Chiang et al. identify GPR18 as a novel receptor for resolvin D2, and show that activation of this receptor in human and mouse phagocytes stimulates phagocytic clearance during bacterial infections and promotes organ protection
Abstract
Endogenous mechanisms that orchestrate resolution of acute inflammation are essential in host defense and the return to homeostasis. Resolvin (Rv)D2 is a potent immunoresolvent biosynthesized during active resolution that stereoselectively stimulates resolution of acute inflammation. Here, using an unbiased G protein–coupled receptor-β-arrestin–based screening and functional sensing systems, we identified a receptor for RvD2, namely GPR18, that is expressed on human leukocytes, including polymorphonuclear neutrophils (PMN), monocytes, and macrophages (MΦ). In human MΦ, RvD2-stimulated intracellular cyclic AMP was dependent on GPR18. RvD2-stimulated phagocytosis of Escherichia coli and apoptotic PMN (efferocytosis) were enhanced with GPR18 overexpression and significantly reduced by shRNA knockdown. Specific binding of RvD2 to recombinant GPR18 was confirmed using a synthetic 3H-labeled-RvD2. Scatchard analysis gave a Kd of ∼10 nM consistent with RvD2 bioactive concentration range. In both E. coli and Staphylococcus aureus infections, RvD2 limited PMN infiltration, enhanced phagocyte clearance of bacteria, and accelerated resolution. These actions were lost in GPR18-deficient mice. During PMN-mediated second organ injury, RvD2’s protective actions were also significantly diminished in GPR18-deficient mice. Together, these results provide evidence for a novel RvD2–GPR18 resolution axis that stimulates human and mouse phagocyte functions to control bacterial infections and promote organ protection.
Inflammation is a vital and normally protective response to defend the host against infection and injury. When excessive, acute inflammation can progress to chronic inflammation, scarring, and fibrosis (Cotran et al., 1999; Nathan, 2012; Tabas and Glass, 2013; Deutschman and Tracey, 2014). Acute inflammatory responses are ideally self-limited, leading to catabasis and complete resolution. With the structure elucidation of endogenous antiinflammatory and pro-resolving mediators and their functional characterization, it is becoming apparent that resolution of inflammation is an active biosynthetic process with specialized mediators that govern the key steps in resolution. Resolution of self-limited inflammation is governed by lipid mediator (LM) class switching from production of proinflammatory prostaglandins and leukotrienes in the initiation phase to biosynthesis of antiinflammatory and pro-resolving local mediators, such as lipoxins (LXs) in the resolution phase (Serhan, 2014). More recently, new families of resolution-phase mediators have been identified, and their structures have been elucidated, including eicosapentaenoic acid (EPA)–derived E-series resolvins (Rv) and docosahexaenoic acid (DHA)-derived D-series Rvs, protectins, and maresins (Serhan, 2014). Together with the LXs, they are agonists of resolution and constitute a genus of potent endogenous mediators termed the specialized pro-resolving mediators (SPMs).
Eicosanoids exhibit their actions by cell surface receptors, which belong to the G protein–coupled receptor (GPCR) superfamily (Shimizu, 2009). SPMs also interact with cell-surface GPCR on leukocytes to limit further PMN infiltration and stimulate phagocyte resolution programs (Serhan and Chiang, 2013). For example, LXA4 and RvD1 each directly activate lipoxin A4 receptor (ALX) and GPR32, denoted DRV1. RvE1 also activates two separate receptors—ChemR23/ERV1 to stimulate macrophage (MΦ) phagocytosis and BLT1 to limit and redirect polymorphonuclear neutrophil (PMN) signals (El Kebir et al., 2012; Serhan and Chiang, 2013).
RvD2 was identified and isolated from murine self-resolving exudates during the resolution phase of self-limited acute inflammation in vivo (Serhan et al., 2002). In human leukocytes, the precursor DHA is converted via 17-lipoxygenation to 17S-hydro(peroxy)-DHA, an intermediate that is enzymatically transformed by 5-lipoxygenase (LOX) to a unique 7(8)epoxide-containing intermediate, followed by enzymatic hydrolysis to form RvD2. The complete structure and stereochemistry of RvD2 is 7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-DHA (Spite et al., 2009). RvD2 is produced during human MΦ efferocytosis, e.g., engulfment of apoptotic PMN (Dalli and Serhan, 2012), in human adipose tissue (Clària et al., 2013) and peripheral blood from healthy donors (Mas et al., 2012), as well as in pulmonary tuberculosis patients (Frediani et al., 2014). In addition, RvD2 levels were increased in healthy donors with n-3 supplementation (Colas et al., 2014). In animal models of diseases, RvD2 stereoselectively reduces excessive PMN trafficking to inflammatory loci, stimulates PMN phagocytosis of Escherichia coli, and controls polymicrobial sepsis (Spite et al., 2009). Using a microfluidic chamber able to monitor single cells, RvD2 restores chemotactic response of PMN, and improves survival after a second septic insult post-burn in rats (Bohr et al., 2013; Kurihara et al., 2013). In addition, RvD2 reduces inflammatory pain (Park et al., 2011) and inflammation in experimental colitis and fibromyalgia (Bento et al., 2011; Klein et al., 2014). Together, these results indicate potent roles for RvD2 in regulating resolution of bacterial infections and sterile inflammation.
Herein, we present evidence for a specific cell surface GPCR for RvD2, namely GPR18. RvD2 activates recombinant human GPR18 in a receptor- and ligand-dependent manner. Using radiolabeled RvD2 ([10,11-3H]-7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-DHA), we obtained evidence for direct binding of RvD2 to recombinant GPR18 with a Kd value within the bioactive concentration range of RvD2. We investigated GPR18’s contribution to RvD2 pro-resolving actions in stimulating phagocytosis of microbes and apoptotic PMN, accelerating resolution of bacterial infections, and organ protection with overexpression or knockdown of GPR18.
RESULTS
RvD2 receptor candidates and GPCR screening
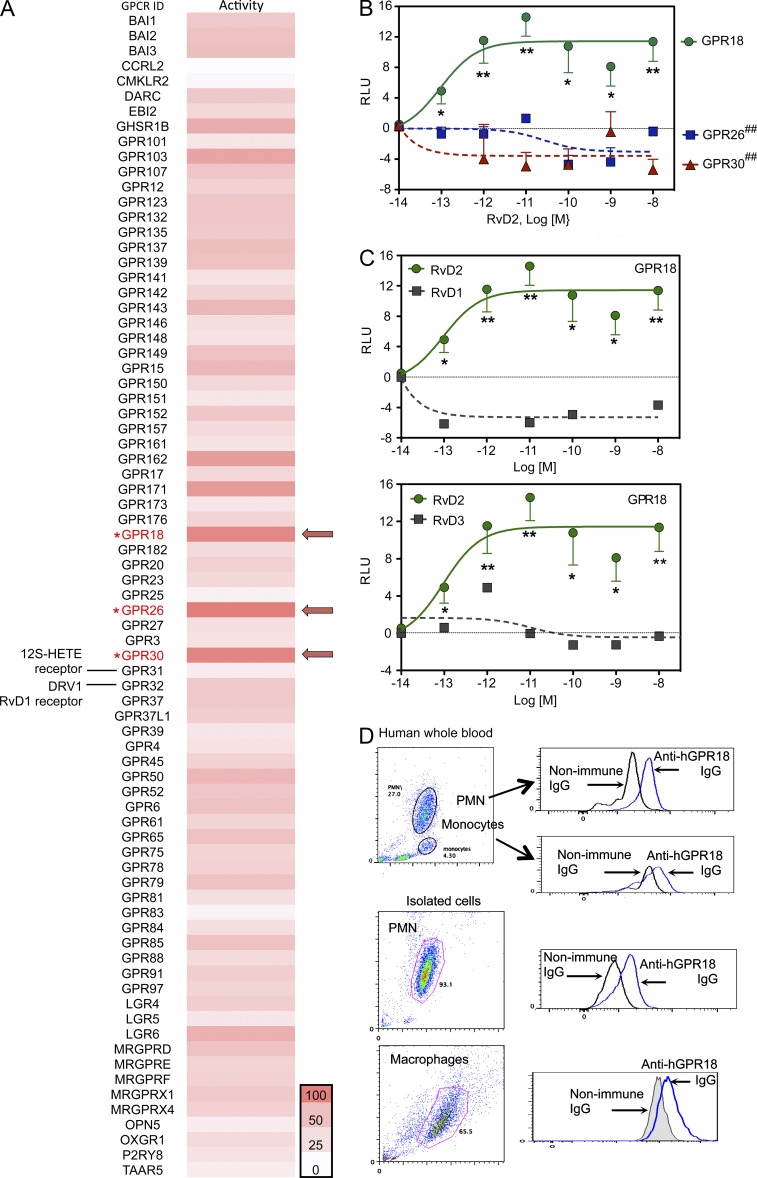
To identify receptors for RvD2, we used an unbiased GPCR-β-arrestin–coupled custom commercial screening system to monitor RvD2–receptor interactions (Krishnamoorthy et al., 2010). Among 77 orphan human GPCRs, three receptors gave the strongest signals increasing chemiluminescence in response to 10 nM of RvD2 (normalized as 100% in the heatmap that was equivalent to [positive signals − baseline/baseline in the absence of ligand]; see Materials and methods), namely GPR18, GPR26, and GPR30 (Fig. 1 A). RvD2 did not activate GPR32, a human receptor for RvD1 (Krishnamoorthy et al., 2010), or GPR31, a recently reported receptor for 12S-hydroxy-eicosatetraenoic acid, which is a product of 12-LOX and arachidonic acid (Fig. 1 A; Guo et al., 2011). We further evaluated human GPR18, GPR26, and GPR30 using this β-arrestin–based ligand receptor interaction system. RvD2 (10−13–10−8 M) significantly increased chemiluminescence signals (RLU) with GPR18-expressing cells compared with vehicle controls in a dose-dependent manner with EC50 values of ∼2.0 × 10−13 M (Fig. 1 B). In contrast, RvD2 (10−13–10−8 M) did not increase chemiluminescence in cells overexpressing either GPR26 or GPR30. RvD1 and RvD3 at equimolar concentrations (10−14–10−8 M) did not activate GPR18 (Fig. 1 C). These results suggested that RvD2 selectively activated human GPR18. Because RvD2 exerts potent actions with leukocytes (Spite et al., 2009; Serhan, 2014), we assessed GPR18 expression on human leukocytes using flow cytometry. GPR18 was expressed in human peripheral blood PMN; anti-GPR18 IgG gave 4.1 ± 1.7 and 3.7 ± 2.3 fold increases of MFI in whole blood PMN and isolated PMN, respectively, compared with the nonimmune IgG control. Peripheral blood monocytes and monocyte-differentiated MΦ also expressed GPR18 (Fig. 1 D).
Figure 1.
Identification of RvD2 receptor candidates. (A) A panel of orphan GPCRs was screened using β-arrestin PathHunter GPCR system in the presence of 10 nM of RvD2 or vehicle control (0.1% ethanol). Results were expressed as a heatmap. The receptors that gave highest chemiluminescence signal in response to RvD2 (see Materials and methods for the screening methodology) were taken as 100% in the heatmap (indicated by arrows). (B) Receptor specificity. Ligand (RvD2)–receptor interaction was monitored using a β-arrestin system overexpressing GPR18 (circle), GPR26 (square), or GPR30 (triangle). Results are mean ± SEM from 3 independent experiments and 4 replicates each experiment. ##, P < 0.01 versus GPR18 (one-way ANOVA with Tukey’s Multiple Comparison test). *, P < 0.05; **, P < 0.01, RvD2 versus vehicle controls (unpaired Student’s t test). RLU, relative luminescence units. (C) Ligand specificity. RvD2 (circle), RvD1, or RvD3 (square) interaction with GPR18 were monitored using β-arrestin system overexpressing GPR18. Results are from 3 (RvD2) or 2 (RVD1 and RvD3) independent experiments and 4 replicates each experiment. *, P < 0.05; **, P < 0.01 RvD2 versus vehicle controls (unpaired Student’s t test). (D) GPR18 expression. Human whole blood (50 µl), isolated PMN (106 cells), or GM-CSF–differentiated MΦ (106 cells) were incubated with rabbit anti-GPR18 IgG or nonimmune rabbit IgG (1:50 dilutions, 30 min), followed by PE-anti-rabbit IgG (1:200 dilutions, 30 min). GPR18 expression was monitored by flow cytometry. Results are representative of 4 independent experiments using 4 separate healthy donors. Results with whole blood and isolated PMN were obtained from the same donor.
