 based on A600nm readings and 2 μl of 1:5 serial dilutions were spotted onto SD−Leu and YPG plate media and incubated at the specified temperatures for 3 days.
The TS19 clone was sequenced to determine the mutations that caused the temperature-sensitive (ts) phenotype. Five mutations were detected in TS19: Adenine−12→Guanine (a−12g), Adenine−93༠Guanine (a−93g), Glu55→Gly (E55G), Arg107→Gly (R107G), and Gln256→Leu (Q256L). The first two mutations are upstream of the COQ9 ORF and the remaining three are within the COQ9 ORF. To identify the amino acids that were critical for Coq9 function, coq9 alleles were generated that contained single, double, or triple mutations. The plasmids generated were then transformed into the yeast Δcoq9 null mutant (W303ΔCOQ9) and subjected to plate dilution assay. Serial dilutions were plated on SD−Leu to confirm the presence of the plasmid. WT was not transformed with any plasmid, so it had no growth on SD−Leu, but it grew well on YPG at both permissive and non-permissive temperatures. W303ΔCOQ9 harboring empty vector (EV) showed no growth on YPG at either temperature as expected. W303ΔCOQ9 harboring wild-type COQ9 (COQ9) was included to provide a positive control for growth on YPG at the different temperatures. Yeast Δcoq9 mutants harboring plasmids containing single mutations, E55G and Q256L, did not show altered growth at the non-permissive temperature, but the presence of the R107G single mutation did produce slightly defective growth at 37 °C (Fig. 4A). Yeast Δcoq9 mutants harboring plasmids containing double mutations in combination with R107G had defective growth at 37 °C, but not with E55G Q256L (Fig. 4B). The combination of three amino acid substitution mutations E55G R107G Q256L recapitulated the TS19 phenotype (Fig. 4C). The presence of two mutations upstream of COQ9 start codon had no effect on yeast growth at non-permissive temperature (Fig. 4). Therefore, the full temperature sensitive phenotype of Δcoq9:TS19 requires the presence of E55G, R107G, and Q256L mutations.
3.3 Temperature-sensitive mutations in COQ9 lead to the destabilization of other Coq polypeptides at non-permissive temperature
Deletion of the yeast COQ9 gene leads to the decreased steady state of other Coq polypeptides, especially Coq4 and Coq7 [6]. To determine whether the Coq9-ts19 polypeptide impacts other Coq polypeptide levels, we grew wild type (WT) and Δcoq9:TS19 (Δ9:TS19) yeast in YPGal for 18.5 hours at 25 °C or 37 °C, and then isolated mitochondria. The steady-state levels of Coq9, Coq4, Coq7, Coq5 and Coq6 in isolated mitochondria were analyzed by immunoblotting (Fig. 5). In wild-type mitochondria, Coq9, Coq4, and Coq7 levels were increased at 37 °C (Fig. 5A, B and C). Expression of certain genes in S. cerevisiae can be induced or repressed in response to environmental changes, such as heat shock [29]. Therefore, the increased steady-state levels of Coq polypeptides at the non-permissive temperature might be a stress response. Coq5 and Coq6 levels were not changed at higher temperature (Fig. 5D and E). In the Δcoq9:TS19 mutant, steady state polypeptide levels of Coq9-ts19 were increased at the non-permissive temperature (Fig. 4A), while Coq4, Coq7, Coq5, and Coq6 tended to be decreased at 37 °C (Fig. 5B, C, D, and E). The results suggest that at high temperature the expression of Coq9-ts19 causes destabilization of the Coq polypeptide complex.
Fig. 5. Expression of Coq9-ts19 polypeptide affects steady state levels of other yeast Coq polypeptides in response to different growth temperatures.
