Kang et al. show that genetic or pharmacological inactivation of the enzyme phospholipase D1 (PLD1) disrupts colitis-associated intestinal tumorigenesis by suppressing the self-renewal capacity of colon cancer stem cells.
Abstract
Expression of the Wnt target gene phospholipase D1 (PLD1) is up-regulated in various carcinomas, including colorectal cancer (CRC). However, the mechanistic significance of its elevated expression in intestinal tumorigenesis remains unknown. In this study, we show that genetic and pharmacological targeting of PLD1 disrupts spontaneous and colitis-associated intestinal tumorigenesis in ApcMin/+ and azoxymethane/dextran sodium sulfate mice models. Intestinal epithelial cell–specific PLD1 overexpression in ApcMin/+ mice accelerated tumorigenesis with increased proliferation and nuclear β-catenin levels compared with ApcMin/+ mice. Moreover, PLD1 inactivation suppressed the self-renewal capacity of colon cancer–initiating cells (CC-ICs) by decreasing expression of β-catenin via E2F1-induced microRNA (miR)-4496 up-regulation. Ultimately, low expression of PLD1 coupled with a low level of CC-IC markers was predictive of a good prognosis in CRC patients, suggesting in vivo relevance. Collectively, our data reveal that PLD1 has a crucial role in intestinal tumorigenesis via its modulation of the E2F1–miR-4496–β-catenin signaling pathway. Modulation of PLD1 expression and activity represents a promising therapeutic strategy for the treatment of intestinal tumorigenesis.
Colorectal cancer (CRC) is one of the leading causes of cancer deaths. Most human CRC involves somatic mutations in the adenomatous polyposis coli (APC) tumor suppressor gene, which leads to activation of Wnt signaling via β-catenin stabilization. Accumulated β-catenin then translocates into the nucleus, where it binds and activates T cell factor (TCF) transcription factor (Fearon and Vogelstein, 1990). Aberrant trans-activation of a certain set of target genes by β-catenin and TCF4–LEF (lymphoid enhancing factor) complexes is believed to be crucial to the initiation of intestinal carcinogenesis. However, it has become increasingly evident that the temporal- and cell-specific expression of known Wnt/β-catenin/TCF target genes does not explain the pervasiveness and complexity of Wnt/β-catenin biology in the context of CRC (Niehrs and Acebron, 2012), which raises the possibility that factors other than β-catenin/TCF contribute to Wnt-induced biological responses. Indeed, a variety of cancer-relevant molecules and signaling pathways intersect with the Wnt pathway and may modulate it. Accordingly, identification of critical effectors regulating the multifaceted cross talk between signaling pathways will lead to nomination of new candidates for creation of targeted therapies useful to CRC management.
Some cancers are hierarchically organized and sustained by a relatively rare population of so-called cancer-initiating cells (C-ICs) or cancer stem cells (CSCs; Al-Hajj and Clarke, 2004). Although the capacity to initiate tumors upon serial transplantation is a hallmark of all C-ICs, little is known about the genes that control this process. The Wnt/β-catenin pathway is a major regulator of colon C-IC (CC-IC) genesis, as well as the self-renewal of normal cells (Brabletz et al., 2009). We recently demonstrated that the genes encoding the lipid enzyme phospholipase D1/2 (PLD1/2) comprise a new transcriptional target of β-catenin/TCF4 (Kang et al., 2010, 2011a). Moreover, PLD1 promotes Wnt signaling by increasing the β-catenin–TCF-4 interaction, revealing bidirectional cross talk between the PLD1 and Wnt/β-catenin pathways. PLD1 and its related isoform PLD2 hydrolyze phosphatidylcholine to generate bioactive lipid phosphatidic acid (PA) and are elevated in various human cancers, including CRC (Selvy et al., 2011). Although PLD has been implicated in tumor growth and metastasis (Su et al., 2009; Chen et al., 2012; Henkels et al., 2013), the potential pathogenetic role of PLD in CRC has yet to be explored. Specifically, the molecular mechanisms linking oncogenic Wnt signaling and the PLD1 pathway in CRC, as well as their pathophysiological significance, have not been addressed. In this study, we explore the role of PLD1 in spontaneous and colitis-associated intestinal tumorigenesis by generating gain- and loss-of-function mouse models with genetically manipulated PLD1 expression. We investigate the role and mechanism of action of PLD1 as a key modulator of the cancer-relevant signaling networks and CC-IC self-renewal capacity that contributes to intestinal tumorigenesis.
RESULTS
PLD1 inactivation retards spontaneous and colitis-associated intestinal tumorigenesis
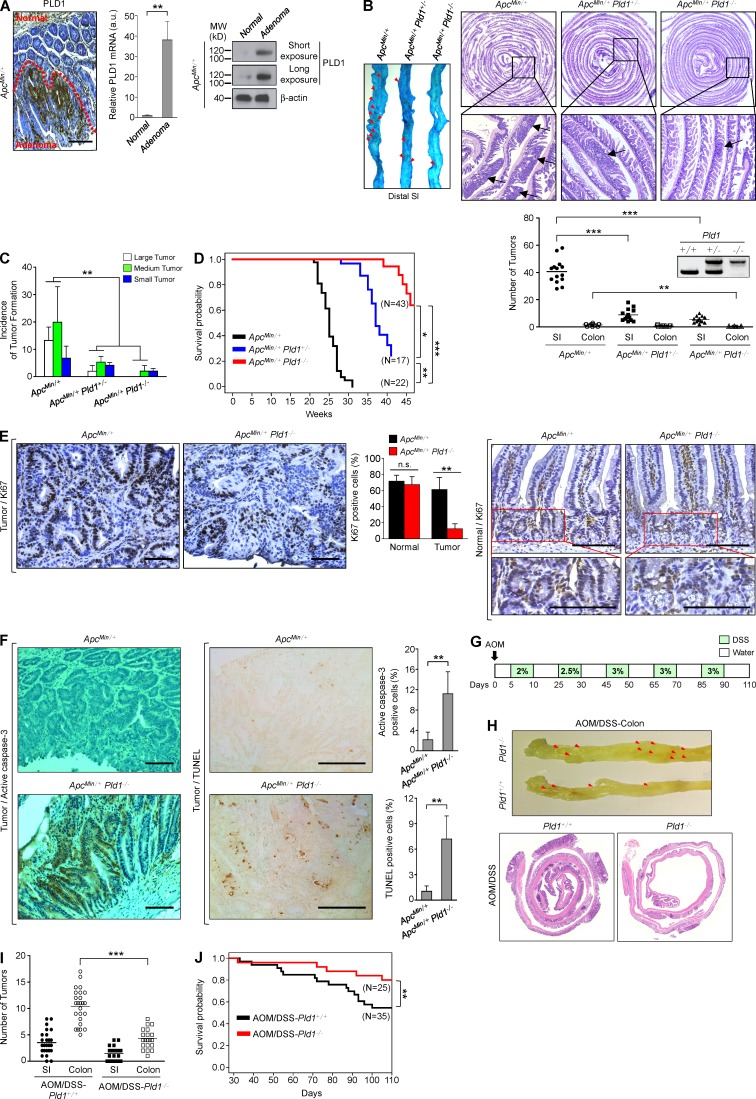
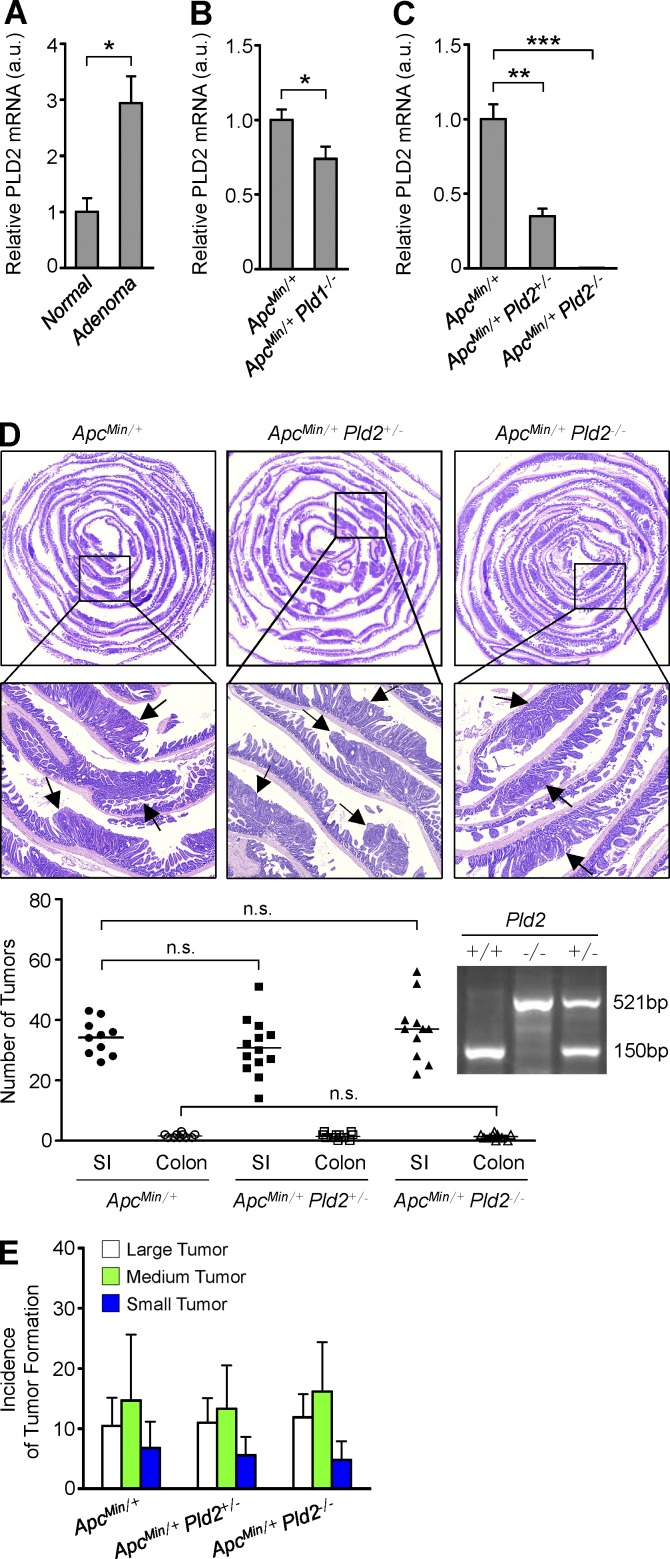
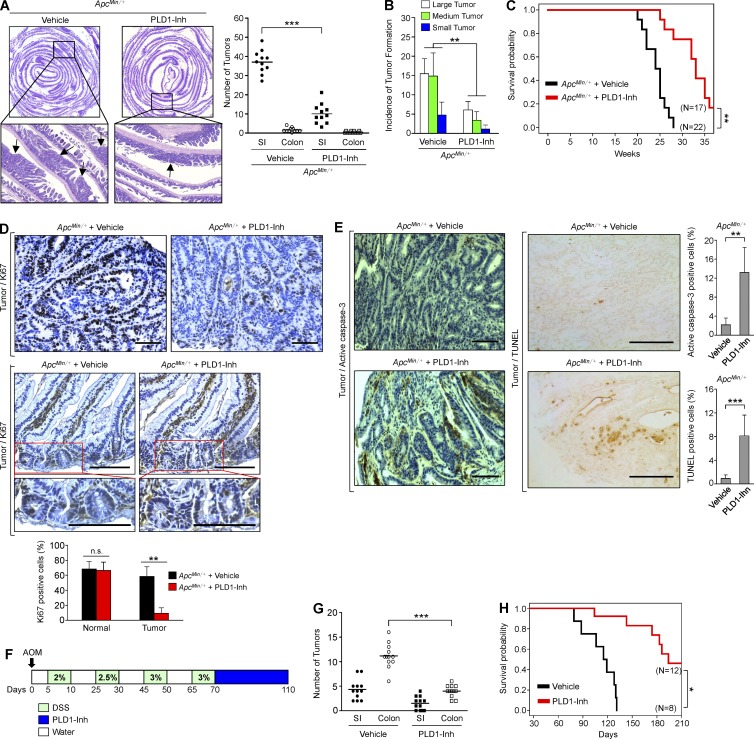
Because Wnt signaling is a well-known oncogenic pathway and PLD1 activates the Wnt signaling pathway, we assessed the function of PLD1 in intestinal tumorigenesis using the ApcMin/+ mouse model. ApcMin/+ mice contain a germline mutation at codon 850 of the Apc gene that results in activation of the Wnt/β-catenin pathway and spontaneous development of numerous adenomatous polyps in the intestine (Kennell and Cadigan, 2009). Expression of PLD1 was dramatically increased in the intestinal adenomas of ApcMin/+ mice relative to normal intestinal tissues, in which the level of PLD1 was very low (Fig. 1 A and see Fig. 4 G). Therefore, we generated ApcMin/+ mice with WT, heterozygous, or homozygous Pld1 (Dall’Armi et al., 2010). The number of intestinal polyps in 16-wk-old ApcMin/+Pld1−/− or ApcMin/+Pld1+/− mice was significantly lower than in ApcMin/+ control mice, and the polyps that were present at the proximal and distal small intestine (SI) were smaller than those present in age-matched ApcMin/+ mice (Fig. 1, B and C). In addition, the mortality of ApcMin/+Pld1−/− or ApcMin/+Pld1+/− mice was significantly reduced relative to ApcMin/+ littermate controls (Fig. 1 D). Immunohistochemical staining (IHC) using antibodies to Ki67 revealed that tumors from ApcMin/+Pld1−/− mice showed lower proportions of proliferating cells than those from control mice (Fig. 1 E). Ki67 in ApcMin/+ and ApcMin/+Pld1−/− mice was expressed at the bottom of the crypts in the normal intestinal area, and the number of Ki67+ cells in the normal crypts and tumors was quantified (Fig. 1 E). Thus, it seems that the animal would not succumb as a result of the intestinal loss. Moreover, tumors from ApcMin/+Pld1−/− showed higher proportions of apoptotic cells than control mice, as analyzed by IHC using antibodies to active caspase-3 and by TUNEL assay (Fig. 1 F). The levels of caspase-3–and TUNEL-positive cells were quantified (Fig. 1 F). Furthermore, we investigated whether PLD1 inactivation inhibits colitis-associated cancer using an azoxymethane (AOM)/dextran sodium sulfate (DSS)–induced mouse colon cancer model (Neufert et al., 2007). For the AOM/DSS model, mice were given a single i.p. injection of the mutagen AOM, after which they received drinking water containing 2–3% DSS in several 5-d periods that were interspersed with periods in which they received normal water (Fig. 1 G). The number of intestinal polyps and the mortality in Pld1-deficient AOM/DSS mice was reduced significantly relative to control