Abstract
Catharanthus roseus is an important medicinal plant, which produces a variety of indole alkaloids of significant pharmaceutical relevance. In the present study, we aimed to investigate the potential stress-induced increase of indole alkaloid biosynthesis in C. roseus using proteomic technique. The contents of the detectable alkaloids ajmalicine, vindoline, catharanthine, and strictosidine in C. roseus were significantly increased under binary stress. Proteomic analysis revealed that the abundance of proteins related to tricarboxylic acid cycle and cell wall was largely increased; while, that of proteins related to tetrapyrrole synthesis and photosynthesis was decreased. Of note, 10-hydroxygeraniol oxidoreductase, which is involved in the biosynthesis of indole alkaloid was two-fold more abundant in treated group compared to the control. In addition, mRNA expression levels of genes involved in the indole alkaloid biosynthetic pathway indicated an up-regulation in their transcription in C. roseus under UV-B irradiation. These results suggest that binary stress might negatively affect the process of photosynthesis in C. roseus. In addition, the induction of alkaloid biosynthesis appears to be responsive to binary stress.
Keywords: biosynthesis pathway, Catharanthus roseus, dark incubation, indole alkaloids, gel-free proteomics, UV-B irradiation
Introduction
Catharanthus roseus (C. roseus) produces a wide range of indole alkaloids, many of which are pharmaceutically important compounds (Ganapathi and Kargi, 1990). Vinblastine and vincristine are frequently used as cancer chemotherapeutics (Gidding et al., 1999). Aajmalicine was shown to have anti-hypertensive and anti-arrhythmias activities (Almagro et al., 2011). Vindoline and catharanthine displayed anti-bacterial activities, anti-diabetic properties, and diuretic actions (Chen et al., 2013). Given these biological activities, the indole alkaloids have been comprehensively investigated in the last two decades (Irie et al., 1987; O'Connor and Maresh, 2006), with C. roseus as an important and attractive resource for research of anticancer drugs.
Since most of the active components accumulate as secondary metabolites, these compounds are often present in trace quantities in plants, which hinders industrial-scale purification. The insufficiency of indole alkaloids from the available natural sources has led to the development of a number of alternative methods for their synthesis and production, including chemical synthesis (total synthesis or semi-synthesis), metabolic engineering, plant cell, and tissue culture (Hughes and Shanks, 2002). However, there are some major flaws in all of the current alternative methods.
Although chemical synthesis of alkaloids from C. roseus had been reported, it was not applicable for industrial-scale production due to its low productivity and high cost. In recent decades, various intermediates of indole alkaloids were identified (St-Pierre et al., 1999), and a series of key enzymes involved in the biosynthesis of indole alkaloids were cloned and characterized (Han et al., 2007). In addition, the metabolic network of indole alkaloids has been vigorously studied (Richer et al., 2006), making it possible to improve the content of alkaloids in C. roseus by metabolic engineering (van der Heijden et al., 2004). For example, overexpression of ORCA3 and G10H genes in C. roseus largely increased the accumulation of vindoline, catharanthine and ajmalicine (Pan et al., 2012). Using transgenic plants for the production of alkaloids has also been explored, however, genetic instability is a major hindrance in its application for commercial production (Koprek et al., 2001). Although large-scale C. roseus cell culture can also be used to produce the alkaloids, the levels of production were still insufficient for commercial production (Mujib et al., 2012). Moreover, due to the absence of certain enzymes, several important alkaloids such as the bisindole alkaloids, cannot be produced in cell cultures (Kutchan, 1995).
The rate of biosynthesis of secondary metabolites in plants is usually influenced by abiotic stress (Ramakrishna and Ravishankar, 2011). The content of catharanthine in C. roseus plant cell suspension was increased by exposure to a low dose of ultraviolet-B (UV-B) irradiation (Ramani and Chelliah, 2007). When C. roseus hairy roots were exposed to UV-B light for 20 min, the concentrations of lochnericine, serpentine, and ajmalicine were increased by 60, 20, and 50%, respectively (Binder et al., 2009). The reported results verified that UV-B was an elicitor of indole alkaloids. Nevertheless, the effect of UV-B irradiation on the content of indole alkaloids in C. roseus at the whole plant scale, which is the sole source for commercial production of alkaloids, has not yet bee investigated.
Recently, comparative proteomics has been widely applied in the investigation of biosynthetic mechanisms in plants. For C. roseus, the proteomic analysis of suspension cell had been carried out using two-dimensional electrophoresis, two-dimensional fluorescence difference gel electrophoresis, and gel-free approaches (Jacobs et al., 2005; Champagne et al., 2012). The results indicated that gel-based proteomic technique had the limitation to detect low-abundant proteins. Gel-free quantitative proteomics has since been rapidly developed, and is more useful for the identification of a large number of proteins (Neilson et al., 2011). This approach provides significantly more detailed proteome information on low-abundant proteins and might be more suitable for the investigation of proteins involved in secondary metabolism in plants.
In this study, the effects of UV-B irradiation and dark incubation on alkaloid contents, mRNA expression, and protein abundance were analyzed to investigate the influence of binary stress on the regulation of alkaloid biosynthesis. The content of the detectable alkaloids, ajmalicine, vindoline, catharanthine, and strictosidine as well as various physiological parameters were analyzed in the leaves of C. roseus after UV-B irradiation and dark incubation. Subsequently, the physiological parameters of C. roseus leaves were analyzed. To better understand the UV responsive mechanism in C. roseus, gel-free/label-free proteomics was performed. Moreover, the expression patterns of key genes involved in alkaloid biosynthesis were analyzed to gain further insights into the molecular mechanism of increased indole alkaloid biosynthesis in C. roseus.
Material and methods
Plant materials and treatments
Catharanthus roseus was provided from College of Pharmacy, Zhejiang University (Hangzhou, China). The plants were cultivated in a greenhouse for 45 days after sowing with temperature maintained at 25–28°C and 80% relative humidity. For UV-B irradiation, the light intensity of UV-B (1345.00 μW·cm−2) was measured from 275 to 320 nm with an UV radiometer (Beijing Normal University photoelectric instrument factory, Beijing, China). For the analysis of alkaloid content, the plants were exposed to UV-B for 1, 2, and 3 h, and then cultured for 72 h in darkness. The leaves were dried at 50°C in a drying oven and ground into a coarse powder for 1 min using a high-speed mixer. The coarse powder was screened into fine powder using a mesh with a 0.43 mm aperture. For the analysis of physiological parameters, the plants were exposed to UV-B for 1 h followed by a 72 h dark incubation prior to harvesting the leaves. For proteomic analysis, the plants were exposed to UV-B for 1 h followed by a 72 h dark incubation. Leaves were then collected, frozen in liquid nitrogen, and stored at −80°C. For quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis, the plants were exposed to UV-B for 0, 15, 30, and 60 min without dark incubation. The fresh leaves were frozen in liquid nitrogen and stored at −80°C. The control groups of all experiments were incubated in the dark for 72 h without prior UV-B irradiation. For all experiments, three independent experiments were performed as biological replicates (Supplementary Figure 1).
Quantitative analysis of alkaloids
Samples (1 g dry weight) were extracted in 100 mL of methanol using an ultrasonic extraction method (Tanabe et al., 1999) for three times (for 1 h each). The methanol extracts were dried in a rotary evaporator at 50°C. The residue was dissolved in 5 mL of methanol, and then filtered through a 0.45-μm filter (Millipore, Bullerica, MA, USA) for high performance liquid chromatography (HPLC) analysis. Several standard compounds, which consisted of strictosidine, vindoline, catharanthine, and ajmalicine were provided by Zhejiang Institute for Food and Drug Control (Hangzhou, China). For quantification, a calibration curve was constructed using the standard solutions diluted in methanol at six different concentrations: strictosidine (0.125, 0.250, 0.375, 0.500, 0.625, and 0.750 mg·mL−1), vindoline (0.181, 0.362, 0.543, 0.724, 0.905, and 1.086 mg·mL−1), catharanthine (0.098, 0.196, 0.294, 0.392, 0.490, and 0.588 mg·mL−1), and ajmalicine (0.035, 0.070, 0.105, 0.140, 0.175, and 0.209 mg·mL−1). For HPLC analysis, 20 μL of the standard solutions and samples were used.