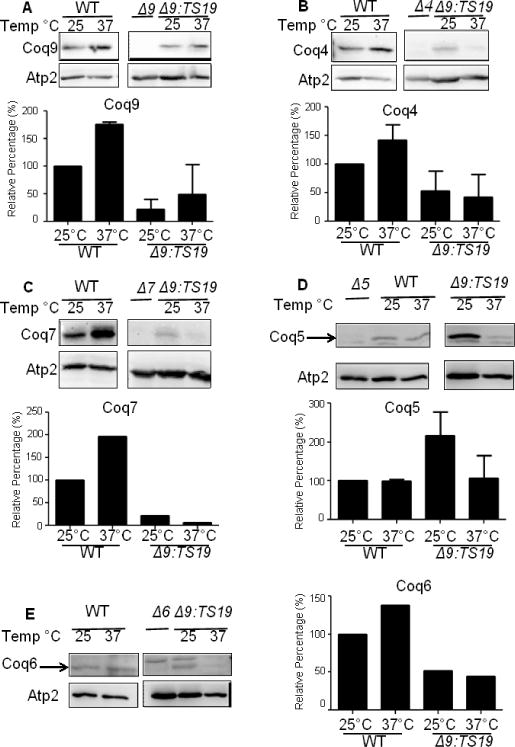
The mitochondria of W3031B (WT) and W303ΔCOQ9 harboring the temperature-sensitive plasmid TS19 (Δ9:TS19) were isolated after yeast were grown for 18.5 hours at either 25°C or 37°C. Mitochondria were also isolated from the null control strains W303ΔCOQ9 (Δ9), W303ΔCOQ7 (Δ7), and W303ΔCOQ4 (Δ4) after yeast were grown overnight at 30 °C. Purified mitochondria (15 μg protein) were separated by SDS-PAGE and analyzed by immunoblot. Immunoblots were performed with antibodies against the designated polypeptides: Coq4, Coq5, Coq6, Coq7, Coq9 and Atp2. Images presented within a given panel were derived from the same gel, and thus the band intensities corresponding to the same antisera can be directly compared. Densitometry quantification was conducted with the software ImageJ (NIH) and plotted as graphs. The signals of COQ4, COQ5, COQ6, COQ7, and COQ9 were each normalized to the signals of ATP2. Normalized values were then compared to WT at 25 °C to get the relative percentage. The immunoblot depicted is representative of two independent blots performed with two different preparations of purified mitochondria. (A, B, D) Bars designate the average signal + S.D. (n=2); (C, E) Each bar represents one measurement (n=1) because signals for Coq6 and Coq7 were detectable at 37 °C in only one set of the immunoblots.
3.4 Changes in COQ RNA levels do not correspond to the observed changes in Coq polypeptide levels
To investigate whether the change of Coq polypeptides at different temperatures is a response at gene expression level or protein level, we analyzed the mRNA levels of COQ4, COQ5, COQ6, COQ7 and COQ9 in WT and Δ9:TS19 yeast grown at either the permissive (25 °C) or non-permissive temperatures (37 °C). The mRNA levels of COQ4, COQ5, COQ6, COQ7 and COQ9 were not changed in the Δ9:TS19 mutant at different temperatures (Fig. 6). This was also the case for the mRNA levels of COQ5, COQ7 and COQ9 in wild type (Fig. 6A, C and D). The mRNA levels of COQ4 and COQ6 in wild-type yeast were decreased at non-permissive temperature (Fig. 6B and E). Thus, the increase observed in the steady state polypeptide levels of Coq9, Coq4, Coq7, and Coq6 in wild-type mitochondria at 37°C (Fig. 5A, B and C) cannot be attributed to corresponding changes in mRNA content. Therefore, it seems most likely that changes in steady state Coq polypeptide levels observed in Fig. 5 are instead due to the stabilization or destabilization of the CoQ-synthome.
Fig. 6. The observed changes in Coq polypeptide levels with temperature do not correspond to alterations in COQ RNA content.
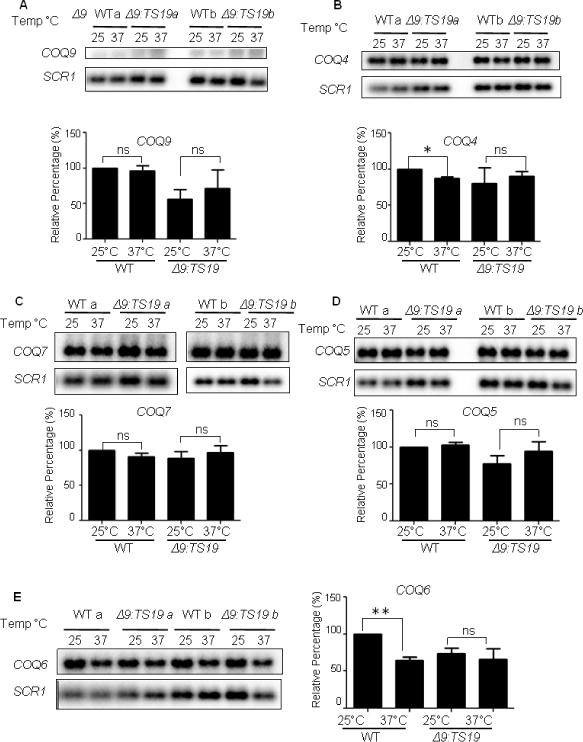
The total RNA of W3031B (WT) and W303ΔCOQ9 harboring the temperature-sensitive plasmid TS19 (Δ9:TS19) were extracted after yeast were grown for 18.5 hours at 25°C or 37°C. RNA were also extracted from the null control strains W303ΔCOQ9 (Δ9), W303ΔCOQ7 (Δ7), and W303ΔCOQ4 (Δ4) after yeast were grown overnight at 30 °C. Aliquots of RNA (5 μg) were separated by 1.2% agarose gel and analyzed by Northern blot. Hybridizations were performed with probes against the designated RNA: COQ4, COQ5, COQ6, COQ7, COQ9 and SCR1. Northern blot assay signals were quantified with the Quantity One software from the Bio-Rad FX Plus Phosphorimaging System. The quantified signals of COQ4, COQ5, COQ6, COQ7, and COQ9 were each normalized by the signals of SCR1. Normalized values were then compared to WT at 25°C to get the relative percentage. Each bar represents a total of two measurements from two independent samples (n=2). Significant changes in the amounts of mRNA at different temperatures were determined with the Student’s two-tailed t-test. The *symbols represent mRNA in samples at 37 °C compared to mRNA at 25 °C; *, p<0.05, **, p<0.01, ***, p<0.001.
