Zou et al. identify a Golgi-associated factor, TRAF3-interacting protein 3 (TRAF3IP3), as a crucial mediator of thymocyte development regulating TCR-stimulated ERK signaling in the Golgi.
Abstract
Generation of T lymphocytes in the thymus is guided by signal transduction from the T cell receptor (TCR), but the underlying mechanism is incompletely understood. Here we have identified a Golgi-associated factor, TRAF3-interacting protein 3 (TRAF3IP3), as a crucial mediator of thymocyte development. TRAF3IP3 deficiency in mice attenuates the generation of mature thymocytes caused by impaired thymocyte-positive selection. TRAF3IP3 mediates TCR-stimulated activation of the mitogen-activated protein kinase (MAPK) extracellular signal-regulated kinase (ERK) and its upstream kinase mitogen/extracellular signal-regulated kinase (MEK). Interestingly, TRAF3IP3 exerts this signaling function through recruiting MEK to the Golgi and, thereby, facilitating the interaction of MEK with its activator BRAF. Transgenic expression of a constitutively active MEK rescues the T cell development block in Traf3ip3 knockout mice. These findings establish TRAF3IP3 as a novel regulator of T cell development and suggest a Golgi-specific ERK signaling mechanism that regulates thymocyte development.
The development of T cells in the thymus involves sequential progression of thymocytes through different stages, which can be defined based on their surface expression of CD4 and CD8 coreceptors (Germain, 2002). During the early CD4−CD8− double-negative (DN) stages, DN1–DN4, the gene encoding the TCRβ chain undergoes rearrangement, leading to the formation of a pre-TCR that drives the further differentiation of the cells into CD4+CD8+ double-positive (DP) stage (Michie and Zúñiga-Pflücker, 2002). Subsequently, rearrangement of the TCRα gene occurs in the CD4+CD8+ DP stage, leading to formation of surface αβ TCR. DP thymocytes are then subject to positive and negative selections by complexes of self-peptide and MHC displayed on thymic epithelial cells and other antigen-presenting cells (Klein et al., 2014). The majority of DP thymocytes die of “neglect” caused by expression of TCRs that fail to recognize self-peptide–MHC complexes, and those that bind self-peptide–MHC complexes with high affinity die of negative selection. Only the thymocytes with TCRs that bind self-peptide–MHC complexes with intermediate strength are positively selected to become mature CD4+ or CD8+ single-positive (SP) thymocytes (Germain, 2002; Klein et al., 2014).
Signal transduction from the TCR plays a crucial role in the progression of thymocytes from the DP to SP stages (Starr et al., 2003; Zamoyska and Lovatt, 2004; Wang et al., 2010). In particular, the MAPK signaling cascade is critically required for the development and maturation of thymocytes (Pagès et al., 1999; Alberola-Ila and Hernández-Hoyos, 2003; Fischer et al., 2005; Kortum et al., 2013). This signaling cascade involves sequential activation of the small G protein Ras and its downstream kinases RAF, mitogen/extracellular signal-regulated kinase (MEK), and extracellular signal-regulated kinase (ERK; Kortum et al., 2013). Among the RAF members, BRAF is particularly important for TCR-stimulated MEK-ERK activation and thymocyte development (Tsukamoto et al., 2008; Dillon et al., 2013). TCR stimulation leads to the activation of BRAF, but not RAF1, in thymocytes (Dillon et al., 1991). Consistently, BRAF is required for TCR-stimulated ERK activation and DP thymocyte positive selection; however, how BRAF mediates activation of MEK and ERK in thymocytes is incompletely understood (Tsukamoto et al., 2008; Dillon et al., 2013).
In the present study, we identified TRAF3-interacting protein 3 (TRAF3IP3) as a novel regulator of MAPK signaling and thymocyte development. TRAF3IP3, also called TRAF3-interacting JNK-activating modulator (T3JAM), was originally identified as a protein that interacts with TRAF3 and synergizes with TRAF3 to activate JNK under overexpression conditions (Dadgostar et al., 2003). TRAF3IP3 mRNA is specifically expressed in lymphoid organs, although its physiological role has not been investigated (Dadgostar et al., 2003). We found that TRAF3IP3 is highly expressed in DP thymocytes and associated with the Golgi apparatus. Using newly generated Traf3ip3 KO mice, we demonstrated a crucial role for TRAF3IP3 in mediating the thymocyte development from DP to SP stages. The TRAF3IP3 deficiency specifically attenuates TCR-stimulated activation of MEK and its downstream kinase ERK. TRAF3IP3 mediates MEK activation by recruiting MEK to the Golgi and, thereby, facilitating MEK interaction with its upstream kinase BRAF. These findings establish TRAF3IP3 as a novel regulator of thymocyte development and suggest a compartmentalized BRAF-MEK-ERK signaling mechanism regulated by TRAF3IP3.
RESULTS
TRAF3IP3 is required for thymocyte development
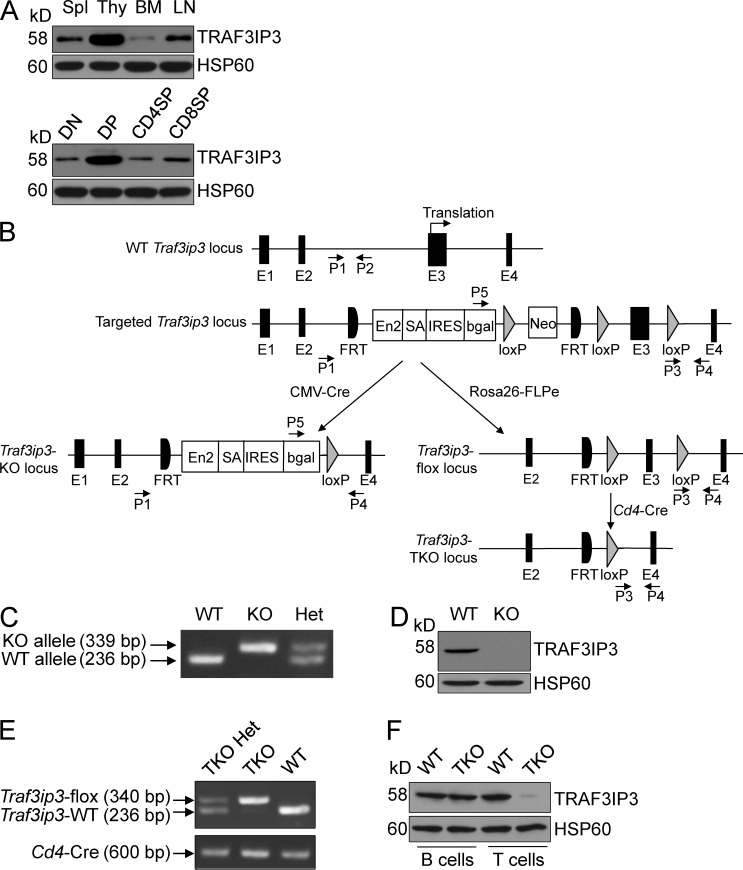
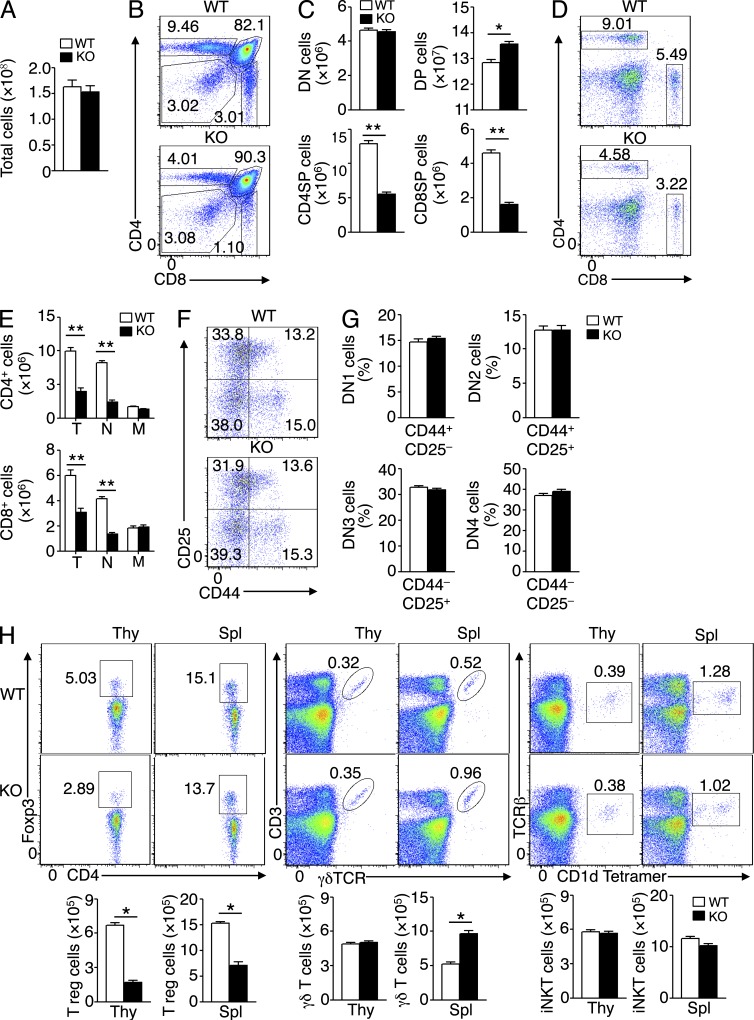
To assess the function of TRAF3IP3 in the immune system, we analyzed its expression at the protein level and found that TRAF3IP3 was highly expressed in the thymocytes, most abundantly in the CD4+CD8+ DP thymocytes (Fig. 1 A). This finding prompted us to examine the role of TRAF3IP3 in thymocyte development. We generated germline Traf3ip3 KO mice by crossing the Traf3ip3-targeted mice with CMV-Cre mice (Fig. 1, B–D; Schwenk et al., 1995). We also crossed the Traf3ip3-targetd mice with FRT deleter (Rosa26-FLPe) mice to generate Traf3ip3-flox mice, which were further crossed with Cd4-Cre mice to produce Traf3ip3 T cell–conditional KO (TKO) mice (Fig. 1, B, E, and F). The Traf3ip3-KO mice were healthy and fertile (not depicted), and these mutant mice and their WT control mice had similar numbers of total thymocytes (Fig. 2 A). However, the Traf3ip3-KO mice had considerably lower frequencies of CD4+ and CD8+ SP thymocytes, and concomitantly higher frequencies of DP thymocytes, than the WT mice (Fig. 2, B and C). The Traf3ip3-KO mice also had a reduced frequency and number of splenic CD4+ and CD8+ T cells (Fig. 2, D and E). In addition, the Traf3ip3-KO mice had fewer naive T cells than WT mice, whereas the number of memory T cells in both genotypes was normal (Fig. 2 E). In contrast, the TRAF3IP3 deficiency did not affect the thymocyte development at the DN stages, DN1–DN4 (Fig. 2, F and G). These data suggest that TRAF3IP3 is required for thymocyte development during the DP to SP transition. We also examined the development of regulatory T (T reg) cells, γδ T cells, and invariant NKT (iNKT) cells in the Traf3ip3-KO mice. The Traf3ip3-KO mice had a reduced number of thymic and splenic T reg cells and an increased number of splenic γδ T cells, whereas the number of iNKT cells was comparable between the KO and WT mice (Fig. 2 H). The defect of the Traf3ip3-KO mice in T reg cell development was consistent with their impaired production of SP thymocytes, and how TRAF3IP3 regulated the peripheral γδ T cells was unclear.