HPLC analysis was performed on a Waters 2695 Alliance HPLC system (Waters, Milford, MA, USA) equipped with a photodiode array detector. The compounds were separated using reversed phase high performance liquid chromatography. A C18 column (250 × 4.6 mm, Waters) was used with a flow rate of 1 mL·min−1 at 40°C. Gradient elution was employed for qualitative and quantitative analyses using mobile phase of 0.025 M ammonium acetate solution and acetonitrile: 0–40 min (30–60% acetonitrile) and 41–50 min (60–80% acetonitrile). Spectra were measured at a wavelength of 220 nm, and peaks were determined by comparing the retention time and UV spectra with those of the standards.
Analysis of the physiological parameters
Photosynthetic pigment content was determined according to the procedures of Lichtenthaler and Wellburn (1983). Malondialdehyde (MDA) content was assayed (Heath and Packer, 1968) with minor modifications. In order to determine the MDA content, a portion (0.3 g fresh weight) of leaves was ground in 3 mL of 10% trichloroacetic acid and centrifuged at 5000 × g for 10 min. Then, 2 mL of 0.6% thiobarbituric acid was added to 2 mL of the obtained supernatant. The mixture was kept in boiling water for 15 min, and the absorbance of the solution was measured at wavelengths of 532, 450, and 600 nm.
The contents of soluble sugar and soluble protein were determined by the anthrone method (Fales, 1951) and the Lowry method (Lowry et al., 1951), respectively. The proline content was measured according to published method (Bates et al., 1973). Briefly, a portion (0.3 g) of leaves was homogenized in 3% aqueous sulfosalicylic acid. Then it was centrifuged at 5000 × g for 10 min. A reaction mixture consisting of 2 mL of supernatant, 2 mL of ninhydrin and 2 mL of glacial acetic acid was boiled at 100°C for 1 h. The resulting reaction mixture was extracted with 5 mL of toluene and the absorbance of the proline-ninhydrin chromophore was measured at 520 nm.
The superoxide dismutase (SOD) activity was assayed using the SOD assay kit-WST (Dojindo Molecular Technologies, Gaithersburg, MD, USA) according to the manufacturer's protocol. Briefly, the formation of a formazan dye upon reduction of the tetrazolium salt WST-1 (2-(4-iodophenyl)-3-(4-nitrophenly)-5-(2, 4-disulfophenyl)-2H-tetrazolium) with superoxide anions was measured. The mixture of the supernatant and enzyme working solution was incubated for 20 min at 37°C and the absorbance was measured at 450 nm.
Peroxidase (POD) activity was assayed using the POD assay kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's protocol. Briefly, a portion (0.3 g) of leaves was homogenized in 5 mL of 0.1 M phosphate buffer (pH 6.8) and then centrifuged at 15,000 × g for 15 min. The supernatant was collected as enzyme extract. A mixture of 0.125 mM phosphate buffer (pH 6.8), 0.05 mM pyrogallol, 0.05 mM hydrogen peroxide, and 1 mL of enzyme extract was incubated for 5 min at 25°C and the absorbance was measured at 420 nm.
The glutathione content was determined using a Reduced Glutathione Detection Kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's protocol. Briefly, a portion (0.3 g) of leaves was homogenized in 3 mL of 10% trichloroacetic acid containing 5 mM EDTA and was then centrifuged at 12,000 × g for 20 min. The supernatant was collected. Then, 1 mL of the obtained supernatant were added to the tubes containing 1 mL of 0.1 M phosphate buffer (pH 7.7), 20 μL of 40 mM 5, 5-dithiobis (2-nitrobenzoic acid), 20 μL of 10 mM NADPH. The reaction was started by the addition of 20 μL glutathione reductase. After mixing, the absorbance was monitored for 5 min at 412 nm.
Protein extraction
Samples (0.5 g fresh weight) were ground to powder in liquid nitrogen and then transferred into a polypropylene tube containing an acetone solution of 10% trichloroacetic acid and 0.07% 2-mercaptoethanol. The resulting mixture was vortexed and then sonicated for 5 min at 4°C. The suspension was incubated for 1 h at −20°C and vortexed every 15 min. It was then centrifuged at 9000 × g for 20 min at 4°C. The supernatant was discarded and the pellet was washed twice with 0.07% 2-mercaptoethanol in acetone. The final pellet was dried and re-suspended in lysis buffer, consisting of 7 M urea, 2 M thiourea, 5% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate, and 2 mM tributylphosphine, by vortexing for 1 h at 25°C. The suspension was then centrifuged at 20,000 × g for 20 min at room temperature until a clean supernatant was obtained. The protein concentrations were determined using the Bradford method (Bradford, 1976) with bovine serum albumin as the standards.
Protein purification and digestion for mass spectrometry analysis
Proteins (100 μg) were purified with methanol and chloroform to remove any detergent from the sample solutions. Briefly, 400 μL of methanol was added to the sample, and the resulting solution was mixed. Subsequently, 100 μL of chloroform and 300 μL of water were added to the samples, which were mixed and centrifuged at 20,000 × g for 10 min to achieve phase separation. The upper phase was discarded, and 300 μL of methanol was added slowly to lower phase. The samples were centrifuged at 20,000 × g for 10 min, supernatants were discarded, and pellets were dried. The dried pellets were resuspended in 50 mM NH4HCO3. The proteins were reduced with 50 mM dithiothreitol for 1 h at 56°C and alkylated with 50 mM iodoacetamide for 1 h at 37°C in the dark. Alkylated proteins were digested with trypsin and lysyl endopeptidase at 1:100 enzyme/protein concentrations for 16 h at 37°C. The resulting peptides were acidified with formic acid to pH < 3, and the resulting solution was centrifuged at 20,000 × g for 10 min. The obtained supernatant was collected and analyzed by nanoliquid chromatography (LC)-mass spectrometry (MS).
Nanoliquid chromatography-tandem mass spectrometry analysis
Peptides were analyzed on a nanospray LTQ XL Orbitrap MS (Thermo Fisher Scientific, San Jose, CA, USA) operated in data-dependent acquisition mode with the installed XCalibur software (version 2.0.7, Thermo Fisher Scientific). Using an Ultimate 3000 NanoLC system (Dionex, Germering, Gemany), peptides in 0.1% formic acid were loaded onto a C18 PepMap trap column (300 μm ID × 5 mm, Dionex). The peptides were eluted from the trap column with a linear acetonitrile gradient (8–30% for 120 min) in 0.1% formic acid and at a flow rate of 200 nL·min−1. The peptides that were eluted from the trap column were separated on a C18 capillary tip column (75 μm ID × 120 mm, Nikkyo Technos, Tokyo, Japan) and ionized at a spray voltage of 1.5 kV. Full-scan mass spectra were acquired in the LTQ Orbitrap mass spectrometer over 400–1500 m/z with a resolution of 30,000. A lock mass function was used for high mass accuracy (Olsen et al., 2005). The 10 most intense precursor ions were selected for collision-induced fragmentation in the linear ion trap at normalized collision energy of 35%. Dynamic exclusion was employed within 90 s to prevent repetitive selection of peptides (Zhang et al., 2009).
Protein identification from acquired mass spectrometry data
Identification of proteins was performed using the Mascot search engine (version 2.5.1; Matrix Science, London, UK) with the NCBInr-Viridiplantae database (2, 564, 738 sequences) obtained from the NCBI database (http://www.nibi.nlm.nih.gov/protein). The acquired raw data files were processed using Proteome Discoverer (version 1.4.0.288; Thermo Fisher Scientific). The parameters used in Mascot searches were as follows: carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine was set as a variable modification. Trypsin was specified as the proteolytic enzyme and one missed cleavage was allowed. Peptide mass tolerance was set at 10 ppm, fragment mass tolerance was set at 0.8 Da, and the peptide charge was set at +2, +3, and +4. An automatic decoy database search was performed as part of the search. Mascot results were filtered with Mascot Percolator to improve the accuracy and sensitivity of peptide identification (Brosch et al., 2009). False discovery rates for peptide identification of all search strategies were < 1.0%. Peptides with a percolator ion score of more than 13 (p < 0.05) were used for protein identification. The acquired Mascot results were exported into the SIEVE software (version 2.1.377; Thermo Fisher Scientific) for differential analysis.