RvD2 activates human recombinant GPR18: ligand–receptor dependency
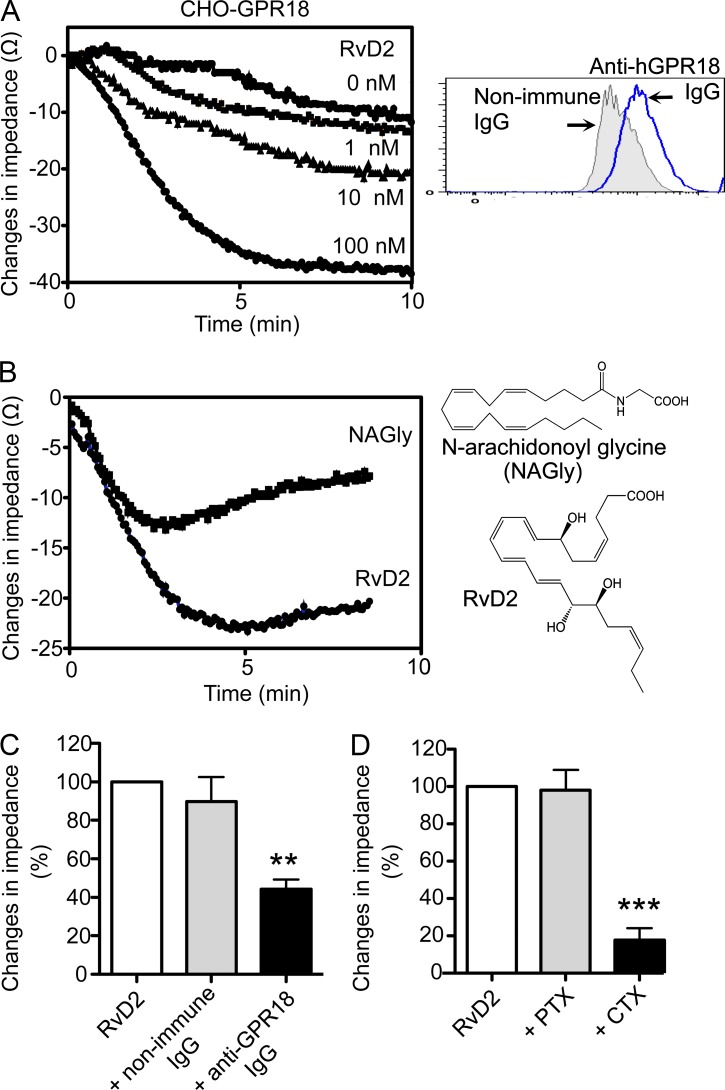
To examine ligand–receptor interactions, we performed electrical cell substrate impedance sensing (ECIS), monitoring changes in impedance upon ligand binding to receptors (Peters and Scott, 2009). In this system, RvD2 (1–100 nM) dose-dependently elicited rapid changes in impedance, with CHO cells overexpressing recombinant human GPR18 (CHO-hGPR18; Fig. 2 A). Because N-arachidonyl glycine (NAGly) was recently identified as a ligand for GPR18 (Kohno et al., 2006), we compared these ligands and found that equimolar concentrations of NAGly and RvD2 both elicited impedance changes activating CHO-hGPR18 (Fig. 2 B). RvD2-initiated changes in impedance were significantly reduced when cells were incubated with anti-hGPR18 antibody before addition of RvD2 (Fig. 2 C; 15.0 ± 1.7 Ω with anti-GPR18 IgG vs. 30.5 ± 4.3 Ω with nonimmune IgG). To test whether GPR18 activation by RvD2 is mediated via G proteins in CHO cells, we incubated CHO-hGPR18 cells with either cholera toxin (CTX) or pertussis toxin (PTX) before RvD2 addition. PTX did not change RvD2-initiated impedance changes, suggesting that with RvD2, GPR18 did not couple to Gαi-like proteins in CHO-hGPR18 cells. CTX markedly inhibited RvD2-initiated impedance changes (Fig. 2 D; 7.0 ± 3.5 Ω with RvD2 plus CTX vs. 16.3 ± 3.5 Ω with RvD2 alone), suggesting that RvD2 triggered GPR18 coupling to Gαs-like proteins in CHO-hGPR18 cells.
Figure 2.
RvD2-dependent activation of GPR18. (A) Dose response. CHO-GPR18 cells were incubated with RvD2 (1–100 nM) or vehicle alone (control). Impedance changes across CHO cell monolayers were continuously recorded in real-time for 10 min (inset). Representative histograms of GPR18 expression. (B) Ligand specificity. Tracings were CHO-GPR18 cells incubated with RvD2 or NAGly (100 nM each; chemical structures depicted on the right). Time 0 denotes the addition of compounds. (C) CHO-GPR18 cells were incubated with anti-GPR18 Ab (1:50) or nonimmune rabbit IgG for 30 min, followed by addition of 100 nM RvD2. (D) CHO-GPR18 cells were treated with CTX (1 µg/ml, 2 h) or PTX (1 µg/ml, 16 h) followed by addition of 100 nM RvD2. Results are expressed as (A and B) changes in impedance (Ω); mean of 4 separate tracings from 4 independent experiments or (C and D) percentage of changes in impedances. RvD2-initiated impedance changes were taken as 100%; mean ± SEM from 4 separate tracings from 4 independent experiments; **, P < 0.01; ***, P < 0.001 versus RvD2 plus nonimmune IgG (C) or RvD2 alone (D) using one-way ANOVA with Tukey’s multiple comparison test.
GPR18 mediates signals and actions of RvD2 in human MΦ
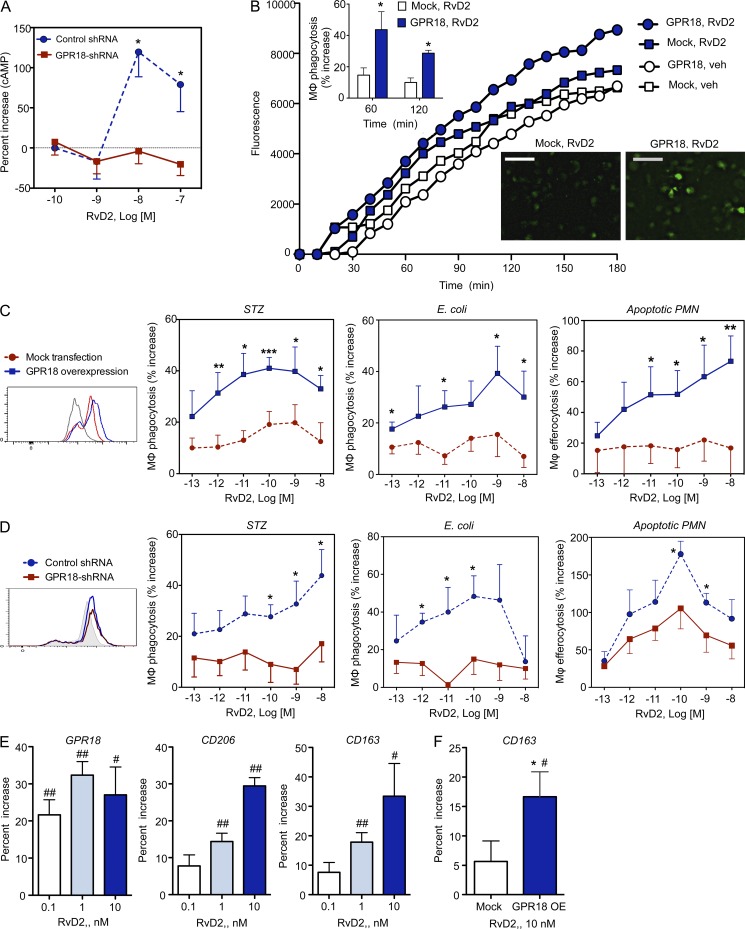
Given that RvD2 activation of GPR18 in CHO cells is likely mediated via a Gαs-like protein, we next determined whether RvD2 regulates cAMP, a second messenger downstream of Gαs, in human MΦ (Rossi et al., 1998). RvD2 at 10–100 nM significantly increased cAMP with human MΦ. This action was diminished when MΦ were transfected with shRNA targeting GPR18 (Fig. 3 A).
Figure 3.
Human GPR18-mediated RvD2 actions in MΦ phagocytosis. (A) Human GPR18 was knocked down with GPR18 shRNA in human MΦ. MΦ (0.4 × 106 cells) were incubated with indicated concentrations of RvD2 for 2 min (37°C), and cAMP was measured. Results are mean ± SEM of four separate experiments and duplicates in each experiment. *, P < 0.05 obtained with unpaired Student’s t test for GPR18 shRNA (solid red line) versus control scrambled shRNA (dashed blue line) transfected MΦ. (B) Human MΦ were transfected with human GPR18 (circle) or mock (square) plasmids; 72 h later, MΦ were plated onto chamber (0.1 × 106 cells/well) incubated with RvD2 at 10−9 M (blue) or vehicle control (white) for 15 min at 37°C, followed by addition of BacLight Green-labeled E. coli to initiate phagocytosis. Fluorescent images were then recorded every 10 min for 180 min. (top inset) Percent increase in phagocytosis by RvD2 in mock (white) or GPR18 (blue) transfected MΦ. (bottom) Representative fluorescent images at 180 min. Bars, 50 µm. Three separate experiments were performed. In each experiment, 4 fields (20×) per condition (per well) were recorded. Results are mean fluorescence of four fields/well from one representative experiment. (C and D) Human GPR18 was overexpressed (C) or knocked down with shRNA (D) in human MΦ and verified by flow cytometry (insets). MΦ were incubated with RvD2 (10−13 to 10−8 M) or vehicle control for 15 min, followed by addition of FITC-zymosan, BacLight Green-labeled E. coli, or CFDA-labeled apoptotic PMN to initiate phagocytosis. Results are percent increases of phagocytosis above vehicle. (C) Mean ± SEM from 5 or (D) mean ± SEM from 3 independent experiments with separate donors and triplicates in each experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001, obtained with unpaired Student’s t test for GPR18 overexpression (solid blue lines) versus mock transfection (dashed red lines) in C and GPR18 shRNA (solid red lines) versus control scrambled shRNA (dashed blue lines) in D. (E and F) GPR18, CD206, and CD163 expression. (E) Human MΦ (0.5 × 106 cells) or (F) human MΦ overexpressing GPR18 (GPR18-OE) or mock plasmids were incubated with vehicle or RvD2 (0.1, 1, or 10 nM) for 24 h. GPR18, CD206, and CD163 were monitored using flow cytometry. Results are percent increase above vehicle; mean ± SEM from 3 independent experiments with 3 separate donors. #, P < 0.05; ##, P < 0.01 RvD2 versus vehicle; *, P < 0.05, GPR18 overexpression versus mock transfection using unpaired Student’s t test.