Figure 1.
Traf3ip3 gene targeting. (A) Immunoblot analysis of TRAF3IP3 and HSP60 using lysates of splenocytes (Spl), thymocytes (Thy), BM cells, and LN cells (top) or flow cytometrically sorted subpopulations of thymocytes (bottom). (B) Schematic picture of Traf3ip3 gene targeting strategy using an FRT-LoxP vector, with genotyping primers indicated (P1–P5). Chimeric mice were crossed with a CMV-Cre mouse to generate germline Traf3ip3-KO mice or with an FRT deleter (Rosa26-FLPe) mouse to generate Traf3ip3-flox mice, which were subsequently crossed with a Cd4-Cre mouse to produce Traf3ip3-TKO mice. (C) Genotyping PCR using P1/P2 primer pair for the WT allele and P5/P4 primer pair for the KO allele. (D) Immunoblot analysis of TRAF3IP3 and Actin using thymocytes of WT and Traf3ip3-KO (KO) mice. (E) Genotyping PCR to amplify the Traf3ip3-flox (using P3/P4 primer pair) and WT (using P1/P2 primer pair) alleles (top) or the Cre cDNA (bottom). (F) Immunoblot analysis of TRAF3IP3, showing T cell–specific TRAF3IP3 ablation. PCR primer sequences are listed in Table S1. Data are representative of four independent experiments with at least six mice of each genotype (A, D, and F).
Figure 2.
Impaired T cell development in Traf3ip3-KO mice. (A) Total number of thymocytes from 4-wk-old WT and Traf3ip3-KO (KO) mice. (B and C) Flow cytometric analysis of the percentage of thymic subpopulations from 4-wk-old WT and Traf3ip3-KO mice, presented as a representative FACS plot (B) or summary graph (C). (D and E) Flow cytometric analysis of the frequency (D) and absolute number (E) of CD4+ and CD8+ T cells in splenocytes of 4-wk-old Traf3ip3+/+ and Traf3ip3-KO mice. T indicates total CD4+ or CD8+ T cells, N indicates naive T cells (CD44−CD62L+), and M indicates memory T cells (CD44+CD62L−). (F and G) Flow cytometry analysis of CD4−CD8− DN thymocytes (based on expression of CD25 and CD44) from 4-wk-old Traf3ip3+/+ and Traf3ip3-KO mice. Data are presented as a representative FACS plot (F) and summary graph of DN1–DN4 stages (G). (H) Flow cytometry analysis of T reg cells, γδ T cells, and iNKT cells in the thymus (Thy) and spleen (Spl) from 4-wk-old WT and Traf3ip3-KO mice. Data are representative of four (A–G) or three (H) independent experiments with at least five mice per group. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05; **, P < 0.01).
TRAF3IP3 has a cell-intrinsic role in regulating thymocyte development
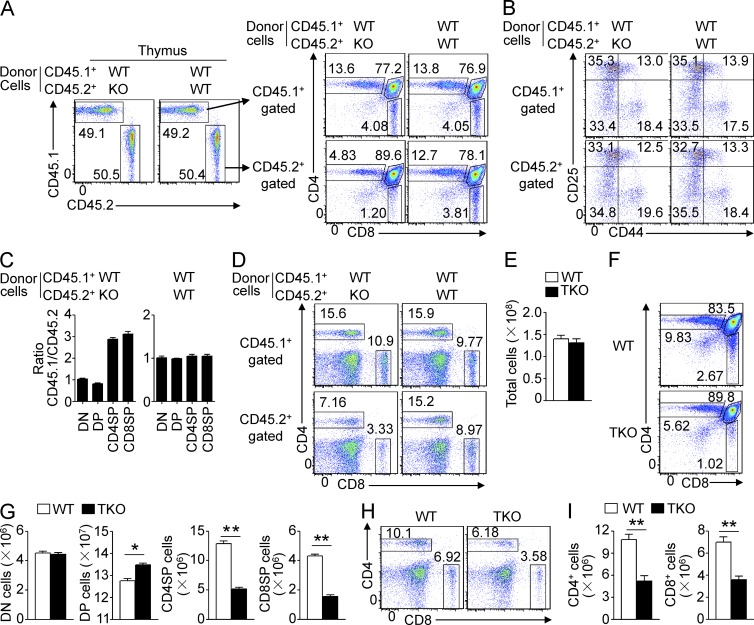
To examine whether the thymocyte development defect of Traf3ip3-KO mice is cell intrinsic, we generated mixed BM chimeric mice by transferring BM cells derived from WT or Traf3ip3-KO mice (CD45.2+), along with BM cells derived from WT B6.SJL mice (CD45.1+), into the lymphocyte-deficient Rag1-KO mice. The chimeric mice had a similar percentage of thymocytes derived from the CD45.1+ and CD45.2+ BM cells (Fig. 3 A). Furthermore, the CD45.1+ and CD45.2+ WT thymocytes displayed a similar profile of development (Fig. 3 A). However, under the same conditions, the CD45.2+ Traf3ip3-KO BM cells yielded a substantially lower frequency of CD4+ and CD8+ SP thymocytes and of splenic T cells than the WT BM cells, although the KO and WT BM cells produced a similar frequency of DN cells (Fig. 3, A–D). This result suggested a cell-intrinsic role for TRAF3IP3 in regulating thymocyte development.
Figure 3.
Cell-intrinsic defect of TRAF3IP3-deficient mice in thymocyte development. (A and B) Flow cytometric analysis of total thymocytes (A) and DN thymocytes (B) in Rag1-KO recipient mice adoptively transferred (for 4 wk) with BM cells derived from WT or Traf3ip3-KO mice (CD45.2+) along with BM cells derived from B6.SJL mice (CD45.1+), gating on CD45.1+ or CD45.2+ cells. (C) The CD45.1/CD45.2 ratio on DN, DP, and SP subsets in the BM chimera mice of A. (D) Flow cytometric analysis of splenocytes in the BM chimera mice of A. (E) Total number of thymocytes from 4-wk-old WT and Traf3ip3-TKO mice. (F and G) Flow cytometric analysis of the percentage of thymic subpopulations from 4-wk-old WT and Traf3ip3-TKO mice, presented as a representative FACS plot (F) or summary graph (G). (H and I) Flow cytometric analysis of the frequency (H) and absolute number (I) of CD4+ and CD8+ T cells in splenocytes of 4-wk-old WT and Traf3ip3-TKO mice. Data are representative of three (A–D) or four (E–I) independent experiments with at least four mice in each group. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05; **, P < 0.01).