mice, suggesting that PLD1 loss also suppresses colitis-associated intestinal tumorigenesis (Fig. 1, H–J). Considering that PLD2 has a close expression pattern to PLD1, we wondered whether the PLD2 level is altered via compensation in the setting of PLD1 loss. PLD2 expression was increased in the intestinal adenomas of ApcMin/+ mice relative to normal intestinal tissues (Fig. 2 A). Moreover, PLD2 expression was somewhat decreased in ApcMin/+Pld1−/− compared with ApcMin/+ (Fig. 2 B). Thus, to examine whether PLD2 loss plays a role in intestinal tumorigenesis, we generated ApcMin/+ mice with WT, heterozygous, or homozygous Pld2. PLD2 expression was decreased or ablated in ApcMin/+Pld2+/− or ApcMin/+Pld2−/− mice, respectively (Fig. 2 C). Contrary to PLD1-deleted ApcMin/+ mice, ApcMin/+ mice with heterozygous or homozygous Pld2 showed a marginal difference in the number and size of intestinal polyps, suggesting that PLD2 deletion does not retard intestinal tumorigenesis (Fig. 2, D and E). Thus, PLD1-dependent signaling can contribute to the regulation of intestinal tumorigenesis. Moreover, it is possible that the biological effects may be through other means such as nonenzymatic protein–protein interactions. We further examined whether a PLD1 inhibitor (VU0155069) known to selectively inhibit PLD1 (Scott et al., 2009) affects intestinal tumorigenesis. Moreover, PLD1 inhibitor–treated ApcMin/+ mice (10 mg/kg, three times a week for 4 wk) also significantly suppressed the number and size of intestinal polyps and increased the mortality relative to vehicle-treated ApcMin/+ mice, which are results comparable to those of ApcMin/+Pld1−/− mice (Fig. 3, A–C). The tumors from PLD1 inhibitor–treated ApcMin/+ mice showed lower proportions of proliferating cells as analyzed by IHC using antibodies to Ki67 (Fig. 3 D). As an internal control, Ki67 in both PLD1 inhibitor– and vehicle-treated ApcMin/+ mice was expressed at the bottoms of the crypts in the normal intestinal area (Fig. 3 D). The number of Ki67+ cells in the normal crypts and tumors was quantified (Fig. 3 D). Additionally, PLD1 inhibition in ApcMin/+ mice increased higher proportions of apoptotic cells (Fig. 3 E). The levels of caspase-3– and TUNEL-positive cells were quantified (Fig. 3 E). These findings indicate that PLD1-dependent signaling contributes substantially to the severe mortality and tumor growth caused by inactivation of APC. Furthermore, PLD1 inhibitor–treated AOM/DSS mice also generated results comparable to those observed in response to PLD1 ablation (Fig. 3, F–H). These data indicate that PLD1 is closely associated with colitis-associated cancer. Collectively, our findings reveal that PLD1 is a crucial mediator of intestinal tumorigenesis that may represent a therapeutic target in intestinal tumors.
Figure 1.
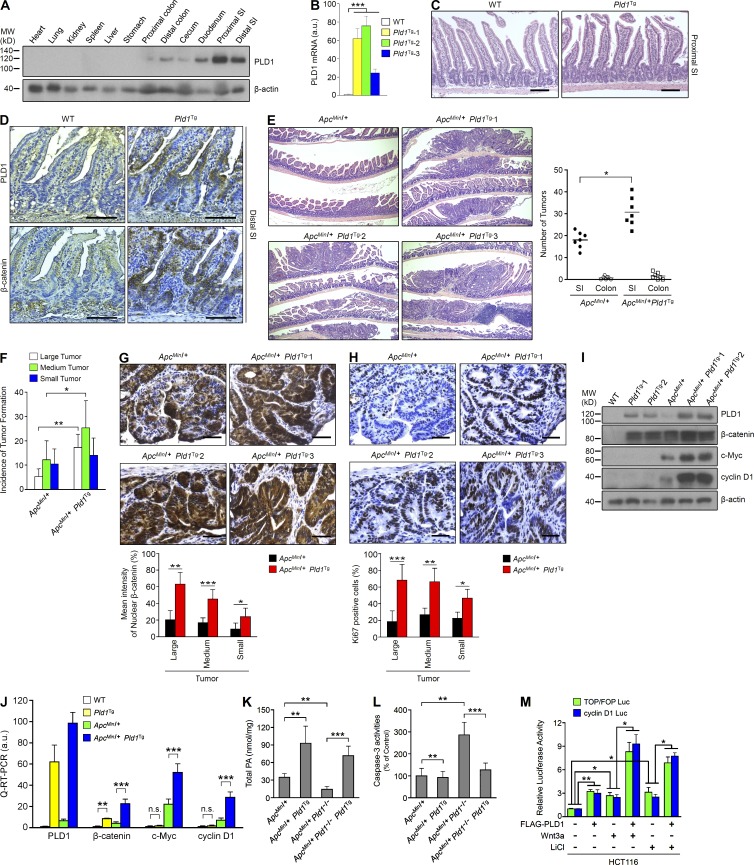
Loss of PLD1 attenuates intestinal tumorigenesis in ApcMin/+ and AOM/DSS mice models. (A) IHC for PLD1 in normal epithelia and adenomas from Apcmin/+ mice (left). q-RT-PCR (middle) and IB (right) analysis of PLD1 in normal SI of WT mice or adenomas of Apcmin/+ mice. n = 6 per group. Results are representative of at least three independent experiments and are shown as mean ± SEM. Student’s t test was used. (B and C) ApcMin/+ (n = 14), ApcMin/+Pld1+/− (n = 13), or ApcMin/+Pld1−/− (n = 13) mice were sacrificed at 16 wk. (B) Methylene blue staining of the representative distal SI of the indicated mice (left). Arrows indicate polyps. H&E staining of the entire SI Swiss roll (top right). The number of visible polyps in the SI and colon intestine (bottom). Student’s t test was used. (C) Size distribution of polyps in the SI of the indicated mice (small, <0.5 mm; medium, 0.5–1.5 mm; large, >1.5 mm). Results are representative of three independent experiments. ANOVA F-test was used. (D) The indicated mice were followed for long-term survival. Survival probability was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. (E) IHC for Ki67 in tumor tissues (left) and normal mucosa (right) of ApcMin/+ and ApcMin/+Pld1−/− mice. Representative images were selected from at least six different fields. The expression of Ki67 in the tumor was quantified (middle). Results are representative of three independent experiments and are shown as mean ± SEM. n = 11 per group; five tumors per mouse. n.s., not significant. (F) IHC for active caspase-3 (left) and TUNEL assay (middle) in tumor tissues of ApcMin/+ and ApcMin/+Pld1−/− mice. The levels of caspase-3– and TUNEL-positive cells were quantified (right). Results are representative of at least two independent experiments and are shown as mean ± SEM. n = 8 per group; five tumors per mouse. IHC staining results were analyzed using a Chi-square test. (G) Schematic view of the colitis-associated cancer model (AOM/DSS) in Pld1+/+ and Pld1−/− mice and AOM/DSS-Pld1+/+ (n = 25) or AOM/DSS-Pld1−/− (n = 20) mice, which were sacrificed at 110 d. (H) The dissected colon intestine was photographed by an optical camera (top). Arrows indicate polyps. H&E staining of the entire colon intestine Swiss roll (bottom). (I) The number of visible polyps in the SI and colon intestine. Student’s t test was used. (J) The indicated mice were followed for long-term survival. Survival probability was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. MW, molecular weight; a.u., arbitrary units. Bars, 100 µm.
Figure 4.
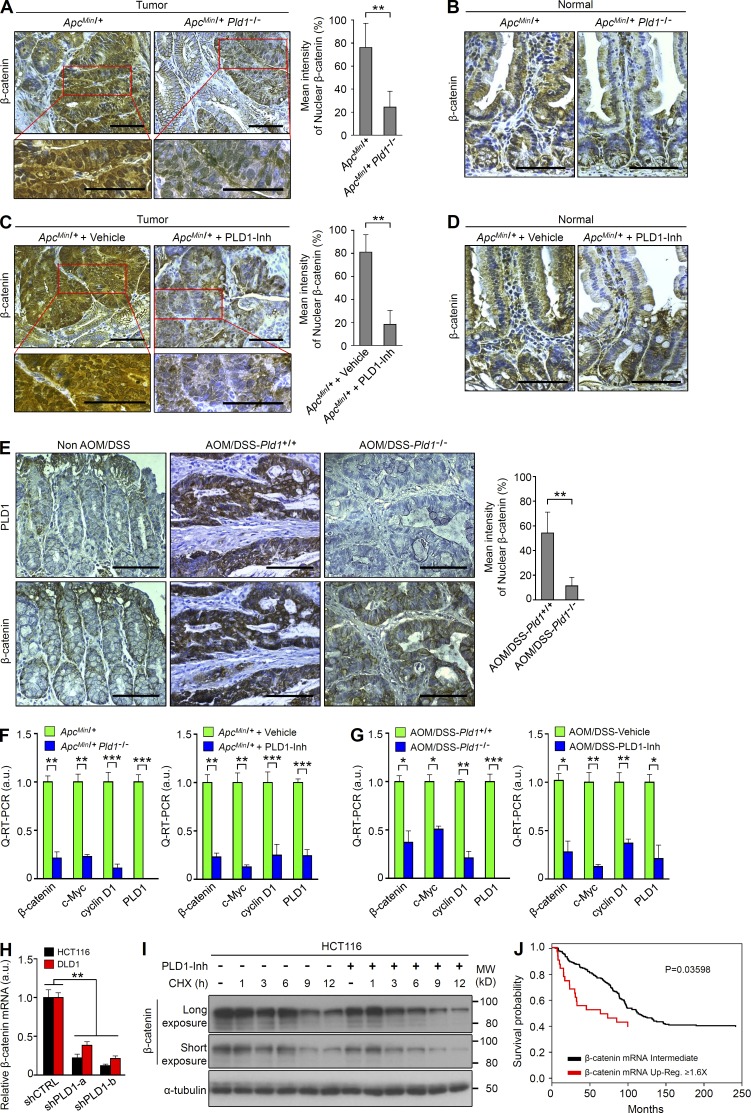
PLD1 inactivation down-regulates expression of β-catenin. (A–D) IHC for β-catenin in tumor tissues (A) and normal mucosa (B) of ApcMin/+ and ApcMin/+Pld1−/− mice and in tumor tissues (C) and normal mucosa (D) of ApcMin/+/vehicle and ApcMin/+/PLD1-Inh mice. The nuclear β-catenin in the tumor was quantified (A and C, right). n = 21 per group; five tumors per mouse. (E) IHC for PLD1 (top) and β-catenin (bottom) in normal colon mucosa of non-AOM/DSS mice (n = 4) and tumor tissues of AOM/DSS-Pld1+/+ and AOM/DSS-Pld1−/− mice. The nuclear β-catenin in the tumor was quantified (right). n = 9 per group; four tumors per mouse. (A–E) Results are representative of at least three independent experiments and are shown as mean ± SEM. A Chi-square test was used. (F) q-RT-PCR analysis of the indicated mRNAs in tumor tissues of ApcMin/+ and ApcMin/+Pld1−/− mice (left) or ApcMin/+/vehicle and ApcMin/+/PLD1-Inh mice (right). (G) q-RT-PCR analysis of the indicated mRNAs in tumor tissues of AOM/DSS-Pld1+/+ and AOM/DSS-Pld1−/− mice (left) or vehicle- and PLD1 inhibitor–treated AOM/DSS mice (right). (F and G) Results are representative of at least two independent experiments and are shown as mean ± SEM. n = 8 per group. A Student’s t test was used. (H) q-RT-PCR analysis of β-catenin in PLD1-depleted stable cells. Results are representative of three independent experiments and are shown as mean ± SEM. A Student’s t test was used. (I) Effect of 10 µM PLD1 inhibitor on the protein stabilization of β-catenin in cells treated with 50 µM cycloheximide (CHX) for the indicated times. Results are representative of at least three independent experiments. (J) β-catenin mRNA expression is inversely correlated with survival in 174 CRC patient specimens. Survival probability was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. MW, molecular weight; a.u., arbitrary units. Bars, 100 µm.
