Abstract
While crystal and NMR structures exist of the influenza A M2 protein, there is disagreement between models. Depending on the requirements of the technique employed, M2 has been studied in a range of membrane mimetics including detergent micelles and membrane bilayers differing in lipid composition. The use of different model membranes complicates the integration of results from published studies necessary for an overall understanding of the M2 protein. Here we show using site-directed spin-label EPR spectroscopy (SDSL-EPR) that the conformations of M2 peptides in membrane bilayers are clearly influenced by the lipid composition of the bilayers. Altering the bilayer thickness or the lateral pressure profile within the bilayer membrane changes the M2 conformation observed. The multiple M2 peptide conformations observed here, and in other published studies, optimistically may be considered conformations that are sampled by the protein at various stages during influenza infectivity. However, care should be taken that the heterogeneity observed in published structures is not simply an artifact of the choice of the model membrane.
Keywords: influenza A M2 protein, model membrane, site-directed spin labeling, SDSL-EPR, hydrophobic mismatch, lateral pressure, phosphatidylethanolamine
INTRODUCTION
The M2 protein is a 97-amino-acid multifunctional protein that is assembled into a tetramer which spans the viral membrane.1 The most extensively studied function of the M2 protein is its proton channel activity that is crucial for uncoating of virions when viruses enter cells via endosomes.2 In addition to acting as a proton channel, M2 has been shown to play a critical role in viral assembly and budding.3
As a hydrophobic membrane-bound protein, M2 presents challenges in terms of protein preparation, reconstitution into membranes and structure determination of large peptide/lipid complexes. A series of biophysical methods have been employed to study the conformation and dynamics of the M2 protein.4-7 Depending on the requirements of the technique employed, a range of membrane mimetics have been used, including detergent micelles and membrane bilayers composed of a range of different lipids. The use of different peptide constructs and different model membranes complicates the integration of results from published studies necessary for an overall understanding of the M2 protein.
In this study, we have probed the conformation and dynamics of two different M2 peptide constructs in different lipid bilayer membranes using site-directed spin-label electron paramagnetic spectroscopy (SDSL-EPR). SDSL-EPR is an information-rich method and is not limited by size of the protein/lipid complex.8 Therefore, SDSL-EPR offers the valuable opportunity to compare how the membrane mimetic used in structure determination impacts the M2 conformation observed.
Using SDSL-EPR, we previously published a study demonstrating that the conformation of the pore region of the M2 proton channel depended on the lipid composition of the membrane bilayers.9 The peptide used in that study was referred to as M2TM and contained transmembrane residues 22-46 (Figure 1). M2TM peptides form a homotetrameric proton channel. In that study we attached a nitroxide spin label to the N-terminus of the M2TM peptide and observed the N-termini of the M2TM peptides moved closer together within the tetramer as the membrane thickness increased, consistent with a conformational change in response to hydrophobic mismatch. We also noted an intriguing finding in this earlier work. Hydrophobic matching could not account for all our data without considering the lateral pressure profiles of the lipid bilayers.
FIGURE 1.
M2 peptide sequences used for SDSL-EPR studies. Sequences correspond to the M2 protein from influenza strain A/Udorn/72 (H3N2). M2TM peptides contain residues 22-46 and are spin-labeled at the N-termini. The M2TMC peptides contain residues 23-60. M2TMC R45C/C50S peptides are spin labeled at a single cysteine site (underlined) and have the WT C50 site changed to a serine. M2TMC C50S peptides do not have a spin-label attached and are used for dilute-labeled spectra.
Here we expand on our previous SDSL-EPR studies and use a longer M2 peptide, called M2TMC, which consists of residues 23-60 and includes both the transmembrane domain and the first 14 residues of the C-terminal domain (Figure 2). We demonstrate that M2TMC peptides mirror the behavior of the shorter M2TM peptides in their response to changes in hydrophobic thickness of the membrane bilayers. Furthermore we probe the role of membrane lateral pressure10 by studying M2TM peptides in lipid bilayers with varying amounts of phosphatidylethanolamine (PE) and demonstrate there are significant changes in the conformation of M2TM.
FIGURE 2.