To further confirm the cell-intrinsic function of TRAF3IP3 in thymocyte development, we used the Traf3ip3-TKO mice generated by crossing the Traf3ip3-flox mice with Cd4-Cre mice, which is known to delete loxP-flanked genes from the very late DN stage of thymocyte development (Fig. 1 B; Wolfer et al., 2002). The Traf3ip3-TKO and WT control mice had comparable numbers of total thymocytes (Fig. 3 E). However, as seen with the Traf3ip3-KO mice, the Traf3ip3-TKO mice had a significantly reduced frequency of CD4+ and CD8+ SP thymocytes and a concomitantly increased frequency of DP thymocytes (Fig. 3, F and G). Traf3ip3-TKO mice also had fewer splenic CD4+ and CD8+ T cells than WT mice (Fig. 3, H and I). These data further emphasize a cell-intrinsic defect of TRAF3IP3-deficient thymocytes in the transition from DP to SP stages.
TRAF3IP3 regulates thymocyte positive selection
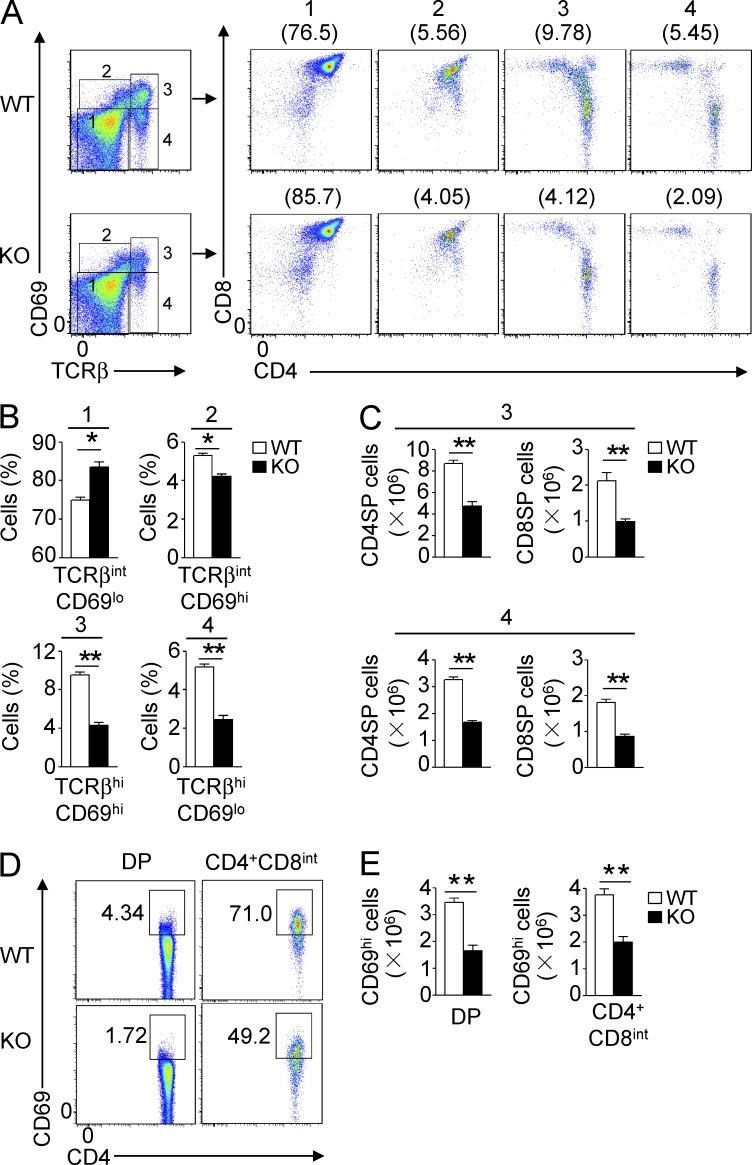
The development of thymocytes from DP to SP stages involves a process of positive selection, which is guided by TCR stimulation by self-peptide–MHC complexes (Alberola-Ila and Hernández-Hoyos, 2003). A hallmark of positive selection is up-regulation of surface TCRβ and CD69 (Swat et al., 1993; Yamashita et al., 1993; Vanhecke et al., 1997; Nakayama et al., 2002). To more precisely define the stage of developmental block in the Traf3ip3-KO thymocytes, we analyzed the thymocyte populations based on the expression of CD69 and TCRβ. This method could separate the thymocytes into four populations, representing preselection DP thymocytes (population 1; TCRloCD69lo), a transitional population initiating positive selection (population 2; TCRintCD69hi), a population immediately after positive selection (population 3; TCRhiCD69hi), and a more mature postselection SP thymocyte population (population 4; TCRhiCD69lo; Hu et al., 2012). Compared with their Traf3ip3+/+ littermates, the Traf3ip3-KO mice had an increased frequency of preselection DP thymocytes (population 1) and concomitant reduction in the subsequent populations, most dramatically the postselection population 3 and population 4 cells (Fig. 4, A and B). Accordingly, the Traf3ip3-KO mice had significantly fewer mature SP thymocytes in populations 3 and 4 than WT mice (Fig. 4 C). Further analysis revealed a substantially reduced frequency and absolute number of CD69hi cells within the total DP thymocyte population and the CD4+CD8int transitional thymocyte population of Traf3ip3-KO mice (Fig. 4, D and E). The similar results were obtained from thymocyte analysis of the Traf3ip3-TKO mice (not depicted). These results suggested a defect of the TRAF3IP3-deficient thymocytes in positive selection.
Figure 4.
Impaired positive selection in Traf3ip3-KO mice. (A) Flow cytometry analysis of the surface expression of CD69 and TCRβ on thymocytes from 4-wk-old WT and Traf3ip3-KO mice, showing four subpopulations of thymocytes. Numbers in parentheses indicate percentage of the individual subpopulations. (B) Summary graph of the thymocyte subpopulations from A. (C) The absolute number of CD4+ SP or CD8+ SP cells in the population of 3 or 4 from A. (D and E) Flow cytometry analysis of CD69 expression on DP and CD4+CD8int thymocytes of 4-wk-old Traf3ip3+/+ and Traf3ip3-KO mice, as shown of percentage (D) and absolute number (E). Data are representative of five (A–C) or three (D and E) independent experiments with at least four mice per group. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05; **, P < 0.01).
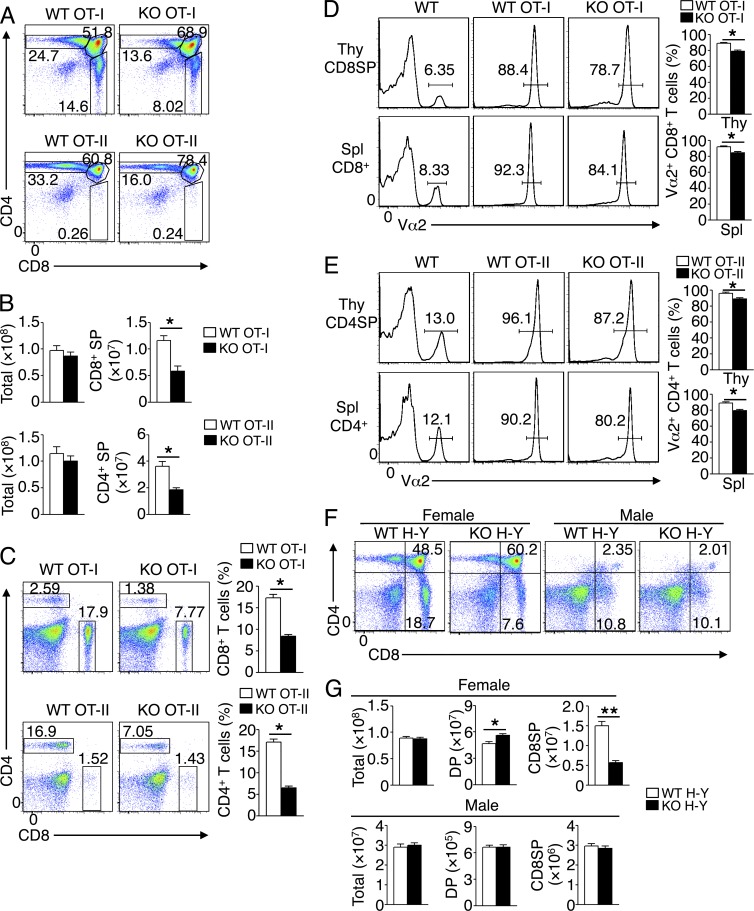
To further examine the role of TRAF3IP3 in regulating thymocyte positive selection, we used TCR transgenic mice expressing an OVA-specific TCR, restricted by MHC class I (OT-I) or MHC class II (OT-II; Hogquist et al., 1994; Barnden et al., 1998). The OT-I and OT-II TCRs positively select OVA-specific CD8+ and CD4+ thymocytes, respectively. We crossed Traf3ip3-KO mice with OT-I or OT-II mice to generate Traf3ip3-KO OT-I or Traf3ip3-KO OT-II mice. The Traf3ip3-KO OT-I and Traf3ip3-KO OT-II mice had a substantially reduced frequency and number of CD8+ and CD4+ SP thymocytes, respectively, than their corresponding control (Traf3ip3+/+ OT-I and Traf3ip3+/+ OT-II) mice (Fig. 5, A and B). Consistently, the Traf3ip3-KO OT-I and Traf3ip3-KO OT-II mice also had a reduced frequency of CD8+ and CD4+ splenic T cells, respectively (Fig. 5 C). Flow cytometry analysis for the transgenic TCRα chain Vα2 indicated that nearly all CD8+ SP thymocytes and splenic CD8+ T cells expressed the OVA-specific TCR in WT and Traf3ip3-KO OT-I mice, although lower Vα2 levels could be detected in the TRAF3IP3-deficient CD8+ SP thymocytes and splenic CD8+ T cells (Fig. 5 D). Similar results were obtained in the analysis of Va2 expression in CD4+ SP thymocytes and splenic CD4+ T cells of the WT and Traf3ip3-KO OT-II mice (Fig. 5 E).