Figure 2.
PLD2 ablation does not reduce intestinal tumorigenesis in ApcMin/+ mice. (A) q-RT-PCR analysis of PLD2 in normal SI of WT mice or adenomas of Apcmin/+ mice. Results are representative of three independent experiments and are shown as mean ± SEM. n = 8 per group. Student’s t test was used. (B) q-RT-PCR analysis of PLD2 in tumor tissues of ApcMin/+ (n = 7) and ApcMin/+Pld1−/− (n = 12) mice, which were sacrificed at 16 wk. Results are representative of three independent experiments and are shown as mean ± SEM. Student’s t test was used. (C) q-RT-PCR analysis of PLD2 in normal SI tissues of ApcMin/+, ApcMin/+Pld2+/−, and ApcMin/+Pld2−/− mice, which were sacrificed at 12 wk (mean ± SEM). n = 7 per group. Student’s t test was used. (D and E) ApcMin/+ (n = 10), ApcMin/+Pld2+/− (n = 13), and ApcMin/+Pld2−/− (n = 11) mice were sacrificed at 16 wk. (D) Arrows indicate polyps. H&E staining of the entire SI Swiss roll (top). The number of visible polyps in the SI and colon intestine (bottom). n.s., not significant. (E) Size distribution of polyps in the SI of the indicated mice. Results are representative of at least two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. a.u., arbitrary units.
Figure 3.
PLD1 inhibition attenuates intestinal tumorigenesis in the ApcMin/+ and AOM/DSS mice model. (A and B) 12-wk-old male ApcMin/+ mice were treated i.p. with either vehicle or 10 mg/kg PLD1 inhibitor three times a week for 4 wk. ApcMin/+/vehicle (n = 11) and ApcMin/+/PLD1-Inh (n = 11) mice were sacrificed at 16 wk. (A) Arrows indicate polyps. H&E staining of representative whole SI Swiss roll (left). The number of visible polyps in the SI and colon intestine (right). Student’s t test was used. (B) Size distribution of polyps in the SI of the indicated mice. Results are representative of at least two independent experiments. ANOVA F-test was used. (C) The indicated mice were followed for long-term survival. Survival probability was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. (D) IHC for Ki67 in tumor tissues (top) and normal mucosa (middle) of vehicle or PLD1 inhibitor–treated ApcMin/+ mice. Representative images were selected from at least six different fields. The expression of Ki67 in the tumor was quantified (bottom). Results are representative of three independent experiments and are shown as mean ± SEM. n = 6 per group; five tumors per mouse. n.s., not significant. (E) IHC for active caspase-3 (left) and TUNEL assay (middle) in tumor tissues of ApcMin/+ and ApcMin/+Pld1−/− mice. The levels of caspase-3– and TUNEL-positive cells were quantified (right). Results are representative of at least two independent experiments and are shown as mean ± SEM. n = 6 per group; five tumors per mouse. IHC staining results were analyzed using the Chi-square test. (F) AOM/DSS-induced mice were treated i.p. with either vehicle or 10 mg/kg PLD1 inhibitor three times a week for 6 wk and AOM/DSS-vehicle (n = 12), or AOM/DSS–PLD1-Inh (n = 12) mice were sacrificed at 110 d. (G) The number of visible polyps in the SI and colon intestine. Student’s t test was used. (H) The indicated mice were followed for long-term survival. Survival probability was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 100 µm.
PLD1 inactivation down-regulates expression of β-catenin
Surprisingly, loss of PLD1 in ApcMin/+ mice greatly decreased the level of nuclear β-catenin in adenoma at 16 wk of age relative to control mice (Fig. 4 A). Membrane-associated β-catenin was detected in both in the nontransformed areas of the ApcMin/+ intestines and in the normal areas of ApcMin/+Pld1−/− (Fig. 4 B). Additionally, PLD1 inhibitor–treated ApcMin/+ mice showed a significant reduction of β-catenin levels, which is comparable to those of PLD1 ablation (Fig. 4 C). As an internal control, membrane-associated β-catenin was detected in both in the nontransformed areas of the vehicle- or PLD1 inhibitor–treated ApcMin/+ intestines (Fig. 4 D). Moreover, the levels of nuclear β-catenin and PLD1 were increased in the AOM/DSS-induced tumors but decreased in PLD1-depleted AOM/DSS mice (Fig. 4 E). These results support the idea that the antitumoral effect of PLD1 deletion in this system is mechanistically equivalent. The level of nuclear β-catenin was quantified (Fig. 4, A, C, and E). To date, it has been believed that β-catenin is mainly regulated at the protein level. However, ablation and inhibition of PLD1 in ApcMin/+ and AOM/DSS mice led to a significant decrease in the mRNA levels of β-catenin and its target genes (Fig. 4, F and G). We further examined whether chronic suppression of PLD1 using two types of shPLD1 (short hairpin to PLD1)-transfected stable CRC cells affected expression of β-catenin. Chronic depletion of PLD1 dramatically decreased the mRNA level of β-catenin in HCT116 and DLD1 cells (Fig. 4 H). However, PLD1 inhibition did not affect the protein stability of β-catenin (Fig. 4 I). Collectively, PLD1 inactivation down-regulates β-catenin at the mRNA level. Interestingly, the mRNA level of β-catenin is correlated with a poorer prognosis of CRC patients on the basis of The Cancer Genome Atlas database (http://www.cbioportal.org/public-portal/; Fig. 4 J), suggesting physiological relevance.
Intestinal epithelial cell (IEC)–specific PLD1 overexpression accelerates tumorigenesis in ApcMin/+ mice with increased proliferation and β-catenin expression
To further study the role of PLD1 in intestinal tumorigenesis, we produced PLD1-Villin transgenic (Tg) mice that overexpress PLD1 specifically in IECs. The Tg mice, but not WT mice, exhibited high PLD1 expression in the duodenum, SI, colon, and cecum, but not in other organs (Fig. 5 A). PLD1 mRNA expression was increased in three Tg lines compared with WT mice (Fig. 5 B). Although the intestinal phenotype in Pld1Tg mice showed no significant lesion (Fig. 5 C), IEC-specific PLD1 overexpression greatly increased the level of β-catenin in SI at 12 wk of age relative to WT control (Fig. 5 D). The expression of PLD1 in sequential sections was mainly detected in the areas where β-catenin is supposed to be overexpressed (Fig. 5 D). β-catenin in SI from Pld1Tg mice was mainly localized in the membrane and cytoplasm, but not in the nucleus. To explore the impact of PLD1 overexpression within an environment that predisposes to neoplasia, we crossed the Pld1Tg mice with the ApcMin/+ mice, generating ApcMin/+Pld1Tg mice. ApcMin/+Pld1Tg mice increased the number and incidence of tumor formation (Fig. 5, E and F). IEC-specific PLD1 overexpression in ApcMin/+ mice also increased nuclear β-catenin levels and proportions of KI67-positive cells with proliferation compared with ApcMin/+ mice (Fig. 5, G and H). ApcMin/+Pld1Tg mice showed increased levels of PLD1, β-catenin, and its target genes (cyclin D1 and c-Myc) compared with ApcMin/+ or Pld1Tg mice (Fig. 5, I and J). The difference in β-catenin target gene expression between ApcMin/+Pld1Tg and Pld1Tg mice might be a result of differential localization of β-catenin in SI from two strains of mice. Because PLD1 is very highly expressed in the intestine from Pld1Tg, ApcMin/+, and ApcMin/+Pld1Tg mice compared with that of WT mice, it seems that PLD1 is not expressed in the intestinal tissues from WT mice, although PLD1 is expressed at a very low level in normal epithelial cells (Fig. 1 A and Fig. 5 I). ApcMin/+Pld1Tg mice showed increased intestinal PA generation, and ApcMin/+Pld1−/− mice showed decreased intestinal PA formation, which was recovered by overexpression of IEC-specific PLD1 in PLD1-depleted mice (ApcMin/+Pld1−/−Pld1Tg; Fig. 5 K). Moreover, PLD1 ablation in ApcMin/+ mice induced apoptosis, which was protected in ApcMin/+Pld1−/−Pld1Tg mice, as analyzed by casapase-3 activity assay (Fig. 5 L). Furthermore, treatment of Wnt3a or LiCl (GSK3β inhibitor) in PLD1 overexpressed cells significantly increased TCF trans-activation and cyclin D1 promoter activity in HCT116 cells relative to either PLD1 overexpression or Wnt (LiCl) treatment (Fig. 5 M). Collectively, these results suggest that IEC-specific PLD1 overexpression promotes tumor development in ApcMin/+ mice, probably through increased IEC proliferation and up-regulation of β-catenin and its target genes.
Figure 5.
IEC-specific PLD1 overexpression accelerates tumorigenesis in ApcMin/+ mice. (A) PLD1 expression in various tissues from IEC-specific PLD1Tg mice. Results are representative of at least four independent experiments. (B) q-RT-PCR analysis in SI from WT and Pld1Tg mice. n = 5 per group. (C and D) WT (n = 5) and Pld1Tg (n = 6) mice were sacrificed at 12 weeks for histology analysis. H&E staining (C) and IHC for PLD1 and β-catenin (D). (E) The histology of representative tumors (left) and incidence of tumor formation (right; n = 7 per group). (B–D) A Student’s t test was used. (F) Size distribution of polyps in the SI of the indicated mice. n = 7 per group. ANOVA F-test was used. (G and H) IHC for β-catenin (G) and Ki67 (H). Representative images were selected from at least three different fields. The expressions of nuclear β-catenin (G) and Ki67 (H) were quantified in size distribution of polyps. n = 5 per group; 10 tumors per mouse. A Chi-square test was used. (I) IB analysis from tissues of the indicated mice. (J) q-RT-PCR analysis from the indicated mice. n = 3–7 per group. (K and L) Total PA levels (K) and caspase-3 activities (L) in lysates from tumor tissues of ApcMin/+ (n = 7), ApcMin/+Pld1Tg (n = 5), ApcMin/+Pld1−/− (n = 9), and ApcMin/+Pld1−/−Pld1Tg (n = 12) mice sacrificed at 18 wk. Results are representative of three independent experiments. (M) Effect of PLD1 and/or Wnt3a (mimics; LiCl) on TCF trans-activation and cyclin D1 promoter activity. (J–M) A Student’s t test was used. All data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant; MW, molecular weight; a.u., arbitrary units. Bars, 100 µm.