Differential analysis of proteins using mass spectrometry data
Proteins that contained common peptides were grouped. Both unique and shared peptides were analyzed for protein quantification. Peptides that are shared between proteins were only included in the quantitative information for the protein that contained the most number of assigned peptides to ensure that a peptide was only used once for quantification. The chromatographic peaks detected by MS were aligned, and the peptide peaks were detected as frames on all parent ions scanned by MS/MS using 5 min of frame time width and 10 ppm of frame m/z width. In the differential analysis of protein abundance, total ion current was used for normalization. Chromatographic peak areas within a frame were compared for each sample, and the ratios between samples were determined for each frame. The frames detected in the MS/MS scan were matched to the imported Mascot results. The peptide ratios between samples were determined from the variance-weighted average of the ratios in frames that matched the peptides in the MS/MS spectrum. The ratios of peptides were further integrated to determine the ratios of the corresponding proteins. For identification of differentially changed proteins, the minimum requirements for the identification of differentially changed proteins were: at least six peptide sequence matches above the identity threshold with more than 10% sequence coverage; the number of matched peptides with more than two peptides; and a fold change of more or less than 1.2 times in protein quantities in the treated samples against the control with a significant difference (p < 0.05).
RNA extraction and quantitative reverse transcription-polymerase chain reaction analysis
Samples (0.1 g fresh weight) were ground to powder in liquid nitrogen using a sterilized mortar and pestle. Total RNA was extracted from the tissue powder using a Quick RNA Isolation Kit (Huayueyang Biotechnology, Beijing, China). RNA was reverse-transcribed using a Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer's protocol. Primers were designed using the Primer Premier 5.0 (Supplementary Table 1). The qRT-PCR was performed in a 10 μL reaction using a QuantiFast SYBR Green PCR Kit (Qiagen, Hilden, Germany) on an IQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The relative quantification method (2−ΔΔCT) was used to evaluate quantitative variation between treatments. 40S Ribosomal protein S9 (RPS9) was used as a single reference gene (Menke et al., 1999).
Functional categorization of identified proteins
Protein functions were categorized using MapMan bin codes (http://mapman.gabipd.org/), as previously described (Usadel et al., 2005). A small-scale prediction of the identified proteins from C. roseus was performed by transferring annotations from the Arabidopsis genome and consideration of orthologous genes.
Statistical analysis
SPSS 19.0 (IBM, Chicago, IL, USA) statistical software was used for the statistical evaluation of the results. One-Way ANOVA followed by Tukey's multiple comparison post-hoc tests and the Student's t-test was also performed when only two groups were compared. All results were shown as mean ± SD from three independent biological replicates. A p value of less than 0.05 was considered statistically significant.
Results
The contents of indole alkaloids in C. roseus leaf increased under binary stress
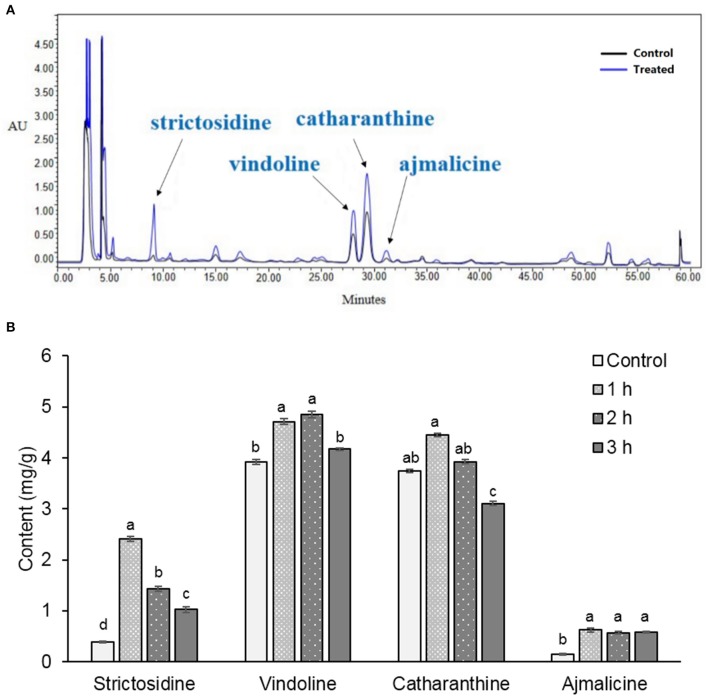
To profile changes in indole alkaloids biosynthesis in C. roseus plant under stresses, the indole alkaloids in C. roseus leaves were analyzed. Methanol extracts of C. roseus leaves were examined by HPLC analysis. Based on retention times and ultraviolet absorbance of the standards, the four alkaloids containing strictosidine, ajmalicine, vindoline, and catharanthine were identified in leaves. The contents of alkaloids in C. roseus leaves increased after 1 h of UV-B irradiation followed by 72 h of dark incubation (Figure 1A). Time-course quantitative analysis of the induced alkaloids demonstrated that 1 h of UV-B irradiation coupled with 72-h of dark incubation were the optimal induction conditions, under which the total contents of the four compounds were at maximum (Figure 1B). Compared with the control, the contents of strictosidine, ajmalicine, vindoline, and catharanthine in C. roseus leaves after 1 h of UV-B irradiation and 72 h of dark incubation increased by 527.9, 321.6, 20.1, and 19.0%, respectively (Supplementary Table 2).
Figure 1.
Pattern of metabolites in leaves of C. roseus. Methanol extracts from leaves collected from 45-day-old C. roseus plants were analyzed by HPLC (A). Arrowheads show the positions of the identified alkaloids. Control: C. roseus plants were only incubated for 72 h in the dark (black line). Treated: C. roseus plants were irradiated by UV-B for 1 h and incubated for 72 h in the dark (blue line). The content of the alkaloids in C. roseus under 1, 2, and 3 h UV-B irradiation times were calculated from standard curves (B). Data were shown as means ± SD from three independent biological replicates. Means with the same letter are not significantly different according to One-Way ANOVA test (p < 0.05). Control, plants with only 72 h of dark incubation.
Quantitative proteomic analysis reveals identities of proteins that were responsive to binary stress
Gel-free proteomic technique was employed to reveal changes in protein abundance that accounted for the increase in alkaloid contents in C. roseus leaves. The leaves that were treated with or without 1 h of UV-B irradiation followed by 72 h of dark incubation were collected. Proteins were extracted, reduced, alkylated, and digested. The tryptic peptides were collected. Principle component analysis of the peptide profile for total proteins was performed for quality assessment of the experimental inputs (Supplementary Figure 2). A total of 90 proteins with more than 2 matched peptides were identified as being differentially changed (fold change > 1.2 and p < 0.05). Out of these 90 proteins, the accumulation level of 24 proteins was decreased and that of 66 proteins was increased significantly in C. roseus leaves upon UV-B irradiation and dark incubation (Table 1, Supplementary Table 3). These differential proteins were categorized into 18 groups (Figure 2). The abundance of proteins related to oxidative pentose phosphate, tricarboxylic acid (TCA) cycle, cell wall, glycolysis, transport, amino acid metabolism, secondary metabolism, C1-metabolism, major CHO metabolism, mitochondrial electron transport, and signaling were increased after UV-B irradiation and dark incubation. In contrast, the abundance of proteins related to tetrapyrrole synthesis were decreased. The abundance of proteins related to photosynthesis, redox ascorbate/glutathione metabolism, stress, and protein synthesis followed either an increasing or decreasing trend. Additionally, the abundance of 10-hydroxygeraniol oxidoreductase (10-HGO) in C. roseus leaves, which is involved in the pathway of indole alkaloid biosynthesis, was 2 times higher than that of the control group.
Table 1.
Total proteins changed between control and UV-B treatment by gel-free proteomics.