Figure 5.
T cell development in Traf3ip3-KO mice expressing OT-I, OT-II, or H-Y TCR. (A and B) Flow cytometry analysis of thymocytes from 4-wk-old WT or Traf3ip3-KO mice crossed to OT-I or OT-II TCR transgenic mice, presented as a representative FACS plot (A) or summary graph of total or CD4+ and CD8+ SP thymocyte numbers (B). (C) Flow cytometry analysis of CD4+ and CD8+ T cells in splenocytes from 4-wk-old WT and Traf3ip3-KO OT-I or OT-II TCR transgenic mice, presented as a representative FACS plot (left) and a summary graph (right). (D and E) Flow cytometry analysis of Vα2 expression in CD8+ SP thymocytes (Thy CD8SP) and splenic CD8+ T cells (Spl CD8+) of 4-wk-old WT and Traf3ip3-KO OT-I mice (D) or in CD4+ SP thymocytes (Thy CD4SP) and splenic CD4+ T cells (Spl CD4+) of 4-wk-old WT and Traf3ip3-KO OT-II TCR transgenic mice (E), presented as a representative FACS plot (left) or summary graph of percentage (right). WT mice with no transgenic TCR were included as control. (F and G) Flow cytometry analysis of thymocytes from 4-wk-old WT and Traf3ip3-KO H-Y TCR transgenic female or male mice, showing a representative FACS plot (F) and summary graphs of total, SP, and DP thymocytes (G). Data are representative of five (A–E) or three (F and G) independent experiments with at least four mice per group. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05; **, P < 0.01).
To examine the role of TRAF3IP3 in thymocyte negative selection, we crossed the Traf3ip3-KO mice with H-Y mice expressing a transgenic TCR specific for the MHCI-restricted male antigen H-Y (Kisielow et al., 1988). In male H-Y mice, CD4+CD8+ DP thymocytes undergo negative selection induced by endogenous H-Y antigen (Kisielow et al., 1988). The male WT and Traf3ip3-KO H-Y TCR transgenic mice displayed similar frequencies and absolute numbers of total, DP, and CD8+ SP thymocyte populations (Fig. 5, F and G). Analysis of the female mice, in which thymocytes were positively selected, revealed a substantial reduction in the CD8+ SP thymocytes (Fig. 5, F and G), thus further confirming the requirement of TRAF3IP3 for positive selection. Together, these data demonstrate a critical role for TRAF3IP3 in regulating thymocyte development at the stage of positive selection.
TRAF3IP3 mediates TCR-stimulated MEK-ERK activation
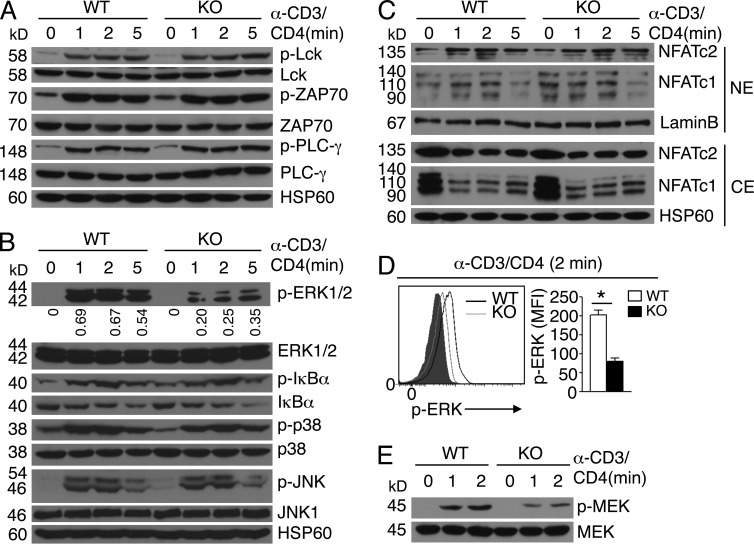
Because thymocyte positive selection is critically dependent on TCR signaling (Alberola-Ila and Hernández-Hoyos, 2003), we examined the role of TRAF3IP3 in regulating DP thymocyte signaling after TCR cross-linking with anti-CD3 plus anti-CD4. The Traf3ip3+/+ and Traf3ip3-KO DP thymocytes had comparable phosphorylation of several TCR-proximal factors, Lck, ZAP70, and PLC-γ, as well as the downstream MAPKs p38 and JNK and the IκB kinase (IKK) substrate IκBα (Fig. 6, A and B). The TRAF3IP3 deficiency also did not influence the activation of the calcium-dependent transcription factor NFAT (Fig. 6 C). However, the Traf3ip3-KO DP thymocytes had a severe defect in the induction of ERK phosphorylation (Fig. 6 B). This result was not caused by reduced frequency of CD69hi thymocytes in the Traf3ip3-KO mice because impaired ERK phosphorylation was also detected in purified CD69-negative Traf3ip3-KO DP thymocytes (Fig. 6 D). The TRAF3IP3 deficiency also attenuated the inducible phosphorylation of the ERK kinase MEK (Fig. 6 E). Thus, TRAF3IP3 has an important role in mediating TCR-stimulated activation of the MEK-ERK signaling pathway.
Figure 6.
Attenuation of TCR-stimulated MEK-ERK activation in Traf3ip3-KO DP thymocytes. (A and B) Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in extracts of sorted WT and Traf3ip3-KO DP thymocytes unstimulated (0 min) or stimulated with anti-CD3 plus anti-CD4 for the indicated time periods. Densitometry-quantified protein bands are presented as p-ERK/ERK values in B. (C) Immunoblot analysis of the indicated proteins using nuclear extracts (NE) or cytoplasmic extracts (CE) of sorted WT and Traf3ip3-KO DP thymocytes unstimulated (0) or stimulated with anti-CD3 and anti-CD4 for the indicated time periods. (D) Intracellular staining and flow cytometry analyses of phosphorylated ERK in WT and Traf3ip3-KO CD4+CD8+CD69− thymocytes stimulated as indicated. Data are presented as a representative FACS plot (left) and a summary graph of mean fluorescence intensity (MFI; right, MFI in the graph calculated after subtracting the background staining of unstimulated cells). Gray area, unstimulated CD4+CD8+CD69− thymocytes. (E) Immunoblot analysis of phosphorylated and total MEK in extracts of sorted WT and Traf3ip3-KO DP thymocytes stimulated as indicated. Data are representative of three (A, B, and D) or four (C and E) independent experiments with at least four mice in each group. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05).
