Abstract
A healthy and independent life requires skeletal muscles to maintain optimal function throughout the lifespan, which is in turn dependent on efficient activation of processes that regulate muscle development, homeostasis, and metabolism. Thus, identifying mechanisms that modulate these processes is of crucial priority. Noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), have emerged as a class of previously unrecognized transcripts whose importance in a wide range of biological processes and human disease is only starting to be appreciated. In this review, we summarize the roles of recently identified miRNAs and lncRNAs during skeletal muscle development and pathophysiology. We also discuss several molecular mechanisms of these noncoding RNAs. Undoubtedly, further systematic understanding of these noncoding RNAs' functions and mechanisms will not only greatly expand our knowledge of basic skeletal muscle biology, but also significantly facilitate the development of therapies for various muscle diseases, such as muscular dystrophies, cachexia, and sarcopenia.
1. Introduction
One of the biggest surprises from the human genome project is that, in contrast to large predicted gene numbers, our genome contains only 20,000~25,000 protein-coding genes, which account for merely ~1.5% of the whole genome [1]. Over the last decade, it has been gradually accepted that the remaining genomic information, originally considered “noise” or “dark matter,” is not “junk DNA” after all. Through comprehensive analyses of mammalian transcriptomes, a vast amount of non-protein-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), have recently been identified. They are emerging as integral components of the gene regulatory networks in a broad range of biological processes, and dysregulation of their expression has been implicated in many human diseases [2, 3]. miRNAs are a class of small noncoding RNAs (approximately 22 nucleotides long) that are evolutionarily conserved from plants to mammals. Generally, miRNAs negatively regulate their targets at the posttranscriptional level by promoting mRNA degradation and/or repressing translation [2]. On the other hand, lncRNAs are transcripts normally longer than 200 nucleotides in length, which do not appear to have protein-coding potential [3]. Only recently discovered, they represent a new class of ncRNAs that have also been implicated in a large spectrum of biological processes.
Skeletal muscle consists of about one-third of our body mass and is the largest tissue in our body. Attached to bones through tendons, skeletal muscle is not only responsible for generating voluntary movement, but also very adaptive and actively participates in metabolism of the whole body. Skeletal muscle is also known for its remarkable ability to regenerate after injury through the use of satellite cells, the endogenous muscle stem cells [4–6]. Satellite cells usually stay quiescent in resting muscle; however, once activated, they can reenter the cell cycle, proliferate, and initiate the myogenic differentiation program. Over the past few decades, a wealth of knowledge has been accumulated regarding the molecular regulatory networks of muscle development and pathophysiology [4, 7, 8]. However, we are still at the beginning of understanding the roles of ncRNAs in skeletal muscle biology. Although aberrant expression of both miRNAs and lncRNAs has been associated with various muscle disorders, such as muscular dystrophies, the functions and mechanisms of these ncRNAs remain unclear [9–12]. Therefore, understanding the functions of miRNAs and lncRNAs during skeletal muscle development and under pathophysiological conditions will greatly expedite the development of therapeutic treatments for many muscle disorders. Here, we briefly summarize the known functions of miRNAs and lncRNAs in skeletal muscle development and pathophysiology.
2. miRNAs, Small Yet Mighty Regulators
2.1. miRNA Biogenesis and Function
More than 1,000 miRNAs are encoded in the human genome. They are abundantly expressed in many cell types and are estimated to target roughly 60% of mammalian genes [2]. miRNAs are either transcribed independently from their own transcriptional units or cotranscribed with the host genes in which they are embedded. The biogenesis pathway of miRNAs has been well documented and is evolutionarily conserved. Briefly, immature primary transcripts containing a stem-loop structure (pri-miRNAs) are initially transcribed by RNA polymerase II [13]. Pri-miRNAs are processed by the Drosha/DGCR8 endonuclease complex into ~70 nucleotides precursor miRNAs (pre-miRNAs) inside the nucleus. The pre-miRNAs are then exported to the cytoplasm, where they are further processed by the Dicer/TRBP endonuclease complex, resulting in imperfect RNA duplexes [14–16]. The mature miRNA duplexes are then separated and incorporated into the RNA-induced silencing complex (RISC), where they bind to the 3′ untranslated regions (UTRs) of their target mRNAs [17, 18]. Thus, miRNAs contribute to posttranscriptional regulation of gene expression by translational inhibition and/or target mRNA degradation. Generally, nucleotides 2–8 of miRNAs, termed the “seed sequence,” are essential for target specificity and binding. Many miRNAs are clustered into families based on their seed sequence; thus, miRNAs with the same seed sequence may target the same set of genes, providing the possibility of functional redundancy and cooperation among different miRNAs [19–21].