| Protein ID | Orthologa | Description | M.P.b | Ratioc | Functional categoryd |
|---|---|---|---|---|---|
| INCREASE | |||||
| GI:30693656 | At3g52560 | Ubiquitin-conjugating enzyme E2 variant 1D [Arabidopsis thaliana] | 2 | 2.16 | Protein |
| GI:159471081 | At3g02230 | UDP-Glucose:protein transglucosylase [Chlamydomonas reinhardtii] | 2 | 2.03 | Cell wall |
| GI:33519154 | – | 10-hydroxygeraniol oxidoreductase [Camptotheca acuminata] | 2 | 1.98 | Secondary metabolism |
| GI:357471289 | At-105710 | Aconitate hydratase [Medicago truncatula] | 2 | 1.96 | TCA |
| GI:21536725 | At5g41670 | 6-phosphogluconate dehydrogenase [Arabidopsis thaliana] | 4 | 1.94 | OPP |
| GI:448872690 | At3g02230 | Alpha-1,4-glucan-protein synthase [Elaeis guineensis] | 3 | 1.93 | Cell wall |
| GI:402228002 | At4g37990 | Cinnamyl alcohol dehydrogenase [Fragaria × ananassa] | 2 | 1.88 | Secondary metabolism |
| GI:508706435 | At5g09590 | Mitochondrial HSO70 2 isoform 2 [Theobroma cacao] | 2 | 1.87 | Stress |
| GI:525507286 | At3g52880 | Monodehydroascorbate reductase, seedling isozyme [Cucumis sativus] | 2 | 1.86 | Redox |
| GI:195633817 | At4g21280 | Oxygen-evolving enhancer protein 3-1 [Zea mays] | 2 | 1.83 | Photosynthesis |
| GI:473888752 | At5g03290 | Isocitrate dehydrogenase [NAD] catalytic subunit 5 [Triticum urartu] | 2 | 1.82 | TCA |
| GI:297807587 | At5g15650 | Reversibly glycosylated polypeptide_3 [Arabidopsis lyrata subsp. lyrata] | 2 | 1.77 | Cell wall |
| GI:351727547 | At3g61440 | OAS-TL3 cysteine synthase [Glycine max] | 3 | 1.77 | Amino acid metabolism |
| GI:162463414 | At5g15650 | Alpha-1,4-glucan-protein synthase [UDP-forming] [Zea mays] | 2 | 1.76 | Cell wall |
| GI:297802428 | At4g35220 | Cyclase family protein [Arabidopsis lyrata subsp. lyrata] | 2 | 1.74 | DNA |
| GI:351722265 | At3g02360 | 6-phosphogluconate dehydrogenase [Glycine max] | 2 | 1.73 | OPP |
| GI:213493066 | At1g65930 | NADP-dependent isocitrate dehydrogenase [Passiflora edulis] | 2 | 1.72 | TCA |
| GI:285309965 | At4g35830 | Aconitate hydratase 1 [Citrus clementina] | 3 | 1.72 | TCA |
| GI:213959111 | At3g12580 | Heat shock protein 70 [Oryza sativa Japonica Group] | 2 | 1.72 | Stress |
| GI:508707706 | At5g13420 | Aldolase-type TIM barrel family protein isoform 2, partial [Theobroma cacao] | 2 | 1.72 | OPP |
| GI:357491797 | At1g65930 | Isocitrate dehydrogenase [Medicago truncatula] | 2 | 1.63 | TCA |
| GI:18414289 | At4g14710 | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Arabidopsis thaliana] | 2 | 1.63 | Amino acid metabolism |
| GI:285309969 | At2g05710 | Aconitate hydratase 2 [Citrus clementina] | 2 | 1.62 | TCA |
| GI:475618083 | At5g15650 | Alpha-1,4-glucan-protein synthase (UDP-forming) [Aegilops tauschii] | 3 | 1.61 | Cell wall |
| GI:197717740 | At1g09210 | Calreticulin [Nicotiana tabacum] | 2 | 1.59 | Signaling |
| GI:297800722 | At4g14880 | O_acetylserine (thiol) lyase (oas_tl) isoform A1 [Arabidopsis lyrata] | 2 | 1.55 | Amino acid metabolism |
| GI:508778858 | At3g12580 | Heat shock protein 70B [Theobroma cacao] | 2 | 1.53 | Stress |
| GI:15218869 | At1g65930 | Isocitrate dehydrogenase [Arabidopsis thaliana] | 3 | 1.51 | TCA |
| GI:315440254 | At3g60100 | Mitochondrial citrate synthase [Pyrus pyrifolia] | 4 | 1.51 | TCA |
| GI:285309967 | At4g26970 | aconitate hydratase 3 [Citrus clementina] | 3 | 1.46 | TCA |
| GI:474119554 | At4g38510 | V-type proton ATPase subunit B 1 [Triticum urartu] | 2 | 1.45 | Transport |
| GI:357439825 | At4g13430 | 3-isopropylmalate dehydratase [Medicago truncatula] | 2 | 1.44 | TCA |
| GI:357445271 | At5g49910 | Heat shock protein [Medicago truncatula] | 2 | 1.43 | Stress |
| GI:351726325 | At3g09640 | L-ascorbate peroxidase 2 [Glycine max] | 2 | 1.43 | Redox |
| GI:308808864 | At1g78900 | Vacuolar H__ATPase V1 sector_ subunit A (ISS) [Ostreococcus tauri] | 2 | 1.42 | Transport |
| GI:390098820 | At2g11270 | Citrate synthase [Capsicum annuum] | 2 | 1.41 | TCA |
| GI:37903393 | At4g09000 | 14-3-3-like protein [Saccharum hybrid cultivar CP65-357] | 2 | 1.41 | Signaling |
| GI:381145559 | At3g53230 | cell division cycle protein 48 [Camellia sinensis] | 2 | 1.4 | Cell |
| GI:297819360 | At5g01530 | chlorophyll a/b_binding protein [Arabidopsis lyrata subsp. lyrata] | 2 | 1.38 | Photosynthesis |
| GI:374433978 | At3g14420 | glycolate oxidase [Wolffia australiana] | 3 | 1.38 | Photosynthesis |
| GI:18409740 | At3g52990 | pyruvate kinase [Arabidopsis thaliana] | 2 | 1.38 | Glycolysis |
| GI:214010947 | At2g13360 | serine glyoxylate aminotransferase 3 [Glycine max] | 2 | 1.37 | Photosynthesis |
| GI:308812360 | At3g12110 | ACT_COLSC Actin (ISS) [Ostreococcus tauri] | 2 | 1.37 | Cell |
| GI:7547401 | At5g40810 | cytochrome c1 precursor [Solanum tuberosum] | 2 | 1.37 | mitoETC |
| GI:302850070 | At2g07698 | F1F0 ATP synthase_ subunit alpha_ mitochondrial [Volvox carteri f. nagariensis] | 2 | 1.37 | mitoETC |
| GI:15231937 | At3g08580 | ADP,ATP carrier protein 1 [Arabidopsis thaliana] | 2 | 1.34 | Transport |
| GI:297807205 | At5g11670 | NADP_malic enzyme 2 [Arabidopsis lyrata subsp. lyrata] | 2 | 1.32 | TCA |
| GI:508710478 | At5g14780 | Formate dehydrogenase [Theobroma cacao] | 2 | 1.32 | C1-metabolism |
| GI:302127766 | At1g53310 | C3 phosphoenolpyruvate carboxylase [Panicum bisulcatum] | 3 | 1.32 | Glycolysis |
| GI:42416979 | At4g33010 | glycine dehydrogenase P protein [Oryza sativa Indica Group] | 4 | 1.31 | Photosynthesis |
| GI:474012573 | At5g02500 | Heat shock cognate 70 kDa protein 1 [Triticum urartu] | 4 | 1.3 | Stress |
| GI:940877 | At5g14590 | Isocitrate dehydrogenase (NADP+) [Solanum tuberosum] | 2 | 1.29 | TCA |
| GI:195605636 | At3g55440 | triosephosphate isomerase, cytosolic [Zea mays] | 2 | 1.29 | Glycolysis |
| GI:15229784 | At3g43810 | calmodulin 7 [Arabidopsis thaliana] | 2 | 1.28 | Signaling |
| GI:15226854 | At2g02010 | glutamate decarboxylase 4 [Arabidopsis thaliana] | 2 | 1.27 | Amino acid metabolism |
| GI:4239891 | At1g79750 | NADP-malic enzyme [Aloe arborescens] | 2 | 1.26 | TCA |
| GI:473968259 | At1g32060 | Phosphoribulokinase, chloroplastic [Triticum urartu] | 2 | 1.26 | Photosynthesis |
| GI:226500740 | At2g19860 | hexokinase-1 [Zea mays] | 2 | 1.26 | Major CHO metabolism |
| GI:162134229 | AtMg01190 | ATP synthase F1 subunit alpha [Trebouxia aggregata] | 2 | 1.25 | mitoETC |