Constitutively active MEK rescues the thymocyte development defect
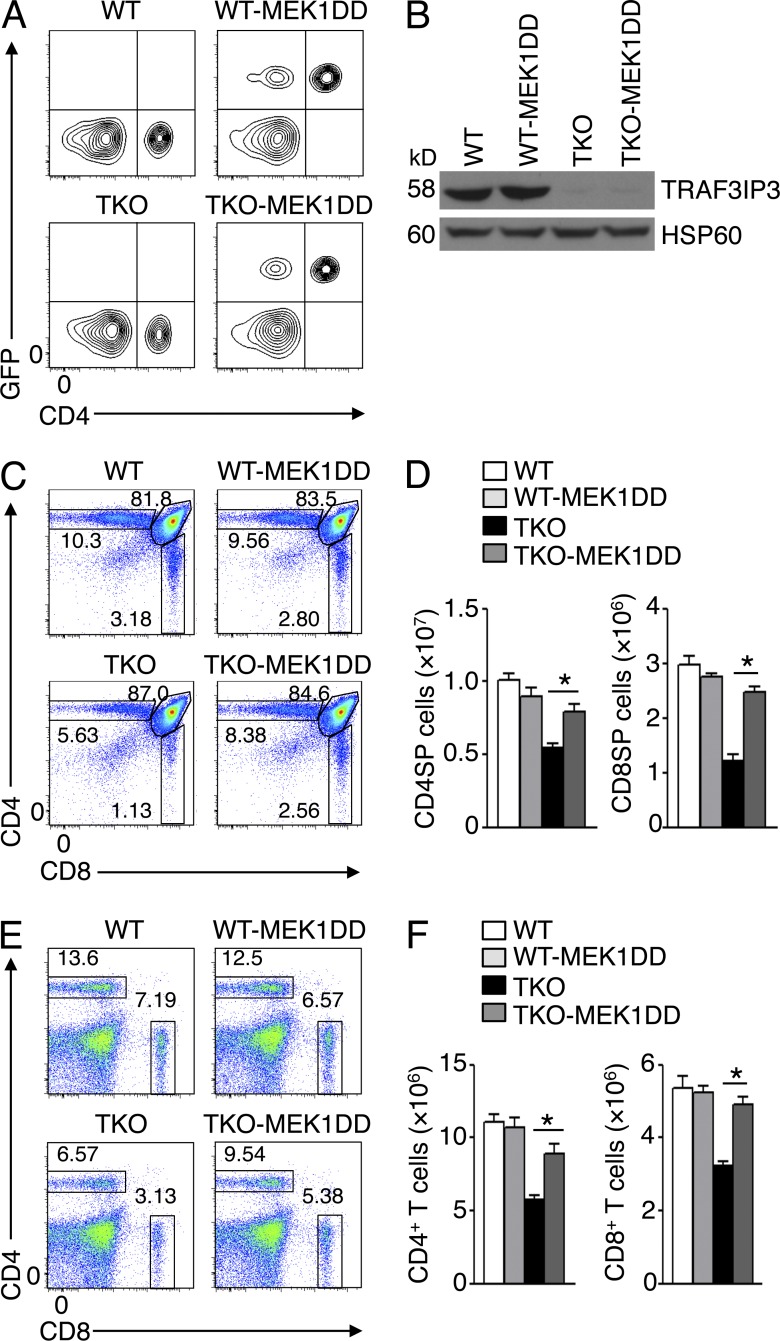
The signaling cascade leading to activation of MEK and ERK is required for thymocyte development (Pagès et al., 1999; Alberola-Ila and Hernández-Hoyos, 2003; Fischer et al., 2005; Kortum et al., 2013). To directly examine the functional significance of attenuated MEK-ERK phosphorylation in TRAF3IP3-deficient thymocytes, we rescued these mutant thymocytes with a constitutively active form of MEK (MEK1DD) by crossing the Traf3ip3-TKO mice with R26StopFLMEK1DD mice expressing MEK1DD under the control of Cre (Srinivasan et al., 2009). MEK1DD transgene was expressed in CD4+ and CD8+ T cells in the progeny mice (Fig. 7 A and not depicted), and the deletion of TRAF3IP3 was confirmed in the CD4+ SP thymocytes (Fig. 7 B). The Cd4-Cre–directed expression of MEK1DD in WT mice did not significantly alter the frequency or number of SP thymocytes (Fig. 7, C and D). However, expression of this constitutively active MEK in Traf3ip3-TKO mice largely restored the population of SP thymocytes (Fig. 7, C and D), as well as splenic T cells (Fig. 7, E and F). These findings emphasized a role for TRAF3IP3-mediated ERK signaling in mediating thymocyte development.
Figure 7.
Constitutively active MEK rescues the defect of TRAF3IP3-deficient mice in T cell development. (A) Flow cytometric analysis of GFP and CD4 expression in PBMCs of WT and Traf3ip3-TKO mice or the same mice crossed with R26StopFLMEK1DD (MEK1DD) mice. GFP was used as a marker for MEK1DD expression because GFP and MEK1DD are coexpressed in T cells via Cd4-Cre–mediated deletion of the STOP cassette in R26StopFLMEK1DD mice. (B) Immunoblot analysis of TRAF3IP3 and HSP60 using lysates of flow cytometrically sorted CD4SP thymocytes from the mice of A. (C and D) Flow cytometric analysis of thymocytes in the indicated mouse strains (4 wk old), presented as a representative plot (C) and a summary graph of CD4+ and CD8+ SP thymocyte number (D). (E and F) Flow cytometric analysis of CD4+ and CD8+ T cells in splenocytes of the indicated mouse strains (4 wk old), presented as a representative plot (E) and a summary graph of CD4+ and CD8+ T cell number (F). Data are representative of three (A and B) or four (C–E) independent experiments with at least four mice of each genotype. Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05).
TRAF3IP3 mediates BRAF-MEK interaction and MEK activation in Golgi
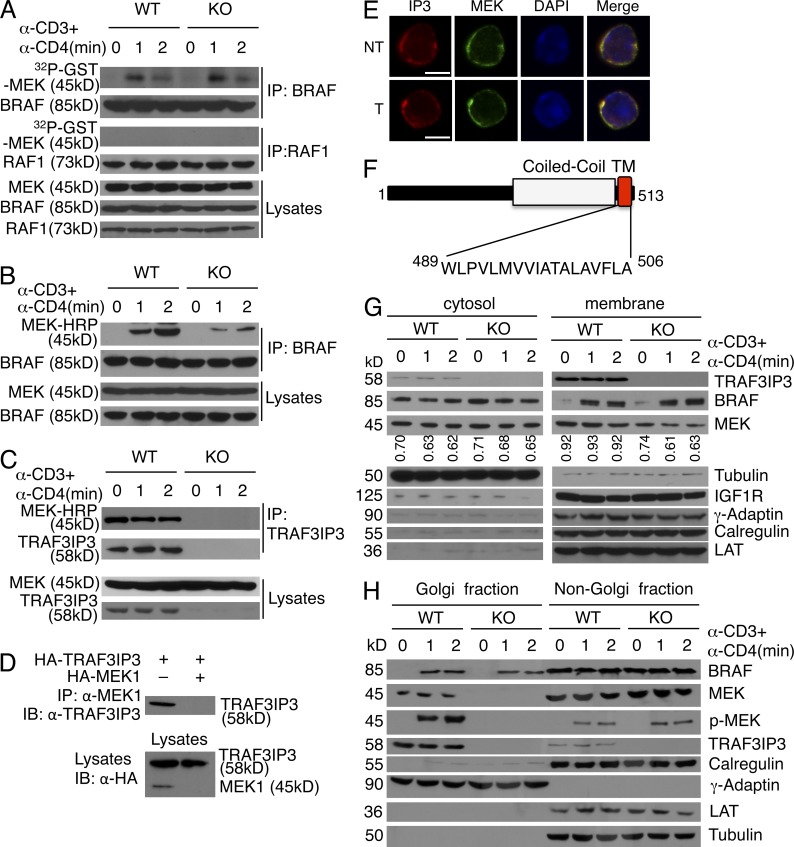
To assess the molecular mechanism by which TRAF3IP3 regulates ERK signaling, we analyzed the upstream signaling events. Consistent with a prior study (Tsukamoto et al., 2008), TCR cross-linking by anti-CD3 plus anti-CD4 stimulated the activation of BRAF but not RAF1 in DP thymocytes (Fig. 8 A). Surprisingly, the TRAF3IP3 deficiency did not affect the catalytic activation of BRAF (Fig. 8 A). This unexpected result prompted us to examine whether TRAF3IP3 regulated the interaction of BRAF with MEK. In response to TCR stimulation, BRAF rapidly bound MEK in WT DP thymocytes; however, this molecular interaction was severely attenuated in the Traf3ip3-KO DP thymocytes (Fig. 8 B). These results suggest that TRAF3IP3 mediates TCR-stimulated MEK-ERK activation by facilitating the inducible interaction of BRAF with MEK.
Figure 8.
TRAF3IP3-mediated Golgi recruitment of MEK and BRAF-MEK interaction. (A) In vitro kinase assays for BRAF and RAF1 isolated from sorted WT and Traf3ip3-KO DP thymocytes stimulated with anti-CD3 and anti-CD4. Protein expression level was analyzed by immunoblot (bottom). (B and C) Coimmunoprecipitation (top) and immunoblot (bottom) analyses to detect BRAF-MEK interaction (B) or TRAF3IP3-MEK interaction (C) in sorted WT and Traf3ip3-KO DP thymocytes stimulated with anti-CD3 and anti-CD4. (D) TRAF3IP3-MEK coimmunoprecipitation (top) and direct immunoblot (bottom) assays using HEK293 cells transfected with the indicated expression vectors. (E) Confocal microscopy analysis of TRAF3IP3 (IP3), MEK, DAPI (nuclear staining), and merged picture in nontreated (NT) or anti-CD3/anti-CD4–stimulated (T) WT DP thymocytes. Bars, 5 µm. (F) Schematic picture of mouse TRAF3IP3 structure, showing a coiled-coil domain and a transmembrane (TM) domain. (G) Immunoblot analysis of the indicated proteins in the cytosol and membrane fractions of WT and Traf3ip3-KO DP thymocytes treated as indicated. The purity of cytosol and membrane fractions was confirmed by using antibodies for Tubulin (a cytosolic marker), IGF1R (a membrane marker), LAT (a plasma membrane marker), Calregulin (an endoplasmic reticulum marker), and γ-adaptin (a Golgi marker). MEK bands were quantified by densitometry and presented as MEK/Tubulin (left) or MEK/IGF1R (right) ratio. (H) Immunoblot analysis of the indicated proteins in Golgi or non-Golgi extracts of sorted WT and Traf3ip3-KO DP thymocytes stimulated as indicated. The purity of Golgi fraction was confirmed using antibodies for Calregulin (an endoplasmic reticulum marker) and γ-adaptin (a Golgi marker). Data are representative of three (A and E) or five (B, C, G, and H) independent experiments with at least four mice per group. Data in D are representative of three independent experiments.