MΦ phagocytosis of apoptotic cells, microbes, and debris is a cellular hallmark of tissue resolution of acute inflammation (Cotran et al., 1999). Because RvD2 enhances MΦ phagocytosis of serum-treated zymosan (STZ), as well as live E. coli (Spite et al., 2009), we next examined whether this action was dependent on GPR18. Human MΦ were differentiated from peripheral blood monocytes (see Materials and methods) and transfected with either human GPR18 or a mock plasmid. Phagocytosis of fluorescently labeled E. coli was monitored in real-time using microscopy (Fig. 3 B). RvD2 (1 nM) increased uptake of E. coli with mock-transfected MΦ (∼15% increase above vehicle control; 1 h), an action that was further enhanced with hGPR18 transfection (>40% increase at 1 h and ∼30% increase at 2 h with 1 nM RvD2).
To further investigate the role of GPR18 in RvD2-stimulated phagocytosis with human MΦ, we transfected MΦ with hGPR18 (overexpression) or specific shRNA targeting hGPR18 (knockdown). Overexpression and knockdown of hGPR18 were verified by flow cytometry (Fig. 3, C and D, insets). Phagocytosis of fluorescently labeled STZ, E. coli, and apoptotic PMN were performed and fluorescence was monitored using a fluorescence plate reader. RvD2 (0.01–1 nM) increased human MΦ phagocytosis of FITC-zymosan in mock-transfected MΦ, and this action was further enhanced with hGPR18 overexpression (Fig. 3 C; ∼40% increase above vehicle in MΦ-hGPR18 vs. 15–20% increase in MΦ-mock; 1 h). hGPR18 overexpression also led to increased phagocytosis of E. coli in response to RvD2 (e.g., 1 nM RvD2 produced a ∼40% increase with MΦ-hGPR18 vs. ∼15% with MΦ-mock). In comparison, RvD2-mediated (10 pM–10 nM) efferocytosis of apoptotic PMN was also increased with hGPR18 overexpression compared with mock transfected MΦ (Fig. 3 C). In separate sets of experiments, knockdown of endogenous MΦ GPR18 using shRNA significantly abolished RvD2-stimulated phagocytosis of STZ, E. coli, and apoptotic PMN (Fig. 3 D). Together, these results demonstrated that human GPR18 contributed to RvD2’s pro-resolving actions in stimulating MΦ ingestion of yeast zymosan particles, live E. coli, and apoptotic human PMN.
We next examined whether RvD2 can regulate MΦ mannose receptor CD206 and CD163, which are phagocytic receptors and markers of the antiinflammatory and efferocytic M2 phenotype (Plüddemann et al., 2011; Zizzo et al., 2012). With human MΦ, RvD2 (0.1–10 nM; 24 h) dose-dependently increased expression of CD206 and CD163 was monitored by flow cytometry (Fig. 3 F). MΦ overexpressing GPR18 increased CD163 expression in response to RvD2 when compared with mock-transfected MΦ (Fig. 3 E). These results suggested that these phagocytic receptors CD163 and CD206 (Plüddemann et al., 2011) contribute to RvD2-enhanced MΦ phagocytosis. In addition, RvD2 (0.1–10 nM; 24 h) up-regulated GPR18 expression (Fig. 3 E). Together, these results suggest a positive feed-forward amplification mechanism for RvD2 actions in promoting MΦ phagocytosis.
RvD2–GPR18 interaction in vivo reduces exudate PMN and stimulates efferocytosis
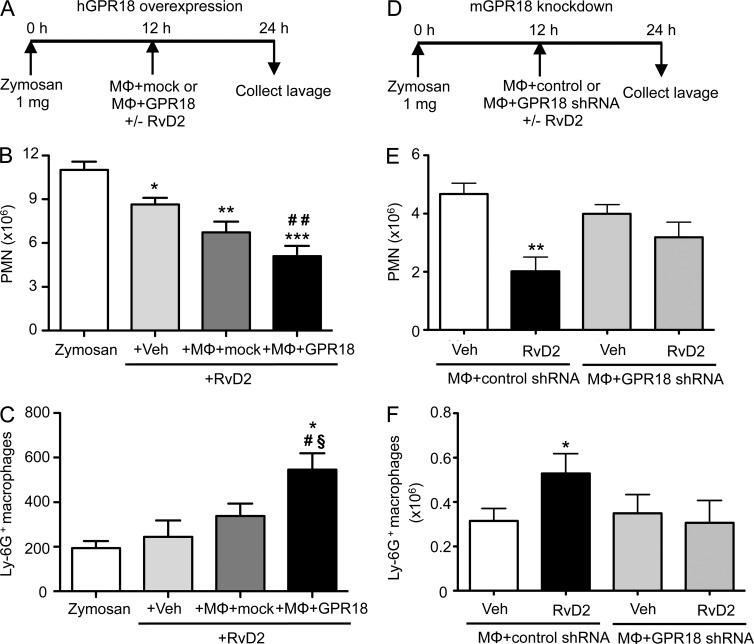
We examined whether overexpression of human GPR18 can enhance RvD2’s action in vivo using a self-limited murine peritonitis. We collected naive peritoneal MΦ and transfected them ex vivo with either GPR18 or mock plasmids. Peritonitis was initiated, transfected MΦ were injected with or without RvD2 (10 ng/mouse, i.p.) at peak of inflammation as monitored by maximal PMN infiltration (12 h after zymosan administration), and exudates collected at 24 h (Fig. 4 A). Administration of RvD2 (10 ng) alone at 12 h significantly reduced PMN numbers at 24 h (Fig. 4 B; 8.6 ± 0.5 × 106 vs. 11.0 ± 0.6 × 106 PMN with zymosan alone). GPR18-transfected MΦ further enhanced this RvD2 action (5.1 ± 0.7 × 106 PMN). In vivo efferocytosis was evaluated using Ly-6G+ MΦ in exudates. Low-dose RvD2 (10 ng) alone did not increase efferocytosis, whereas administration of RvD2 and mock-transfected MΦ gave significantly higher Ly-6G+ MΦ (Fig. 4 C; 339 ± 54 vs. 194 ± 31 Ly-6G+MΦ). Moreover, administration of RvD2 and GPR18-tranfected MΦ further enhanced efferocytosis (545 ± 74 Ly-6G+ MΦ).
Figure 4.
RvD2 in vivo actions were enhanced by overexpression and reduced by knockdown of GPR18. (A–C) Naive peritoneal MΦ were collected and transfected ex vivo (106 cells) with either GPR18 (5 µg) or mock plasmids for 48 h. Zymosan (1 mg) was injected into peritoneum to initiate peritonitis. 12 h later, transfected MΦ (1.5 × 105/mouse) and/or RvD2 (10 ng) was injected i.p. (A) Timeline. (B) PMN numbers (Ly6G+ CD11b+) and (C) efferocytosis (Ly6G+ F4/80+) were determined using flow cytometry. Results are expressed as mean ± SEM from 2 independent experiments and 6 mice/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001, versus zymosan (zym) alone. #, P < 0.05, ##, P < 0.01, versus zym+veh+RvD2 (one-way ANOVA with Dunnett’s multiple comparison test). §, P < 0.05, versus zym+MΦ-mock+RvD2 (unpaired Student’s t test). (D–F) Naive peritoneal MΦ (106 cells) were transfected ex vivo with either GPR18 shRNA (5 µg) or control-scrambled shRNA. Zymosan (1 mg) was injected to initiate peritonitis. 12 h later, transfected MΦ (2 × 105/mouse) and/or RvD2 (20 ng) was injected i.p. Inflammatory exudates were collected at 24 h. (D) Timeline. (E) PMN numbers (Ly6G+CD11b+) and (F) efferocytosis (Ly6G+F4/80+) were determined using flow cytometry. Results are expressed as mean ± SEM from 2 independent experiments and 6 mice/group. *, P < 0.05; **, P < 0.01, obtained with unpaired Student’s t test for vehicle versus RvD2 in MΦ + control shRNA group.
In a second set of experiments, we collected peritoneal resident MΦ and knocked down endogenous mouse GPR18 using targeted shRNA (Fig. 4 D). RvD2 (20 ng/mouse, i.p.) significantly reduced PMN numbers (>50%) and increased efferocytosis (>60%) in mice given control shRNA-transfected MΦ. In contrast, RvD2’s actions were abolished in mice that received GPR18 shRNA-transfected MΦ (Fig. 4, E and F). Collectively, these results indicated that, in acute peritonitis, RvD2 limited PMN infiltration and enhanced efferocytosis in a GPR18-dependent manner.
3H-RvD2–specific binding
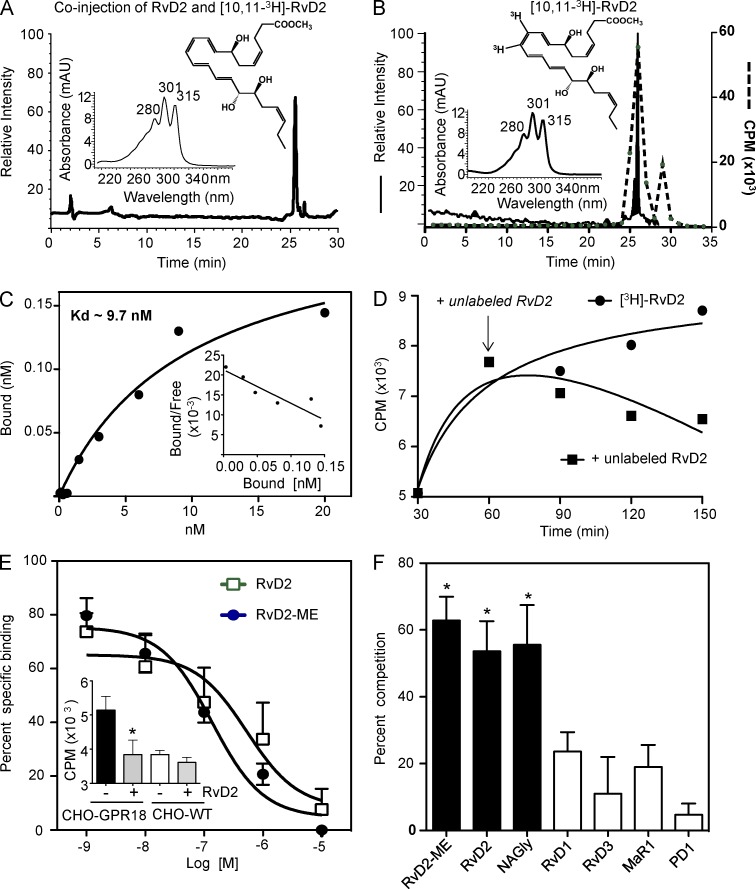
Because GPR18 mediated RvD2’s potent pro-resolving actions in vitro and in vivo, we next determined whether RvD2 directly binds to recombinant human GPR18 using radiolabeled ligand binding. To this end, we prepared tritium-labeled [10,11-3H]-RvD2 methyl ester (ME) by catalytic hydrogenation of synthetic precursor 10,11-acetylenic RvD2-ME (Fig. 5). After hydrogenation with tritium, [3H]-RvD2-ME was obtained and its integrity was confirmed after HPLC isolation. Fig. 5 (A and B) shows the chromatographic tracing of [10,11-3H]-RvD2-ME that coeluted with authentic synthetic RvD2-ME standard, and matched radioactivity peak. RvD2 has a tetraene structure with characteristic UV bands of absorbance at λmax 301 nm with shoulders at 280 and 315 nm (insets, Fig. 5, A and B). The qualified [3H]-RvD2-ME was then used for saturation binding with recombinant hGPR18 expressed in CHO cells in the absence or presence of 10 μM unlabeled RvD2-ME. Specific binding was obtained and Scatchard plot analysis produced a Kd of 9.6 ± 0.9 nM (Fig. 5 C). This value is within the bioactive range of RvD2 (Fig. 3). To test whether RvD2-ME can displace [3H]-RvD2-ME binding, CHO-hGPR18 cells were incubated with [3H]-RvD2-ME for 60 min, followed by addition of unlabeled RvD2-ME, which time-dependently displaced [3H]-RvD2-ME binding to CHO-hGPR18 (Fig. 5 D).
Figure 5.