| GI:15219234 | At1g78900 | V-type proton ATPase catalytic subunit A [Arabidopsis thaliana] | 7 | 1.24 | Transport |
| GI:15233115 | At3g54890 | light-harvesting complex I chlorophyll a/b binding protein 1 [Arabidopsis thaliana] | 3 | 1.23 | Photosynthesis |
| GI:297812157 | At5g19760 | dicarboxylate/tricarboxylate carrier [Arabidopsis lyrata subsp. lyrata] | 2 | 1.23 | Transport |
| GI:195658441 | At1g78900 | vacuolar ATP synthase catalytic subunit A [Zea mays] | 3 | 1.23 | Transport |
| GI:240254125 | At1g20260 | V-type proton ATPase subunit B3 [Arabidopsis thaliana] | 5 | 1.23 | Transport |
| GI:474369193 | At1g23490 | ADP-ribosylation factor 2 [Triticum urartu] | 2 | 1.22 | Protein |
| GI:224127848 | At4g10340 | light-harvesting complex II protein Lhcb5 [Populus trichocarpa] | 2 | 1.21 | Photosynthesis |
| DECREASE | |||||
| GI:18411711 | At3g60750 | transketolase [Arabidopsis thaliana] | 2 | 0.83 | Photosynthesis |
| GI:297794397 | At5g66190 | ferredoxin_NADP_ reductase [Arabidopsis lyrata subsp. lyrata] | 2 | 0.82 | Photosynthesis |
| GI:78675163 | AtCg00270 | photosystem II protein D2 [Lactuca sativa] | 2 | 0.82 | Photosynthesis |
| GI:307136095 | At5g66190 | ferredoxin–NADP reductase [Cucumis melo subsp. melo] | 3 | 0.82 | Photosynthesis |
| GI:297840973 | AtCg00340 | photosystem I P700 chlorophyll a apoprotein A2 [Arabidopsis lyrata subsp. lyrata] | 2 | 0.81 | Photosynthesis |
| GI:511348322 | AtCg00130 | ATP synthase CF0 subunit I (chloroplast) [Catharanthus roseus] | 2 | 0.81 | Photosynthesis |
| GI:27448410 | AtCg00490 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Selaginella wrightii] | 2 | 0.79 | Photosynthesis |
| GI:374975197 | AtCg00490 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Neoregelia carolinae] | 4 | 0.79 | Photosynthesis |
| GI:413968524 | At1g31330 | photosystem I subunit III precursor (chloroplast) [Solanum tuberosum] | 2 | 0.77 | Photosynthesis |
| GI:89280643 | AtCg00490 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Solanum lycopersicum] | 2 | 0.76 | Photosynthesis |
| GI:428697696 | AtCg00490 | ribulose 1_5_bisphosphate carboxylase/oxygenase large subunit [Pellia endiviifolia] | 2 | 0.76 | Photosynthesis |
| GI:443329645 | AtCg00490 | ribulose 1_5_bisphosphate carboxylase/oxygenase large subunit [Equisetum hyemale] | 3 | 0.76 | Photosynthesis |
| GI:18412632 | At4g04640 | ATP synthase gamma chain 1 [Arabidopsis thaliana] | 2 | 0.75 | Photosynthesis |
| GI:224179477 | AtCg00490 | ribulose_1_5_bisphosphate carboxylase/oxygenase large subunit [Monomastix sp.] | 4 | 0.75 | Photosynthesis |
| GI:139389650 | AtCg00490 | large subunit of riblose-1,5-bisphosphate carboxylase/oxygenase [Arabis hirsuta] | 3 | 0.7 | Photosynthesis |
| GI:508781449 | At4g04640 | ATPase, F1 complex, gamma subunit protein [Theobroma cacao] | 2 | 0.66 | Photosynthesis |
| GI:508777181 | At1g62750 | Translation elongation factor EFG/EF2 protein [Theobroma cacao] | 2 | 0.57 | Protein |
| GI:474036467 | At5g45930 | Magnesium-chelatase subunit chlI, chloroplastic [Triticum urartu] | 3 | 0.55 | Tetrapyrrole synthesis |
| GI:148763638 | At5g45930 | Magnesium chelatase 40-kDa subunit [Hordeum vulgare subsp. vulgare] | 2 | 0.52 | Tetrapyrrole synthesis |
| GI:297804102 | At4g20360 | Chloroplast elongation factor tub [Arabidopsis lyrata subsp. lyrata] | 2 | 0.42 | Protein |
| GI:300681420 | At3g55800 | sedoheptulose-1,7-bisphosphatase, chloroplast precursor, expressed [Triticum aestivum] | 2 | 0.36 | Photosynthesis |
Ortholog, indicates AGI E.C.code.
M.P., indicates number of matched peptide, the proteins with >2 matched peptides and p-value < 0.05 were considered.
Ratio, indicates ratio of quantities of protein (UV-B treated group/control group), the proteins with ratio 1.2/0.8 were considered.
“Functional category” indicates protein function categorized using MapMan bin codes.
Figure 2.
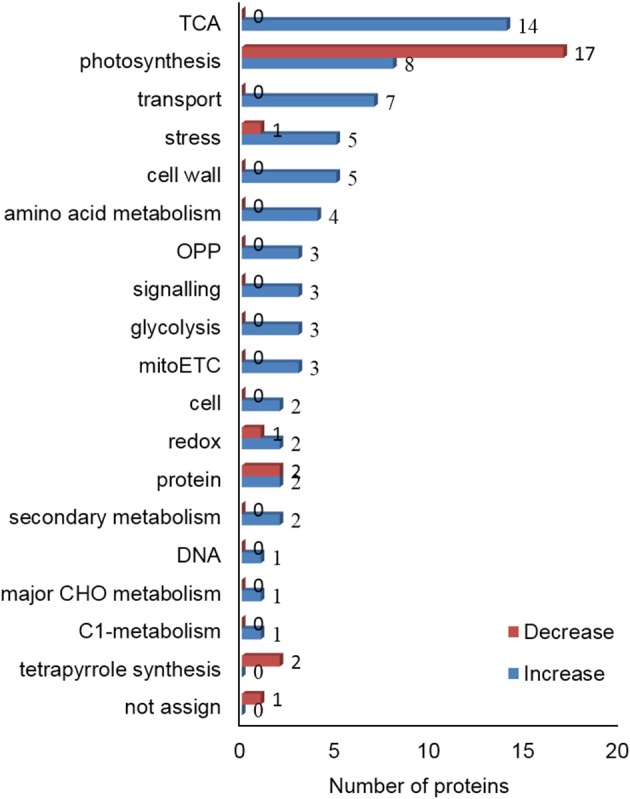
Functional categorization of differential proteins in leaves of C. roseus treated with UV-B and dark stress. C. roseus plants were irradiated with or without 1 h of UV-B irradiation and 72 h of dark incubation. Leaves were collected and proteins were then extracted, digested and analyzed by nanoLC-MS/MS. The identified proteins were categorized using MapMan bin codes. Numbers of categorized proteins are shown in the graph. Blue bars and red bars indicate increased and decreased levels of proteins. The abbreviations are as follows: TCA, tricarboxylic acid cycle; OPP, oxidative pentose phosphate; mitoETC, mitochondrial electron transport chain; cell, cell organization/vesicle transport; redox, redox ascorbate/glutathione metabolism; protein, protein synthesis/posttranslational modification/folding/degradation/activation; DNA, DNA synthesis/repair; CHO, carbohydrates; and C1-metabolism, carbon 1-metabolism.
Expression of genes involved in the biosynthesis of indole alkaloid in C. roseus was up-regulated in response to binary stress
To uncover the molecular mechanism of increased indole alkaloid in C. roseus upon exposure to binary stress, qRT-PCR analysis was performed. Forty five-day-old C. roseus were treated with UV-B irradiation for 0, 15, 30, and 60 min and RNAs extracted from leaves were analyzed by qRT-PCR. The nine genes and one transcription factor containing tryptophan decarboxylase (tdc), geraniol-10-hydroxylase (g10h), 10-hydroxygeraniol oxidoreductase (10hgo), secologanin synthase (sls), strictosidine synthase (str), strictosidine β-glucosidase (sgd), tabersonine 16-hydroxylase (t16h), deacetoxyvindoline 4-hydroxylase (d4h), 6-17-O-deacetylvindoline O-acetyltransferase (dat), and Octadecaniod-derivative Responsive Catharanthus AP2-domain Protein 3 (ORCA3), which are involved in the biosynthesis pathway of the alkaloids, were selected for further analysis of mRNA expression levels (Supplementary Figure 3). RPS9 was selected as a reference gene (Supplementary Table 1). Among the examined genes, the mRNA expression levels of dat, t16h, d4h, ORCA3, str, g10h, and 10-hgo were up-regulated in leaves under UV-B stress. In addition, the expression levels of t16h, ORCA3, and str in C. roseus leaf after UV-B irradiation were four times higher than those of the control. The levels of sgd, sls, and tdc were up-regulated after 30-min of UV-B irradiation, but down-regulated after 60 min of UV-B irradiation (Figure 3).
Figure 3.