3.5 Incubation at the non-permissive temperature leads to decreased Q6 and the accumulation of nitrogen-containing intermediates in the yeast Δcoq9 mutant harboring TS19
The results in Figs. 4 and 5 suggest that the Coq9 polypeptide harboring the TS19 mutations (Coq9-ts19) is functionally impaired at the non-permissive temperature, 37 °C. To gain further insight into the nature of the Coq9-ts19 temperature sensitive defects, we analyzed the de novo synthesis of Q6 and Q6-intermediates in the Δcoq9:TS19 mutant. Wild type (WT) and coq9 null mutant harboring empty vector (Δcoq9: EV) were included as controls. Yeast were grown in selective liquid media for 18.5 hours at 25 °C or 37 °C followed by labeling with 10 μg/ml 13C6-pABA for five hours at 25 °C or 37 °C. At 37 °C there was a two-fold decrease in de novo synthesized 13C6-Q6 in WT, but there was not significant change of 12C-Q6. In contrast, there was a marked decrease of both 12C-Q6 and 13C6-Q6 in Δcoq9:TS19 at the non-permissive temperature (Fig.7A). Therefore, both the content of 12C-Q6 and the synthesis of de novo 13C6-Q6 are dramatically decreased by high-temperature incubation in the Δcoq9:TS19 mutant.
Fig. 7. Yeast strains expressing Coq9-ts19 have decreased Q6 content and accumulate higher levels of nitrogen-containing Q-intermediates at the restrictive temperature.
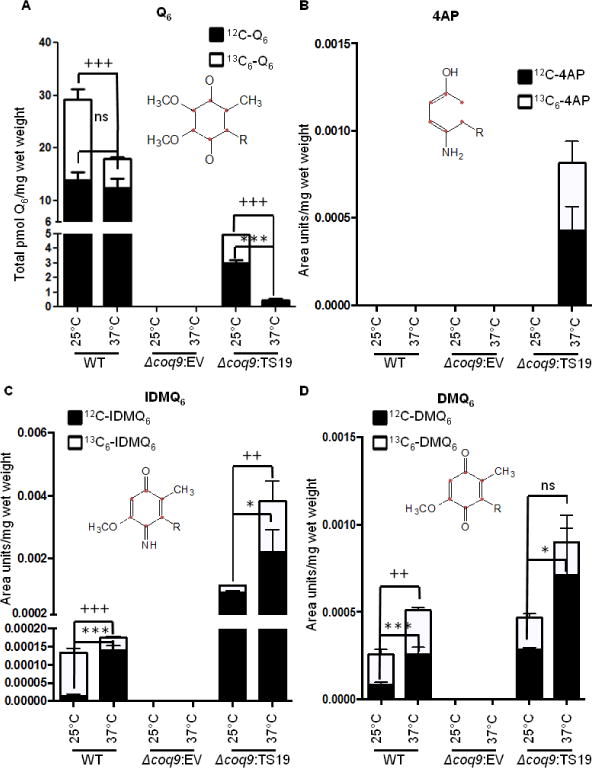
Yeast W3031B (WT), W303ΔCOQ9 harboring pRS315 empty vector (Δcoq9: EV), and W303ΔCOQ9 harboring the temperature-sensitive plasmid TS19 (Δcoq9: TS19) were seeded in 20 ml of SD-Complete (WT) or SD−Leu (Δcoq9: EV and Δcoq9: TS19) at 0.01 A600nm/ml and grown for 18.5 hours at 25 °C or 37 °C. 13C6-pABA (10 μg/ml) was added to yeast cultures and incubations were continued at either 25 °C or 37 °C. After labeling for 5 h, yeast cells (30 A600nm) were collected as cell pellets from which lipids were extracted and analyzed by RP-HPLC-MS/MS. Each bar represents a total of four measurements from two independent samples each with two injections. Black bars represent the amount of the designated 12C-compounds and white bars represent the amount of 13C6-compounds. For Q6, IDMQ6, and DMQ6, the total amounts of the 12C- and 13C6-compounds represent the sum of reduced and oxidized forms; 12C-AP and 13C6-4AP, were present only in the reduced form. Significant changes in the amounts of Q6 and Q6 intermediates at different temperatures were determined with the Student’s two-tailed t-test. The *symbols represent 12C-compounds in samples at 37 °C compared to 12C-compounds at 25 °C; *, p<0.05, **, p<0.01, ***, p<0.001. The + symbols represent 13C6-labeled compounds in samples at 37°C were compared to samples at 25°C; +, p<0.05, ++, p<0.01, +++, p<0.001; ns designates “non-significant”.