PLD1 is highly expressed in the CC-IC population and regulates in vitro sphere-forming capacity
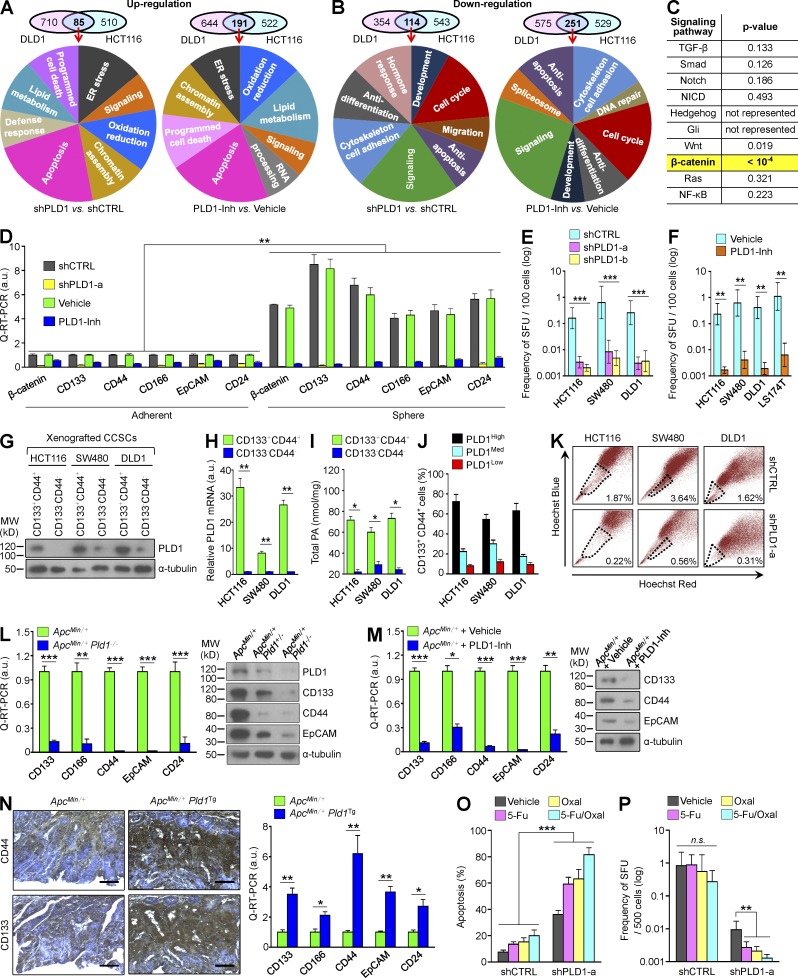
To identify potential effectors responsible for antitumorigenesis by targeting PLD1, we performed a microarray in DLD1 and HCT116 cells and then analyzed the transcriptional profile of genes that were differently expressed by inhibition and depletion of PLD1 under serum-deprived conditions. The genes commonly up- or down-regulated in PLD1 inhibitor–treated or PLD1-depleted CRC cells were subsequently classified according to gene ontology–based functional categories (Fig. 6, A and B). PLD1 inactivation up-regulated expression of genes involved in apoptosis, lipid metabolism, oxidation/reduction, ER stress, and chromatin assembly, but a large portion of up-regulated genes was associated with apoptosis. The positive regulators of apoptosis (Bim, NOXA, p27KIP1, p73, p14ARF, CHOP, Sos2, GCLC, and DR5) increased significantly in response to inhibition and depletion of PLD1 (Tables S1 and S6). Genes down-regulated by targeting PLD1 were primarily associated with signaling, cell cycle, antiapoptosis, antidifferentiation, and cytoskeleton/cell adhesion (Fig. 6 B). We further analyzed the Gene Set Enrichment Analysis signaling pathway to determine whether targeting of PLD1 regulates the main pathways associated with colorectal tumorigenesis. The results revealed that targeting PLD1 significantly represses the β-catenin signaling pathway (P < 10−4), but not other pathways (Fig. 6 C). Moreover, targeting of PLD1 down-regulated expression of β-catenin and its target genes (LEF1, ID3, ENC1, c-Myc, cyclin D1, CDC25A, and MET) with C-IC markers (CD44, CD133, EpCAM, CD24, ID1, and CD166; Tables S2 and S7). A rare subpopulation of cells with special surface markers within CRC has the potential to initiate and sustain tumor growth. These C-ICs, associated with tumor relapse and progression, are considered to be responsible for the poor outcome of CRC. The subpopulation of stem or progenitor cells in heterogeneous CRC cells is correlated with their sphere-forming abilities under serum-free culture conditions (Moon et al., 2011). Thus, we examined whether PLD1 is involved in the self-renewal of C-ICs. Xenografted shcontrol or shPLD1-transfected stable CRC cells were cultured under adherent or sphere conditions. Expression of β-catenin, PLD1, and C-IC markers was significantly increased, especially under sphere culture conditions, when compared with adherent conditions (Fig. 6 D). Inhibition and deletion of PLD1 dramatically abolished expression of β-catenin and the C-IC markers under both adherent and sphere culture conditions (Fig. 6 D). To determine whether PLD1 affected the capacity of serial sphere formation, experiments were conducted in which clonally derived primary cells were replated at limited dilution into secondary sphere-forming assays. Targeting PLD1 significantly decreased the sphere-forming capacity of several CRC cells (Fig. 6, E and F). The CD133+CD44+ population sorted from xenografted CRC cells by flow cytometry showed dramatically increased expression and activity of PLD1 when compared with the CD133–CD44– population (Fig. 6, G–I). Additionally, a high level of PLD1 expression was detected in the population of CD133+CD44+ CRC cells (Fig. 6 J). ATP-binding cassette (ABC) transporter participates in tumor resistance by actively transporting drugs across the cell membrane, which protects cells from chemotherapeutic agents (Donnenberg and Donnenberg, 2005). Stem cells are frequently identified as the side population (SP) by flow cytometry based on ABC transporter–mediated efflux of Hoechst dye (Goodell et al., 1996). As most cells accumulate Hoechst 33342, SP cells can be isolated by dual-wavelength flow cytometry based on their ability to efflux this dye. We identified an SP among HCT116, SW480, and DLD1 cells that was generally small, and depletion of PLD1 greatly reduced the SP from 1.87, 3.64, or 1.62% to 0.22, 0.56, or 0.31%, respectively (Fig. 6 K). Moreover, ablation and inhibition of PLD1 in ApcMin/+ mice significantly decreased the expression of C-IC markers (Fig. 6, L and M). These phenomena were also observed in AOM/DSS mice (unpublished data). IEC-specific PLD1 overexpression in ApcMin/+ mice significantly increased expression of the C-IC markers relative to ApcMin/+ mice (Fig. 6 N). The chemoresistance of C-ICs has emerged as an important cellular property that enables tumors to regrow after initial cytoreductive therapy. Although treatment of 5-fluorouracil and oxaliplatin, clinically used for CRC therapy, induced apoptosis of DLD1 cells under sphere culture, the drugs did not affect the sphere-replating capacity (Fig. 6, O and P). However, treatment of the drug in PLD1-depleted cells resulted in enhanced chemosensitivity and significantly decreased sphere-replating capacity relative to either PLD1 depletion or drug treatment. Collectively, these findings demonstrate that PLD1 is highly up-regulated in the CC-IC population and involved in the self-renewal capacity of C-ICs.
Figure 6.
PLD1 is highly up-regulated in the CC-IC population and in vitro sphere-forming capacity. (A and B) Venn diagram for transcripts up- (A) and down-regulated (B) by PLD1 inhibition in DLD1 and HCT116 cells. Summary of the functional categories of genes significantly enriched upon treatment with PLD1 inhibitor. Gene ontology groups demonstrated enhanced statistical representation (P < 0.01). (C) Targeting PLD1 significantly represses the β-catenin signaling pathway but not other pathways in the CRC cells. In yellow: over-representation reaches statistical significance only for the β-catenin signatures. (D) q-RT-PCR analysis of β-catenin and C-IC markers under adherent and sphere culture conditions of xenografted DLD1 cells stably transfected with shPLD1 or treated with PLD1 inhibitor. Data are shown as mean ± SEM. ANOVA F-test was used. (E and F) In vitro LDAs of SFUs by PLD1 depletion (E) and PLD1 inhibition (F) in xenografted cells; error bars represent 95% confidence intervals. (G–I) IB (G) and (H) q-RT-PCR (H) analysis of PLD1 expression and cellular PA levels (I) in CD133+CD44+ and CD133–CD44– cells sorted from xenografted CRC spheres. (E, F, H, and I) Data are shown as mean ± SEM. A Student’s t test was used. (J) After the indicated cells were sorted by flow cytometry using antibody to PLD1, the percentage of CD133+CD44+ populations was analyzed by FACS. (K) Effect of PLD1 depletion on SP in CRC cells. (L and M) q-RT-PCR and IB analysis of the indicated C-IC markers in tumor tissues of the indicated mice. (N) IHC and q-RT-PCR analysis of the indicated C-IC markers in ApcMin/+ (n = 7) and ApcMin/+Pld1Tg (n = 6) mice. (O and P) After exposing the vehicle or PLD1-Inh group to 50 µg/ml 5-fluorouracil (5-Fu) and/or 100 µM oxaliplatin (Oxal), the percentage of apoptotic cells (O) and LDA analysis of SFUs (P) was measured. (L–P) Results are representative of at least three independent experiments and are shown as mean ± SEM. A Student’s t test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant; MW, molecular weight; a.u., arbitrary units. Bars, 100 µm.
PLD1 inhibition down-regulates β-catenin expression through E2F1-induced microRNA (miR)-4496 up-regulation
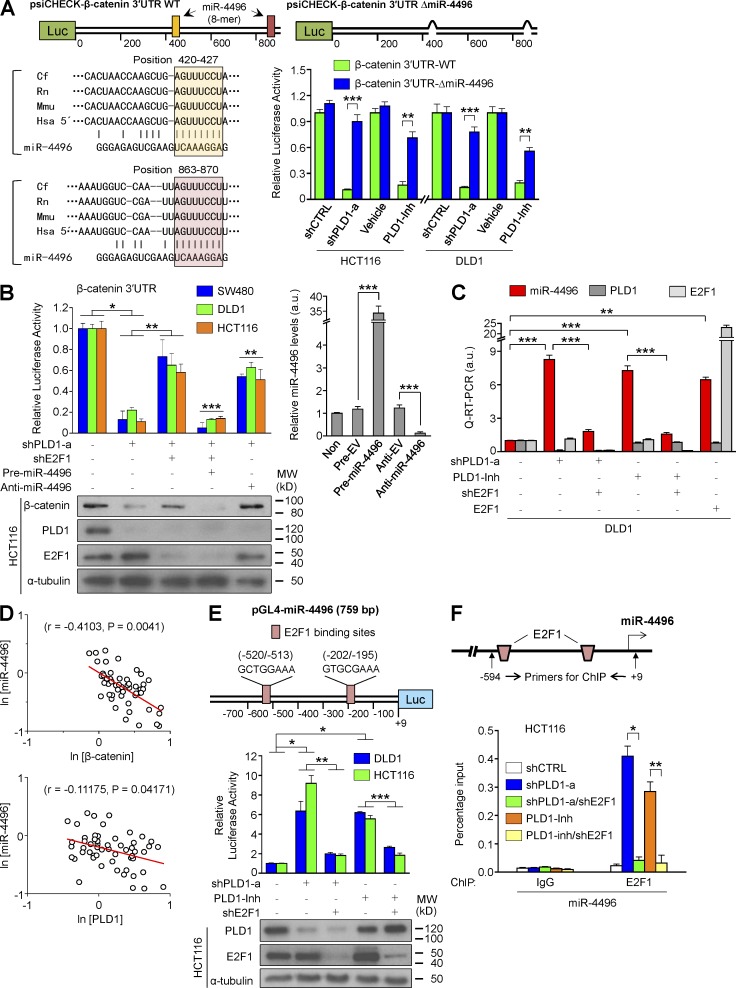
Because PLD1 inactivation decreases β-catenin expression, we examined how PLD1 regulates expression of β-catenin; however, depletion and inhibition of PLD1 did not affect the promoter activity of β-catenin (unpublished data). miRs silence gene expression by binding to the 3′ untranslated regions (UTRs) of target miR, inhibiting their translation or marking them for degradation (Pauli et al., 2011). Thus, it is assumed that PLD1 inhibition down-regulates β-catenin via miR. We next attempted to identify relevant miR targeting a 3′ UTR of β-catenin using an miR array conducted in PLD1 inhibitor–treated DLD1 cells (Table S8). Based on the bioinformatic approach, we found that PLD1 inhibition enhanced the expression of miR-4496, which has a putative target site in the 3′ UTR of β-catenin that is conserved in various species (Fig. 7 A). The luciferase activity of the β-catenin 3′ UTR reporter was decreased remarkably in response to suppression of PLD1 in CRC cells (Fig. 7 A). The deletion of the putative binding sites of miR-4496 and anti–miR-4496 led to significant recovery of the luciferase activity of the β-catenin 3′ UTR that had decreased in response to PLD1 suppression (Fig. 7, A and B), suggesting that β-catenin is a target of miR-4496. It has been reported that E2F1 down-regulates β-catenin in both GSK3β- and caspase-independent manners (Morris et al., 2008). However, it is unknown how E2F1 regulates β-catenin expression in transcriptional or posttranscriptional levels. Thus, we examined whether PLD1 is involved in the regulation of E2F1-mediated β-catenin and miR-4496 expression. Depletion of E2F1 significantly recovered the decrease in the activity of β-catenin 3′ UTR reporter in response to PLD1 depletion, which was suppressed by precursor (pre)miR-4496 and recovered by anti–miR-4496 (Fig. 7 B). Expression of β-catenin protein was also comparable to the results of β-catenin 3′ UTR (Fig. 7 B). Moreover, suppression of PLD1 enhanced expression of miR-4496, which was reduced by E2F1 depletion, whereas E2F1 increased expression of miR-4496 (Fig. 7 C). The expression of miR-4496 was inversely correlated with the expression of β-catenin and PLD1 in 55 CRC tissues (Fig. 7 D), indicating biological relevance. These results demonstrate that PLD1 inhibition down-regulates expression of β-catenin at the miR-4496–mediated posttranscriptional level and that E2F1 might negatively regulate β-catenin levels via up-regulation of miR-4496. Interestingly, we identified putative E2F1 binding sites in the promoter of miR-4496 (Fig. 7 E). Depletion and inhibition of PLD1 significantly increased the promoter activity of miR-4496 and binding of E2F1 to the promoters, which was reduced by E2F1 depletion, suggesting miR-4496 as a new target gene of E2F1 (Fig. 7, E and F). Collectively, these results suggest that PLD1 inhibition down-regulates β-catenin expression via E2F1-induced miR-4496 up-regulation.