[3H]-RvD2 isolation and specific binding with human recombinant GPR18. (A and B) [10,113H]-RvD2-ME characterization and isolation. (A) HPLC chromatographs of [10,113H]-RvD2-ME and unlabeled RvD2-ME co-injection. (B) Chromatographic and radioactive tracing; [10,113H]-RvD2-ME (solid line) and radioactivity (dashed line). (insets) Structure and online UV spectra of RvD2-ME and [10,113H]-RvD2-ME. Results are representative of 5 separate experiments. (C–F) CHO cells were transfected with human GPR18. (C) Saturation binding. GPR18-transfected CHO cells (0.5 × 106 cells in 100 µl PBS2+) were incubated with [3H]-RvD2-ME at indicated concentrations in the presence or absence of 10 µM unlabeled RvD2-ME for 60 min at 4°C. Bound and unbound radioligands were separated by filtration, and specific binding was determined. Results are representative of 4 independent experiments and 2 replicates in each experiment. (D) Displacement binding. CHO-GPR18 cells (0.5 × 106 cells) were incubated with 3 nM of [3H]-RvD2-ME. 1 h later, 1 µM of unlabeled RvD2-ME (square) was added to displace radioligand binding (denoted by an arrow). Radioactivity was determined at indicated time points. Results are representative of 3 independent experiments and 2–3 replicates in each experiment. (E) Competition binding. CHO-GPR18 cells (0.5 × 106 cells) were incubated with 3 nM of [3H]-RvD2-ME in the presence or absence of increasing concentrations of unlabeled RvD2-ME (circle) or RvD2 (square) for 60 min at 4°C. (inset) Specific [3H]-RvD2-ME (3 nM) binding on CHO-GPR18 and CHO-WT cells in the absence or presence of 1 µM of RvD2-ME. (F) Ligand specificity. CHO-GPR18 cells (0.5 × 106 cells) were incubated with 3 nM of [3H]-RvD2-ME in the absence or presence of 100 nM of RvD2-ME, RvD2, NAGly, RvD1, RvD3, MaR1, or PD1 for 60 min at 4°C. (E and F) Results are mean ± SEM from 4 (RvD2-ME, RvD2, NAGly, RvD1) or 2 (RvD3, MaR1, PD1) independent experiments and 2 replicates in each experiment; *, P < 0.05, compared with incubations with cells and [3H]-RvD2-ME in the absence of competing unlabeled compound (one-way ANOVA with Tukey’s multiple comparison test).
To determine the ligand specificity, competition binding was performed. RvD2 and RvD2-ME gave similar affinities to CHO-GPR18 with IC50 ∼100 nM (Fig. 5 E). [3H]-RvD2-ME did not give significant specific binding with CHO-WT (Fig. 5 E, inset). NAGly also competed for [3H]-RvD2-ME binding, with, essentially, equipotency at 100 nM. In contrast, select SPMs, including RvD1, RvD3, maresin 1 (MaR1), and protectin D1 (PD1) did not significantly compete for [3H]-RvD2-ME–specific binding (Fig. 5 F). Collectively, these results indicated high affinity and specific binding of RvD2 with recombinant human GPR18.
GPR18-deficient mice display impaired resolution of infections and diminished responses with RvD2
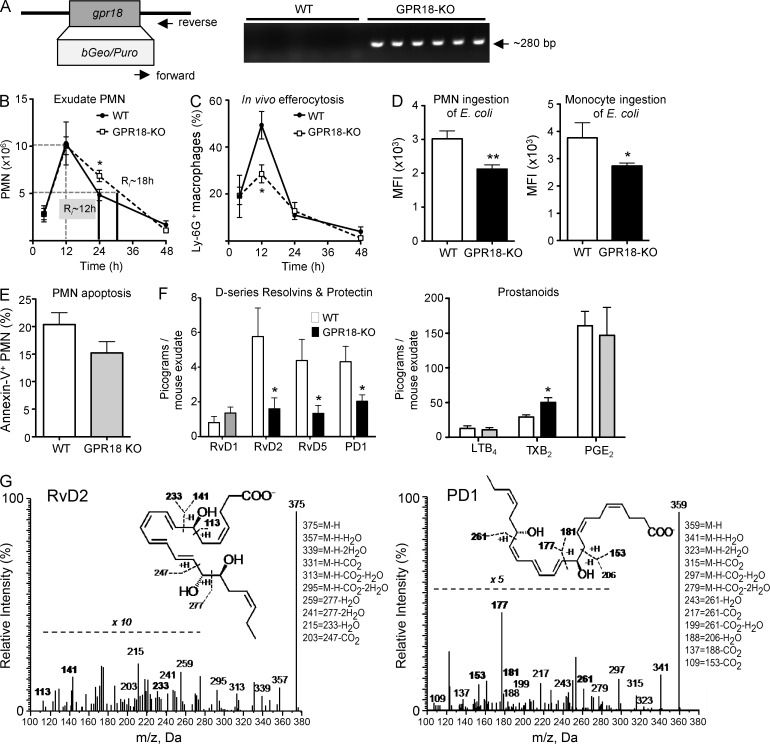
To determine the GPR18-dependent in vivo actions of RvD2, we prepared GPR18-deficient mice (GPR18 knockout; GPR18-KO) by insertion of a bGeo/Puro gene into the coding region of mouse gpr18. Targeted deletion of gpr18 was confirmed by genotyping using PCR (Fig. 6 A). These mice were born without apparent pathological phenotypes. Using GPR18-KO mice and their WT littermates, we investigated whether GPR18 played a role in controlling E. coli infections, a common and urgent health concern worldwide (Mead et al., 1999). Here, we performed a self-resolving E. coli (105 CFU) peritoneal infection. In WT littermates, PMN infiltration into peritoneum reached maximum at 12 h, followed by a decline, giving a resolution interval (Ri) of ∼12 h (Fig. 6 B). In comparison, GPR18-KO mice gave significantly higher PMN at 24 h (6.9 ± 0.6 vs. 5.0 ± 0.6 × 106 PMN in WT mice), leading to a delayed resolution of infection with Ri ∼18 h. These KO mice gave impaired efferocytosis with significantly lower Ly-6G+ MΦ at 12 h after E. coli inoculation (Fig. 6 C). Phagocyte ingestion of E. coli was also determined at 24 h, and significantly fewer intracellular E. coli were found in GPR18 KO in both exudate PMN and monocytes/MΦ, compared with WT littermates (Fig. 6 D). There were no statistical differences in exudate PMN apoptosis (percentage of Annexin V+ PMN) between WT and GPR18-KO mice at 24 h (Fig. 6 E). We performed mass spectrometry (MS)–based metabololipidomics focusing on local acting lipid mediators (LM). Each LM was profiled using multiple reaction monitoring and identified by direct comparison with synthetic and authentic standards using matching criteria including retention time, characteristic fragmentation patterns, and at least six diagnostic ions (Colas et al., 2014). Select SPM, including RvD2, RvD5, and PD1, were present in infectious exudates collected from WT mice at 24 h, and their levels were significantly reduced in GPR18-KO mice (Fig. 6 F). The representative MS-MS spectra of RvD2 and PD1 used for their identification are shown in Fig. 6 G. GPR18-KO gave increased amounts of TXB2, and there were no statistically significant differences in LTB4 and PGE2 between WT and GPR18-KO mice. Together, these results indicated a delayed-resolution phenotype with GPR18 KO mice in E. coli infection with heightened PMN infiltration, reduced SPM, impaired efferocytosis, and E. coli ingestion by phagocytes.
Figure 6.
Targeted deletion of mouse gpr18 delays resolution of E. coli infection. (A) Targeted deletion of mouse gpr18 (NM_182806) was constructed by insertion of bGeo/Puro gene into the coding region of gpr18 (left). Mice tails were collected, genomic DNA was isolated, and PCR was performed using primers specific for KO construct (right). (B–G) GPR18-deficient mice (white) and WT littermates (black) were inoculated with E. coli (105 CFU) by i.p. injection, and peritoneal exudates were collected at indicated time points. (B) PMN numbers were determined, and resolution indices were calculated (see Materials and methods). WT (black) and GPR18-KO (white). (C) In vivo efferocytosis (F4/80+Ly-6G+), (D) intracellular E. coli levels in PMN (Ly-6G+ CD11b+ E. coli+) or monocytes (Ly-6G− CD11b+ E. coli+), and (E) PMN apoptosis (Ly-6G+Annexin V+) were monitored by flow cytometry. MFI, mean fluorescence intensity. (F) d-series resolvins, protectins, and prostanoids in 24 h infectious exudates. (G) Representative MS/MS spectra of RvD2 and PD1. (B–F) Results are expressed as mean ± SEM from 2 independent experiments and 6–7 mice/group. *, P < 0.05; **, P < 0.01, obtained with unpaired Student’s t test for GPR18-KO versus WT.
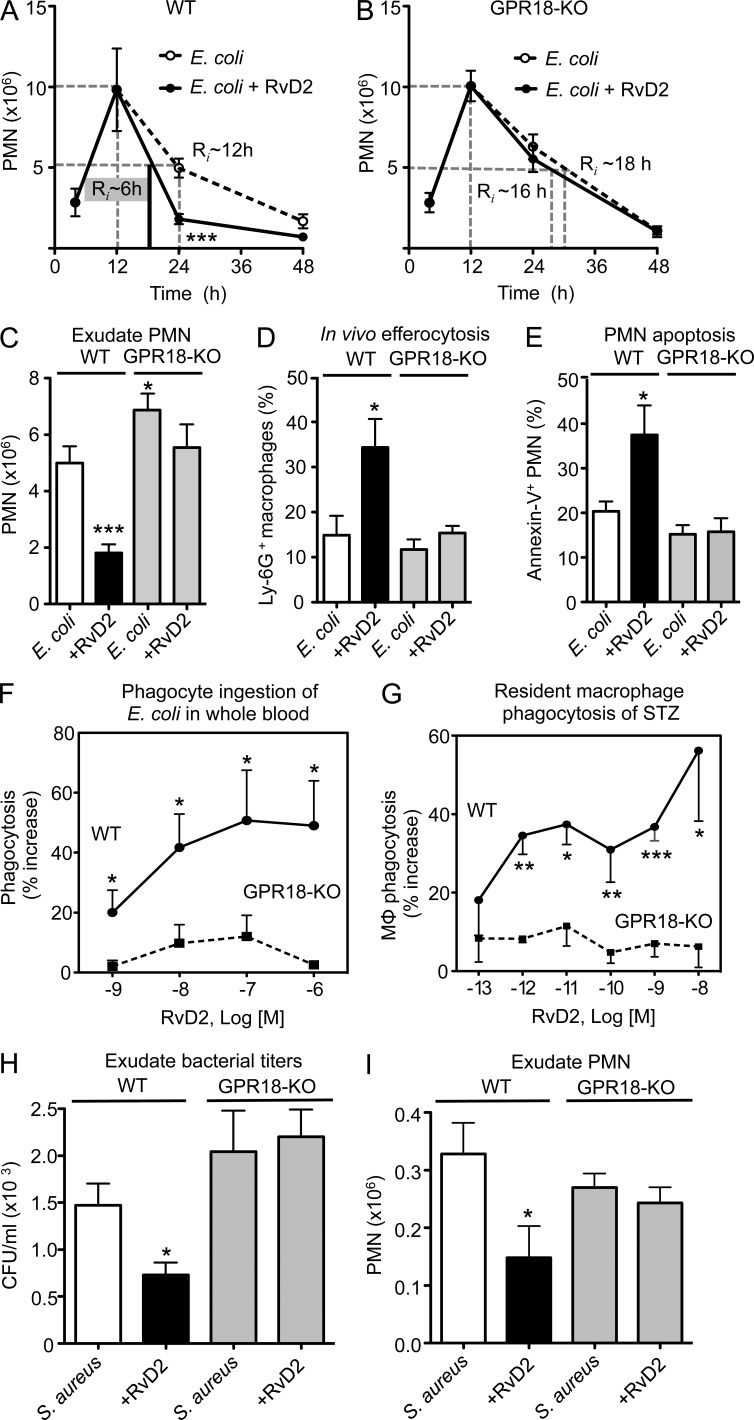
We next assessed whether RvD2 was protective in E. coli infections and if this action was GPR18-dependent. In WT mice, 100 ng RvD2 administered at 12 h (peak of inflammation) after E. coli inoculation lowered PMN numbers by ∼60% (1.9 ± 0.3 vs. 5.0 ± 0.6 × 106 PMN with E. coli alone; P < 0.001) at 24 h, giving a shortened Ri of ∼6 h compared with vehicle control with Ri ∼12 h (Fig. 7 A). This action of RvD2 was diminished in GPR18-KO mice, where RvD2 administration did not significantly alter PMN numbers (Fig. 7, B and C). RvD2 significantly enhanced efferocytosis and PMN apoptosis in WT mice by ∼120 and ∼70%, respectively (Fig. 7, D and E). These actions of RvD2 were diminished in GPR18-KO mice, pointing to the contribution of GPR18 to RvD2’s protective actions in E. coli infection. In comparison, when RvD2 was given at the onset of infection (100 ng/mouse i.p., together with E. coli), it significantly lowered the numbers of exudate PMN at 12 h (0.74 ± 0.18 vs. 1.50 ± 0.29 × 106 PMN with E. coli alone; P = 0.04). At this time point, there were no statistically significant differences in the percentage of apoptotic PMN (19.4 ± 4.3% with E. coli plus RvD2 vs. 13.5 ± 3.6% Annexin V+ PMN with E. coli alone; P = 0.16). These results suggest differential actions of RvD2 in vivo when it’s given at the peak of PMN infiltration (12 h) versus at the onset of inflammation (T0) during E. coli infection.