The nitrogen containing compounds, 4-AP and IDMQ6 accumulated in the Δcoq9:TS19 mutant. For example, 12C- and 13C6-4-AP were uniquely present in the Δcoq9:TS19 mutant at 37 °C (Fig. 7B) and levels of 13C6-IDMQ6 and 12C6-IDMQ6 increased six- and two-fold, respectively, at the non-permissive temperature (Fig. 7C). In contrast, WT had decreased amount of 13C6-IDMQ6 at the non-permissive temperature (Fig. 7C). Because Coq9 appears to be required to convert IDMQ6 to DMQ6, we also measured the amount of DMQ6. We found that both 12C6- and 13C6-DMQ6 were increased significantly in WT at the non-permissive temperature. However the amount of 13C6-DMQ6 was not changed in the Δcoq9:TS19 mutant at 37 °C (Fig. 7D). In conclusion, incubation of the Δcoq9:TS19 mutant at 37 °C led to a dramatic decline in the amount of Q6 and accumulation of the nitrogen containing compounds, 4-AP and IDMQ6, that derive from 13C6-pABA.
4. Discussion
Q plays a crucial role in mitochondrial electron transport and also serves as an important lipid soluble antioxidant. Despite the obvious importance of Q in human health and mitochondrial disease, many questions remain regarding its biosynthesis. Although the Coq9 polypeptide is one of eleven polypeptides essential for Q biosynthesis in yeast and human cells, the functional role Coq9 plays in the Q biosynthetic pathway remains an outstanding question.
In this study we examined the role of Coq9 in removing amino/imino groups from yeast Q-intermediates derived from pABA (Fig. 1). The finding that IDMQ6 accumulates in a yeast coq9 null mutant overexpressing COQ8 [10] suggested that Coq9 is required for the deamination of IDMQ6. Here we identified an earlier and new imino-intermediate in the pathway, IDDMQ6. We discovered this intermediate when the coq5-5 point mutant, defective in the C-methyltransferase step, was fed 13C6-pABA. We showed that the coq5-5 mutant fed 13C6-pABA accumulated both 13C-DDMQ6 and 13C6-IDDMQ6 (Fig. 2A and 2B). We speculated that the deamination of IDDMQ6 would depend on Coq9. Therefore, we analyzed the intermediates that accumulated in the coq5-5, Δcoq9 double mutant over-expressing Coq8 (coq5-5 Δcoq9:mcCOQ8). We found that this yeast strain lacked 13C6-DDMQ6, but still accumulated 13C6-IDDMQ6 (Fig. 2A and 2B). The data are consistent with the idea that Coq9 is required for removal of the nitrogen substituent for IDDMQ6 to form DDMQ6. The results also suggest that normally, the Coq5 C-methyltransferase acts prior to Coq9 and methylates IDDMQ6 to form IDMQ6. In the event that Coq5 activity is slow (or defective) then Coq9 is able to process IDDMQ6 to DDMQ6. There are likely to be profound differences between the yeast and human enzymes at these steps, because human cells are unable to convert pABA to Q [17]
Yeast Coq9 also plays an important role in supporting the activity of Coq6. This is evident because the dysfunction of Coq9 leads to the accumulation of 4-AP (Fig. 2C and 6B), which is an intermediate found in coq6 null yeast mutants over-expressing COQ8 [10]. In both the Δcoq6 and Δcoq9 mutants, the accumulation of 13C6-4-AP depended on the presence of 13C6-pABA and on Coq8 over-expression. Thus, even though Coq9 appears to be essential in removing the nitrogen groups from Q-intermediates, we have not demonstrated that Coq9 is the enzyme that catalyzes the deamination step directly. Indeed, based on the accumulation of 4-AP, it is likely that Coq6 may play an important role in mediating the deamination step(s). It is important to note that there is some Coq6 activity present in the coq9 null mutant over-expressing Coq8, because Q-intermediates accumulate that harbor the Coq6-mediated hydroxyl-group, such as 13C6-IDMQ6. While we have postulated potential pathways linking 4-AP, IDDMQ6 and IDMQ6 to the production of Q6 (Fig. 1 and [7]), none have yet been proved to be productive intermediates in the pathway leading to Q6.