Figure 7.
PLD1 inhibition down-regulates β-catenin expression through E2F1-induced miR-4496 up-regulation. (A) Schematic representation of luciferase constructs of β-catenin 3′ UTR. Luciferase vectors (psiCHECK) were inserted with full-length β-catenin 3′ UTR (930 bp). ΔmiR-4496 (914 bp) were generated by deletions of highly conserved and predicted binding sites for the seed sequences of the miR. Effect of depletion or inhibition of PLD1 on the luciferase activity of β-catenin 3′ UTR reporters. A Student’s t test was used. The pink and peach boxes represent conserved seed pairing. (B) Effect of the pre- and anti-miR on PLD1 or E2F1-mediated β-catenin 3′ UTR luciferase activity and IB analysis of β-catenin (left). The levels of pre- and anti-miR are shown as a control (right). ANOVA F-test was used. (C) q-RT-PCR analysis of miR-4496 under the indicated conditions. A Student’s t test was used. (D) Inverse correlation of β-catenin or PLD1 with miR-4496 mRNA levels in CRC tissues. Spearman’s correlation coefficient (r) is provided with its statistical significance. The red lines represent the best-fit curves. (E) Schematic view of two putative E2F1 binding elements in promoter regions of the human miR-4496 (top). Effect of PLD1 and E2F1 on promoter activity of miR-4496 (middle). IB analysis of PLD1 and E2F1 is shown as a control (bottom). (F) ChIP assays for the binding of E2F1 to the promoter of miR-4496. (E and F) Results are representative of at least three independent experiments and are shown as mean ± SEM. A Student’s t test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001. MW, molecular weight; a.u., arbitrary units.
PLD1 regulates tumor-initiating capacity through the E2F1–miR-4496–β-catenin signaling axis
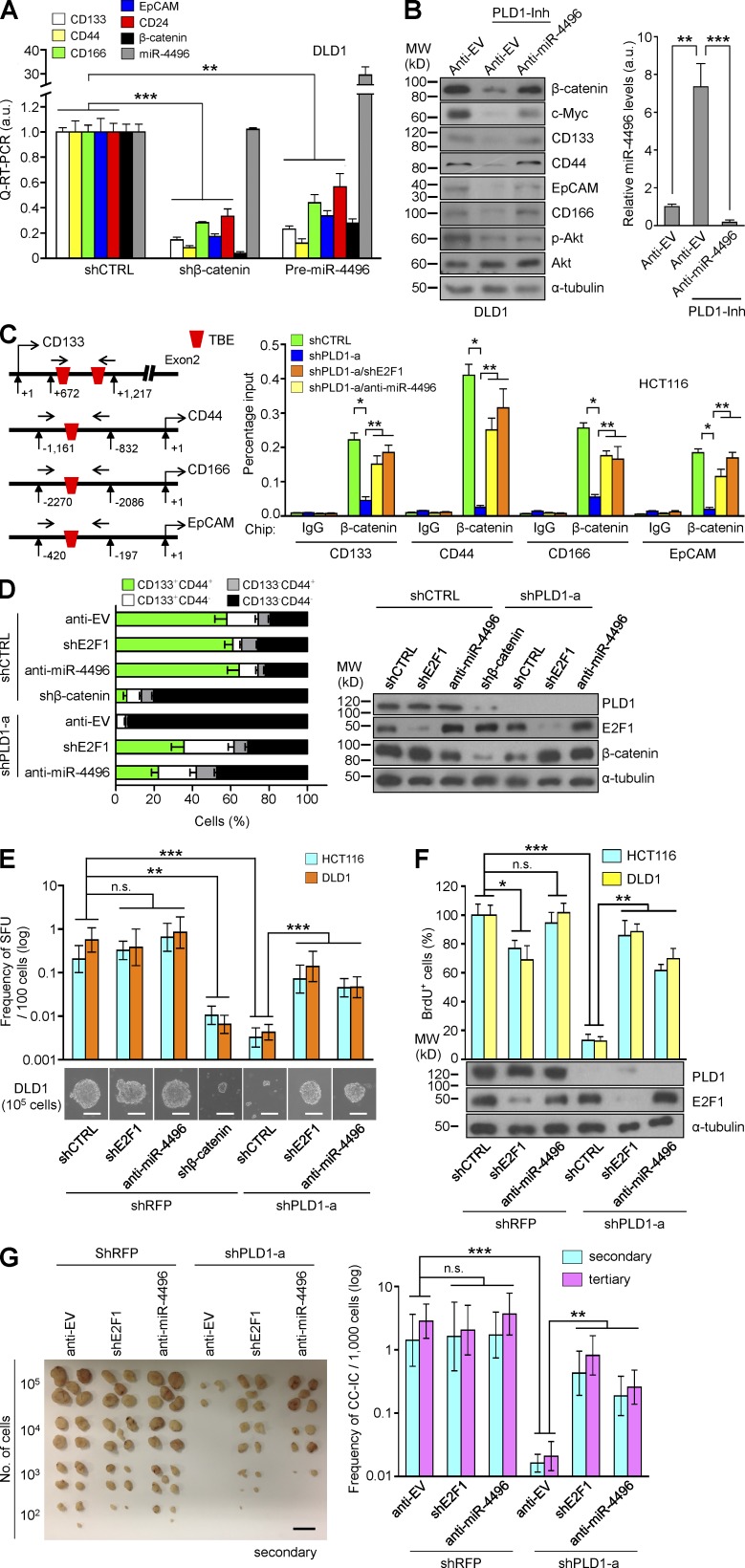
β-Catenin depletion and premiR-4496 significantly decreased expression of the C-IC markers (Fig. 8 A), whereas anti–miR-4496 recovered expression of the β-catenin/TCF target genes and the C-IC markers that had decreased in response to PLD1 inhibition (Fig. 8 B). Furthermore, we identified a TCF binding site or sites in the promoter of the C-IC markers. Depletion of PLD1 under sphere conditions decreased the binding of β-catenin to the promoter of the C-IC markers (CD133, CD44, CD166, and EcCAM), which was recovered by E2F1 depletion and anti–miR-4496 (Fig. 8 C). Depletion of E2F1 recovered the decrease in the expression of β-catenin and the C-IC markers in response to PLD1 knockdown (Table S9). Moreover, depletion of PLD1 greatly reduced the CD44highCD133high population (Fig. 8 D), sphere-forming capacity (Fig. 8 E), and proliferation (Fig. 8 F), which were recovered by E2F1 depletion and anti–miR-4496 (Fig. 8, D–F). To further determine whether the PLD1–E2F1–miR-4496 signaling pathway affects the capacity for serial tumor initiation of C-ICs at the clonal level, in vivo secondary and tertiary transplantation studies were performed in nonobese diabetic (NOD)/severe combined immunodeficiency (SCID) mice (Fig. 7 G). To assess the frequency of tumor-initiating cells in mice, limiting dilution assays (LDAs) were performed by diluting sphere-cultured cells and injecting defined cell doses subcutaneously. Six different groups of DLD1 cells were prepared and injected subcutaneously in the mice: control, E2F1 deletion, anti–miR-4496 overexpression, PLD1 depletion, both E2F1 and PLD1 depletion, and anti–miR-4496 overexpression with PLD1 depletion. To assess for C-IC self-renewal capacity, cells were isolated from xenografts, and secondary LDAs were performed at serial dilution (102, 103, 104, or 105); a minimum of five sites were tested. For all experiments, the number of injection sites containing tumors was counted and analysis was performed. The LDAs were calculated using the limiting dilution function of the Walter and Eliza Hall Institute Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda/index.html). The PLD1-depleted group showed a 200-fold decrease in tumor reinitiation capacity, whereas the capacity was recovered with depletion of E2F1 or miR-4496 (Fig. 8 G). Collectively, these findings demonstrate that PLD1 regulates the in vivo self-renewal capacity of C-ICs through the E2F1–miR-4496–β-catenin axis.
Figure 8.
Targeting PLD1 attenuates tumor-initiating capacity through the E2F1–miR-4496–β-catenin axis. (A) Effect of β-catenin depletion or premiR-4496 on the expression of the indicated genes. (B) Effect of anti–miR-4496 on the expression of the indicated proteins (left). The levels of anti-miR are shown as a control (right). (C) Effect of E2F1 depletion or miR-4496 on the binding of β-catenin/TCF to the promoter of C-IC marker genes. (D) Representative flow cytometric profiles of CD44 and CD133 expression under the indicated conditions (left). IB analysis of PLD1, E2F1, and β-catenin was shown as a control (right). (E) Frequency of secondary SFUs by transduction with anti–miR-4496 or shE2F1 in PLD1-depleted CRC cells, as determined by in vitro LDAs. (F) BrdU incorporation analysis in the indicated cells. (G) Photographs of excised tumors (left) and in vivo serial transplantation assays (right) from NOD/SCID mice (n = 5 per group) injected with DLD1 cells derived from the indicated xenografts. Results are representative of at least three independent experiments and are shown as mean ± SEM. A Student’s t test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant; MW, molecular weight; a.u., arbitrary units. Bars: (E) 50 µm; (G) 1 cm.
Low PLD1 levels coupled with low levels of C-IC markers is associated with increased survival of CRC patients
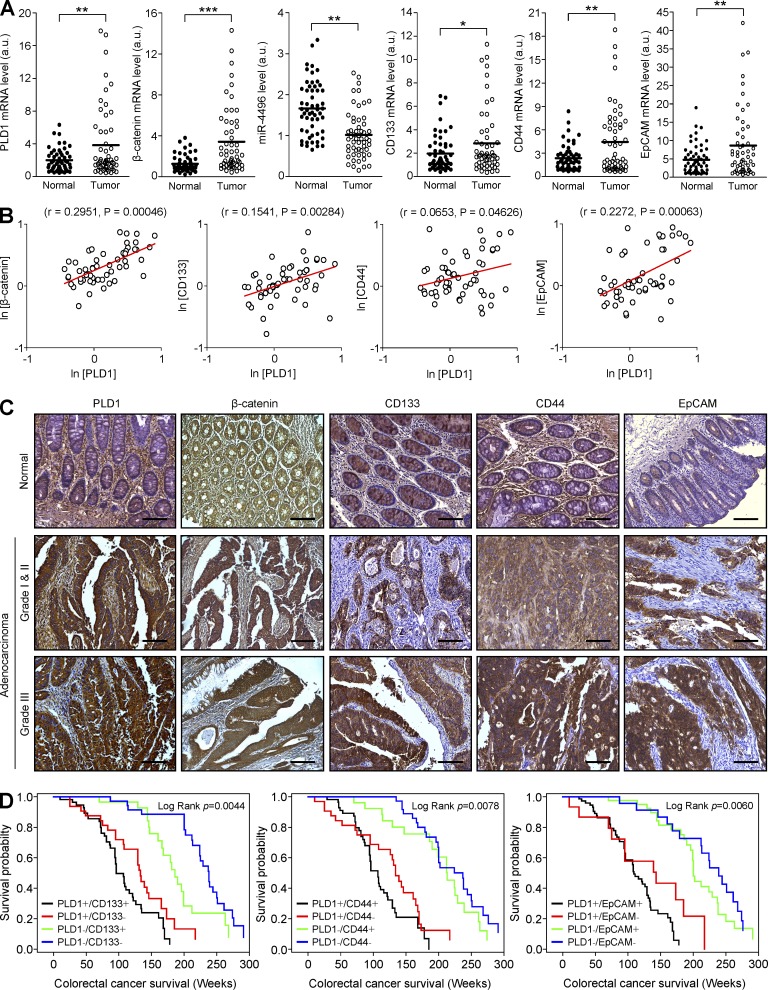
We next attempted to demonstrate the physiological relevance of the relationship of the PLD1 level with the expression of β-catenin and C-IC markers. The mRNA expression in 55 CRC tissues exhibited elevated expression of CD133, CD44, EpCAM, and miR-4496 (Fig. 9 A) when compared with that of normal tissues. Moreover, expression of PLD1 showed a positive correlation with expression of the C-IC markers in CRC tissues as analyzed by quantitative (q)-RT-PCR (Fig. 9 B). IHC analysis using CRC tissue shows the pathological correlation comparable to the results of q-RT-PCR (Fig. 9 C). Further analysis of the prognostic value of combining PLD1 and C-IC marker expression in CRC was conducted. A low level of PLD1 with low levels of C-IC markers (PLD1–/CD133–, PLD1–/CD44–, and PLD1–/EpCAM–) was significantly correlated with higher overall survival of CRC patients, and vice versa (Fig. 9 D). These data support an intimate association of PLD1 expression with levels of the C-IC markers in survival of CRC patients and suggest a potential value of PLD1 and C-IC markers as prognostic biomarkers and therapeutic targets.
Figure 9.