Figure 7.
RvD2-dependent protection is diminished with GPR18 deficiency in mice. (A–E) E. coli peritonitis; GPR18-deficient mice and WT littermates were inoculated with E. coli (105 CFU). 100 ng RvD2 was given by i.p. injection 12 h after E. coli inoculation, and peritoneal exudates collected at indicated time points. (A–C) PMN numbers were determined and resolution indices were calculated. E. coli alone (white), E. coli plus RvD2 (black). Results are expressed as mean ± SEM from 2 independent experiments with 4–5 mice/group (for 4-, 12-, and 48-h time points), or 3 independent experiments with 7–8 mice/group (for 24-h time point). *, P < 0.05; ***, P < 0.001, using unpaired Student’s t test for RvD2 versus vehicle group at 24 h. (D) In vivo efferocytosis (F4/80+Ly-6G+) and (E) PMN apoptosis (Ly-6G+Annexin V+) were monitored by flow cytometry. Results are expressed as mean ± SEM from 2 independent experiments and 5 mice/group; *, P < 0.05; ***, P < 0.001, using unpaired Student’s t test for RvD2 versus vehicle group at 24 h. (F) Mouse peripheral blood was collected from WT (circle) and GPR18 KO (square) mice, incubated with RvD2 (10−9–10−6 M) or vehicle for 15 min, followed by addition of BacLight Green-labeled E. coli for 2 h. RBCs were lysed, and fluorescence associated with phagocytes monitored by flow cytometry. Results are expressed as mean ± SEM from 2 independent experiments, 4 mice/group. *, P < 0.05, using unpaired Student’s t test for WT versus GPR18-KO. (G) Peritoneal MΦ were collected from naive WT (circle) and GPR18 KO (square) mice and incubated with RvD2 (10−13– 10−8 M) or vehicle for 15 min, followed by addition of FITC-zymosan to initiate phagocytosis. Results are mean ± SEM from 2 independent experiments, 4 mice/group and 4 replicates for each experimental condition. *, P < 0.05; **, P < 0.01 using unpaired Student’s t test for WT versus GPR18-KO. (H and I) S. aureus skin infection. Murine dorsal pouches were raised in GPR18-KO mice and WT littermates for 6 d. Live S. aureus (105 CFU) was given together with RvD2 (200 ng) or vehicle by intra-pouch injection, and pouch exudates were collected at 4 h. (H) Bacterial counts (CFU/ml) and (I) exudate PMN numbers were determined. Results are expressed as mean ± SEM from 2 independent experiments and 6–8 mice/group. *, P < 0.05, using unpaired Student’s t test for RvD2+S. aureus versus S. aureus alone.
We examined E. coli phagocytosis with mouse peripheral blood ex vivo and demonstrated the diminished response to RvD2 in GPR18 KO mice when compared with WT littermates (Fig. 7 F). We next investigated phagocytosis with isolated naive peritoneal MΦ collected from GPR18-KO and WT mice. RvD2 gave dose-dependent increases in phagocytosis of STZ with WT MΦ but not KO MΦ (Fig. 7 G). These results indicated that in GPR18-KO mice, phagocyte responses to RvD2 were lost in both isolated and whole blood phagocytes.
To assess the role of RvD2 and GPR18 in Gram-positive bacterial infections, we performed Staphylococcus aureus–initiated infections in murine dorsal skin pouches using GPR18 KO and WT littermates. S. aureus is an emerging cause of various skin infections, and a high percentage of hospital-acquired infections are caused by antibiotic-resistant S. aureus (World Health Organization, 2014). RvD2 (200 ng) administered via intrapouch injection together with S. aureus significantly reduced ∼50% bacterial titers (729 ± 144 vs. 1417 ± 234 CFU with S. aureus alone; P < 0.05) and PMN numbers (0.15 ± 0.07 vs. 0.33 ± 0.16 × 106 PMN with S. aureus alone; P < 0.05) at 4 h in WT mice (Fig. 7, H and I). These actions of RvD2 were abolished in GPR18 KO mice. Thus, these results demonstrated that the endogenous mouse GPR18 contributed to pro-resolving actions of RvD2 in infections, i.e., accelerating resolution of infection by enhancing efferocytosis and phagocyte clearance of bacteria.
PMN-mediated second organ injury initiated by ischemia–reperfusion (I/R): GPR18 and ligand dependency in vivo
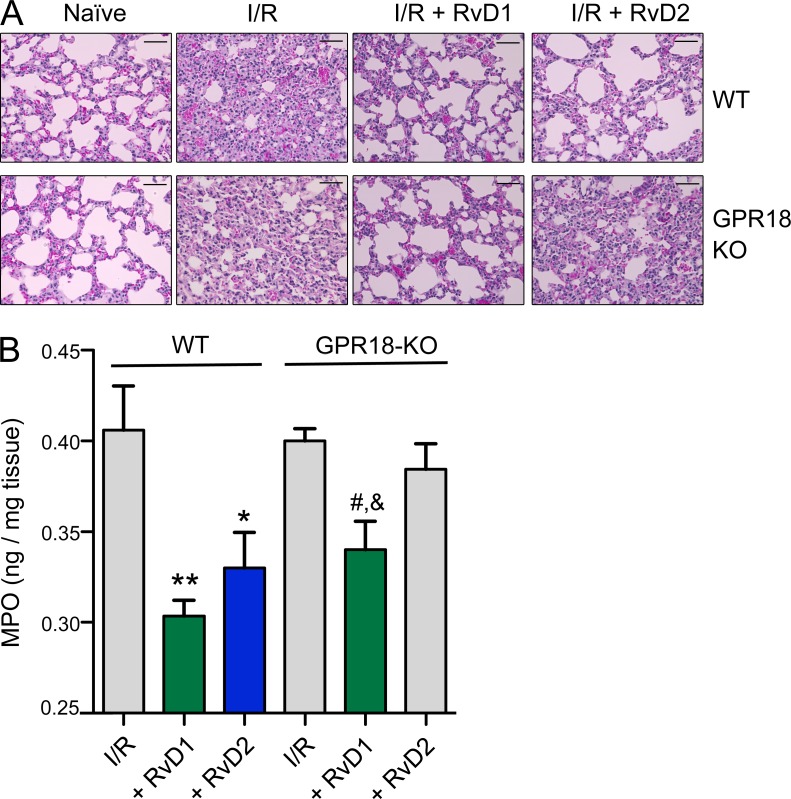
To investigate contribution of GPR18 in sterile injury (i.e., injury from within) and inflammation, we assessed I/R-initiated lung injury. Surgically based clamping procedures are well known to lead to aberrant PMN activation via stasis and vessel occlusion that gives rise to second organ injury that contributes to longer hospitalization (Eltzschig and Eckle, 2011). Here, we used a hind limb I/R (tourniquet occlusion) model of second organ lung injury, an established murine model of operating room surgical insults in humans (Chiang et al., 1999). Ischemia (1 h) followed by reperfusion (2 h) initiated second organ lung injury and PMN infiltration into the lungs in both WT vs. KO mice as illustrated by H&E staining (Fig. 8 A). PMN infiltration was quantified by measuring MPO levels, which had no statistically significant differences between WT and GPR18-KO mice (0.41 ± 0.02 vs. 0.40 ± 0.01 ng MPO/mg tissue; P = 0.76; Fig. 8 B). In WT mice, RvD2 administration (100 ng i.v./mouse) protected WT mice from second organ reflow injury (Fig. 8 A) and significantly lowered PMN infiltration (0.33 ± 0.02 vs. 0.41 ± 0.02 ng MPO/mg tissue; P < 0.05; Fig. 8 B). In contrast, in GPR18 KO mice there were no statistically significant differences in MPO values between RvD2 and vehicle-treated mice (0.38 ± 0.02 vs. 0.40 ± 0.01 ng MPO/mg tissue; P = 0.41; Fig. 8 B), indicating that RvD2-mediated organ protection was diminished in GPR18-KO mice. In comparison, RvD1, which was not a ligand for recombinant GPR18 (Figs. 1 C and 5 F), exerted organ protection in both GPR18 KO and WT mice (Fig. 8, A and B). These results indicated that specific RvD2–GPR18 interactions in vivo controlled PMN-mediated remote organ injury as illustrated herein with hind-limb occlusion-initiated reflow lung injury.
Figure 8.
I/R injury. RvD2 decreases PMN-mediated lung injury in WT but not GPR18-deficient mice. Mice were subjected to hind limb ischemia (60 min). RvD1, 100 ng RvD2, or vehicle control (0.1% ethanol in saline) was then administered i.v., followed by reperfusion (2 h). Mice were sacrificed and lung was collected. (A) Lung tissue histology. Hematoxylin and eosin (H&E) staining of I/R lungs. Bars, 50 µm. (B) Lung PMN infiltration was quantified by myeloperoxidase (MPO). Results are MPO values (nanogram/milligram lung tissue); mean ± SEM from 2 independent experiments and 5–7 mice/group. *, P < 0.05; **, P < 0.01, treatment (RvD1 or RvD2) versus I/R alone in WT group. #, P < 0.05 versus I/R alone; &, P < 0.05 versus RvD2 in GPR18-KO group using one-way ANOVA with Tukey’s multiple comparison post-test.
DISCUSSION
During self-limited inflammation and active resolution, precursor essential polyunsaturated fatty acids (e.g., AA, EPA, DHA) are converted by exudate leukocytes to several chemically and functionally distinct families of SPM that act on specific target cell types to evoke potent stereoselective actions (Serhan, 2014). The defining pro-resolving actions of SPM include limiting further PMN infiltration, stimulating MΦ phagocytosis, and efferocytosis contributing to shortened resolution intervals. In this study, we performed unbiased screening for functional RvD2 GPCRs and identified GPR18. To further assess this candidate receptor, we prepared synthetic labeled RvD2 and demonstrated direct evidence for specific and stereoselective binding with recombinant GPR18. We also obtained functional evidence for RvD2–GPR18 interactions with isolated cells and in GPR18-deficient mice that we prepared for these studies.