miRNA-mediated gene regulation is a complex and well-orchestrated process. Unlike transcriptional factor-mediated regulation of gene expression, which appears to be “on or off,” miRNAs tend to only moderately regulate the overall level of their target's expression, so they are referred to as “fine-tuners” [21]. Although the effect of a single miRNA on a specific gene may appear to be small, the combinatory effect of miRNAs on multiple mRNA targets functioning within the same biological pathway can be synergistic and sometimes dramatic [22, 23]. In addition, a mRNA normally possesses multiple miRNA-binding sites in its 3′ UTR and is likely a target for numerous different miRNAs. Therefore, this reciprocal multiplicity between miRNAs and mRNAs increases not only the complexity but also the robustness of the miRNA regulatory network. This, to some extent, may also explain why knockouts of many miRNAs, when deleted individually in animal models, do not result in apparent defects.
However, it should be noted that this moderate regulation of gene expression does not necessary suggest that miRNAs are dispensable. Perhaps the most convincing evidence supporting the importance of miRNAs in mammals is the embryonic lethal phenotype of Dicer knockout mice [24]. Dicer is an endonuclease, encoded at a single locus in mammals, which is required for generating biologically active mature miRNAs. Dicer-deficient mice die at the gastrulation stage before the embryo body is fully developed, demonstrating that functionally mature miRNAs are critical for early mammalian development [24]. More specifically, the importance of miRNAs in skeletal muscle has also been demonstrated through a Dicer conditional loss-of-function study in skeletal muscle. Using a MyoD-Cre recombinase transgene to remove a conditional Dicer allele in skeletal muscle, Harfe and colleagues found that collective loss of functional miRNAs in skeletal muscle resulted in muscle hypoplasia, increased apoptosis, and perinatal lethality [25], clearly demonstrating the critical role of miRNAs in skeletal muscle. Here we will discuss the function of some known miRNAs in skeletal muscle development (myogenic differentiation), muscle physiology (fiber types, hypertrophy/atrophy), and the pathophysiology of muscle diseases.
2.2. miRNA in Skeletal Muscle Development (Myogenic Differentiation)
Since the first report of a myogenic regulatory miRNA by our and other groups [26–28], increasing numbers of miRNAs have been identified as able to modulate myogenesis. In Table 1, we list the miRNAs that have been implicated in myogenesis, along with their identified target(s). Due to space limitations, here we will only summarize some well-characterized miRNAs with validated targets in myogenic differentiation.
Table 1.
miRNAs and targets implicated in the development of skeletal muscle.