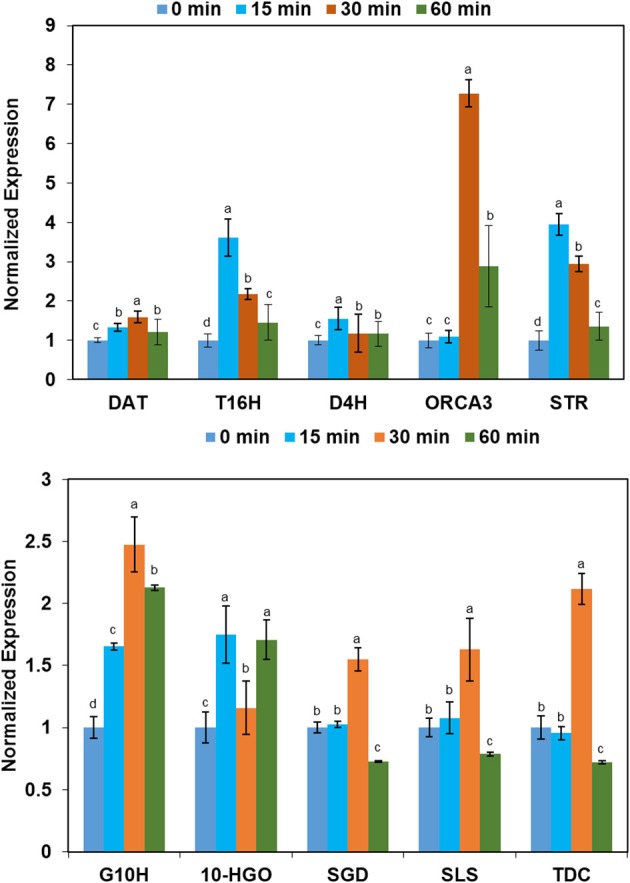
The variation in expression tendency of the selected genes of leaves derived from C. roseus under conditions of different UV-B exposure. The leaves of C. roseus plants were collected at 0, 15, 30, and 60 min UV-B irradiation. Total RNA was extracted from the collected leaves. The transcript abundance of the selected genes was analyzed by qRT-PCR. Data were shown as means ± SD from three independent biological replicates. Means with the same letter are not significantly different according to One-Way ANOVA test (p < 0.05). The abbreviations are follows: the genes tdc, g10h,10-hgo, sls, str, sgd, t16h, d4h, dat, and ORCA3 code for tryptophan decarboxylase,geraniol-10-hydroxylase, 10-hydroxygeraniol oxidoreductase, secologanin synthase, strictosidine synthase, strictosidine β-glucosidase, tabersonine 16-hydroxylase, deacetoxyvindoline 4-hydroxylase, 6-17-O-deacetylvindoline O-acetyltransferase and Octadecaniod-derivative Responsive Catharanthus AP2-domain Protein 3, respectively. The RPS9 gene was used as a reference control gene.
The physiological activity of C. roseus was changed under binary stress
Physiological parameters were determined to reflect the physiological activities of C. roseus leaves after UV-B irradiation and dark incubation. The leaves of C. roseus plants that were treated with or without 1 h of UV-B irradiation and 72 h of dark incubation were collected and homogenized in buffer solution. After centrifugation, the supernatants were collected for the analysis of physiological parameters. After UV-B irradiation and dark incubation, the contents of soluble protein, soluble sugar, MDA, and proline were decreased by 5.6, 7.5, 24, and 19.2%, respectively; while, the content of glutathione was increased by 33%. Compared with the control, the activities of SOD and POD were increased by 21.4 and 124%, respectively. In contrast, the activity of glutathione reductase was decreased by 31.4% (Table 2). However, the content of photosynthetic pigments did not show a significant change after UV-B irradiation and dark incubation (Supplementary Table 4).
Table 2.
Changes of physiological parameters in C. roseus leaves.
| Soluble sugar (mg/g) | Soluble protein (mg/g) | SOD [U/mg(pro)] | GRa [U/g(pro)] | POD [U/(g.min)] | MDA (umol/g) | Proline (ug/g) | GSHb(mg/g) | |
|---|---|---|---|---|---|---|---|---|
| Control | 5.288 ± 0.067 | 8.517 ± 0.058 | 122.361 ± 2.465 | 39.360 ± 2.622 | 1255.093 ± 20.789 | 0.0071 ± 0.0001 | 35.880 ± 1.363 | 0.058 ± 0.002 |
| Treated | 4.990 ± 0.027* | 7.879 ± 0.167* | 148.501 ± 1.671* | 27.009 ± 2.462* | 2810.691 ± 46.463* | 0.0054 ± 0.0004* | 29.005 ± 1.373* | 0.077 ± 0.003* |
Significant at P < 0.05;
GR, glutathione reductase;
GSH, glutathione.
Discussion
The increase in indole alkaloid biosynthesis in C. roseus is induced by binary stress
Although UV-B serves as an adverse environmental factor for the growth and development of plants (Caldwell et al., 2007), it has been shown to induce a variety of bioactive secondary metabolites, such as indole alkaloids, in medicinal plants (Ramani and Chelliah, 2007). Indole alkaloids provide protection against microbial infection, herbivores consumption, and abiotic environmental stresses (Cordell, 2013). In this study, the contents of the indole alkaloids strictosidine, vindoline, catharanthine, and ajmalicine in C. roseus leaves were increased under the binary stress of UV-B irradiation and dark incubation. Both UV-B irradiation and dark incubation are essential stresses for alkaloid induction (Zhang et al., 2014). Additionally, it is known that reactive oxygen species (ROS) are largely released under conditions of high-energy irradiation of UV-B, resulting in oxidative injury and disturbance of metabolism in plants (Apel and Hirt, 2004). It is likely that the alkaloids are accumulated to enhance the scavenging capacity of ROS. Moreover, the enhanced activities of the enzymatic and non-enzymatic antioxidant systems also indicate the accumulation of ROS.
The accumulation of 10-HGO, which is a key enzyme involved in the alkaloid biosynthesis pathway, provides further insights into the induction of the indole alkaloids. 10-HGO also plays a key role in the secoiridoid pathway and catalyzes the synthesis of secologanin which is one of the substrates for strictosidine synthase (Valletta et al., 2010). The other substrate for strictosidine synthase is tryptamine, which is derived from tryptophan metabolism via the shikimate pathway. Strictosidine synthase catalyzes the condensation of secologanin and tryptamine to yield strictosidine, which is the key precursor for the biosynthesis of indole alkaloids in C. roseus (Yamazaki et al., 2003). In the present study, 10-HGO was two-fold more abundant in the treated group compared to the control group, which may explain the increased amount of strictosidine observed after stress treatment.
To verify the mechanism behind the change in the alkaloid at the transcript level, 10 key genes, which are involved in indole alkaloids biosynthesis, were selected for investigation by qRT-PCR. The genes tdc, g10h, 10-hgo, sls, and str encode the key enzymes that catalyze the biosynthesis of strictosidine from tryptophan and geraniol, which is a key intermediate in indole alkaloid biosynthesis (Patra et al., 2013). ORCA3 is a transcription factor that plays a key role in controlling several important key genes involved in indole alkaloid biosynthesis (Goklany et al., 2013). The overexpression of ORCA3 increases the expression of indole alkaloid biosynthetic genes, leading to the accumulation of indole alkaloids (van der Fits and Memelink, 2000). In the present study, the continual up-regulated expression of tdc, g10h, 10-hgo, sls, and str was consistent with the increased content of strictosidine in C. roseus under stress. Similarly, the up-regulated expression of sgd, d4h, dat, ORCA3, and t16h may have contributed to the observed increasing contents of ajmalicine, vindoline, and catharanthine under binary stress. Furthermore, the variation tendency in gene expression under stresses was the same as that of alkaloids. Taken together, our results indicate that the genes involved in the biosynthesis of indole alkaloids are induced under binary stress, which may account for the increased contents of indole alkaloids in C. roseus.
Enhanced tricarboxylic acid cycle in C. roseus might be related to the biosynthesis of secondary metabolites
The abundance of aconitate hydratase, iscocitrate dehydrogenase, and mitochondrial citrate synthase which were involved in the TCA cycle, was increased in C. roseus under UV-B irradiation and dark incubation. The TCA cycle which is composed of a set of eight enzymes can oxidize pyruvate and malate to form CO2 and nicotinamide adenine dinucleotide (NADH) and provide intermediates for metabolic pathways (Nunes-Nesi et al., 2013). Aconitase hydratase which is involved in the TCA cycle plays a vital role in nutrient metabolism and energy supply (Zhang et al., 2003). NADP dependent iscocitrate dehydrogenase is related to the synthesis of 2-oxoglutarate for ammonia assimilation and glutamate in plants (Palomo et al., 1998). Additionally, citrate synthase catalyzes the condensation reaction to form citrate (Mulholland and Richards, 1997). Functioning with the described key enzymes, the TCA cycle provides ATP for the fundamental physiological activities of plants (Fernie et al., 2004). In C. roseus leaves, the enhanced TCA cycle may serve to provide energy and intermediates for secondary metabolism. Furthermore, the increase in abundance of proteins related to primary metabolism, such as acetylserine lyase, cysteine synthase, and aldolase-type barrel family protein isoforms, and the resulting accumulation of the primary metabolites, might promote the biosynthesis of secondary metabolites.