Expression of PLD1 and C-IC markers is associated with survival of CRC patients. (A) Relative expression of the indicated genes and miR-4496 in 55 pairs of tumor and adjacent normal tissues from CRC patients was measured by q-RT-PCR. Data were analyzed using the paired t test. Results are representative of at least two independent experiments and are shown as mean ± SEM. (B) In 55 tumor tissues from CRC patients, there was a positive correlation of PLD1 with β-catenin, CD133, CD44, and EpCAM mRNA expression. Spearman’s correlation coefficient (r) is provided with its statistical significance. The red lines represent the best-fit curves. (C) IHC in tumor and adjacent normal tissues from CRC patients. (D) Correlation with survival when patient specimens are segregated according to immunoreactivity of PLD1 and CC-IC surface markers (CD133, CD44, or EpCAM). Kaplan-Meier survival curves of CRC patients (n = 153) are presented. Results are representative of at least two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. a.u., arbitrary units. Bars, 100 µm.
DISCUSSION
To our knowledge, the role of PLD in intestinal tumorigenesis has not previously been investigated. Here, we show that genetic and pharmacological inhibition of PLD1 disrupts spontaneous and colitis-associated intestinal tumorigenesis in ApcMin/+ and AOM/DSS mice models. Haploinsufficiency with deletion of only a single allele of PLD1 also showed decreased tumorigenesis in ApcMin/+ mice, despite the presence of an enzyme corresponding to a single allele of PLD1. Haploinsufficiency of PLD1 might reduce tumorigenesis because of the relatively decreased enzymatic activity of PLD1. IEC-specific PLD1 overexpression in ApcMin/+ mice accelerated tumorigenesis with increased IEC proliferation and nuclear β-catenin levels compared with ApcMin/+ mice. We recently demonstrated a direct connection between PLD and Wnt signaling pathways (Kang et al., 2010). Genetic and pharmacological targeting of PLD1 in ApcMin/+ and AOM/DSS mice decreased the expression of β-catenin and its target genes but increased the expression of proapoptotic E2F1 target genes, which ultimately attenuated intestinal tumorigenesis (unpublished data). We have identified miR-4496 as a new target of E2F1 that has a crucial node responsible for mediation of the cross talk between E2F1 and Wnt/β-catenin signaling. Our results differ from a previous study in which E2F1 induces the posttranslational degradation of β-catenin (Morris et al., 2008). As AXIN1/2 and SIAH1 are E2F target genes, E2F1 may be involved in the degradation of β-catenin. The components of the β-catenin destruction complex, such as APC and GSK3β, are required for degradation of β-catenin by AXIN1/2 or SIAH1 (Liu et al., 2001; Morris et al., 2008; Li et al., 2012). However, because most CRCs have mutations in APC or β-catenin, it is unlikely that AXIN- or SIAH1-mediated degradation of β-catenin occurs in CRC. Here, we provide evidence that targeting PLD1 reduces expression of β-catenin in the posttranscriptional level via up-regulation of E2F1-induced miR-4496, suggesting dynamic changes in the β-catenin levels by PLD1 and a new pathway by which E2F1 represses β-catenin–TCF activity. We identified miR-4496 as a new miR targeting β-catenin. E2F1 can function as both a tumor suppressor or as an oncogene under different conditions (Trimarchi and Lees, 2002). Unlike other human cancers, in most CRCs, whereas Wnt/β-catenin signaling is activated, E2F1 activity is kept at a low level because there are no mutations in retinoblastoma 1 (RB1; Nevins, 2001). E2F1 in CRC may function as a tumor suppressor (Bramis et al., 2004). It has long been known that RB1 in colon carcinoma is not mutated, but overexpressed, and even amplified (Lai et al., 2006; Firestein et al., 2008). Thus, in the context of CRC, RB is more likely to act as an oncoprotein than a tumor suppressor. In this regard, strategies focused on the control of E2F1 proteins hold particular promise because activation of E2F activity is the ultimate consequence of deregulation of the RB pathway. E2F1 activity is negatively regulated by PLD1; accordingly, retaining RB1 and amplifying PLD1 may enable CRC cells to select for mechanisms that limit the activity of E2F1 and tip the balance toward β-catenin–driven proliferation conditions that suppress E2F1 and enhance the activity of β-catenin. Interestingly, we found that PLD1 inactivation reduced the expression of RB1 as analyzed by a microarray (Tables S2 and S7); thus, down-regulation of RB1 by targeting PLD1 may release active E2F1 and induce E2F1 trans-activation. This is the first evidence that PLD1 regulates the expression of β-catenin at the posttranscriptional level via miR-4496; accordingly, our results demonstrate a new regulatory mechanism for cross talk between Wnt/β-catenin and E2F1 signaling pathways regulated by PLD1. Therapeutic approaches that are not capable of eradicating the C-IC subset are unlikely to be successful because although they might be able to destroy the majority of tumor cells and induce regression of tumor lesions, they fail to prevent disease relapse and metastatic dissemination (Dalerba et al., 2007). Recently, it has been reported that ID1 and ID3 promote the self-renewal capacity of CC-ICs with chemoresistance (O’Brien et al., 2012). Interestingly, the results from the mRNA microarray showed reduced expression of ID1 and ID3 genes in response to PLD1 inhibition. Here, we demonstrate that PLD1 governs self-renewal of CC-ICs through the E2F1–miR-4496–β-catenin axis. PLD1 inactivation decreases the expression of C-IC markers, self-renewal capacity of CC-ICs for serial tumor initiation, and chemoresistance. IEC-specific PLD1 overexpression in the ApcMin/+ mice greatly increased the expression of β-catenin and C-IC markers compared with ApcMin/+ mice. The expression of PLD1 coupled with the levels of C-IC markers is associated with survival in patients with CRC and can thus be considered predictors of patient prognosis. C-ICs interplay closely with the tumor microenvironment, and disrupting this niche microenvironment impairs CSC self-renewal and thereby significantly inhibits the growth of tumors (Boral and Nie, 2012). Metastasis to the lung after introduction of tumor cells intravenously was reduced in Pld1−/− mice, and this finding reflects aberrant interaction of the tumor cells with the Pld1−/− platelets (Chen et al., 2012). Moreover, impairment of vascular endothelial growth factor receptor 2 signaling in the absence of PLD1, which leads to reduced vascular permeability, could potentially normalize tumor blood vessels and improve chemotherapeutic delivery to preestablished and vascularized tumors (Chen et al., 2012). PLD1 may trigger a signal from the platelets to the C-ICs to promote tumor progression, epithelial-mesenchymal transformation/metastasis, and angiogenesis. Collectively, our findings demonstrate the central roles that PLD1 plays in regulating the cross talk among E2F1–miR-4496 and Wnt/β-catenin pathways and the tumor-initiating program of CC-ICs. These results provide a missing link between the networks mediating cancer and may fill the gap in current knowledge regarding how the complexity of the Wnt/β-catenin pathway is delicately regulated in cancer. Considering the highly complex interactions among cancer-relevant pathways, PLD1 might be an attractive target for the development of promising therapeutic drugs for treatment of CRC.
MATERIALS AND METHODS
Mice.
Gender- and age-matched Pld1−/− (Dall’Armi et al., 2010) and Pld2−/− (Oliveira et al., 2010) mice have been backcrossed seven times onto the C57BL/6J backgrounds. Apcmin/+ mice were purchased from The Jackson Laboratory. Apcmin/+Pld1−/− and Apcmin/+Pld2−/− mice were generated by interbreeding mice carrying Pld1−/−, Pld2−/−, and Apcmin/+. For all in vivo and ex vivo experiments, Apcmin/+Pld1+/−, Apcmin/+Pld1−/−, Apcmin/+Pld2+/−, and Apcmin/+Pld2−/− mice and age-matched Apcmin/+ or WT littermate controls were used in this study. Apcmin/+ or AOM/DSS-induced mice were i.p. injected with either vehicle or 10 mg/kg PLD1 inhibitor (VU0155069; Cayman Chemical) three times a week for 4 wk. For the AOM/DSS model, mice were given a single i.p. injection of the mutagen AOM at 12.5 mg/kg, after which they received drinking water containing 2–3% DSS in several 5-d periods that were interspersed with periods in which they received normal water (Fig. 1 G and Fig. 3 F). The cDNA of the human PLD1 gene was cloned into a pBS-Villin vector that contains the mouse villin promoter (Pinto et al., 1999). The pBS-Villin/PLD1 plasmid was verified by sequencing, and the transgene was isolated from the plasmid by restriction enzyme digestion and gel purification. To generate PLD1 mice, the transgene was microinjected into C57BL/6J mouse eggs (Macrogen), and the resulting founders and their progeny were genotyped by PCR amplification of tail DNA using the following primers: 5′-CACTGGCCTAAAGCTCACCA-3′ and 5′-TTGATGCCAAGAGCGAGTTC-3′. Tg mice were congenically rebred to C57BL/6J for seven generations. Three Tg lines (1, 2, and 3) were established from these founders. Apcmin/+Pld1Tg mice were generated by interbreeding mice carrying Pld1Tg lines and Apcmin/+. Apcmin/+Pld1−/−Pld1Tg mice were generated by interbreeding mice carrying Pld1Tg lines and Apcmin/+Pld1−/− mice. NOD/SCID mice (The Jackson Laboratory) were used for tumor xenograft maintenance and in vivo LDAs. Animal studies were approved by the Institutional Animal Care and Use Committee of Pusan National University.
Human primary tumor samples and tissue microarray.
55 human colon tumors and adjacent normal tissues for q-RT-PCR analysis were obtained from Pusan National University Hospital, a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare. All samples derived from the National Biobank of Korea were obtained with informed consent under institutional review board–approved protocols. For IHC analysis, tissue microarray slides were purchased from SuperBioChips and US-Biomax. Each array includes >174 cases of normal, reactive, premalignant, and malignant tissues of the colon (various grades and stages). Overall survival was calculated from the date of treatment start to the date of death or date of sacrifice. Survival curves were constructed using Kaplan-Meier methodology. Log-rank tests were used to assess differences in tumor characteristics.
Histology and immunohistochemistry.
Anti-Ki67 (Abcam), anti–β-catenin (BD), anti-active caspase-3 (Cell Signaling Technology), anti-CD133 (MyBioSource), anti-CD44 (Proteintech), anti–cyclin D1, and anti-EpCAM antibodies (Santa Cruz Biotechnology, Inc.) were used as the primary antibodies. IHC analyses of diaminobenzidine staining were performed using an HRP kit (UltraTek; Scytek). The diaminobenzidine-stained specimens were visualized using a general optical microscope with a camera (AxioCam ICc5; Carl Zeiss). Hematoxylin and eosin (H&E) staining of the entire intestinal Swiss roll was conducted using a MIRAX scan(Carl Zeiss). Images were processed with equivalent parameters using ZEN Light Edition software (Carl Zeiss).
In vitro LDAs.
To determine the number of sphere-forming units (SFUs), cells were cultured in serum-free medium with epidermal growth factor and basic fibroblast growth factor. We plated a defined number of cells per well in a 96-well plate. The highest dose was 100,000, and the lowest dose was 10 cells per well, all plated in a fixed volume of 200 µl per well. There were a minimum of 6 wells per cell concentration. Every sample and each condition was tested by plating cells in this dilution series, down to one cell per well. At the end of the experiment, we scored a binary output: the wells were read as positive or negative. A positive well was defined as having at least one sphere, and the negative wells had no spheres, based on visual inspection. Using the Walter and Eliza Hall Institute Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda/index.html), we were able to calculate a sphere-initiating cell frequency. The methods were such that we never tried to count multiple spheres in one well because this method is inaccurate based on the fact that spheres aggregate, making it impossible to establish the sphere as clonal in bulk culture (Pastrana et al., 2011). Sphere-replating frequency was obtained from repeating the in vitro LDA with 10 individual spheres. The mean number of SFUs counted upon replating of 10 LDAs derived from single spheres constituted the in vitro self-renewal assay.
In vivo LDAs.
To assess the frequency of tumor-initiating cells in mice, LDAs were performed by diluting cells and injecting defined cell doses subcutaneously. To assess for C-IC self-renewal capacity, cells were isolated from xenografts, and secondary LDAs were performed at serial dilution (102, 103, 104, or 105); a minimum of five sites were tested. For all experiments, the number of injection sites containing tumors was counted, and analysis was performed. The LDAs were calculated using the limiting dilution function of the Walter and Eliza Hall Institute Extreme Limiting Dilution Analysis.
miR microarray data analysis.
For control and test RNAs, synthesis of target miR probes and hybridization were performed using an miR Labeling Reagent and Hybridization kit (Agilent Technologies) according to the manufacturer’s instructions. In brief, each 100 ng of total RNA was dephosphorylated with ∼15 U of calf intestine alkaline phosphatase, followed by RNA denaturation with ∼40% DMSO and 10-min incubation at 100°C. Dephosphorylated RNA was ligated with pCp-Cy3 mononucleotide and purified with MicroBioSpin 6 columns (Bio-Rad Laboratories). After purification, labeled samples were resuspended with Gene Expression Blocking Agent (Agilent Technologies) and Hi-RPM Hybridization Buffer (Agilent Technologies) followed by boiling for 5 min at 100°C and chilling on ice for 5 min. Finally, denatured labeled probes were pipetted onto an assembled human miR microarray (Human miRNA Microarray Release 19.0, 8 × 60 K; Agilent Technologies) and hybridized for 20 h at 55°C with 20 rpm rotating in a hybridization oven (Agilent Technologies). The hybridized microarrays were washed according to the manufacturer’s washing protocol. All data normalization and selection of fold-changed probes were performed using GeneSpringGX 7.3 (Agilent Technologies). We performed data transformation (set measurements ≤0.01) and per chip (normalized to the 75th percentile) normalization. Probes that changed >2-fold, 1.25-fold, and 2-fold of ratio in PLD1 inhibitor/vehicle, shPLD1/shCTRL, and shPLD1-shE2F1/shPLD1 groups, respectively, were selected and considered as differentially expressed probes. Microarrays are available in the GEO database under accession no. GSE55724.
mRNA microarray analysis.