| miRNA | miRNA target(s) | Function in myogenic differentiation | References |
|---|---|---|---|
| miR-1 | HDAC4 Pax7 |
Promote differentiation Inhibit proliferation |
[26, 33] |
| miR-133 | SRF UCP2 |
Promote proliferation Promote differentiation |
[26, 110] |
| miR-206 | Polα1, Cx43; Pax3/7, Fstl1, and Utrn; Notch3 and Igfbp5 | Promote differentiation Inhibit proliferation |
[33–35, 41, 63, 64, 111] |
| miR-23a | Myh | Inhibit differentiation | [112] |
| miR-24 | Smad 3 | Promote differentiation | [59] |
| miR-26a | Ezh2 Smad 1 and Smad 4 |
Promote differentiation | [48, 49] |
| miR-27 | Pax3 Myostatin |
Promote satellite cell activation Promote differentiation | [37, 57, 58] |
| miR-29 | HDAC4, YY1, Ring1, Rybp, Akt3, Col, and Lims1 | Promote differentiation, inhibit proliferation, and fibrosis | [50–52, 58, 113, 114] |
| miR-31 | Myf5 | Maintenance of quiescence/stemness | [39] |
| miR-124 | Dlx5 | Inhibit differentiation | [115] |
| miR-125b | IGF-II | Inhibit differentiation | [116] |
| miR-128a | Insr; IRS1 and Pik3r1 | Inhibit proliferation | [117] |
| miR-148a | ROCK1 | Promote differentiation | [118] |
| miR-146b | Smad4, Notch1, and Hmga2 | Promote differentiation | [119] |
| miR-155 | MEF2A | Inhibit differentiation | [120] |
| miR-181 | Hox-A11 | Promote differentiation | [32] |
| miR-186 | Myogenin | Inhibit differentiation | [31] |
| miR-195/497 | Cdc25, Ccnd | Maintenance of quiescence/stemness | [43] |
| miR-199a-3p | IGF-1, mTOR, and RPS6KA6 | Inhibit differentiation | [121] |
| miR-199a-5p | FZD4, JAG1, and WNT2 | Promote proliferation | [99] |
| miR-203 miR-203b | c-JUN, MEF2C, and MyoD | Inhibit proliferation and differentiation | [122, 123] |
| miR-208b miR-499 |
Sox6, Purβ, Sp3, and HP-1β | Fiber type determination | [67] |
| miR-214 | Ezh2 | Promote differentiation | [47, 124] |
| miR-221/222 | p27, MyoD | Inhibit differentiation | [125, 126] |
| miR-322/424 | Cdc25A | Promote differentiation | [44] |
| miR-351 | E2f3 | Promote proliferation | [127] |
| miR-378 | MyoR | Promote differentiation | [128] |
| miR-486 | Pax7 | Promote differentiation | [38] |
| miR-489 | Dek | Maintenance of quiescence/stemness | [129] |
| miR-503 | Cdc25A | Inhibit proliferation | [44] |
| miR-669 | MyoD | Inhibit proliferation | [130] |
| miR-675 | Smad1, Smad5, and Cdc6 | Promote differentiation | [60] |
| miR-682 | Unknown | Promote proliferation | [131] |
| miR-1192 | HMGB1 | Inhibit differentiation | [132] |
| miR-3906 | Homer-1b | Promote differentiation | [133] |
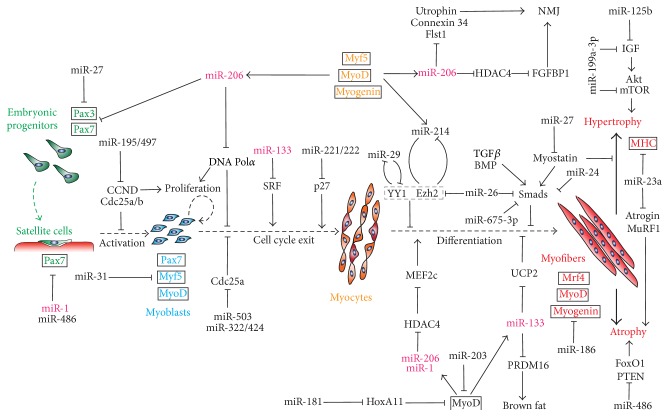
Myogenesis is a complex process that requires coordination of multiple factors governing activation of quiescent satellite cells, proliferation of myoblasts, cell cycle exit, and subsequent terminal differentiation resulting in multinucleated myofibers [4, 5]. Over the past few decades, using C2C12 cells as an in vitro model of myogenesis, together with numerous in vivo studies, the regulatory networks of these factors have been well defined [4, 7, 8, 29]. It is now known that both adult muscle regeneration and embryonic myogenesis share a strikingly similar genetic hierarchy orchestrated by a cascade of myogenic transcription factors, including the paired box (PAX) family of transcription factors (Pax3/7), the basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs) such as Myf5, MyoD, myogenin, and MRF4, and MEF2 and SRF [4, 7, 8, 29]. These transcription factors can act in either synergy or antagonism through feedforward and feedback loops. Recently, miRNAs have been shown to play critical roles in myogenesis through their reciprocal regulatory relationship with these transcription factors (Figure 1). miRNAs