While yeast Coq6 does not function very well in the absence of Coq9, several lines of evidence suggest that Coq7 is completely inactive in the absence of Coq9. Coq7 is a di-iron containing hydroxylase that catalyzes the hydroxylation of DMQ [30, 31]. Mutations in COQ7 result in the accumulation of DMQ in S. cerevisiae, C. elegans and mice [10, 32, 33]. Yeast coq7 and coq9 null mutants over-expressing COQ8 both accumulate DMQ6 when 4-HB is provided as the ring precursor [10]. In the yeast Δcoq9 strain, Coq8 over-expression only slightly increases the steady state level of Coq7, while Coq8 over-expression restored Coq9 polypeptide to wild-type level in Δcoq7. Purification of HA-tagged yeast Coq9 captures the yeast Coq7, Coq4, Coq6 and Coq5 polypeptides [6]. In addition, purification of tagged forms of Coq3 and Coq6 also capture Coq4, Coq5, Coq7, Coq8, and Coq9 [8]. These results indicate that yeast Coq9 and Coq7 are in a complex, together with other polypeptides and Q and Q-intermediates, termed the CoQ-synthome [7, 8]. Such Coq polypeptide biosynthetic complexes also appear to play a role in Q biosynthesis in the mouse. The lack of a functional Coq9 protein in homozygous Coq9 mutant mice causes a severe reduction in the Coq7 protein and accumulation of DMQ9 [34]. Human cells with Coq9 defects accumulate an intermediate slightly more polar than Q10; based on studies in yeast and mice this seems likely to be DMQ10 [35]. Thus it seems likely that the function of Coq9 in enhancing Coq7 function is conserved from yeast to humans.
To gain further insight into the function of Coq9 we created a temperature-sensitive yeast coq9 allele (coq9-ts19). We showed that amino acid substitution mutations E55G, R107G and Q256L recapitulated the YPG growth phenotype of the coq9-ts19 mutant. At non-permissive temperature (37 °C), the presence of the Coq9-ts19 polypeptide led to a trend of decreased steady-state polypeptide levels of Coq4, Coq5, Coq6, and Coq7 in isolated mitochondria (Fig. 5). These observed changes in Coq polypeptide levels did not result from changes in the corresponding COQ RNA levels (Fig. 6). Lipid extracts prepared from the coq9-ts19 mutant grown at non-permissive temperature showed a drastic decrease in Q6 content and the accumulation of intermediates containing the nitrogen group, IDMQ6 and 4-AP (Fig. 7). Taken together, these findings indicate that at the restrictive temperature, the coq9-ts19 mutant loses the ability to support the activities catalyzed by Coq6 and Coq7, and that the CoQ-synthome is destabilized.
In contrast, when wild-type yeast cells were subjected to the same temperature shift, steady state polypeptide levels of the Coq4, Coq7 and Coq6 polypeptides were increased (Fig. 5). Again, these changes were not paralleled by changes in the corresponding COQ RNA levels (Fig. 6). In contrast to the increase in the Coq polypeptide content at high temperature, the level of de novo synthesized Q6 was decreased, while the level of de novo synthesized 13C6-DMQ6 increased (Fig. 7). This observation is consistent with impaired Coq7 function at the elevated temperature. The phosphorylation state of Coq7 affects Q6 biosynthesis and the status of respiratory metabolism can cause Coq7 become dephosphorylated or phosphorylated [36]. It would be interesting to compare the phosphorylation state of Coq7 at permissive and non-permissive temperatures. It is also possible that certain Coq enzyme activities may be sensitive to high temperatures. It was shown that Coq3 homologs from either C. elegans or S. cerevisiae rescue the E. coli ubiG mutant at 30 °C, but not at 37 °C [37].
It is important to note that the recent study by Lohman et al. [38] provides insights into the possible effects of these yeast Coq9-ts19 amino acid substitutions. Lohman et al. solved the structure of human Coq9 and identified it as a member of an ancient protein family TFR (TetR family of regulators) with a canonical amino terminal helix-turn-helix (HTH) domain. Two human Coq9 polypeptides crystallized as a dimer and formed a hydrophobic interface that binds lipids, including phospholipids and Q [38]. Another separate surface patch of Coq9 was shown to be key to the ability to bind human Coq7. Intriguingly, the authors identified key amino acid residues in the human Coq9 polypeptide that affected binding with Coq7. We used the human Coq9 structure to predict the structure of yeast Coq9 with the protein homology/analogy recognition engine (Phyre 2) [39]. The predicted yeast structure is comprised of residues P40 to L231, so only the E55G and R107G can be evaluated (Fig. 8A). In the predicted structure, residue E55 is at the end of α helix one and R107 is in α helix five (Fig. 8A). When compared to the human Coq9 structure, yeast E55 corresponds to human A113, which resides at the C-terminus of α helix one, and is part of the HTH domain (Fig. 8B). The HTH motif is structurally similar to the TFR family of bacterial transcriptional regulators, but is predicted to lack DNA-binding capacity [38]. The presence of the E55G on its own did not affect growth on at the non-permissive temperature (Fig. 4). This is similar to the observation that single mutations introduced into the HTH domain of yeast Coq9 did not affect respiration competence [38]. Yeast R107 aligns with Q165 in α helix four of human Coq9 (Fig 8B). We found that the R107G substitution on its own produced a moderate temperature sensitive phenotype, but the combination or either E55G or Q256L in combination with R107G gave a much more pronounced temperature sensitivity. In the human Coq9 structure, Q165 resides in a region that is neither highly conserved nor is shown to be important in lipid binding or interaction with human Coq7 [38]. Our immunoblot analyses show that at non-permissive temperature, the steady state level of Coq9-ts19 is increased, while other Coq polypeptides are destablized (Fig. 5A).