Fundamental physiology of C. roseus was influenced by binary stress
UV-B can induce plant photomorphogenic development which is characterized by the inhibition of flavonoid accumulation and UV-B stress tolerance (Huang et al., 2014). Low-intensity UV-B radiation can act as entraining signal for the circadian clock (Feher et al., 2011). Photomorphogenic UV-B signaling also controls plant immune response (Demkura and Ballare, 2012). Photosynthesis has been reported as a direct and most affected system in plants upon UV-B stress, which frequently results in reduced abundance of photosynthesis related proteins (Strid et al., 1994; Kanalas et al., 2008). In the present study, the abundance of proteins related to photosynthesis such as sedoheptulose-1, 7-bisphosphatase, chlorophyll a/b binding protein, serine glyoxylate aminotransferase 3, ribulose bisphosphate carboxylase/oxygenase, and light-harvesting complex II protein were decreased in C. roseus leaves. It is known that chlorophyll can be synthesized via a common tetrapyrrole biosynthetic pathway. In our study, the abundance of two tetrapyrrole synthesis proteins were also decreased after UV stress. Moreover, this observation was consistent with the content of the photosynthetic pigment observed in C. roseus leaves. This evidence suggests that binary stress might negatively affect the process of photosynthesis in C. roseus.
Cinnamyl alcohol dehydrogenases is a zinc-dependent dehydrogenase, responsible for catalyzing the reversible conversion of p-hydroxycinnamaldehydes to the corresponding alcohols in lignin biosynthesis (Blanco-Portales et al., 2002). Lignin is one of the most important components in plant cell walls, which protects cellulose in the cell walls of plants by adhering to hydrolytic enzymes (Moura et al., 2010). UDP-glucose: protein transglucosylase (UPTG) catalyzes the synthesis of α-1, 4-glucan, which is related to starch biosynthesis and plays a crucial role in the synthesis of cell wall polysaccharides (Bocca et al., 1999). In addition it has been shown to relate to cell wall compound metabolism and contribute to cell wall thickening (Ge et al., 2007). In this study, the E. guineensis and in Z. mays orthologs of alpha-1,4-glucan-protein synthase were identified (Supplementary Table 3). The abundance of cinnamyl alcohol dehydrogenases and proteins related to cell wall such as alpha-1,4-glucan-protein synthase, reversibly glycosylated polypeptide, and UPTG were increased compared to the control group. These results suggest that the increase of cell wall formation is a response to binary stress in C. roseus.
Furthermore, heat shock proteins (HSP) are produced by cells in response to environmental stress conditions in plants (Wang et al., 2004). HSP-70 is widely distributed in plants and is responsible for preventing aggregation and assisting refolding of non-native proteins under stressful conditions (Frydman, 2001). Some studies found that HSP 70 participates in photo-protection and the process of photosystem II repair (Schroda et al., 1999). The up-regulated Hsp70 level may be involved in the modulation of signal transduction and maintenance of key functions of basic cellular proteins after UV-B exposure and dark stress conditions such as damaged photosystem II centers from UV-B irradiation.
Concluding remarks
In conclusion, our data indicated that the increase of indole alkaloids production in C. roseus leaves under binary stress was a consequence of metabolic pathways alteration. Comparative gel-free proteomics revealed significant proteome difference in C. roseus leaves between control and treated groups. The proteins related to cell wall and secondary metabolism in C. roseus leaves might be parts of the defense system against injuries resulted from binary stress. The alteration in secondary metabolism, might account for the increase in indole alkaloids observed in C. roseus leaves after stress exposure. In addition, the enhanced TCA cycle might affect the biosynthesis of indole alkaloid in C. roseus. Finally, the data derived from this study is highly applicable to other medicinal plant and provides important insights for effectively improving the contents of alkaloids in medicinal plants for industrial production.
Accession codes
The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaino et al., 2013) with the dataset identifier PXD001937.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. L. Sun at Zhejiang University for her valuable discussions. This work was supported by the National Science Foundation of China (Grant No. 81274033), the Research Project of Chinese Ministry of Education (Grant No. 113037A), and the Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ14H280001).
Glossary
Abbreviations
- HPLC
high performance liquid chromatography
- MDA
Malondialdehyde
- POD
peroxidase
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- TCA
tricarboxylic acid
- 10-HGO
10-hydroxygeraniol oxidoreductase.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00582
References
- Almagro L., Perez A. J. L., Pedreno M. A. (2011). New method to enhance ajmalicine production in Catharanthus roseus cell cultures based on the use of cyclodextrins. Biotechnol. Lett. 33, 381–385. 10.1007/s10529-010-0430-6 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Bates L. S., Waldren R. P., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF0001806020688380 [DOI] [Google Scholar]
- Blanco-Portales R., Medina-Escobar N., Lopez-Raez J. A., Gonzalez-Reyes J. A., Villalba J. M., Moyano E., et al. (2002). Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase from strawberry. J. Exp. Bot. 53, 1723–1734. 10.1093/jxb/erf029 [DOI] [PubMed] [Google Scholar]
- Binder B. Y. K., Peebles C. A. M., Shanks J. V., San K. Y. (2009). The effects of UV-B Stress on the production of terpenoid indole alkaloids in Catharanthus roseus hairy roots. Biotechnol. Prog. 25, 861–865. 10.1002/btpr.97 [DOI] [PubMed] [Google Scholar]
- Bocca S. N., Kissen R., Rojas-Beltran J. A., Noel F., Gebhardt C., Moreno S., et al. (1999). Molecular cloning and characterization of the enzyme UDP-glucose: protein transglucosylase from potato. Plant Physiol. Biochem. 37, 809–819. 10.1016/S0981-9428(99)00117-5 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of proteindye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brosch M., Yu L., Hubbard T., Choudhary J. (2009). Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome Res. 8, 3176–3181. 10.1021/pr800982s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell M. M., Bornman J. F., Ballare C. L., Flint S. D., Kulandaivelu G. (2007). Terrestrial ecosystems, increased solar ultraviolet irradiation, and interactions with bother climate change factors. Photochem. Photobiol. Sci. 6, 252–266. 10.1039/B700019G [DOI] [PubMed] [Google Scholar]
- Champagne A., Rischer H., Oksman-Caldentey K. M., Boutry M. (2012). In-depth proteome mining of cultured Catharanthus roseus cells identifies candidate proteins involved in the synthesis and transport of secondary metabolites. Proteomics 12, 3536–3547. 10.1002/pmic.201200218 [DOI] [PubMed] [Google Scholar]
- Chen Q. H., Zhang W. P., Zhang Y. L., Chen J., Chen Z. L. (2013). Identification and quantification of active alkaloids in Catharanthus roseus by liquid chromatography-ion trap mass spectrometry. Food Chem. 139, 845–852. 10.1016/j.foodchem.2013.01.088 [DOI] [PubMed] [Google Scholar]
- Cordell G. A. (2013). Fifty years of alkaloid biosynthesis in Phytochemistry. Phytochemistry 91, 29–51. 10.1016/j.phytochem.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Demkura P. V., Ballare C. L. (2012). UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 5, 642–652. 10.1093/mp/sss025 [DOI] [PubMed] [Google Scholar]
- Fales F. W. (1951). The assimilation and degradation of carbohydrates by yeast cells. J. Biol. Chem. 193, 113–124. [PubMed] [Google Scholar]
- Feher B., Kozma-Bognar L., Kevei E., Hajdu A., Binkert M., Davis S. J., et al. (2011). Functional interaction of the circadian clock and UV resistance locus 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67, 37–48. 10.1111/j.1365-313X.2011.04573.x [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Carrari F., Sweetlove L. J. (2004). Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261. 10.1016/j.pbi.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Frydman J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647. 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- Ganapathi B., Kargi F. (1990). Recent advances in indole alkaloid production by Catharanthus-Roseus (Periwinkle). J. Exp. Bot. 41, 259–267. 10.1093/jxb/41.3.25916716935 [DOI] [Google Scholar]