GPR18 gene is localized to human chromosome 13q32, encoding an open reading frame (ORF) of 993 bp. Human GPR18 ORFs is highly conserved with canine and mouse orthologues, sharing ∼89% and ∼83% nucleotide identity, respectively (Gantz et al., 1997; Samuelson et al., 1996). The GPR18 gene is abundantly expressed in testis and spleen, as well as several other tissues associated with endocrine and immunological/hematologic functions (Gantz et al., 1997). The pattern of expression is consistent with information obtained from BioGPS (http://biogps.org) showing that GPR18 is highly expressed in testis and immune systems, including bone marrow, lymph nodes, and tonsil. Within immune cells, the highest expression was found with leukocytes. Along these lines, our results demonstrated GPR18 expression in peripheral blood PMNs and monocytes, as well as in monocyte-derived MΦ (Fig. 1 D). These are target cell types for RvD2’s pro-resolving actions, e.g., limiting PMN and stimulating MΦ functions (Spite et al., 2009; Serhan, 2014; Fig. 3).
NAGly was shown earlier to activate GPR18-transfected cells, suggesting that NAGly is a ligand for GPR18 (Kohno et al., 2006). In this study, we confirmed that NAGly activated recombinant GPR18 using the ECIS system (Fig. 2 B). In addition, NAGly enhances PMN apoptosis in murine peritonitis, and NAGly treatment of HEK-GPR18 cells increases the production of the pro-resolving mediator LXA4, suggesting a role for NAGly-GPR18 in promoting resolution of acute inflammation via SPM production (Burstein et al., 2011).
In I/R-initiated sterile injury, PMN infiltration to the lungs was not statistically significantly different between WT and GPR18-KO mice 2 h after reperfusion (Fig. 8 B). In comparison, in S. aureus skin infection, GPR18-KO did not have impaired ability to clear bacteria or heightened PMN infiltration 4 h after initiation of infection (Fig. 7, H and I). These results suggest that in the acute phase of infection and sterile lung injury, it is possible that endogenous RvD2 was not produced and/or did not play an essential protective role at these initial time intervals. Nevertheless, exogenous administration of RvD2 in WT mice rendered marked protection, which was lost in GPR18-KO mice. RvD1’s protection in I/R-initiated acute lung injury was not diminished in these KO mice (Fig. 8 and vide infra). Of note, in E. coli infection, GPR18-KO mice showed characteristics of resolution deficit, including heightened PMN infiltration, impaired efferocytosis, and reduced E. coli ingestion by phagocytes in the resolution phase, i.e., 12–24 h (Fig. 6, B–D), suggesting a critical role for endogenous RvD2, the GPR18 ligand in controlling inflammation and infection in the resolution phase. Along these lines, RvD2 and other SPMs, including RvD5 and PD1, were present in 24 h infectious exudates and significantly higher in WT mice compared with GPR18-KO (Fig. 6 F). Together, these results pointed to a positive-feedback loop initiated by endogenous RvD2–GPR18 interactions, leading to further increases of select SPMs and enhanced phagocyte pro-resolving functions during bacterial infection.
It is customary to treat infections with antibiotics that directly target bacteria. The present results with RvD2–GPR18 axis, together with our earlier findings with RvD1, RvD5, and PD1 in bacterial infections (Chiang et al., 2012), underscore a potential new option, namely directly treating the host via stimulating innate host responses with SPM to enhance phagocytosis and killing of microbes, and to accelerate resolution of infections. Thus, these findings support the potential for host-directed SPM treatments together with traditional antibiotic therapy. In this regard, SPM are immunoresolvents, and not immunosuppressive at the bioactive range demonstrated in the present study (picomolar to nanomolar in vitro and nanogram doses in vivo in mice). These findings may add new opportunities for host-directed therapy in treating infections, a concept that is also supported by recent results with viral infections of H5N1 influenza and with Mycobacterium tuberculosis infections (Baillie and Digard, 2013; Morita et al., 2013; Mayer-Barber et al., 2014).
With synthetic 3H-RvD2, we report specific binding of 3H-RvD2 to recombinant GPR18, with a Kd ∼10 nM (Fig. 5 C). This value is commensurate with RvD2 bioactions. For example, in human MΦ, RvD2 at 10 nM increases cAMP and stimulated phagocytosis of STZ, E. coli, and apoptotic PMN in a GPR18-dependent manner (Fig. 3). By comparison, RvD1–ALX receptor interactions in MΦ also activate cAMP intracellular signaling (Lee and Surh, 2013). Other SPMs, including RvD1, did not compete for 3H-RvD2 binding to GPR18 (Fig. 5 F). This is consistent with the findings in GPR18 KO mice, where RvD1 retains its organ protective actions in hind limb I/R-initiated and PMN-mediated second organ reflow injury in the lungs, while the response with RvD2 is lost in the GPR18-KO mice (Fig. 8). These results lend support for specific RvD2–GPR18 interactions in recombinant systems and in vivo.
Recently, GPR18 was found to be abundantly expressed in mouse intestinal intraepithelial lymphocytes (IELs; Wang et al., 2014). In catfish, GPR18 is protective in Aeromonas hydrophila infection. Transfection of GPR18 in catfish gill cells offers significant protection against A. hydrophila (Pridgeon and Klesius, 2013). The ligands activating GPR18 were not investigated. Along these lines, several species of fish, including rainbow trout and Atlantic salmon, produce endogenous SPM, including RvD2 (Rowley, 1991; Sharp et al., 1992; Hong et al., 2005; Raatz et al., 2011). These earlier findings, together with our present results that the RvD2–GPR18 axis stimulates human and mouse phagocyte clearance of microbes and limits excessive PMN infiltration in vivo, suggest a potential evolutionarily conserved role for GPR18 and RvD2 in regulating phagocyte responses to protect the host during infections.
Results of the present experiments provide direct evidence to support GPR18 as a GPCR mediating pro-resolving actions of RvD2 with human and mouse phagocytes. Moreover, they illustrate a novel endogenous resolution mechanism with the RvD2–GPR18 axis regulating bacterial infections and intrinsic organ protection.
MATERIALS AND METHODS
GPCR screening.
A panel of 77 orphan GPCRs was screened using the PathHunter β-arrestin enzyme fragment complementation (EFC) technology with β-galactosidase (Discoverx). In this system, β-galactosidase was split into two inactive fragments, enzyme acceptor and enzyme donor. ProLink-tagged proteins were then generated; in this panel, an enzyme acceptor was fused to β-arrestin and an enzyme donor was fused to the orphan GPCRs. Activation of GPCR recruited β-arrestin to the receptor, bringing two inactive fragments in close proximity and restoring β-galactosidase activity, which was monitored via chemiluminescent signals using PathHunter detection reagents (Discoverx). Custom GPCR screening was performed with RvD2 10 nM or vehicle control (0.1% ethanol) using the agonist format; RvD2 was incubated with cells expressing the orphan panel of GPCRs at 37°C for 90 min. Negative controls measured potential constitutive activity in the absence of ligand. This custom screening was performed in duplicate and mean chemiluminescence was used for analysis as follows. For agonist format, the percentage of activity was calculated using the following formula: percent activity = 100% × (mean RLU of test sample – mean RLU of vehicle control)/(mean RLU of vehicle control). RLU, relative luminescence units.
GPCR–β-arrestin system.
Ligand–receptor interactions were monitored using the Beta Arrestin PathHunter eXpress system (Discoverx) and performed essentially as in Krishnamoorthy et al. (2010), with CHO cells stably overexpressing recombinant human GPR18, GPR26, or GPR30 receptors. In brief, cells were plated in 96-well plates 48 h before experiments. Test compounds were incubated with cells for 1 h at 37°C, and receptor activation was determined by measuring chemiluminescence using the PathHunter detection kit (Discoverx).
ECIS system.
Ligand–receptor interactions were determined by measuring impedance across CHO cell monolayers using an ECIS system (Applied Biophysics; Peters and Scott, 2009), and performed essentially as in Krishnamoorthy et al. (2010). In brief, select GPCR or mock-transfected CHO cells were plated at 105 per well of an 8-well ECIS array (8W10E+). Test compounds were added to the chambers in serum-free medium, and real-time impedance changes were monitored (0–10 min, 37°C). For antibody incubations, anti-GPR18 Ab (Imgenex) or nonimmune rabbit IgG was incubated with cells in the ECIS chambers at 1:50 dilutions for 30 min before addition of compounds. For CTX and PTX treatment, CTX (1 µg/ml, 2 h) or PTX (1 µg/ml, 24 h) were incubated with CHO-GPR18, and cells were washed with HAM F-12 serum-free media, followed by addition of RvD2.
Human leukocyte isolation and MΦ differentiation.
Human peripheral blood was drawn from healthy volunteers, who denied taking medications at least 2 wk before donation, by venipuncture in a heparinized syringe (Partners Human Research Committee Protocol No. 1999-P-001297). PMN and monocytes were isolated (Krishnamoorthy et al., 2010). MΦ were differentiated by culturing freshly isolated monocytes in RPMI media supplemented with 10% FBS and recombinant human GM-CSF (10 ng/ml; R&D Systems) for 7 d.
Receptor expression.
Human peripheral blood, isolated PMN, CHO cells or MΦ were incubated with rabbit anti-human GPR18 or rabbit nonimmune IgG (1:50 Imgenex) for 30 min, followed by PE-donkey anti–rabbit IgG (1:200) for 20 min. Flow cytometry was performed using FACSCantoII (BD). CD163 and CD206 expression on MΦ were monitored by flow cytometry using PE-conjugated anti–human CD163 IgG and APC-conjugated anti–human CD206 IgG (R&D Systems).
GPR18 transfection, phagocytosis, and cAMP.
For overexpression of GPR18, MΦ (5 × 106 cells in a 10-cm Petri dish) were transfected with a mock vector or with expression vector for human GPR18 (5 µg; Origene) using Jet-Pei transfection reagent following manufacturer’s instruction (Polyplus-Transfection SA). For knockdown of GPR18, MΦ (5 × 106 cells) were transfected with shRNA plasmids for GPR18 (QIAGEN) or with negative scrambled controls (5 µg) using Jet-Pei transfection reagent. For real-time imaging, MΦ were plated onto 8-well chamber slides (50,000 cells/well in PBS2+) 48 h after transfection. Imaging was then performed 24 h after replating. Chamber slides were kept in a Stage Top Incubation system for microscopes equipped with a built-in digital gas mixer and temperature regulator (TOKAI HIT model INUF-K14). RvD2 was added to MΦ (1 nM, 15 min) followed by BacLight Green-labeled E. coli (2.5 × 106 CFU). Images were then acquired every 10 min for 3 h (37°C) with Keyence BZ-9000 (BIOREVO) inverted fluorescence phase-contrast microscope (20X objective) equipped with a monochrome/color switching camera using BZ-II Viewer software (Keyence). Green fluorescence intensity was quantified using BZ-II Analyzer. For dose–response studies, MΦ were plated onto 96-well plates (50,000 cells/well in PBS2+), and phagocytosis was performed 24 h after replating. RvD2 (0.1 pM-10 nM) was incubated with MΦ for 15 min at 37°C, followed by incubation with FITC-labeled zymosan particles at 10:1 ratio (zymosan: MΦ), CFDA-labeled apoptotic PMN at 5:1 ratios or fluorescent-labeled E. coli (BacLight Green; Molecular Probes) at 50:1 ratio for 60 min at 37°C. Plates were gently washed, extracellular fluorescence was quenched by Trypan blue, and phagocytosis was determined by measuring total fluorescence (Ex 493/Em535 nm) using SpectraMax M3 plate reader (Molecular Probes).
For cAMP measurements, GPR18 shRNA or control scrambled shRNA-transfected human MΦ (0.4 × 106 cells) were incubated with the RvD2 (1–100 nM) for 2 min (37°C). After the incubation, 50 µl of 5% Triton-X 100 was added to stop incubations and cells were homogenized. cAMP levels were measured by ELISA following manufacturer’s instruction (Elite cAMP ELISA Assay kit; eEnzyme).