Fig. 8. A homology model of yeast Coq9.
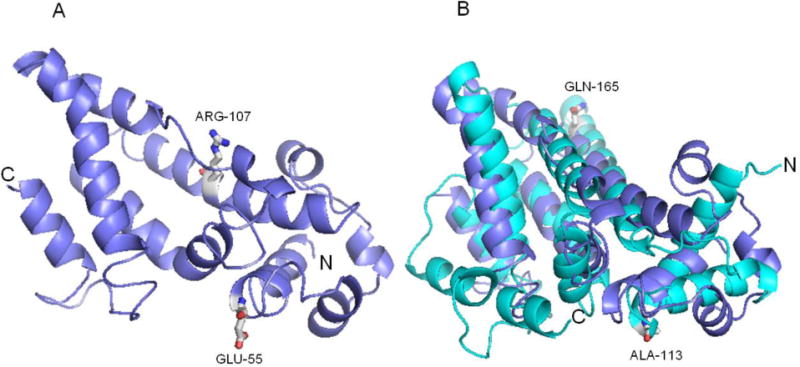
(A) The yeast Coq9 structure was predicted based on the crystal structure of human Coq9 by protein homology/analogy recognition engine (Phyre 2). There are 163 aligned residues, 13% identity and 99.54% confidence. The positions of two of the temperature sensitive amino acid substitution mutations are shown: E55 in α helix one, and R107 in α helix five. (B) The predicted yeast Coq9 structure (purple) is shown aligned with human Coq9 structure (cyan) [38]. Yeast E55 corresponds to human A113 (both are in a helix one in the HTH domain) and yeast R107 aligns with Q165 in α helix four of human Coq9.
According to a model proposed by Gonzalez-Mariscal, Coq7 is recruited to the precomplex to catalyze the conversion of DMQ6 to Q6 [40]. They proposed that Coq9 plays an important structural role to stabilize the Q-biosynthetic complex, and is recruited to form a 700 kDa precomplex as part of the nucleation process initiated by Coq4 binding to HHB [40]. Hence it seems plausible that the temperature-sensitive mutations cause misfolding of yeast Coq9-ts19 at high temperature and prevent its proper function or interaction with other Coq polypeptides and lead to the destabilization of the precomplex. There might be a small amount of functional Coq9-ts19, so some of the 700 kDa precomplex is able to form and produces DMQ6 [40]. Although their model did not depict the interaction of Coq9 and Coq7 as the means of Coq7’s recruitment, the results presented here, and the Coq9 structure by Lohman et al suggest that this may be the case. There is a large amount of DMQ6 accumulated in the temperature sensitive mutant at non-permissive temperature, but very little Q6 was produced (Fig. 7). In this scenario, the mutations in Coq9-ts19 disrupt the interaction of Coq7 and Coq9, so Coq7 cannot bind to the pre-complex and perform its function. Lohman et al. [38]noted that several residues predicted to affect the interaction of yeast Coq9 with Coq7 resulted in decreased Q6 and increased DMQ6. It would be interesting to determine the effect of these mutations on the accumulation of IDMQ6.
In conclusion, we found that yeast Coq9 is required for the deamination of 4-AP, IDDMQ6 and IDMQ6. At the non-permissive temperature, the coq9-ts19 mutant has low steady state levels of Coq4, Coq5, Coq6 and Coq7 polypeptides, shows defective growth on non-fermentable carbon source and a drastic decrease in the content of Q6, and accumulates imino/amino Q-intermediates The results presented here identify Coq9 as a multi-functional protein that is required for the function of Coq6 and Coq7, for removal of the nitrogen substituent from pABA-derived Q-intermediates, and is an essential component of the CoQ synthome.
Supplementary Material
Highlights.
For the first time, a temperature sensitive coq9 mutant, coq9-ts19 was isolated.
Coq4, Coq5, Coq6, and Coq7 polypeptides were destabilized in coq9-ts19 at 37 °C.
The coq9-ts19 has decreased Q6 and elevated imino-/amino-Q-intermediates at 37°C.
Coq9 is required to remove the nitrogen group from pABA-derived Q intermediates.
Coq9 is required for the function of Coq6 and Coq7 in coenzyme Q biosynthesis.