can target several myogenic transcription factors, thus affecting myogenesis; on the other hand, their own expression is sometimes directly controlled by these transcription factors [10, 30]. This intricate relationship between miRNAs and myogenesis regulatory factors supports a view of miRNAs as an integral part of the regulatory network of myogenesis (Figure 1). Indeed, several miRNAs have been identified as directly suppressing myogenic transcription factors and are thus able to control the progression of myogenic differentiation. For example, miR-186 targets myogenin and thus inhibits terminal muscle differentiation [31]. Similarly, miR-181 promotes MyoD activity by repressing its negative regulator HoxA11 in myogenic cells [32]. It could be envisioned that further studies will identify novel miRNAs controlling myogenesis by directly regulating myogenic transcription factors.
Figure 1.
miRNAs in myogenesis. The diagram shows various miRNAs and targets that regulate the quiescence and activation of satellite cells, proliferation of myoblasts, and their subsequent cell cycle exit and terminal differentiation into myofibers. The myogenic transcription factors play a central role by governing the expression of several muscle-enriched miRNAs such as miR-1, miR-206, and miR-133 (magenta). The myogenic transcription factors characteristic of each stage of the myogenic process are marked in corresponding colors in a black box. The listed targets include paired box genes 3 and 7 (Pax3 and Pax7), serum response factor (SRF), DNA polymerase catalytic subunit (DNA polα), cell-division cycle protein 25A (Cdc25A), homeobox A11 (HoxA11), Yin and Yang 1 (YY1), enhancer of zeste homolog 2 (Ezh2), histone deacetylase 4 (HDAC4), insulin-like growth factor (IGF), uncoupling protein 2 (UCP2), and PR domain containing 16 (PRDM16).
Satellite cells are muscle stem cells that have the capacity to differentiate into muscle fibers upon muscle damage, yet maintain a quiescent status under normal conditions. Two paired box transcription factors, Pax3 and Pax7, are essential for maintaining this muscle stem cell status through their antidifferentiation capacity, owing to their negative regulation of MyoD [4, 6, 8]. Others and we have found that the miR-1/206 family, comprised of miR-1-1, miR-1-2, and miR-206, is capable of promoting myogenesis, in part by inhibiting Pax3/7 in embryonic muscle precursors and satellite cells [33–35]. The miR-1/206 family is transcribed from three different chromosomal loci in the form of bicistronic transcripts with miRNAs in the miR-133 family (miR-133a-1, miR-133a-2, and miR-133b) [28, 33, 36]. miR-1-1 and miR-1-2 are expressed in both skeletal and cardiac muscles, while miR-206 is specific for skeletal muscle [28, 30, 33, 36]. Similarly, miR-27b has been found to regulate Pax3 levels in embryonic myotomes and activate satellite cells to stimulate myogenic robustness during embryonic myogenesis [37]. In addition, miR-486 can target the Pax7 3′ UTR in satellite cells [38]. Therefore, most miRNAs modulate satellite cells' quiescent status and/or myogenic activation by directly targeting Pax3/7, the master transcription factors that maintain satellite cell quiescence. Alternatively, the recently identified function of miR-31 represents a new mode of miRNA action for regulating satellite cell myogenic activation. miR-31 helps satellite cells maintain their quiescence state by sequestering Myf5 mRNA in mRNP granules in the cell nucleus. Upon satellite cell activation, downregulation of miR-31 in these cells results in rapid release of Myf5 mRNA for protein translation and subsequent myogenesis initiation [39].