Murine zymosan-initiated peritonitis, overexpression, and knockdown of GPR18.
Mice were anesthetized with isoflurane and experiments performed with male FVB mice (6–8 wk old; Charles River Labs; laboratory diet containing essential fatty acids from supplier) in accordance with the Harvard Medical Area Standing Committee on Animals (protocol no. 02570). For overexpression of GPR18, peritoneal MΦ (2 × 106 cells) were collected and transfected with either GPR18 (5 µg) or mock plasmids for 3 d. For knockdown experiments, shRNA plasmids for mouse GPR18 (5 µg; Origene) or with negative scrambled controls using Jet-Pei transfection reagent following manufacturer’s instruction (Polyplus-transfection SA). Zymosan (1 mg) was injected i.p. to initiate peritonitis. 12 h later, transfected MΦ (2.0 × 105 cells) and/or RvD2 were injected i.p. Inflammatory exudates were collected at 24 h (see timeline). Total leukocytes were enumerated and PMN (Ly6G+) and efferocytosis (Ly6G+F4/80+ representing MΦ with ingested PMN) were determined using flow cytometry with FACSCantoII (BD).
Preparation of [3H]-RvD2-ME and radioligand binding.
Synthetic precursor 10,11-acetylenic RvD2-ME was prepared by total organic synthesis (purchased as a custom order from Cayman Chemicals), and custom tritiation was performed with S. Gupta (American Radiolabeled Chemicals, St. Louis, MO) by catalytic hydrogenation to give tritiated [10,11-3H]-RvD2-ME (ME of [10,11-3H]-7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-DHA). Integrity of the radioligand was confirmed and isolated using RP-UV-HPLC (1100 Series; Agilent Technologies) using an Eclipse Plus C18 column (100 mm × 4.6 mm × 1.8 µm; Agilent Technologies) coupled with a DAD (G1315B; Agilent Technologies). A gradient of methanol/water of 55:45 (vol/vol) was ramped to 63:37 (vol/vol) over 22 min and then to 98:2 (vol/vol) for the next 8 min. The flow rate was maintained at 0.5 ml/min.
[3H]-RvD2-ME binding was performed with CHO cells transfected with GPR18. Cells were suspended in Dulbecco’s phosphate buffered saline with CaCl2 and MgCl2 (DPBS2+). For saturation binding, cells (0.5 × 106 cells/0.1 ml) were incubated with 0.1–20 nM of [3H]-RvD2-ME, specific activity ∼80 Ci/mmol in the presence or absence of 10 µM of unlabeled RvD2-ME for 60 min at 4°C. For competition binding, cells (0.5 × 106 cells/0.1 ml) were incubated with ∼3 nM of [3H]-RvD2-ME in the absence or presence of increasing concentrations of unlabeled RvD2 (1 nM–10 µM) or related compounds for 60 min at 4°C. RvD1 and RvD2 were purchased from Cayman Chemicals. RvD3, MaR1, and PD1 were prepared by total organic synthesis and complete stereochemistries were determined (Serhan and Petasis, 2011) for National Institutes of Health Program Project (P01-GM095467, CNS). The bound and unbound radioligands were separated by filtration through Whatman GF/C glass microfiber filters (Thermo Fisher Scientific). Filters were washed 2 times with 5 ml ice-cold DPBS. The radioactivity retained on the filter was determined using a scintillation counter (Beckman Coulter). Nonspecific binding was determined in the presence of 10 µM of unlabeled RvD2.
GPR18-deficient mice.
Targeted deletion of mouse gpr18 (NM_182806) was constructed by Lexicon Pharmaceuticals. A bGeo/Puro gene was inserted into the coding region of gpr18 (see Fig. 6 A for target strategy). Gpr18-deficient lines were created by injecting 129/Sleeves cells into C57BL/6 blastocysts. Resulting mice were then bred with B6129FI hybrids to maintain a 129/SvEv-C57B/6 mixed background. Mouse tails (1 cm) were collected, genomic DNA isolated and knockdown of gpr18 was validated by PCR using oligonucleotide primers: forward, 5′-GAGGAAATTGCATCGCATTGTCT-3′; and reverse, 5′-GACCTTGGGCTTCAGCTTAGA-3′, which amplify a DNA fragment of 280 bps.
Microbial-initiated peritonitis.
Mice were anesthetized with isoflurane, and experiments were performed with male GPR18-deficient mice or WT littermates (8–10 wk old). In brief, mice were anesthetized, 100 ng RvD2 or vehicle controls was injected into the peritoneal cavity together with live E. coli (105 CFU). At designated points, mice were euthanized (overdose of isoflurane) and peritoneal exudate was collected by ravaging with 5 ml PBS. Aliquots of exudate cells were incubated with anti–mouse CD16/32 blocking antibody (0.5 µg/0.5 × 106 cells, 5 min), and then incubated (20 min, 4°C) with individual or a combination of fluorescently labeled antibodies including anti–mouse CD14 (clone rmC5-3) for mononuclear cells and anti–mouse F4/80 antibody (clone BM8) for MΦ or anti–mouse Ly6G (clone RB6-8C5) for PMN, to determine leukocyte sub-types by flow cytometry (FACS Canto II). Antibodies were purchased from BD and eBioscience. Intracellular E. coli levels were determined using a FITC-conjugated anti-E. coli antibody (GTX40856; Genetics). Cells were incubated with anti-mouse CD16/32 blocking antibody and labeled with FITC-conjugated anti–mouse F4/80 Ab for 20 min at 4°C, followed by permeabilization (Cytofix/Cytoperm solution kit; BD). Next, permeabilized cells were labeled with PE-conjugated anti–mouse Ly6G antibody and F4/80+Ly-6G+ MΦ population was determined by flow cytometry.
Mouse phagocyte phagocytosis.
For peritoneal MΦ phagocytosis, resident peritoneal MΦ were collected from naive WT and GPR18 KO mice and plated onto 96-well plates (50,000 cells/well). RvD2 (0.1 pM–10 nM) was incubated with MΦ for 15 min at 37°C, followed by incubation with FITC-labeled zymosan particles at 10:1 ratio (zymosan: MΦ) for 60 min at 37°C. Plates were gently washed, extracellular fluorescence was quenched by Trypan blue, and phagocytosis was determined by measuring total fluorescence (Ex 493/Em535 nm) using a fluorescent plate reader (Molecular Probes). For phagocytosis in whole blood, heparinized peripheral blood (50 µl) of WT and KO mice was incubated with BacLight Green–labeled E. coli (∼2 × 107 CFU) at 37°C for 60 min. RBCs were then lysed, and fluorescence associated with PMNs and monocytes were monitored by flow cytometry.
Murine dorsal skin pouches.
Pouches were raised for 6 d (Wynyard and Willoughby, 2003). Mice were given RvD2 (200 ng) or vehicle control with live S. aureus (serotype [b]c1; 105 CFU) by intra-pouch injection. 4 h later, mice were euthanized, intra-pouch exudates were collected, and leukocyte counts were determined. For bacterial titers, aliquots of lavage were used for serial dilution, plated onto LB agar plates, and cultured overnight at 37°C.
I/R induced second organ reflow injury.
Bilateral hind limb ischemia was initiated using tourniquets placed on each hind limb. Mice were subjected to ischemia for 60 min, followed by tail vein injection of 100 ng RvD2, and then tourniquets were removed to initiate reperfusion for 2 h. At this interval, mice were euthanized, and lungs were harvested and stored at −80° C or in 10% (vol/vol) buffered formalin for histology assessment by the Histology Core of Boston Children’s Hospital. PMN infiltrations into lungs were quantified using increments of lung myeloperoxidase (MPO). In brief, the frozen lungs were homogenized and centrifuged, and tissue levels of MPO were determined with ELISA (R&D Systems).
LM metabololipidomics.
All samples for LC-MS/MS analysis were extracted using SPE columns. In brief, two volumes of methanol with deuterium-labeled internal standards (d4-PGE2, d4-LTB4, d5-LXA4, and d5-RvD2; 500 pg. each) were added to the lavages to facilitate quantification of sample recovery. Samples were kept at −20°C for 45 min to allow protein precipitation. Samples were then placed into an automated extraction system (RapidTrace; Biotage) and products extracted as follows. Solid-phase C18 cartridges were equilibrated with 3 ml of methanol and 6 ml of H2O. 9 ml H2O (pH 3.5, HCl) were then added to the samples, and the acidified solutions were rapidly loaded onto the conditioned C18 columns that were washed with 4 ml of H2O to neutralize the acid. Next, 5 ml hexane was added, and products were eluted with 9 ml of methyl formate. Products were brought to dryness using the automated evaporation system (TurboVap LV; Biotage) and immediately suspended in methanol-water (50:50 vol/vol) for LC-MS/MS automated injections. The LC-MS/MS system consisted of a QTrap 5500 (ABSciex) equipped with a Shimadzu LC-20AD HPLC and a Shimadzu SIL-20AC autoinjector (Shimadzu Corp.). An Agilent Poroshell 120 EC-C18 column (100 mm × 4.6 mm × 2.7 µm) was kept at 50°C and LM were eluted with a mobile phase consisting of methanol–water–acetic acid (50:50:0.01, vol/vol/vol) that was ramped to 80:20:0.01 (vol/vol/vol) from 2 to 11 min, maintained till 14.5 min, and then rapidly ramped to 98:2:0.01 (vol/vol/vol) for the next 0.1 min. This was subsequently maintained at 98:2:0.01 (vol/vol/vol) for 5.4 min, and the flow rate was maintained at 0.5 ml/min. To monitor and quantify the levels of targeted LM, a multiple reaction monitoring (MRM) method was devised with signature ion fragments for each molecule. Identification was conducted using published criteria (Dalli and Serhan, 2012) using retention times and at least six diagnostic ions. Calibration curves were obtained using synthetic and authentic LM mixtures, including d4-LTB4, d5-LXA4, d4-PGE2, d5-RvD2, RvD1, RvD2, RvD5, PD1, MaR1, RvE1, RvE2, LXA4, LXB4, PGE2, PGD2, PGF2α, TXB2, and LTB4 at 1.56, 3.12, 6.25, 12.5, 25, 50, 100 and 200 pg. Linear calibration curves for each were obtained with r2 values of 0.98–0.99. Quantification was performed based on peak area of the MRM transition and the linear calibration curve obtained with authentic standard for each compound.
Statistical analysis.
Results are expressed as mean ± SEM. Statistical analysis were performed using nonparametric tests including Student’s t test for two-group comparisons and one-way analysis of variance (ANOVA) for multiple group comparisons with post-hoc analysis using Tukey’s or Dunnett’s Multiple Comparison test (Prism; GraphPad). P < 0.05 was considered to be significant.
Acknowledgments
The authors thank Mary Small for assistance with manuscript preparation, Dr. Masakazu Shinohara for carrying out cAMP measurements while a member of this laboratory, and Chien-Yee Cheng for technical assistance.
This work was supported in part by National Institutes of Health grants R01GM38765 and a Mérieux Research Grant from the Institut Mérieux (C.N. Serhan).