Acknowledgments
We thank Dr. Carla Koehler (UCLA) for her advice on generating the temperature-sensitive mutant plasmid, TS19, and for the Atp2 antibody. We thank Dr. A. Tzagoloff (Columbia University) for the original yeast coq mutant strains. We thank Kevin Roy for advice and assistance with RNA analyses, and the members of the Clarke lab for their helpful discussions and feedback on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by the National Science Foundation Grant MCB-1330803 (C.F.C.), and the National Institutes of Health Grant T32 GM 008496 and the Whitcome Pre-doctoral Training Program (UCLA Molecular Biology Institute) supported C. H. H. The authors thank and the General Biochemistry Laboratory course students (fall 2013 and spring 2013) at California State University Fullerton for their technical assistance in contributing towards the generation of some of the mutants used in this study.
The abbreviations used are: 4-AP, 3-hexaprenyl-4-aminophenol; DDMQ6, demethyl-demethoxy; DMQ6, demethoxy-Q6; HAB, 3-hexaprenyl-4-aminobenzoic acid; 4HB, 4-hydroxybenzoic acid; HHB, 3-hexaprenyl-4-hydroxybenzoic acid; 4-HP, 3-hexaprenyl-4-hydroxyphenol; IDDMQ6, imino-demethyldemethoxy Q6; IDMQ6, imino-demethoxy-Q6; mcCOQ8, multi-copy COQ8; pABA, para-aminobenzoic acid; RP-HPLC-MS/MS; reverse phase-HPLC-MS/MS; Q, Coenzyme Q.
References
- 1.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Crane F, Barr R. Chemical structure and properties of coenzyme Q and related compounds, Coenzyme Q biochemistry, bioenergetics and clinical applications of ubiquinone. John Wiley & Sons; Chichester, United Kingdom: 1985. pp. 1–37. [Google Scholar]
- 3.Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Mühlenhoff U, Ozeir M, Lill R, Fontecave M. Involvement of Mitochondrial Ferredoxin and para-Aminobenzoic Acid in Yeast Coenzyme Q Biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Allan CM, Hill S, Morvaridi S, Saiki R, Johnson JS, Liau WS, Hirano K, Kawashima T, Ji Z, Loo JA, Shepherd JN, Clarke CF. A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim Biophys Acta. 2013;1831:776–791. doi: 10.1016/j.bbalip.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marbois B, Xie LX, Choi S, Hirano K, Hyman K, Clarke CF. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh EJ, Gin P, Gulmezian M, Tran UC, Saiki R, Marbois BN, Clarke CF. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007;463:19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He CH, Xie LX, Allan CM, Tran UC, Clarke CF. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allan CM, Awad AM, Johnson JS, Shirasaki DI, Wang C, Blaby-Haas CE, Merchant SS, Loo JA, Clarke CF. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J Biol Chem. 2015;290 doi: 10.1074/jbc.M114.633131. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7(Supplement):S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie LX, Ozeir M, Tang JY, Chen JY, Jaquinod SK, Fontecave M, Clarke CF, Pierrel F. Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J Biol Chem. 2012;287:23571–23581. doi: 10.1074/jbc.M112.360354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauche A, Krause-Buchholz U, Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 12.Xie LX, Hsieh EJ, Watanabe S, Allan CM, Chen JY, Tran UC, Clarke CF. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim Biophys Acta. 2011;1811:348–360. doi: 10.1016/j.bbalip.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefely JA, Reidenbach AG, Ulbrich A, Oruganty K, Floyd BJ, Jochem A, Saunders JM, Johnson IE, Minogue CE, Wrobel RL, Barber GE, Lee D, Li S, Kannan N, Coon JJ, Bingman CA, Pagliarini DJ. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Molecular cell. 2015;57:83–94. doi: 10.1016/j.molcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler B, Jia Z. Preparation and characterization of human ADCK3, a putative atypical kinase. Protein expression and purification. 2015;108:13–17. doi: 10.1016/j.pep.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- 16.Padilla S, Tran UC, Jimenez-Hidalgo M, Lopez-Martin JM, Martin-Montalvo A, Clarke CF, Navas P, Santos-Ocana C. Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell Mol Life Sci. 2009;66:173–186. doi: 10.1007/s00018-008-8547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie LX, Williams KJ, He CH, Weng E, Khong S, Rose TE, Kwon O, Bensinger SJ, Marbois BN, Clarke CF. Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J Lipid Res. 2015;56 doi: 10.1194/jlr.M057919. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Block A, Widhalm JR, Fatihi A, Cahoon RE, Wamboldt Y, Elowsky C, Mackenzie SA, Cahoon EB, Chapple C, Dudareva N, Basset GJ. The Origin and Biosynthesis of the Benzenoid Moiety of Ubiquinone (Coenzyme Q) in Arabidopsis. The Plant cell. 2014;26:1938–1948. doi: 10.1105/tpc.114.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 2000. [Google Scholar]
- 20.Barkovich RJ, Shtanko A, Shepherd JA, Lee PT, Myles DC, Tzagoloff A, Clarke CF. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J Biol Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- 21.Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh EJ, Dinoso JB, Clarke CF. A tRNA(TRP) gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochem Biophys Res Commun. 2004;317:648–653. doi: 10.1016/j.bbrc.2004.03.096. [DOI] [PubMed] [Google Scholar]