- Ge C. L., Wan D. Z., Wang Z. G., Ding Y., Wang Y. L., Shang Q., et al. (2007). Response of rice roots to 1, 2, 4-trichlorobenzene stress. Acta Agronom. Sin. 33, 1991–2000. 10.3321/j.issn:0496-3490.2007.12.012 [DOI] [Google Scholar]
- Gidding C. E. M., Kellie S. J., Kamps W. A., de Graaf S. S. N. (1999). Vincristine revisited. Crit. Rev. Oncol. Hematol. 29, 267–287. 10.1016/S1040-8428(98)00023-7 [DOI] [PubMed] [Google Scholar]
- Goklany S., Rizvi N. F., Loring R. H., Cram E. J., Lee-Parsons C. W. T. (2013). Jasmonate-dependent alkaloid biosynthesis in Catharanthus roseus hairy root cultures is correlated with the relative expression of Orca and Zct transcription factors. Biotechnol. Progr. 29, 1367–1376. 10.1002/btpr.1801 [DOI] [PubMed] [Google Scholar]
- Han M., Zhao B., An Z. G., Rausch T., Zu Y. G. (2007). Cloning and expression of cDNA encoding key enzymes (DXR, SLS, G10H and STR) in terpene indole alkaloids biosynthesis pathway from Catharanthus roseus. B. Bot. Res. 27, 564–568. 10.1007/s00425-012-1712-0 [DOI] [Google Scholar]
- Heath R. L., Packer L. (1968). Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. 10.1016/0003-9861(68)90654-1 [DOI] [PubMed] [Google Scholar]
- Huang X., Yang P., Ouyang X., Chen L., Deng X. W. (2014). Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 10:e1004218. 10.1371/journal.pgen.1004218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E. H., Shanks J. V. (2002). Metabolic engineering of plants for alkaloid production. Metab. Eng. 4, 41–48. 10.1006/mben.2001.0205 [DOI] [PubMed] [Google Scholar]
- Irie K., Hagiwara N., Tokuda H., Koshimizu K. (1987). Structure-activity studies of the indole alkaloid tumor promoter teleocidins. Carcinogenesis 8, 547–552. [PubMed] [Google Scholar]
- Jacobs D. I., Gaspari M., van der Greef J., van der Heijden R., Verpoorte R. (2005). Proteome analysis of the medicinal plant Catharanthus roseus. Planta 221, 690–704. 10.1007/s00425-004-1474-4 [DOI] [PubMed] [Google Scholar]
- Kanalas P., Veres S., Levai L., Sarvari E. (2008). Interactive effects of UV-B irradiation and water stress on leaf water relations and photosynthesis of sessile oak (Quercus petraea L.). Cereal Res. Commun. 36, 303–306. 10.1556/CRC.36.2008.Suppl.1 [DOI] [Google Scholar]
- Koprek T., Rangel S., McElroy D., Louwerse J. D., Williams-Carrier R. E., Lemaux P. G. (2001). Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barely. Plant Physiol. 125, 1354–1362. 10.1104/pp.125.3.1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchan T. M. (1995). Alkaloid biosynthesis-The basis for metabolic engineering of medicinal plants. Plant Cell 7, 1059–1070. 10.1105/tpc.7.7.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. K., Wellburn A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 603, 591–592. [Google Scholar]
- Lowry O. H., Rosenbrough N. J., Farr A. L., Randall R. J. (1951). Protein measurement with folin phenol reagent. J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- Menke F. L. H., Parchmann S., Mueller M. J., Kijne J. W., Memelink J. (1999). Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol. 119, 1289–1296. 10.1104/pp.119.4.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujib A., Ilah A., Aslam J., Fatima S., Siddiqui Z. H., Maqsood M. (2012). Catharanthus roseus alkaloids: application of biotechnology for improving yield. Plant Growth Regul. 68, 111–127. 10.1007/s10725-012-9704-4 [DOI] [Google Scholar]
- Mulholland A. J., Richards W. G. (1997). Acetyl-CoA enolization in citrate synthase: a quantum mechanical/molecular mechanical (QM/MM) study. Proteins 27, 9–25. 10.1002/(SICI)1097-0134(199701) [DOI] [PubMed] [Google Scholar]
- Moura J. C. M. S., Bonine C. A. V., Viana J. D. F., Dornelas M. C., Mazzafera P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. 10.1111/j.1744-7909.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Neilson K. A., Ali N. A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., et al. (2011). Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11, 535–553. 10.1002/pmic.201000553 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araujo W. L., Obata T., Fernie A. R. (2013). Regulation of the mitochondrial tricarboxylic acid cycle. Curr. Opin. Plant Biol. 16, 335–343. 10.1016/j.pbi.2013.01.004 [DOI] [PubMed] [Google Scholar]
- O'Connor S. E., Maresh J. J. (2006). Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep. 23, 532–547. 10.1039/B512615K [DOI] [PubMed] [Google Scholar]
- Olsen J. V., de Godoy L. M. F., Li G. Q., Macek B., Mortensen P., Pesch R., et al. (2005). Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021. 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- Palomo J., Gallardo F., Suarez M. F., Canovas F. M. (1998). Purification and characterization of NADP-linked isocitrate dehydrogenase from scots pine. Plant Physiol. 118, 617–626. 10.1104/pp.118.2.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q. F., Wang Q., Yuan F., Xing S. H., Zhao J. Y., Choi Y. H., et al. (2012). Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-Metabolomics. PLoS ONE 7:e43038. 10.1371/journal.pone.0043038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B., Schluttenhofer C., Wu Y. M., Pattanaik S., Yuan L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 1829, 1236–1247. 10.1016/j.bbagrm.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Ramakrishna A., Ravishankar G. A. (2011). Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 6, 1720–1731. 10.4161/psb.6.11.17613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani S., Chelliah J. (2007). UV-B-induced signaling events leading to enhanced-production of catharanthine in Catharanthus roseus cell suspension cultures. BMC Plant Biol. 7:61. 10.1186/1471-2229-7-61corde [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer H., Oresic M., Seppanen-Laakso T., Katajamaa M., Lammertyn F., Ardiles-Diaz W., et al. (2006). Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl. Acad. Sci. U.S.A. 103, 5614–5619. 10.1073/pnas.0601027103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Wollman F. A., Beck C. F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165–1178. 10.1105/tpc.11.6.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B., Vazquez-Flota F. A., De Luca V. (1999). Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11, 887–900. 10.1105/tpc.11.5.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strid A., Chow W. S., Anderson J. M. (1994). UV-B damage and protection at the molecular level in plants. Photosynth. Res. 39, 475–489. 10.1007/BF00014600 [DOI] [PubMed] [Google Scholar]
- Tanabe A., Suzuki S., Hanada Y. (1999). Evaluation of an ultrasonic extraction method compared with the soxhlet extraction method for the determination of polyaromatic hydrocarbons and n-alkanes in airborne particulate matter. Bunseki Kagaku 48, 939–944. 10.2116/bunsekikagaku.48.939 [DOI] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138, 1195–1204. 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta A., Trainotti L., Santamaria A. R., Pasqua G. (2010). Cell-specific expression of tryptophan decarboxylase and 10-hydroxygeraniol oxidoreductase, key genes involved in camptothecin biosynthesis in Camptotheca acuminate Decne (Nyssaceae). BMC Plant Biol. 10:69 10.1186/1471-2229-10-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. 10.1126/science.289.5477.295 [DOI] [PubMed] [Google Scholar]
- van der Heijden R., Jacobs D. I., Snoeijer W., Hallared D., Verpoorte R. (2004). The Catharanthus Alkaloids: pharmacognosy and biotechnology. Curr. Med. Chem. 11, 607–628. 10.2174/0929867043455846 [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., et al. (2013). The proteomics identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41, D1063–D1069. 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. X., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–253. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Sudo Y., Yamazaki M., Aimi N., Saito K. (2003). Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: cloning, characterization, and differential expression in tissues and by stress compounds. Plant Cell Physiol. 44, 395–403. 10.1093/pcp/pcg051 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhu W., Zhang Y., Yang B., Fu Z., Li X., et al. (2014). Proteomics analysis of Mahonia bealei leaves with induction of alkaloids via combinatorial peptide ligand libraries. J. Proteomics 110, 59–71. 10.1016/j.jprot.2014.07.036 [DOI] [PubMed] [Google Scholar]
- Zhang W. L., Shen W. B., Ye M. B., Xu L. L. (2003). Aconitase and its physiological roles in plants. Plant Physiol. Commun. 39, 391–398. 10.13592/j.cnki.ppj.2003.04.040 [DOI] [Google Scholar]
- Zhang Y., Wen Z., Washburn M. P., Florens L. (2009). Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 81, 6317–6326. 10.1021/ac9004887 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.