Total RNA was extracted with TRIzol (Invitrogen) and purified using RNeasy columns (QIAGEN) according to the manufacturers’ instructions. Microarray analysis was performed by Macrogen. In brief, biotinylated complementary RNAs were amplified and purified using an RNA amplification kit (Illumina; Ambion) according to the manufacturer’s instructions. Labeled cRNA samples were hybridized to each human HT12 expression version 4 bead array for 16–18 h at 58°C, according to the manufacturer’s instructions (Illumina). Detection of array signal was performed using streptavidin-Cy3 (FluoroLink; GE Healthcare). Arrays were scanned with a bead array reader confocal scanner (Illumina), and the scanned images were analyzed using BeadStudio version 3.1.3 software (Gene Expression Module version 3.3.8; Illumina). Probe signal value was transformed by logarithm and normalized by quantile method. Gene enrichment and functional annotation analysis for a significant probe list was performed using DAVID (http://david.abcc.ncifcrf.gov) and Gene Set Enrichment Analysis (http://www.broadinstitute.org/gsea/msigdb/collections.jsp). Microarrays are available in the GEO database under accession no. GSE55771.
Apoptosis.
Apoptotic cell death was measured by APC-conjugated anti–Annexin V Apoptosis Detection kit I (BD). Apoptotic cell death in tumor tissue lysates of Apcmin/+, Apcmin/+Pld1Tg, Apcmin/+Pld1−/−, and Apcmin/+Pld1−/−Pld1Tg mice (18 wk; n = 5–8 for each group) was measured by use of a colorimetric assay (Caspase-3 kit; BioVision) according to the manufacturer’s instructions.
TUNEL assay.
The TUNEL assay was performed using the In Situ Cell Death Detection kit, POD (Roche), according to the manufacturer’s protocol. The percentage of apoptotic cells in the tumor tissue in each section was counted in 10 different microscopic fields.
Immunoprecipitation (IP) and Western blotting.
Cell lysates were analyzed by IP and/or immunoblot (IB) as described previously (Kang et al., 2008). Enhanced chemiluminescence was used for detection of the signal. The following antibodies were used: anti–α-tubulin, anti–β-actin, anti–TCF-4, anti–c-Myc, anti–cyclin D1, anti-EpCAM, anti-CD44, anti-CD24, anti-GFP, anti–mouse-IgG, and anti–rabbit-IgG (Santa Cruz Biotechnology, Inc.); CD133 (Miltenyi Biotec); anti–β-catenin (BD); and anti-E2F1, anti-FLAG, anti-CD44, and anti-active caspase-3 (Cell Signaling Technology). The monoclonal anti-PLD1 and anti-PLD2 antibodies were generated as described previously (Kang et al., 2013).
SP analysis.
Single-cell suspensions of cells were detached from dishes with trypsin-EDTA (Invitrogen) and suspended at 106 cells/ml in HBSS supplemented with 3% FCS and 10 mM Hepes. These cells were then incubated at 37°C for 90 min with 20 µg/ml Hoechst 33342 (Sigma-Aldrich), either alone or in the presence of 100 nM/ml verapamil (Sigma-Aldrich), an inhibitor of the verapamil-sensitive ABC transporter. After 90-min incubation, the cells were centrifuged immediately for 5 min at 300 g and 4°C and resuspended in ice-cold HBSS. The cells were kept on ice to inhibit efflux of the Hoechst dye, and 1 µg/ml propidium iodide (Sigma-Aldrich) was added to discriminate dead cells. Finally, these cells were filtered through a 40-µm cell strainer (Falcon; BD) to obtain single-suspension cells. Cell dual-wavelength analysis and purification were performed on a dual-laser FACS (AriaIII; BD). Hoechst 33342 was excited at 355 nm UV light and emitted blue fluorescence with a 450/20 band-pass filter and red fluorescence with a 670-nm edge filter long-pass. Propidium iodide–positive (dead) cells were excluded from the analysis.
FACS and flow cytometry.
The phycoerythrin (PE)-conjugated anti-CD133 (Miltenyi Biotec), allophycocyanin-conjugated anti-CD44 (BD), PE-conjugated anti-CD24 (BD), or FITC–anti-EpCAM (EMD Millipore) antibodies were used for FACS analysis. To obtain CD133+CD44+ or CD133–CD44– cells, xenografted tumor cells stained with PE-conjugated anti-CD133 and APC-conjugated anti-CD44 antibody (both 1:40) were subjected to cell sorting performed by the FACS. For cell cycle analyses, cells were incubated with 20 µM BrdU (Sigma-Aldrich) for 2 d, prepared as previously described (Tardat et al., 2007), and incubated for 30 min with PE-conjugated anti-BrdU antibody (R&D Systems).
PLD activity assay.
PA production was measured using the Total Phosphatidic Acid Assay kit (Cayman Chemical) for in vivo and in vitro. Tumor tissues (10–15 mg) of Apcmin/+, Apcmin/+Pld1Tg, Apcmin/+Pld1−/−, and Apcmin/+Pld1−/−Pld1Tg mice (18 wk; n = 5–8 for each group) or colonospheres were washed in ice-cold PBS, adding 1.5 ml of ice-cold methanol and 1 M NaCl (2.25 ml)/chloroform (2.5 ml), and were centrifuged at 1,500 g for 10 min at 4°C. The lower chloroform phase was washed twice with 2 ml preequilibrated upper phase and centrifuged at 1,500 g for 10 min at 4°C. The lower chloroform phase was transferred to a 12 × 75–mm glass tube, dried under N2, and resuspended in 500 µl of 1% Triton X-100. To obtain the PA standard curve, PA from egg yolk lecithin was used. The extracted lipids were used to determine PA content using the total PA assay kit, according to the manufacturer’s protocol.
Promoter reporter constructs.
For measurement of TCF activity, TOPflash (eight WT TCF binding sites) and FOPflash (eight mutant-type TCF binding sites) luciferase plasmids were purchased from Addgene (plasmids #12457 and #12457). For construction of the cyclin D1 (–1,748 to +133), β-catenin (–2,645 to +93), CD133 (+461 to +1,639), CD44 (–1,171 to +17), CD166 (–2,392 to –60), EpCAM (–612 to +52), and hsa–miR-4496 (–743 to +9), promoter reporter plasmids (relative to transcription start site or first nucleotide of human miRNA stem loop) were amplified from human normal genomic DNA (Invitrogen) and cloned downstream of the Firefly luciferase gene in pGL4.14b (Promega). The listed primer sets were used in promoter cloning of the indicated genes (Table S3).
3′ UTR reporter constructs and site-directed mutagenesis.
For construction of the human β-catenin (+1 to +930), 3′ UTR reporter plasmids (relative to translation last nucleotide) were amplified from cDNA synthesized from the human normal colon RNA (Invitrogen) and cloned into the XhoI or NotI sites downstream of the Renilla luciferase gene in psiCHECK-2 (Promega). The indicated miRNA seed sequence binding sites in the β-catenin 3′ UTR were deleted completely (miR-4496) using the QuikChange Site-Directed Mutagenesis kit II (Agilent Technologies). The primers for 3′ UTR cloning of the indicated genes are β-catenin forward, 5′-AAACTCGAGCAATCAGCTGGCCTGGTTTGATAC-3′, and reverse, 5′-GCGGCCGCCAATAGTTTTTGATCAAAAACATGAAATAGATC-3′.
Plasmids.
The shRNA lentiviral constructs against PLD1 (two types) or E2F1 in the pLKO vector were purchased from Sigma-Aldrich. premiR-4496 lentiviral constructs in the pLenti-III-miR-GFP were purchased from ABM. Anti–miR-4496 lentiviral constructs in the pEZX-mcherry were purchased from GeneCopoeia.
Viral production and infection.
The shCTRL, shPLD1-a, shPLD1-b, FLAG-PLD1, shE2F1, HA-E2F1, and all pre- and anti-miR lentivirus were produced by using lentivirus packaging mix (Invitrogen). 293FT cells were seeded at 105 cells per 35-mm dish in DMEM/10% FBS. After 18 h, cells were transfected as follows: 10 µl Lipofectamine 2000 (Invitrogen) was added to 100 µl DMEM and incubated for 20 min. 1 μg of viral vector along with 0.9 mg of the appropriate gag/pol expression vector and 0.1 mg VSVG expression vector were then added to the DMEM/Lipofectamine 2000 mixture. The mixture was incubated for 30 min and then added to 293FT cells overnight. The next day, fresh media were added to the transfected 293FT cells. Viral supernatant was harvested 48 and 72 h after transfection, filtered, and added to the recipient cell lines with 6 µg/ml Polybrene (Sigma-Aldrich) for 12-h infection. Human PLD1 were cloned into the pAdeono-IRES-GFP vector. PLD1 adenovirus was produced by using the Fast-Trap adenovirus purification kit (EMD Millipore). After 24 h of incubation in the presence of viral particles, the medium was changed, and cells were cultured for an additional 24 h.
Reagents.
Recombinant Wnt3a (R&D Systems), LiCl (Sigma-Aldrich), oxaliplatin (Oxal; Sigma-Aldrich), 5-fluorouracil (Sigma-Aldrich), and cycloheximide (Sigma-Aldrich) were commercially obtained. PLD1 inhibitor was purchased from Cayman Chemical. Dual-luciferase assay kits were purchased from Promega.
Transient transfection and reporter gene assay.
Following the manufacturer’s instructions, luciferase reporter plasmids, expression plasmids, or siRNAs were transiently transfected into cells with Lipofectamine 2000, Lipofectamine Plus (Invitrogen), Fugene 6 (Roche), or Polyethylenimine (Sigma-Aldrich) reagents. Transfection and luciferase assays were performed as previously described (Kang et al., 2008, 2011b). Relative luciferase activity was obtained by normalization of firefly and Renilla luciferase activity.
Cell lines.
HEK293 and human CRC cells (HCT116, DLD1, SW480, and LS174T) were obtained from ATCC. Adherent cells were grown in DMEM/F12, McCoy’s, or RPMI 1640 (Gibco) containing 1–10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C. Xenografted tumors of HCT116, SW480, DLD1, and LS174T cell lines were minced and enzymatically dissociated with 1 mg/ml collagenase D (Roche) and 1 mg/ml DNase I (Roche) for 1 h at 37°C and then sequentially filtered through 100- and 70-mm cell strainers (BD). After the lysis of RBCs with Red Blood Cell Lysis Solution (Miltenyi Biotec), the filtered cells were grown in B27 (Invitrogen) supplemented with 20 µg/ml of basic fibroblast growth factor and 20 µg/ml epidermal growth factor (Sigma-Aldrich) and penicillin/streptomycin on ultra-low attachment culture dishes (Corning) as a sphere culture condition. For serial passage, sphere cells were dissociated into single cells with Accutase (Invitrogen) once for 4–7 d and incubated under the previously described culture conditions.
Establishment of stable cell lines.
The PLD1 shRNA vectors were generated by ligation of vector pCMV-RFP. To establish shRFP, shPLD1-a, and shPLD1-b stable cell lines, xenografted DLD1, HCT116, and SW480 cells were transfected using control or two types of shRNA for PLD1 for 72 h under sphere cultured conditions, and then cells were selected with 500 µg/ml neomycin (Sigma-Aldrich) for 14 d. To avoid clonal variation, the stable cell lines were established from the mixed population of multiple neomycin-resistant clones. RFP-positive cells were sorted by using flow cytometry (FACS). shPLD1-a plus shE2F1, anti-EV, or anti–miR-4496 stable cell lines were established by transduction of their lentiviruses with 6 µg/ml polybrene (Sigma-Aldrich) and 500 µg/ml neomycin/4 µg/ml puromycin double selection.
Real-time q-PCR.
Total RNA was extracted by using TRIzol reagent (Invitrogen). 3 µg RNA was reverse transcribed to cDNA using the Reverse Transcription Master kit (Invitrogen) according to the manufacturer’s instructions. Real-time q-PCR reactions were performed in triplicate, and the final results were found by using a relative standard curve. The primer sets used in q-RT-PCR for measuring gene expression relative to β-actin or 18S are listed in Table S4.
Quantification of mature miR.
A TaqMan MicroRNA RT kit (Applied Biosystems) was used in combination with the miR-4496–specific reverse transcription primers followed by PCR using the indicated TaqMan-specific primers and TaqMan 2X Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). RNU6B was used as a normalization control.