X-band EPR spectra of M2TMC spin-labeled at position 45 in DLPC/DLPG 4:1, DMPC/DMPG 4:1 and POPC/POPG 4:1. Peptide lipid molar ratio 1:200. Dilute-labeled spectra are shown in grey and fully labeled spectra are shown in black. Addition of M2TMC C50S was used for dilute-labeled spectra. Dilute-labeled samples have one or less spin label per tetramer. All spectra have been normalized to the same number of spins.
MATERIALS AND METHODS
Synthesis, spin labeling and purification of peptides
The 25-residue M2TM peptides (Figure 1) were prepared by solid phase Fmoc synthesis, spin-labeled at the N-terminus with 2,2,5,5-tetramethyl-3-pyrrolin-1-oxyl-3-carboxylic acid N-hydroxysuccinimide ester and purified as described previously.9 The 38-residue M2TMC peptides (Figure 1) were synthesized, spin-labeled with 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate spin label (MTSSL) and purified as previously described.11 Electrophysiology experiments indicate that a cysteine modification at the labeling site (45) used for the M2TMC peptide does not significantly perturb channel function.12 The identities of peptides were confirmed using matrix-assisted laser desorption ionization mass spectrometry.
Sample Preparation
Samples were prepared using the following lipids: 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dilauroyl-sn-glycero-3-phosphatidylethanolamine (DLPE), 1,2-dimyristoyl-sn-glycero-3-phosphatidylethanolamine (DMPE), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanol- amine (DOPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE), 1,2-dilauroyl -sn-glycero-3-[phospho-rac-(1-glycerol)] (DLPG), 1,2-dimyristoyl -sn-glycero-3-[phospho-rac-(1-glycerol)] (DMPG) and 1-palmitoyl-2-oleyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG). All lipids were purchased from Avanti Polar Lipids (Alabaster, AL). All PC/PE lipid mixtures were miscible and in the liquid crystalline phase at 300 K which was the collection temperature for EPR spectroscopy.13, 14 The M2TM reconstitution procedure9 and M2TMC reconstitution procedures7, 11 were described previously using a peptide to lipid ratio of 1:200. Although M2TM peptides can be reconstituted into pure phosphatidylcholine (PC) lipid bilayers, we found that addition of phosphatidylglycerol (PG) lipids was necessary for the reproducibility and stability of bilayer reconstituted M2TMC peptides. All samples were collected in 50 mM Tris, 100 mM KCl, 1 mM EDTA, pH 8.6 buffer.
EPR Spectroscopy
CW X-band continuous wave EPR spectra were collected on a Bruker EMX spectrometer. All spectra were collected were collected in glass capillary tubes with 1.0 mm ID at 300 K. Each spectrum was collected with 2 mW incident power, 100 kHz modulation frequency and 1 G modulation amplitude. For comparison of CW line shapes, each spectrum was double integrated and normalized to the same number of spins. Dilute-labeled samples (with one or less spin label per tetramer) were compared with fully labeled samples (four spin labels per tetramer). Broadening in the fully labeled samples with respect to the dilute-labeled samples is due to spin-spin interactions. Due to the tetrameric geometry of the M2 channel, spin-spin coupling originates from interactions between both lateral neighbors and diagonally related subunits. Under these conditions, a qualitative estimate of magnitude of spin-spin interactions (Ω) can be obtained from the ratio of the amplitudes of the central resonance line (M=0) between the dilute-labeled and fully labeled samples, both normalized to the total number of spins in the samples.16 At large spin-spin distances, Ω is approximately one (no spin-spin coupling) but increases as spin labels approach each other. Calculation and comparison of Ω values has been previously used to characterize the structural rearrangements that occur during the gating of other homooligomeric channels including a potassium channel17 and a mechanosensitive channel.18
RESULTS AND DISCUSSION
Conformation of M2TMC in Membrane Bilayers of Differing Hydrophobic Thickness
Previously we showed the conformation of the pore region of the M2 proton channel formed by the M2TM peptides depends on hydrophobic thickness of the membrane.9 In that study we attached a nitroxide spin label to the N-terminus of M2TM peptides and observed the N-termini of the peptides moved closer together within the tetramer as the membrane thickness increased, consistent with conformational change in response to hydrophobic mismatch.13 If the hydrophobic mismatch hypothesis is relevant for M2 we should also be able to observe changes in conformation in response to changes in hydrophobic thickness of the membrane for the longer M2TMC construct.