- 23.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 24.Padilla-Lopez S, Jimenez-Hidalgo M, Martin-Montalvo A, Clarke CF, Navas P, Santos-Ocana C. Genetic evidence for the requirement of the endocytic pathway in the uptake of coenzyme Q6 in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1788:1238–1248. doi: 10.1016/j.bbamem.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayani S, Chanfreau GF. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA. 2012;18:1563–1572. doi: 10.1261/rna.033779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Hadid Q, Roy K, Munroe W, Dzialo MC, Chanfreau GF, Clarke SG. Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly. Molecular and cellular biology. 2014;34:2903–2916. doi: 10.1128/MCB.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba SW, Belogrudov GI, Lee JC, Lee PT, Strahan J, Shepherd JN, Clarke CF. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J Biol Chem. 2004;279:10052–10059. doi: 10.1074/jbc.M313712200. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen TPT, Casarin A, Desbats MA, Doimo M, Trevisson E, Santos-Ocana C, Navas P, Clarke CF, Salviati L. Molecular characterization of the human COQ5 C-methyltransferase in Coenzyme Q biosynthesis. Biochim Biophys Acta. 2014;1841:1628–1638. doi: 10.1016/j.bbalip.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behan RK, Lippard SJ. The aging-associated enzyme CLK-1 is a member of the carboxylate-bridged diiron family of proteins. Biochemistry. 2010;49:9679–9681. doi: 10.1021/bi101475z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu TT, Lee SJ, Apfel UP, Lippard SJ. Aging-associated enzyme human clock-1: substrate-mediated reduction of the diiron center for 5-demethoxyubiquinone hydroxylation. Biochemistry. 2013;52:2236–2244. doi: 10.1021/bi301674p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu AY, Poon WW, Shepherd JA, Myles DC, Clarke CF. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Hekimi S. Molecular genetics of ubiquinone biosynthesis in animals. Crit Rev Biochem Mol Biol. 2013;48:69–88. doi: 10.3109/10409238.2012.741564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Corzo L, Luna-Sanchez M, Doerrier C, Garcia JA, Guaras A, Acin-Perez R, Bullejos-Peregrin J, Lopez A, Escames G, Enriquez JA, Acuna-Castroviejo D, Lopez LC. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum Mol Genet. 2013;22:1233–1248. doi: 10.1093/hmg/dds530. [DOI] [PubMed] [Google Scholar]
- 35.Duncan AJ, Bitner-Glindzicz M, Meunier B, Costello H, Hargreaves IP, Lopez LC, Hirano M, Quinzii CM, Sadowski MI, Hardy J, Singleton A, Clayton PT, Rahman S. A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am J Hum Genet. 2009;84:558–566. doi: 10.1016/j.ajhg.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Montalvo A, Gonzalez-Mariscal I, Padilla S, Ballesteros M, Brautigan DL, Navas P, Santos-Ocana C. Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p. Biochemical J. 2011;440:107–114. doi: 10.1042/BJ20101422. [DOI] [PubMed] [Google Scholar]
- 37.Gomez F, Saiki R, Chin R, Srinivasan C, Clarke CF. Restoring de novo coenzyme Q biosynthesis in Caenorhabditis elegans coq-3 mutants yields profound rescue compared to exogenous coenzyme Q supplementation. Gene. 2012;506:106–116. doi: 10.1016/j.gene.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sanchez M, Jochem A, Lew S, Seetharaman J, Xiao R, Wang H, Westphall MS, Wrobel RL, Everett JK, Mitchell JC, Lopez LC, Coon JJ, Tong L, Pagliarini DJ. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci U S A. 2014;111:E4697–4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Mariscal I, Garcia-Teston E, Padilla S, Martin-Montalvo A, Pomares-Viciana T, Vazquez-Fonseca L, Gandolfo-Dominguez P, Santos-Ocana C. Regulation of coenzyme Q biosynthesis in yeast: a new complex in the block. IUBMB life. 2014;66:63–70. doi: 10.1002/iub.1243. [DOI] [PubMed] [Google Scholar]
- 41.Hsu AY, Do TQ, Lee PT, Clarke CF. Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim Biophys Acta. 2000;1484:287–297. doi: 10.1016/s1388-1981(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 42.Marbois BN, Clarke CF. The COQ7 gene encodes a protein in saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 43.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 44.Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol Biol Cell. 2008;19:5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 46.Belogrudov GI, Lee PT, Jonassen T, Hsu AY, Gin P, Clarke CF. Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch Biochem Biophys. 2001;392:48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- 47.Gin P, Hsu AY, Rothman SC, Jonassen T, Lee PT, Tzagoloff A, Clarke CF. The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J Biol Chem. 2003;278:25308–25316. doi: 10.1074/jbc.M303234200. [DOI] [PubMed] [Google Scholar]
- 48.Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. The Journal of biological chemistry. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