Chromatin IP (ChIP) assay.
ChIP assay was performed as previously described (Kang et al., 2013). The cells were used for cross-linking with 2.5% paraformaldehyde in PBS for 10 min. Cells were scraped and collected by centrifugation. Cells were lysed in lysis buffer (50 mM Hepes, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, and 1.0 mM protease inhibitor cocktail) and sonicated for 20 s three times. IP reaction was performed at 4°C overnight. Immunocomplexes were extracted three times with 1% SDS and 0.1 M NaHCO3, and cross-linking was reversed by incubation at 65°C overnight. The saved chromatin input fraction was also processed in the same manner. Samples were then digested with DNase- and RNase-free proteinase K at 50°C for 4 h, followed by extraction with phenol-chloroform-isoamyl alcohol. About 1/20 of the immunoprecipitated DNA was used in each q-RT-PCR. For the sequences of the promoter-specific primers used in q-RT-PCR, see Table S5.
Mice genotype.
The following primer sets were used in PCR. Apc (WT) forward, 5′-GCCATCCCTTCACGTTAG-3′, Apc (Min) forward, 5′-TTCTGAGAAAGACAGAAGTTA-3′, and Apc (WT/Min) reverse, 5′-TTCCACTTTGGCATAAGGC-3′; Pld1 (WT) forward, 5′-GAGCTACAGAAAAGCATAGAAAC-3′, Pld1 (MUT) forward, 5′-GAGTAGCTGCAGGGGTCCTC-3′, and Pld1 (WT/MUT) reverse, 5′-GGACAGCAGCTGCGTATGTT-3′; Pld2 (WT) forward, 5′-GCACGGGGCCCTCCCACATC-3′, Pld2 (MUT) forward, 5′-GGGAATCTGAGGCTTCAAGACTGGG-3′, and Pld2 (WT/MUT) reverse, 5′-GCTGGTGTTTTGAGGATGCTTG-3′.
Statistical analysis.
Data were analyzed using the paired t test, and correlation coefficients were calculated using Spearman’s r. CRC patient survival probability, defined as the time from colon resection to death or date of last follow-up, was analyzed using Kaplan-Meier, and differences were evaluated using the log-rank test. IHC results were analyzed using a Chi-square test. Statistical analysis was performed using Origin 8.0 and Prism 5.0 (GraphPad).
Accession numbers.
The GEO accession nos. for the mRNA and miR microarray data reported in this paper are GSE55724 and GSE55771, respectively.
Online supplemental material.
Table S1 shows gene counts for all genes commonly up-regulated by shPLD1 in CRC cells. Table S2 shows gene counts for all genes commonly down-regulated by shPLD1 in CRC cells. Tables S3, S4, and S5 provide primer sets for promoter cloning, gene expression, and ChIP assay used in this study, respectively. Table S6 shows gene counts for all genes commonly up-regulated by PLD1 inhibitor in CRC cells. Table S7 shows gene counts for all genes commonly down-regulated by PLD1 inhibitor in CRC cells. Table S8 shows gene counts for all miR up-regulated or down-regulated by PLD1 inhibitor in CRC cells. Table S9 shows gene counts for some genes up-regulated by shE2F1 among all the genes down-regulated by shPLD1 in CRC cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20141254/DC1.
Supplementary Material
Acknowledgments
This study was supported by a National Research Foundation of Korea grant funded by the Korean government (MEST; 2012002009), a Translational Research Center for Protein Function Control grant (National Science Foundation 2009-0083522), and a National R&D Program for Cancer Control grant from the Ministry for Health, Welfare, and Family Affairs (Republic of Korea; 0920050).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ABC
- ATP-binding cassette
- AOM
- azoxymethane
- APC
- adenomatous polyposis coli
- CC-IC
- colon C-IC
- C-IC
- cancer-initiating cell
- ChIP
- chromatin IP
- CRC
- colorectal cancer
- DSS
- dextran sodium sulfate
- H&E
- hematoxylin and eosin
- IB
- immunoblot
- IEC
- intestinal epithelial cell
- IHC
- immunohistochemical staining
- IP
- immunoprecipitation
- LDA
- limiting dilution assay
- miR
- microRNA
- NOD
- nonobese diabetic
- PA
- phosphatidic acid
- PE
- phycoerythrin
- PLD
- phospholipase D
- RB
- retinoblastoma
- SCID
- severe combined immunodeficiency
- SFU
- sphere-forming unit
- SI
- small intestine
- SP
- side population
- TCF
- T cell factor
- Tg
- transgenic
- UTR
- untranslated region
References
- Al-Hajj M., and Clarke M.F.. 2004. Self-renewal and solid tumor stem cells. Oncogene. 23:7274–7282. 10.1038/sj.onc.1207947 [DOI] [PubMed] [Google Scholar]
- Boral D., and Nie D.. 2012. Cancer stem cells and niche microenvironments. Front Biosci (Elite Ed). 4:2502–2514. 10.2741/E561 [DOI] [PubMed] [Google Scholar]
- Brabletz S., Schmalhofer O., and Brabletz T.. 2009. Gastrointestinal stem cells in development and cancer. J. Pathol. 217:307–317. 10.1002/path.2475 [DOI] [PubMed] [Google Scholar]
- Bramis J., Zacharatos P., Papaconstantinou I., Kotsinas A., Sigala F., Korkolis D.P., Nikiteas N., Pazaiti A., Kittas C., Bastounis E., and Gorgoulis V.G.. 2004. E2F-1 transcription factor immunoexpression is inversely associated with tumor growth in colon adenocarcinomas. Anticancer Res. 24:3041–3047. [PubMed] [Google Scholar]
- Chen Q., Hongu T., Sato T., Zhang Y., Ali W., Cavallo J.A., van der Velden A., Tian H., Di Paolo G., Nieswandt B., et al. . 2012. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci. Signal. 5:ra79 10.1126/scisignal.2003257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Cho R.W., and Clarke M.F.. 2007. Cancer stem cells: models and concepts. Annu. Rev. Med. 58:267–284. 10.1146/annurev.med.58.062105.204854 [DOI] [PubMed] [Google Scholar]
- Dall’Armi C., Hurtado-Lorenzo A., Tian H., Morel E., Nezu A., Chan R.B., Yu W.H., Robinson K.S., Yeku O., Small S.A., et al. . 2010. The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 1:142 10.1038/ncomms1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg V.S., and Donnenberg A.D.. 2005. Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J. Clin. Pharmacol. 45:872–877. 10.1177/0091270005276905 [DOI] [PubMed] [Google Scholar]
- Fearon E.R., and Vogelstein B.. 1990. A genetic model for colorectal tumorigenesis. Cell. 61:759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I., Freed E., Ligon A.H., Vena N., Ogino S., et al. . 2008. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 455:547–551. 10.1038/nature07179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell M.A., Brose K., Paradis G., Conner A.S., and Mulligan R.C.. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183:1797–1806. 10.1084/jem.183.4.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkels K.M., Boivin G.P., Dudley E.S., Berberich S.J., and Gomez-Cambronero J.. 2013. Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene. 32:5551–5562. 10.1038/onc.2013.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.W., Park M.H., Lee Y.J., Kim H.S., Kwon T.K., Park W.S., and Min S.. 2008. Phorbol ester up-regulates phospholipase D1 but not phospholipase D2 expression through a PKC/Ras/ERK/NFκB-dependent pathway and enhances matrix metalloproteinase-9 secretion in colon cancer cells. J. Biol. Chem. 283:4094–4104. 10.1074/jbc.M707416200 [DOI] [PubMed] [Google Scholar]
- Kang D.W., Lee S.H., Yoon J.W., Park W.S., Choi K.Y., and Min S.. 2010. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/β-catenin/TCF signaling axis. Cancer Res. 70:4233–4242. 10.1158/0008-5472.CAN-09-3470 [DOI] [PubMed] [Google Scholar]
- Kang D.W., Choi K.Y., and Min S.. 2011a. Phospholipase D meets Wnt signaling: a new target for cancer therapy. Cancer Res. 71:293–297. 10.1158/0008-5472.CAN-10-2463 [DOI] [PubMed] [Google Scholar]
- Kang D.W., Park M.H., Lee Y.J., Kim H.S., Lindsley C.W., Alex Brown H., and Min S.. 2011b. Autoregulation of phospholipase D activity is coupled to selective induction of phospholipase D1 expression to promote invasion of breast cancer cells. Int. J. Cancer. 128:805–816. 10.1002/ijc.25402 [DOI] [PubMed] [Google Scholar]
- Kang D.W., Park M.K., Oh H.J., Lee D.G., Park S.H., Choi K.Y., Cho M.L., and Min S.. 2013. Phospholipase D1 has a pivotal role in interleukin-1β-driven chronic autoimmune arthritis through regulation of NF-κB, hypoxia-inducible factor 1α, and FoxO3a. Mol. Cell. Biol. 33:2760–2772. 10.1128/MCB.01519-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell J., and Cadigan K.M.. 2009. APC and β-catenin degradation. Adv. Exp. Med. Biol. 656:1–12. [DOI] [PubMed] [Google Scholar]
- Lai P.S., Cheah P.Y., Kadam P., Chua C.L., Lie D.K., Li H.H., Eu K.W., Seow-Choen F., and Lee A.S.. 2006. Overexpression of RB1 transcript is significantly correlated with 13q14 allelic imbalance in colorectal carcinomas. Int. J. Cancer. 119:1061–1066. 10.1002/ijc.21945 [DOI] [PubMed] [Google Scholar]
- Li V.S., Ng S.S., Boersema P.J., Low T.Y., Karthaus W.R., Gerlach J.P., Mohammed S., Heck A.J., Maurice M.M., Mahmoudi T., and Clevers H.. 2012. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 149:1245–1256. 10.1016/j.cell.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Liu J., Stevens J., Rote C.A., Yost H.J., Hu Y., Neufeld K.L., White R.L., and Matsunami N.. 2001. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell. 7:927–936. 10.1016/S1097-2765(01)00241-6 [DOI] [PubMed] [Google Scholar]
- Moon B.S., Kim H.Y., Kim M.Y., Yang D.H., Lee J.M., Cho K.W., Jung H.S., and Choi K.Y.. 2011. Sur8/Shoc2 involves both inhibition of differentiation and maintenance of self-renewal of neural progenitor cells via modulation of extracellular signal-regulated kinase signaling. Stem Cells. 29:320–331. 10.1002/stem.586 [DOI] [PubMed] [Google Scholar]
- Morris E.J., Ji J.Y., Yang F., Di Stefano L., Herr A., Moon N.S., Kwon E.J., Haigis K.M., Näär A.M., and Dyson N.J.. 2008. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature. 455:552–556. 10.1038/nature07310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufert C., Becker C., and Neurath M.F.. 2007. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2:1998–2004. 10.1038/nprot.2007.279 [DOI] [PubMed] [Google Scholar]
- Nevins J.R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699–703. 10.1093/hmg/10.7.699 [DOI] [PubMed] [Google Scholar]
- Niehrs C., and Acebron S.P.. 2012. Mitotic and mitogenic Wnt signalling. EMBO J. 31:2705–2713. 10.1038/emboj.2012.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C.A., Kreso A., Ryan P., Hermans K.G., Gibson L., Wang Y., Tsatsanis A., Gallinger S., and Dick J.E.. 2012. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell. 21:777–792. 10.1016/j.ccr.2012.04.036 [DOI] [PubMed] [Google Scholar]
- Oliveira T.G., Chan R.B., Tian H., Laredo M., Shui G., Staniszewski A., Zhang H., Wang L., Kim T.W., Duff K.E., et al. . 2010. Phospholipase d2 ablation ameliorates Alzheimer’s disease-linked synaptic dysfunction and cognitive deficits. J. Neurosci. 30:16419–16428. 10.1523/JNEUROSCI.3317-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E., Silva-Vargas V., and Doetsch F.. 2011. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 8:486–498. 10.1016/j.stem.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Rinn J.L., and Schier A.F.. 2011. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12:136–149. 10.1038/nrg2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Robine S., Jaisser F., El Marjou F.E., and Louvard D.. 1999. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J. Biol. Chem. 274:6476–6482. 10.1074/jbc.274.10.6476 [DOI] [PubMed] [Google Scholar]
- Scott S.A., Selvy P.E., Buck J.R., Cho H.P., Criswell T.L., Thomas A.L., Armstrong M.D., Arteaga C.L., Lindsley C.W., and Brown H.A.. 2009. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5:108–117. 10.1038/nchembio.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvy P.E., Lavieri R.R., Lindsley C.W., and Brown H.A.. 2011. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111:6064–6119. 10.1021/cr200296t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Chen Q., and Frohman M.A.. 2009. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 5:1477–1486. 10.2217/fon.09.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M., Murr R., Herceg Z., Sardet C., and Julien E.. 2007. PR-Set7–dependent lysine methylation ensures genome replication and stability through S phase. J. Cell Biol. 179:1413–1426. 10.1083/jcb.200706179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., and Lees J.A.. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11–20. 10.1038/nrm714 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.