We collected X-band CW EPR spectra of a M2TMC peptide with a site-specific label at an introduced cysteine residue placed close to the C-terminal end of the transmembrane helix. Based on the geometry of the bundle, labels at the helix ends should experience the most significant changes in spin-coupling distance with helix tilt. Spin labels located near the middle of the helix might interfere with helix-helix contacts essential to oligomerization and should have only small, likely undetectable changes in spin coupling with helix tilt. Three bilayers with different acyl chain hydrophobic thicknesses were tested, DLPC (19.5 Å), DMPC (23.0 Å) and POPC (26.5 Å). The 19 residue hydrophobic stretch of the transmembrane helix of M2 has been estimated bê28.5 Å.9
Two spectra are shown for each different bilayer composition in Figure 2. The higher amplitude grey spectra are dilute-labeled and the superimposed broader black spectra are fully labeled. The dilute-labeled samples have one or less spin label per tetramer whereas the fully labeled samples contain four spin labels per tetramer. Broadening in the fully labeled samples with respect to the dilute-labeled samples is due to spin-spin interactions. Due to the tetrameric geometry of the M2 channel, spin-spin coupling originates from interactions between both lateral neighbors and diagonally related subunits. Under these conditions, a qualitative estimate of magnitude of spin-spin interactions (Ω) can be obtained from the ratio of the amplitudes of the central resonance line (M=0) between the dilute-labeled and fully labeled samples, both normalized to the total number of spins in the samples.
As shown in Figure 3B, the observed spin-spin interaction increases for the spin-labeled M2TMC peptides as the bilayers thicken, consistent with a conformational change that brings the spin labels closer together to best match the peptide hydrophobic region to that of the hydrophobic lipid bilayer. Previously, we proposed that M2TM helices could adapt to the hydrophobic thickness of the membrane (Figure 3A) either by adjusting their tilt angle with respect to the membrane normal and/or by changing the ordering of the helical bundle from a looser tetramer, where helices make some contacts with each other, to a tighter tetramer, where helix-helix associations are maximized.9 These two mechanisms are not mutually exclusive and they may occur in concert with one another. Several previously models have shown that a kink can form in the M2 transmembrane helix.19 For simplicity, models in Figure 3 show a simple helix tilt mechanism.
FIGURE 3.
(A) Spin coupling (Ω) data from our previous SDSL-EPR study using N-terminus labeled M2TM peptides in PC bilayers9 and the current study (B) using M2TMC data from Figure 2. Both data sets provide support for a conformation change due to a change in bilayer thickness. For simplicity, only two of four peptides in the tetramer are shown in the hypothetical cartoon models shown in C and D. M2 peptides could adapt to the hydrophobic thickness of the membrane either by adjusting their tilt angle with respect to the membrane normal and/or by changing the ordering of the helical bundle from a looser tetramer, where helices make some contacts with each other, to a tighter tetramer, where helix-helix associations are maximized. These two mechanisms are not mutually exclusive and they may occur in concert with one another. For simplicity, the models shown here show a simple helix tilt mechanism.
Another possibility to consider is that a monomer-tetramer equilibrium could contribute to our results. The presence of spin-labeled monomers would produce a reduction in the spin-coupling values that would be indistinguishable from an increase in the distance between probes due to conformational change. A previous study of M2TM demonstrated that the peptides were essentially fully tetrameric in bilayers composed of POPC, DMPC and DLPC lipids15 suggesting that a monomer-tetramer equilibrium is not likely to be relevant for the studies presented here. However, the lipid bilayers used in that study are not the same as those used here, which include PG lipids, and it has not yet been demonstrated that M2TMC peptides are fully tetrameric in PC/PG lipid mixtures.