We surmised that TRAF3IP3 might function as a scaffold protein facilitating the BRAF-MEK interaction. Coimmunoprecipitation assays revealed constitutive binding of TRAF3IP3 with MEK in both DP thymocytes and transfected HEK293 cells (Fig. 8, C and D), and this molecular interaction was also indicated by the colocalization of TRAF3IP3 and MEK in confocal microscopy (Fig. 8 E). However, we were unable to detect appreciable binding between TRAF3IP3 and BRAF (not depicted), suggesting that TRAF3IP3 might not function as a simple adaptor. Domain analysis of TRAF3IP3 using the SMART program (Schultz et al., 1998) revealed a transmembrane domain located at its C terminus (Fig. 8 F). Subcellular fractionation assays confirmed the predominant localization of TRAF3IP3 in the membrane fraction (Fig. 8 G). MEK1 was also partially localized to the membrane fraction, and the level of membrane-associated MEK1 was reduced in the Traf3ip3-KO cells (Fig. 8 G). Furthermore, in response to TCR/CD28 stimulation, a proportion of BRAF was transferred to the membrane in a TRAF3IP3-independent manner.
MAPK signaling is known to occur in the membrane of intracellular organelles, particularly the Golgi, and the Golgi-specific MAPK signaling has been implicated in the regulation of thymocyte positive selection (Daniels et al., 2006; Mayinger, 2011). Interestingly, subcellular fractionation studies readily revealed the association of TRAF3IP3 with the Golgi (Fig. 8 H). Moreover, MEK was located to the Golgi in a TRAF3IP3-dependent manner and became phosphorylated within the Golgi in response to TCR stimulation (Fig. 8 H). The MEK phosphorylation was associated with TCR-stimulated translocation of BRAF to the Golgi, although this latter event was independent of TRAF3IP3 (Fig. 8 H). Collectively, these results suggest that TRAF3IP3 may recruit MEK to the Golgi, thereby facilitating TCR-stimulated BRAF-MEK interaction and MEK phosphorylation in the Golgi.
The transmembrane domain of TRAF3IP3 is required for its function
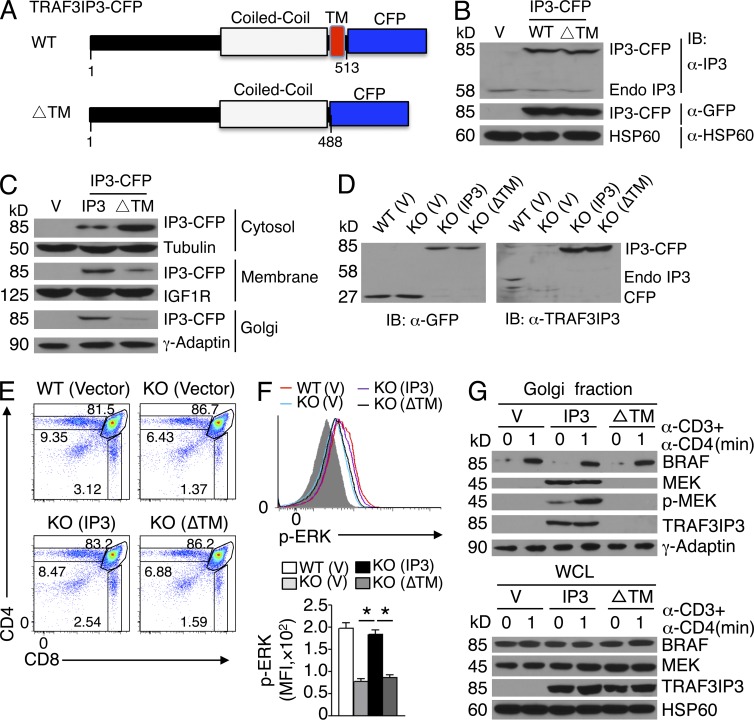
We next examined the role of the transmembrane domain of TRAF3IP3 in mediating its subcellular localization and signaling function. We constructed retroviral vectors encoding WT TRAF3IP3 or its mutant lacking the transmembrane domain (ΔTM) fused with CFP (Fig. 9 A). When stably expressed in Jurkat T cells (Fig. 9 B), the WT TRAF3IP3 was located in the membrane, suggesting that the CFP tag did not affect its subcellular localization (Fig. 9 C). In contrast to the WT TRAF3IP3, the ΔTM mutant was predominantly in the cytosol, thus confirming the function of the transmembrane domain (Fig. 9 C). To study the function of the TRAF3IP3 transmembrane domain in thymocytes, we transduced the Traf3ip3-KO BM cells with WT or ΔTM TRAF3IP3 and adoptively transferred the BM cells into Rag1-KO mice. Although TRAF3IP3 and its ΔTM mutant were comparably expressed in the thymocytes of the BM chimeric mice (Fig. 9 D), expression of WT TRAF3IP3, but not ΔTM, rescued the defect in thymocyte development (Fig. 9 E) and ERK phosphorylation (Fig. 9 F). Importantly, the WT TRAF3IP3, but not its ΔTM mutant, located to the Golgi and recruited MEK to the Golgi (Fig. 9 G). Thus, the transmembrane domain of TRAF3IP3 is required for its Golgi localization and mediating compartmentalized MEK activation during thymocyte development.
Figure 9.
Transmembrane domain of TRAF3IP3 is required for its function. (A) Diagram of WT TRAF3IP3 and its mutant lacking the transmembrane domain (ΔTM) fused with CFP. (B) Immunoblot analysis of TRAF3IP3-CFP fusion protein or endogenous TRAF3IP3 (Endo IP3) in whole-cell lysates of Jurkat T cells stably transduced with a retroviral vector (V) or the same vector encoding WT TRAF3IP3-CFP (WT) or TRAF3IP3ΔTM-CFP (ΔTM) fusion proteins. (C) Immunoblot analysis of the indicated proteins using subcellular extracts of Jurkat cells from B. (D) Immunoblot analysis of TRAF3IP3-CFP fusion protein or endogenous TRAF3IP3 (Endo IP3) using whole-cell lysates of thymocytes derived from Rag1-KO chimeric mice 4 wk after adoptive transfer with WT or Traf3ip3-KO BM cells transduced with pCLXSN-CFP (vector), pCLXSN-TRAF3IP3-CFP (IP3), or pCLXSN-TRAF3IP3ΔTM-CFP (ΔTM). (E) Flow cytometry analysis of CFP-positive donor thymocytes from Rag1-KO recipient mice adoptively transferred as described in D (for 4 wk). (F) Flow cytometry analysis of phosphorylated ERK in CFP-positive DP thymocytes derived from the BM chimeric mice of D, stimulated with anti-CD3 and anti-CD4 for 2 min. Data are presented as a representative FACS plot (top) and a summary graph of mean fluorescence intensity (MFI; bottom, MFI in the graph calculated after subtracting the background staining of unstimulated cells). Gray area, unstimulated CD4+CD8+ thymocytes. (G) Immunoblot analysis in Golgi extracts and whole-cell lysates (WCL) of DP thymocytes derived from the Rag1-KO BM chimeric mice adoptively transferred with retrovirus-transduced Traf3ip3-KO BM cells as described in D. Data are representative of four independent experiments with five mice per group in each experiment (D–G) or three independent experiments (B and C). Error bars show mean ± SEM. Significance was determined by two-tailed Student’s t test (*, P < 0.05).
DISCUSSION
The results described in this paper established TRAF3IP3 as a crucial regulator of thymocyte development. TRAF3IP3 specifically mediated TCR-stimulated activation of MEK and ERK. Consistent with the requirement of the MEK-ERK pathway in thymocyte development, we obtained genetic evidence that the attenuated MEK-ERK signaling in TRAF3IP3-deficient thymocytes critically contributed to their developmental defect.
Our data suggested a role for TRAF3IP3 in mediating the late stages of thymocyte development. Germline ablation of TRAF3IP3 did not affect the DN thymocyte development or the generation of immature DP thymocytes. In fact, the frequency of DP thymocytes was substantially elevated in the Traf3ip3-KO mice, consistent with a block in their progression to the mature SP stage. These results were corroborated by the finding that TRAF3IP3 ablation during DP stage of thymocyte development in the Traf3ip3-TKO mice also caused a defect in SP thymocyte generation. We obtained strong evidence that TRAF3IP3 was involved in positive selection of the DP thymocytes. Based on the analysis of CD69 expression, the TRAF3IP3 deficiency greatly reduced the populations of DP thymocytes that were undergoing, or had completed, positive selection. Similar results were obtained through analysis of another positive selection marker, CD5 (unpublished data). The role of TRAF3IP3 in mediating positive selection was further confirmed by using TCR transgenic mice. This specific function of TRAF3IP3 was correlated with its predominant expression in DP thymocytes.