C.N. Serhan is an inventor on patents [resolvins] assigned to BWH and licensed to Resolvyx Pharmaceuticals. C.N. Serhan was a scientific founder of Resolvyx Pharmaceuticals and owns equity in the company. C.N. Serhan’s interests were reviewed and are managed by the Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- CTX
- cholera toxin
- DHA
- docosahexaenoic acid
- ECIS
- electrical cell substrate impedance sensing
- GPCR
- G protein–coupled receptor
- I/R
- ischemia-reperfusion
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- LM
- lipid mediator
- LOX
- lipoxygenase
- LT
- leukotriene
- LX
- lipoxin
- MΦ
- macrophage
- MaR1
- maresin 1
- NAGly
- N-arachidonyl glycine
- PD1
- protectin D1 (10R,17S-dihydroxy-4Z,7Z,11E,13E,15Z,19Z-DHA)
- PTX
- pertussis toxin
- Rv
- resolvin
- RvD2
- resolvin D2 (7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-DHA)
- [3H]-RvD2
- [10,11-3H]-7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-DHA
- SPM
- specialized pro-resolving mediator
- STZ
- serum-treated zymosan
References
- Baillie J.K., and Digard P.. 2013. Influenza—time to target the host? N. Engl. J. Med. 369:191–193. 10.1056/NEJMcibr1304414 [DOI] [PubMed] [Google Scholar]
- Bento A.F., Claudino R.F., Dutra R.C., Marcon R., and Calixto J.B.. 2011. Omega-3 fatty acid-derived mediators 17(R)-hydroxy docosahexaenoic acid, aspirin-triggered resolvin D1 and resolvin D2 prevent experimental colitis in mice. J. Immunol. 187:1957–1969. 10.4049/jimmunol.1101305 [DOI] [PubMed] [Google Scholar]
- Bohr S., Patel S.J., Sarin D., Irimia D., Yarmush M.L., and Berthiaume F.. 2013. Resolvin D2 prevents secondary thrombosis and necrosis in a mouse burn wound model. Wound Repair Regen. 21:35–43. 10.1111/j.1524-475X.2012.00853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein S.H., McQuain C.A., Ross A.H., Salmonsen R.A., and Zurier R.E.. 2011. Resolution of inflammation by N-arachidonoylglycine. J. Cell. Biochem. 112:3227–3233. 10.1002/jcb.23245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N., Gronert K., Clish C.B., O’Brien J.A., Freeman M.W., and Serhan C.N.. 1999. Leukotriene B4 receptor transgenic mice reveal novel protective roles for lipoxins and aspirin-triggered lipoxins in reperfusion. J. Clin. Invest. 104:309–316. 10.1172/JCI7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N., Fredman G., Bäckhed F., Oh S.F., Vickery T., Schmidt B.A., and Serhan C.N.. 2012. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. 484:524–528. 10.1038/nature11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clària J., Nguyen B.T., Madenci A.L., Ozaki C.K., and Serhan C.N.. 2013. Diversity of lipid mediators in human adipose tissue depots. Am. J. Physiol. Cell Physiol. 304:C1141–C1149. 10.1152/ajpcell.00351.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas R.A., Shinohara M., Dalli J., Chiang N., and Serhan C.N.. 2014. Identification and signature profiles for pro-resolving and inflammatory lipid mediators in human tissue. Am. J. Physiol. Cell Physiol. 307:C39–C54. 10.1152/ajpcell.00024.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotran R.S., Kumar V., and Collins T., 1999. Robbins Pathologic Basis of Disease. W.B. Saunders Co., Philadelphia: 1425 pp. [Google Scholar]
- Dalli J., and Serhan C.N.. 2012. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 120:e60–e72. 10.1182/blood-2012-04-423525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschman C.S., and Tracey K.J.. 2014. Sepsis: current dogma and new perspectives. Immunity. 40:463–475. 10.1016/j.immuni.2014.04.001 [DOI] [PubMed] [Google Scholar]
- El Kebir D., Gjorstrup P., and Filep J.G.. 2012. Resolvin E1 promotes phagocytosis-induced neutrophil apoptosis and accelerates resolution of pulmonary inflammation. Proc. Natl. Acad. Sci. USA. 109:14983–14988. 10.1073/pnas.1206641109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig H.K., and Eckle T.. 2011. Ischemia and reperfusion—from mechanism to translation. Nat. Med. 17:1391–1401. 10.1038/nm.2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frediani J.K., Jones D.P., Tukvadze N., Uppal K., Sanikidze E., Kipiani M., Tran V.T., Hebbar G., Walker D.I., Kempker R.R., et al. 2014. Plasma metabolomics in human pulmonary tuberculosis disease: a pilot study. PLoS ONE. 9:e108854 10.1371/journal.pone.0108854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz I., Muraoka A., Yang Y.K., Samuelson L.C., Zimmerman E.M., Cook H., and Yamada T.. 1997. Cloning and chromosomal localization of a gene (GPR18) encoding a novel seven transmembrane receptor highly expressed in spleen and testis. Genomics. 42:462–466. 10.1006/geno.1997.4752 [DOI] [PubMed] [Google Scholar]
- Guo Y., Zhang W., Giroux C., Cai Y., Ekambaram P., Dilly A.K., Hsu A., Zhou S., Maddipati K.R., Liu J., et al. 2011. Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)-hydroxyeicosatetraenoic acid. J. Biol. Chem. 286:33832–33840. 10.1074/jbc.M110.216564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Tjonahen E., Morgan E.L., Lu Y., Serhan C.N., and Rowley A.F.. 2005. Rainbow trout (Oncorhynchus mykiss) brain cells biosynthesize novel docosahexaenoic acid-derived resolvins and protectins-Mediator lipidomic analysis. Prostaglandins Other Lipid Mediat. 78:107–116. 10.1016/j.prostaglandins.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Klein C.P., Sperotto N.D., Maciel I.S., Leite C.E., Souza A.H., and Campos M.M.. 2014. Effects of d-series resolvins on behavioral and neurochemical changes in a fibromyalgia-like model in mice. Neuropharmacology. 86:57–66. 10.1016/j.neuropharm.2014.05.043 [DOI] [PubMed] [Google Scholar]
- Kohno M., Hasegawa H., Inoue A., Muraoka M., Miyazaki T., Oka K., and Yasukawa M.. 2006. Identification of N-arachidonylglycine as the endogenous ligand for orphan G-protein-coupled receptor GPR18. Biochem. Biophys. Res. Commun. 347:827–832. 10.1016/j.bbrc.2006.06.175 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy S., Recchiuti A., Chiang N., Yacoubian S., Lee C.-H., Yang R., Petasis N.A., and Serhan C.N.. 2010. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc. Natl. Acad. Sci. USA. 107:1660–1665. 10.1073/pnas.0907342107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Jones C.N., Yu Y.M., Fischman A.J., Watada S., Tompkins R.G., Fagan S.P., and Irimia D.. 2013. Resolvin D2 restores neutrophil directionality and improves survival after burns. FASEB J. 27:2270–2281. 10.1096/fj.12-219519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.N., and Surh Y.J.. 2013. Resolvin D1-mediated NOX2 inactivation rescues macrophages undertaking efferocytosis from oxidative stress-induced apoptosis. Biochem. Pharmacol. 86:759–769. 10.1016/j.bcp.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Mas E., Croft K.D., Zahra P., Barden A., and Mori T.A.. 2012. Resolvins D1, D2, and other mediators of self-limited resolution of inflammation in human blood following n-3 fatty acid supplementation. Clin. Chem. 58:1476–1484. 10.1373/clinchem.2012.190199 [DOI] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Andrade B.B., Oland S.D., Amaral E.P., Barber D.L., Gonzales J., Derrick S.C., Shi R., Kumar N.P., Wei W., et al. 2014. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 511:99–103. 10.1038/nature13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead P.S., Slutsker L., Dietz V., McCaig L.F., Bresee J.S., Shapiro C., Griffin P.M., and Tauxe R.V.. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625. 10.3201/eid0505.990502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S., et al. 2013. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 153:112–125. 10.1016/j.cell.2013.02.027 [DOI] [PubMed] [Google Scholar]
- Nathan C. 2012. Fresh approaches to anti-infective therapies. Sci. Transl. Med. 4:sr2 10.1126/scitranslmed.3003081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.K., Xu Z.Z., Liu T., Lü N., Serhan C.N., and Ji R.R.. 2011. Resolvin D2 is a potent endogenous inhibitor for transient receptor potential subtype V1/A1, inflammatory pain, and spinal cord synaptic plasticity in mice: distinct roles of resolvin D1, D2, and E1. J. Neurosci. 31:18433–18438. 10.1523/JNEUROSCI.4192-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M.F., and Scott C.W.. 2009. Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity. J. Biomol. Screen. 14:246–255. 10.1177/1087057108330115 [DOI] [PubMed] [Google Scholar]
- Plüddemann A., Mukhopadhyay S., and Gordon S.. 2011. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol. Rev. 240:11–24. 10.1111/j.1600-065X.2010.00989.x [DOI] [PubMed] [Google Scholar]
- Pridgeon J.W., and Klesius P.H.. 2013. G-protein coupled receptor 18 (GPR18) in channel catfish: expression analysis and efficacy as immunostimulant against Aeromonas hydrophila infection. Fish Shellfish Immunol. 35:1070–1078. 10.1016/j.fsi.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Raatz S.K., Golovko M.Y., Brose S.A., Rosenberger T.A., Burr G.S., Wolters W.R., and Picklo M.J. Sr. 2011. Baking reduces prostaglandin, resolvin, and hydroxy-fatty acid content of farm-raised Atlantic salmon (Salmo salar). J. Agric. Food Chem. 59:11278–11286. 10.1021/jf202576k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A.G., McCutcheon J.C., Roy N., Chilvers E.R., Haslett C., and Dransfield I.. 1998. Regulation of macrophage phagocytosis of apoptotic cells by cAMP. J. Immunol. 160:3562–3568. [PubMed] [Google Scholar]
- Rowley A.F. 1991. Lipoxin formation in fish leucocytes. Biochim. Biophys. Acta. 1084:303–306. 10.1016/0005-2760(91)90073-Q [DOI] [PubMed] [Google Scholar]
- Samuelson L.C., Swanberg L.J., and Gantz I.. 1996. Mapping of the novel G protein-coupled receptor Gpr18 to distal mouse chromosome 14. Mamm. Genome. 7:920–921. 10.1007/s003359900272 [DOI] [PubMed] [Google Scholar]
- Serhan C.N. 2014. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 510:92–101. 10.1038/nature13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., and Chiang N.. 2013. Resolution phase lipid mediators of inflammation: agonists of resolution. Curr. Opin. Pharmacol. 13:632–640. 10.1016/j.coph.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., and Petasis N.A.. 2011. Resolvins and protectins in inflammation resolution. Chem. Rev. 111:5922–5943. 10.1021/cr100396c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., Hong S., Gronert K., Colgan S.P., Devchand P.R., Mirick G., and Moussignac R.-L.. 2002. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J. Exp. Med. 196:1025–1037. 10.1084/jem.20020760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp G.J., Pettitt T.R., Rowley A.F., and Secombes C.J.. 1992. Lipoxin-induced migration of fish leukocytes. J. Leukoc. Biol. 51:140–145. [DOI] [PubMed] [Google Scholar]
- Shimizu T. 2009. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49:123–150. 10.1146/annurev.pharmtox.011008.145616 [DOI] [PubMed] [Google Scholar]
- Spite M., Norling L.V., Summers L., Yang R., Cooper D., Petasis N.A., Flower R.J., Perretti M., and Serhan C.N.. 2009. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 461:1287–1291. 10.1038/nature08541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., and Glass C.K.. 2013. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science. 339:166–172. 10.1126/science.1230720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sumida H., and Cyster J.G.. 2014. GPR18 is required for a normal CD8αα intestinal intraepithelial lymphocyte compartment. J. Exp. Med. 211:2351–2359. 10.1084/jem.20140646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winyard P.G., and Willoughby D.A., 2003. Inflammation Protocols. Humana, Totowa, NJ: 378 pp 10.1385/1592593747 [DOI] [Google Scholar]
- World Health Organization. 2014. Antibiotics Resistance, Factsheet No. 194. http://www.who.int/mediacentre/factsheets/fs194/en/.
- Zizzo G., Hilliard B.A., Monestier M., and Cohen P.L.. 2012. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J. Immunol. 189:3508–3520. 10.4049/jimmunol.1200662 [DOI] [PMC free article] [PubMed] [Google Scholar]