Conformation of M2TM in Membrane Bilayers with Different Amounts of PE
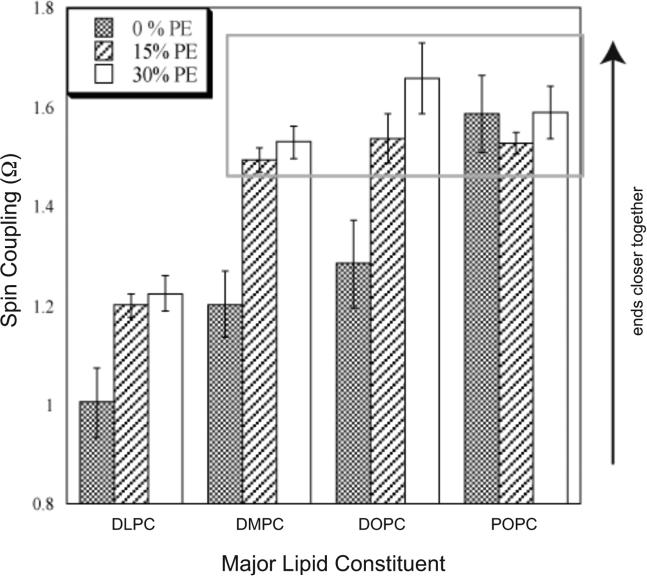
N-terminally spin-labeled M2TM peptides were reconstituted into DLPC(C12:0) , DMPC (C14:0), DOPC (C18:1) and POPC (C16:0-18:1) lipid bilayers, as well as mixed with either 15-mol% or 30-mol% of the cognate phosphatidylethanolamine (PE). The spectra are shown in Figure 4 and the corresponding interaction parameters (Ω) for each lipid environment are shown in Figure 5. A comparison of the grey bars show the M2TM peptides respond to an increase in hydrophobic thickness in pure PC bilayer (no PE added) as demonstrated in our previously published work.9 The observed pattern of spin interaction (Ω) shown in Figure 5 indicates that the spin labels are furthest in DLPC, approximately the same distance apart in DOPC and DMPC and closest in POPC. As the bilayer thickens, the conformation of the M2TM bundle changes to best match the peptide hydrophobic region to that of the hydrophobic lipid bilayer with spin labels getting closer leading to increased spin coupling.
FIGURE 4.
X-band EPR spectra of M2TM in DLPC, DMPC, DOPC, and POPC membranes upon addition of cognate PE lipids. Peptide to lipid molar ration of 1:200. Dilute-labeled spectra are shown in black and fully labeled spectra are shown in red. Dilute-labeled samples have one or less spin label per tetramer. All spectra have been normalized to the same number of spins.
FIGURE 5.
Calculation of spin coupling (Ω) for M2TM reconstituted into PC/PE bilayers. Ω is the ratio of the dilute-labeled spectral peak-to-peak amplitude at the central resonance line (M=0) to that of the fully labeled sample. At large spin-spin distances, Ω is approximately one (no spin-spin coupling) but increases as spin labels approach each other. The grey box indicates the limiting conformation discussed in the text.
In our earlier study, however, we noted an intriguing finding. Although of a similar hydrophobic thickness13 as POPC (26.5 Å), the DOPC (27 Å) bilayer supports a M2TM conformation closer to that found in the thinner DMPC bilayer (23.0 Å). Thus, a hydrophobic matching argument alone does not account for all the data. Peptide-lipid systems are complex and conceivably several mechanisms operate simultaneously to determine equilibrium conformations. DOPC (C18:1), unlike the other lipids tested, has a double bond in each of its acyl chains. The acyl chain unsaturation in DOPC leads to an increase in lateral pressure in the acyl chain region of the bilayer with a decrease in lateral pressure in the head group region as compared to DMPC.20 Thus, despite the differing hydrophobic thicknesses, the lateral pressure profile of a DOPC bilayer could energetically favor a M2TM conformation similar to that found in DMPC.