In thymocytes, BRAF is largely responsible for TCR-stimulated MEK-ERK activation and required for thymocyte positive selection (Dillon et al., 2013). However, how BRAF transduces the TCR signal to the activation of downstream kinases has remained elusive. We found that BRAF rapidly moved to Golgi in response to TCR stimulation, suggesting Golgi-specific activation and/or function of this MEK kinase. Although how BRAF is recruited to the Golgi remains to be further studied, our findings suggest a TRAF3IP3-independent mechanism. TRAF3IP3 is also dispensable for the catalytic activation of BRAF. In contrast, TRAF3IP3 was required for MEK localization to the Golgi and TCR-stimulated MEK-BRAF physical association. It is thus likely that TRAF3IP3 may facilitate the interaction of BRAF with MEK in the Golgi, thereby allowing BRAF to phosphorylate and activate MEK. Indeed, MEK became phosphorylated in the Golgi upon TCR stimulation, which was dependent on TRAF3IP3. Thus, our findings provide an example for how the BRAF-MEK-ERK signaling cascade is activated in a specific subcellular compartment during T cell development. The sequence of TRAF3IP3 predicts a coiled-coil domain and a C-terminal transmembrane domain. We found that the transmembrane domain of TRAF3IP3 was indeed required for its association with membrane and localization to the Golgi apparatus. Importantly, the function of TRAF3IP3 in mediating MEK-ERK activation and thymocyte development also relied on its transmembrane domain. These findings shed additional light onto the mechanism by which TRAF3IP3 mediates TCR signaling in thymocytes.
A prior study suggests that overexpressed TRAF3IP3 induces JNK activation in cooperation with TRAF3 (Dadgostar et al., 2003). This function of TRAF3IP3 may involve its association with TRAF3 and recruitment of TRAF3 to detergent-insoluble cellular compartments (Dadgostar et al., 2003). Our preliminary studies failed to detect appreciable binding of TRAF3IP3 with TRAF3 in thymocytes (unpublished data). Furthermore, the TRAF3IP3 deficiency did not affect TCR-stimulated activation of JNK in thymocytes, although it remains possible that TRAF3IP3 is required for JNK activation in other cell types. Nevertheless, because TRAF3 is dispensable for thymocyte development (Xie et al., 2011), it is unlikely that the function of TRAF3IP3 in thymocytes is mediated by TRAF3. Instead, our data suggest a specific role for TRAF3IP3 in mediating MEK-ERK activation and DP thymocyte positive selection.
MATERIALS AND METHODS
Mice.
Traf3ip3-targeted mice, Traf3ip3tm1a(KOMP)Wtsi (in C57BL/6N background), were generated at Knockout Mouse Project (KOMP) by targeting exon 3 (first coding exon) of Traf3ip3 gene using a FRT-LoxP vector (Fig. 1 B). Germline Traf3ip3-KO mice were generated by crossing the Traf3ip3-targeted mice with CMV-Cre mice (The Jackson Laboratory, C57BL/6 background). Heterozygous (Traf3ip3+/−) mice were bred to generate age-matched WT and homozygous Traf3ip3-KO experimental mice. Traf3ip3-flox mice were generated by crossing the Traf3ip3-targeted mice with FLP deleter mice (Rosa26-FLPe; 129S4/Sv background; The Jackson Laboratory), and the Traf3ip3-flox mice were further crossed with Cd4-Cre mice (The Jackson Laboratory) to generate Traf3ip3-TKO and WT (Traf3ip3+/+Cd4-Cre) mice. Genotyping was performed as indicated in Fig. 1 (C and E) using primers listed in Table S1. B6.SJL mice (expressing the CD45.1 congenic marker), Rag1-KO mice, OT-I and OT-II TCR-transgenic mice, and R26StopFLMEK1DD (Map2k1*) mice were from the Jackson Laboratory. H-Y TCR transgenic mice were provided by M. Demetriou (University of California, Irvine, Irvine, CA). All of these mouse strains were in C57BL/6 background, except the Traf3ip3-TKO mice, which were in B6/129 mixed background. Mice were maintained in a specific pathogen–free facility, and all animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
Plasmids, antibodies, and reagents.
pcDNA-HA-TRAF3IP3 was created by inserting mouse TRAF3IP3 into the pcDNA3-HA vector; pCLXSN-TRAF3IP3-CFP and pCLXSN-TRAF3IP3ΔTM were generated by inserting the mouse TRAF3IP3 and TRAF3IP3ΔTM (lacking the C-terminal 25 amino acids containing a transmembrane domain), fused with CFP, into the pCLXSN retroviral vector (Naviaux et al., 1996). pCLXSN-CFP was generated by inserting CFP cDNA into pCLXSN, and mouse HA-MEK1 plasmid was purchased from Addgene. Antibodies for γ-Adaptin (M-300), ERK (K-23), GFP (B-2), IGF1R (111a9), JNK1 (C-17), NFATc1 (7A6), NFATc2 (4G6-G5), TRAF3IP3 (H-210), T3JAM (E-13, a goat anti-TRAF3IP3 used in the confocal experiment), RAF1 (C-20), BRAF (F-7), Lamin B (C-20), HSP60 (H1), Zap70 (1E7.2), Tubulin (TU-02), p-ERK (E-4), MEK1 (C-18), Lck (3A5), p38α/β (H-147), PLCγ (1249), and Calregulin (N-19) were from Santa Cruz Biotechnology, Inc. HRP-conjugated anti-HA antibody (3F10) was from Roche. Antibodies for phospho-IκBα (Ser32), phospho-JNK, phospho-PLCγ (Tyr319), phospho-MEK1/2 (Ser217/221), phospho-p38 (Thr180/Thr182), and phospho-Zap70 (Tyr319) (2701), as well as HRP-conjugated anti-MEK1/2 antibody, were from Cell Signaling Technology. Other antibodies for cell stimulation, immunoblot, and flow cytometry were as described previously (Zou et al., 2014).
Flow cytometry and intracellular cytokine staining.
Suspensions of thymocytes were stained with the indicated specific conjugated antibodies and subjected to flow cytometry and cell sorting as described previously (Reiley et al., 2007) using LSR II (BD) and FACSAria (BD) flow cytometers, respectively. The data were analyzed using FlowJo software. For intracellular cytokine staining of p-ERK, the anti-CD3/anti-CD4–stimulated thymocytes were fixed in Fix Buffer I (BD) for 10 min, followed by incubation in cold Perm Buffer III (BD) for 30 min, and then subjected to antibody staining and flow cytometry analyses.
Immunoblot, immunoprecipitation, and kinase assays.
Total cell lysates or subcellular extracts were prepared and subjected to immunoblot, coimmunoprecipitation, and in vitro kinase assays as described previously (Sun et al., 1994; Uhlik et al., 1998).
Cell culture and transfection.
HEK293 and Jurkat cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. For retroviral transduction, recombinant viral particles were produced by transiently transfecting HEK293 cells (using calcium method) with the indicated retroviral vectors along with packaging plasmids (pCL-Eco plus VSV-G for mouse BM cells and pCL-Ampho plus VSV-G for Jurkat T cells), as previously described (Cvijic et al., 2003; Nakaya et al., 2014).
BM transduction and adoptive transfer.
BM cells isolated from Traf3ip3-KO or WT mice (CD45.2+) were mixed with BM cells from B6.SJL (CD45.1+) mice (in 4:1 ratio) and adoptively transferred into irradiated (950 rad) Rag1-KO mice.
For retroviral transduction experiments, BM cells were collected from the Traf3ip3-KO and WT mice pretreated with 5-fluorouracil (150 mg/kg) for 48 h (to enhance the frequency of hematopoietic stem cells). After removal of red blood cells and CD90.2+ mature T cells, they were cultured in IMDM supplemented with 15% FBS in the presence of IL-3 (20 ng/ml), IL-6 (20 ng/ml), and stem cell factor (50 ng/ml) for 3 d. These BM cells were mixed with recombinant retroviruses encoding TRAF3IP3-CFP or TRAF3IP3ΔTM (in pCLXSN vector) in the presence of polybrene (8 µg/ml), cultured in the presence of IL-3, IL-6, and stem cell factor for an additional 24 h, and then mixed with viral supernatant again. The efficiency of viral transduction of BM cells was examined by monitoring CFP expression using fluorescence microscopy. Before adoptive transfer, transduced BM cells were harvested and washed once with IMDM without FBS. A total of 106 cells were injected i.v. via a tail vein into lethally irradiated (950 rad) Rag1-KO mice.
Subcellular fractionation.
Cytoplasmic and membrane (including plasma membrane and organelle membrane) fractions were prepared using a membrane protein extraction kit (Thermo Fisher Scientific), and Golgi fraction was isolated using a Golgi isolation kit (Sigma-Aldrich).
Confocal microscopy.
Sorted thymocytes were incubated in serum-free RPMI1640 medium for 30 min and then stimulated for 2 min with anti-CD3 (1 µg/ml) plus anti-CD4 (1 µg/ml) that had been premixed with a secondary cross-linking antibody: goat anti–mouse immunoglobulin G (10 µg/ml; H+L; Jackson ImmunoResearch Laboratories, Inc.). The cells were fixed and permeabilized for 20 min with Cytofix/Cytoperm solution (BD), blocked in 0.5% BSA in PBS, and then stained with primary antibody for 60 min, followed by staining with secondary Alexa Fluor 488 or 647 antibodies (Molecular Probes) for 30 min. Slides were mounted in antifade reagent with DAPI. Pictures were taken with an SP5 RS confocal microscope (Leica) and analyzed by SlideBook 5.0 software.
Statistical analysis.
Statistical analysis was performed using Prism software (GraphPad Software). Two-tailed unpaired Student’s t tests were performed, p-values <0.05 were considered significant, and the level of significance was indicated as follows: *, P < 0.05; **, P < 0.01. In the animal experiments, four mice are required for each group based on the calculation to achieve a 2.3-fold change (effect size) in two-tailed Student’s t test with 90% power and a significance level of 5%.
Online supplemental material.
Table S1 list the primers used for genotyping PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150110/DC1.
Supplementary Material
Acknowledgments
We thank M. Demetriou for H-Y transgenic mice. We also thank the personnel from the National Institutes of Health (NIH)/National Cancer Institute–supported resources (flow cytometry, DNA analysis, and animal facilities) under award number P30CA016672 at The MD Anderson Cancer Center.
This study was supported by grants from the NIH (AI057555, AI064639, GM84459, and AI104519).
The authors declare no competing financial interests.
Author contributions: Q. Zou designed and performed the experiments, prepared the figures, and wrote the manuscript. J. Jin, Y. Xiao, H. Hu, X. Zhou, Z. Jie, X. Xie, and X. Cheng contributed to the performance of the experiments. J.Y.H. Li provided key reagents. S.-C. Sun supervised the work and wrote the manuscript.
Footnotes
Abbreviations used:
- DN
- double negative
- DP
- double positive
- ERK
- extracellular signal-regulated kinase
- iNKT cell
- invariant NKT cell
- MEK
- mitogen/extracellular signal-regulated kinase
- SP
- single positive
- TKO
- T cell–conditional KO
References
- Alberola-Ila J., and Hernández-Hoyos G.. 2003. The Ras/MAPK cascade and the control of positive selection. Immunol. Rev. 191:79–96. 10.1034/j.1600-065X.2003.00012.x [DOI] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., and Carbone F.R.. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Cvijic M.E., Xiao G., and Sun S.C.. 2003. Study of T-cell signaling by somatic cell mutagenesis and complementation cloning. J. Immunol. Methods. 278:293–304. 10.1016/S0022-1759(03)00191-1 [DOI] [PubMed] [Google Scholar]
- Dadgostar H., Doyle S.E., Shahangian A., Garcia D.E., and Cheng G.. 2003. T3JAM, a novel protein that specifically interacts with TRAF3 and promotes the activation of JNK. FEBS Lett. 553:403–407. 10.1016/S0014-5793(03)01072-X [DOI] [PubMed] [Google Scholar]
- Daniels M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G.A., Gascoigne N.R., and Palmer E.. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 444:724–729. 10.1038/nature05269 [DOI] [PubMed] [Google Scholar]
- Dillon N., Kollias G., Grosveld F., and Williams J.G.. 1991. Expression of adult and tadpole specific globin genes from Xenopus laevis in transgenic mice. Nucleic Acids Res. 19:6227–6230. 10.1093/nar/19.22.6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon T.J., Takahashi M., Li Y., Tavisala S., Murray S.E., Moran A.E., Parker D.C., and Stork P.J.. 2013. B-Raf is required for positive selection and survival of DP cells, but not for negative selection of SP cells. Int. Immunol. 25:259–269. 10.1093/intimm/dxs104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.M., Katayama C.D., Pagès G., Pouysségur J., and Hedrick S.M.. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 23:431–443. 10.1016/j.immuni.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Germain R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. 10.1038/nri798 [DOI] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., and Carbone F.R.. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Hu Q., Nicol S.A., Suen A.Y., and Baldwin T.A.. 2012. Examination of thymic positive and negative selection by flow cytometry. J. Vis. Exp. (68):e4269 10.3791/4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P., Blüthmann H., Staerz U.D., Steinmetz M., and von Boehmer H.. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 333:742–746. 10.1038/333742a0 [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum R.L., Rouquette-Jazdanian A.K., and Samelson L.E.. 2013. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 34:259–268. 10.1016/j.it.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayinger P. 2011. Signaling at the Golgi. Cold Spring Harb. Perspect. Biol. 3:a005314 10.1101/cshperspect.a005314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie A.M., and Zúñiga-Pflücker J.C.. 2002. Regulation of thymocyte differentiation: pre-TCR signals and β-selection. Semin. Immunol. 14:311–323. 10.1016/S1044-5323(02)00064-7 [DOI] [PubMed] [Google Scholar]
- Nakaya M., Xiao Y., Zhou X., Chang J.H., Chang M., Cheng X., Blonska M., Lin X., and Sun S.C.. 2014. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 40:692–705. 10.1016/j.immuni.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Kasprowicz D.J., Yamashita M., Schubert L.A., Gillard G., Kimura M., Didierlaurent A., Koseki H., and Ziegler S.F.. 2002. The generation of mature, single-positive thymocytes in vivo is dysregulated by CD69 blockade or overexpression. J. Immunol. 168:87–94. 10.4049/jimmunol.168.1.87 [DOI] [PubMed] [Google Scholar]
- Naviaux R.K., Costanzi E., Haas M., and Verma I.M.. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès G., Guérin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., and Pouysségur J.. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 286:1374–1377. 10.1126/science.286.5443.1374 [DOI] [PubMed] [Google Scholar]
- Reiley W.W., Jin W., Lee A.J., Wright A., Wu X., Tewalt E.F., Leonard T.O., Norbury C.C., Fitzpatrick L., Zhang M., and Sun S.-C.. 2007. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204:1475–1485. 10.1084/jem.20062694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., and Ponting C.P.. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron U., and Rajewsky K.. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L., Sasaki Y., Calado D.P., Zhang B., Paik J.H., DePinho R.A., Kutok J.L., Kearney J.F., Otipoby K.L., and Rajewsky K.. 2009. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 139:573–586. 10.1016/j.cell.2009.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., and Hogquist K.A.. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176. 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- Sun S.-C., Ganchi P.A., Béraud C., Ballard D.W., and Greene W.C.. 1994. Autoregulation of the NF-kappa B transactivator RelA (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc. Natl. Acad. Sci. USA. 91:1346–1350. 10.1073/pnas.91.4.1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swat W., Dessing M., von Boehmer H., and Kisielow P.. 1993. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23:739–746. 10.1002/eji.1830230326 [DOI] [PubMed] [Google Scholar]
- Tsukamoto H., Irie A., Senju S., Hatzopoulos A.K., Wojnowski L., and Nishimura Y.. 2008. B-Raf-mediated signaling pathway regulates T cell development. Eur. J. Immunol. 38:518–527. 10.1002/eji.200737430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik M., Good L., Xiao G., Harhaj E.W., Zandi E., Karin M., and Sun S.-C.. 1998. NF-κB-inducing kinase and IkappaB kinase participate in human T-cell leukemia virus I Tax-mediated NF-κB activation. J. Biol. Chem. 273:21132–21136. 10.1074/jbc.273.33.21132 [DOI] [PubMed] [Google Scholar]
- Vanhecke D., Verhasselt B., De Smedt M., Leclercq G., Plum J., and Vandekerckhove B.. 1997. Human thymocytes become lineage committed at an early postselection CD69+ stage, before the onset of functional maturation. J. Immunol. 159:5973–5983. [PubMed] [Google Scholar]
- Wang L., Xiong Y., and Bosselut R.. 2010. Tenuous paths in unexplored territory: From T cell receptor signaling to effector gene expression during thymocyte selection. Semin. Immunol. 22:294–302. 10.1016/j.smim.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A., Wilson A., Nemir M., MacDonald H.R., and Radtke F.. 2002. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 16:869–879. 10.1016/S1074-7613(02)00330-8 [DOI] [PubMed] [Google Scholar]
- Xie P., Kraus Z.J., Stunz L.L., Liu Y., and Bishop G.A.. 2011. TNF receptor-associated factor 3 is required for T cell-mediated immunity and TCR/CD28 signaling. J. Immunol. 186:143–155. 10.4049/jimmunol.1000290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Nagata T., Tada T., and Nakayama T.. 1993. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol. 5:1139–1150. 10.1093/intimm/5.9.1139 [DOI] [PubMed] [Google Scholar]
- Zamoyska R., and Lovatt M.. 2004. Signalling in T-lymphocyte development: integration of signalling pathways is the key. Curr. Opin. Immunol. 16:191–196. 10.1016/j.coi.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Zou Q., Jin J., Hu H., Li H.S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., et al. 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 15:562–570. 10.1038/ni.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.