To further explore the effects of the membrane lateral pressure profile on the conformation of M2TM, we decided to test lipid bilayers containing PE. The addition of PE to a PC bilayer has been shown to increase the lateral pressure in the acyl chain region and decrease the pressure in the head group region.10 As shown in Figure 5, the addition of 15 mol% PE to PC bilayers leads to an increase in observed spin coupling for all binary lipid mixtures studied with the exception of POPC/POPE. When increasing the proportion of PE from 15-mol% to 30-mol% the observed spin coupling is virtually unchanged for all four binary lipid mixtures studied. The spin coupling values, Ω, are approximately 1.2 for the DLPC/DLPE mixtures and approach 1.6 for all the remaining PE-containing bilayers. Ω reflects the proximity of the spin-labeled ends and thus the conformation of the tetramer. Upon the addition of a PE lipid to DMPC, DOPC, and POPC, the M2 peptides appear to approach a limiting conformer (see grey box in Figure 5). Note that although pure DMPC and pure DOPC do not support the limiting conformation of M2TM, adding 15-mol% PE allows a shift of the equilibrium conformation of the M2 peptide to the limiting conformation. Once the tetramer reaches this conformation, further modulating the lipid-protein interactions (such as increasing PE content from 15 to 30-mol%) has little effect on measured spin-spin couplings.
In the DLPC/DLPE bilayers, the M2 peptide is unable to reach the limiting conformation described above, achieving a spin coupling of only 1.2 upon the addition of PE. It appears the acyl chain region of a DLPC bilayer is not thick enough to accommodate the limiting conformation without the energetically costly effect of exposing hydrophobic residues to the aqueous phase. Thus DLPC/DLPE incorporated M2 peptide cannot reach the conformation seen in the other PC/PE environments regardless of the increase in membrane lateral pressure contributed by PE. However, it is important to note that previous thiol-disulfide exchange experiments indicated that the antiviral drug amantadine binds to M2TM peptides in DLPC bilayers 15 consistent with the peptide forming a tetramer capable of drug binding in DLPC bilayers. Despite that fact, another factor to consider in the interpretation of the DLPC results is the possibility of a monomer-tetramer equilibrium. Spin-labeled monomers would produce a reduction in the spin-coupling values that would be indistinguishable from at increase in the distance between probes due to conformational change. A previous study of M2TM peptides demonstrated that the peptides were essentially fully tetrameric in bilayers composed DLPC lipids.15 However, it has not yet been demonstrated that M2 peptides are fully tetrameric in DLPC/DLPE lipid mixtures.
PE is a major constituent of cell membranes, ranging between 12% and 75% abundance, depending on tissue type.21 Analysis of the lipid composition of influenza A virions indicate that PE is a significant constituent of the virus envelope.22 It has been suggested that one possible reason for the abundance of PE lipids in biological membranes is that PE lipids can provide special packing properties essential for the function of some integral membrane proteins.10
PE lipids have smaller head groups than PC lipids and are cone-shaped. Unlike cylindrically shaped PC, the cone-like shape of PE may allow it to pack against the M2 peptide tetramer in a fashion that maximizes protein-protein contacts within the helical bundle. PE has been co-crystallized with some membrane proteins, suggesting that PE may be necessary to stabilize certain conformations of membrane proteins.23 The addition of PE was shown to stabilize the oligomeric structure of the KcsA potassium channel by increasing the membrane lateral pressure in the acyl chain region.24 The stabilization of distinct conformations of the large mechanosensitive channel for E.coli (MscL) can be achieved by manipulating the nature and extent of lipid-protein interactions.25
SUMMARY AND CONCLUSIONS
We have observed that the conformations of two M2 protein constructs are clearly influenced by the model membrane used. Several different studies have already pointed out the intrinsic plasticity of the truncated constructs of the M2 protein26 and conformational heterogeneity has been observed in previously published work on M2.19, 27 The full-length M2 protein has a more favorable free energy of association than truncated peptides30 and may not be as malleable as the M2 peptides studied here due to additional elements of conformational specificity contained within the full-length protein.
The structural plasticity displayed by M2 in response to membrane composition, as well as mutagenesis,26, 28 may be indicative of functional requirements for conformational changes during proton channel function and viral budding. For example, it is known that there is, at the least, a closed conformation at high pH and an open conformation at low pH for the M2 channel.11, 29 The multiple M2 peptides conformations observed here, and in other published studies, optimistically may be conformations that are sampled by the protein at various stages during influenza infectivity. However, care should be taken that the heterogeneity observed is not simply an artifact of sample design or reconstitution protocol.
ACKNOWLEDGEMENTS
Our work on the M2 protein has benefitted from a long-term and productive collaboration with many members of the DeGrado group past and present. In particular, we sincerely thank Bill DeGrado for being a gracious and generous colleague.
Grant Sponsor: NIH
Grant number: 1R15AI094483-01
REFERENCES
- 1.Pinto LH, Lamb RA. J Biol Chem. 2006;281(14):8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Qiu JX, Soto C, DeGrado WF. Curr Opin Struct Biol. Vol. 21. Elsevier Ltd; England: 2010. pp. 68–80. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossman JS, Lamb RA. Virology. Vol. 411. Elsevier Inc; United States: 2010. pp. 229–36. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claridge JK, Aittoniemi J, Cooper DM, Schnell JR. Biochemistry. 2013;52(47):8420–8429. doi: 10.1021/bi401035m. [DOI] [PubMed] [Google Scholar]
- 5.Liao SY, Fritzsching KJ, Hong M. Protein Sci. 2013;22(11):1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross TA, Ekanayake V, Paulino J, Wright A. J Mag Res. 2014;239:100–109. doi: 10.1016/j.jmr.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomaston JL, Nguyen PA, Brown EC, Upshur MA, Wang J, Degrado WF, Howard KP. Protein Sci. 2013;22(1):65–73. doi: 10.1002/pro.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klug CS, Feix JB. Biophysical Tools for Biologists: Vol 1 in Vitro Techniques. Vol. 84. Elsevier Academic Press Inc; San Diego: 2008. Methods and applications of site-directed spin Labeling EPR Spectroscopy. pp. 617–658. [DOI] [PubMed] [Google Scholar]
- 9.Duong-Ly KC, Nanda V, Degrad WF, Howard KP. Protein Sci. 2005;14(4):856–861. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kruijff B. Curr Opin Chem Biol. 1997;1(4):564–569. doi: 10.1016/s1367-5931(97)80053-1. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, Ma CL, Ohigashi Y, Pinto LH, DeGrado W, Howard KP. Biochemistry. 2008;47(38):9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. 1997;94(21):11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Planque MRR, Killian JA. Molecular Membrane Biology. 2003;20(4):271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 14.Sugar IP, Monticelli G. Biophys. Chem. 1983;18(4):281–289. doi: 10.1016/0301-4622(83)80041-6. [DOI] [PubMed] [Google Scholar]; Ahn T, Yun CH. Archives of Biochemistry and Biophysics. 1999;369(2):288–294. doi: 10.1006/abbi.1999.1376. [DOI] [PubMed] [Google Scholar]
- 15.Cristian L, Lear J, DeGrado W. Proc Natl Acad Sci. 2003;100(25):14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mchaourab HS, Perozo E. Berliner L, Eaton S, Eaton G, editors. Determination of Protein Folds and Conformational Dynamics Using Spin-Labeling EPR Spectroscopy. Biological Magnetic Resonance. Distance Measurements in Biological Systems by EPR. 2000;19:185–247. [Google Scholar]
- 17.Perozo E, Cortes DM, Cuello LG. Science. 1999;285(5424):73–8. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 18.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Nature. 2002;418(6901):942–8. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Cady S, Hong M. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantor RS. Biophysical Journal. 1999;76(5):2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gennis RB. Biomembranes: Molecular Structure and Function. Springer-Verlag; New York: 1989. [Google Scholar]
- 22.Zhang J, Pekosz A, Lamb RA. J Virol. 2000;74(10):4634–44. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opekarova M, Tanner W. Biochim Biophys Acta. 2003;1610(1):11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 24.van den Brink-van der Laan E, Chupin V, Killian JA, de Kruijff B. Biochemistry. 2004;43(14):4240–50. doi: 10.1021/bi036129d. [DOI] [PubMed] [Google Scholar]
- 25.Perozo E, Kloda A, Cortes DM, Martinac B. Nat Struct Biol. 2002;9(9):696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 26.Howard KP, Lear J, DeGrado WF. Proc Natl Acad Sci. 2002;99(13):8568–72. doi: 10.1073/pnas.132266099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi M, Cross T, Zhou H. Proc Natl Acad Sci. 2009;106(32):13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stouffer AL, Nanda V, Lear J, DeGrado WF. J Mol Biol. 2005;347(1):169–79. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 29.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Nature. 2008;451(7178):596–600. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SB, DeGrado WF. Biochemistry. 1999;38(37):11905–13. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]