Upon activation, Pax3/7 expression is immediately reduced, while several bHLH myogenic transcription factors such as Myf5 and MyoD are upregulated in satellite cells [8]. During embryonic myogenesis or muscle regeneration, activated myoblasts first undergo a quick phase of active proliferation that normally produces an ample number of myoblasts before they exit the cell cycle and start terminal differentiation. Interference with either the proliferation or cell cycle exit stage of activated myoblasts will result in premature or delayed differentiation. Therefore, in theory, any miRNA that affects cell proliferation or the cell cycle will be able to control myogenic differentiation. In myoblasts, several highly expressed miRNAs appear to coordinately regulate this process. Activated by MyoD in activated myoblasts, miR-206 not only can repress Pax3/7 expression, thus further stimulating myoblast activation, but also can repress the p180 subunit of DNA polymerase alpha, whose downregulation coincides with cell cycle exit and differentiation of various tissues [40, 41]. Interestingly, although miR-133 is also upregulated during C2C12 differentiation and muscle regeneration, it promotes myoblast proliferation by inhibiting serum responding factor (SRF) [26]. Since SRF is directly involved in the regulation of miR-1/miR-133a transcription [26, 28, 42], miR-133 and SRF form a negative feedback circuit that balances myoblast proliferation and differentiation. In addition, several other miRNAs (miR-195/497, miR-322/424, and miR-503) have been recently identified as capable of regulating myoblast proliferation by targeting Cdc25a/b and CCNDs, all well-known cell cycle regulators [43, 44].
Once myoblasts exit the cell cycle, they become committed myocytes. MyoD and myogenin then coordinately drive their terminal differentiation by activating muscle specific transcription that was originally suppressed by the polycomb group (PcG) comlpex [4, 6, 8]. Enhancer of zeste homolog 2 (Ezh2) is a histone lysine methyltransferase subunit of the PcG complex that mediates MyoD- and myogenin-dependent promoter silencing in myoblasts through H3K4 and H3K27 methylation [45, 46]. Thus, downregulation of the PcG complex, allowing subsequent MyoD binding to activate muscle genes, is required for myoblasts to appropriately progress into myogenic differentiation. Interestingly, upregulation of several miRNAs (miR-214, miR-26, and miR-29) collaboratively represses PcG complex expression and function in myocytes and thereby promotes muscle-specific gene expression and differentiation (Figure 1). In undifferentiated myoblasts, PcG occupies a MyoD enhancer site upstream of miR-214, which is released by MyoD upon differentiation, allowing the activation of miR-214 expression. Therefore, by targeting and inhibiting Ezh2, miR-214 forms a negative feedback loop with the PcG complex [47]. Similarly, miR-26a also targets and suppresses Ezh2 in differentiating myocytes, although it is not clear if MyoD regulates its expression [48, 49]. Like miR-214, miR-29 forms a negative feedback loop with several other critical components of the PcG complex such as Yin Yang 1 (YY1), RING, and Rybp, an YY1 binding protein, by inhibiting their expression [50, 51]. Furthermore, miR-29 can also regulate myogenesis more directly by repressing AKT3 in differentiating myocytes [52].
While both the miR-1/miR-206 family and miR-133 family of miRNAs become enriched in myocytes during differentiation, likely via MyoD- and/or myogenin-dependent transcriptional regulation, their effects and mode of action on myogenesis are different. miR-1/206 inhibits the expression of histone deacetylase 4 (HDAC4), which suppresses the MEF2c transcriptional activity to induce muscle gene expression. Thus, this forms a positive feedforward loop in which MEF2c induces miR-1 expression, causing inhibition of the MEF2c repressor HDAC4, leading to enhancement of MEF2 activity [26]. In contrast, miR-133 inhibits myogenic differentiation and sustains myoblast proliferation by inhibiting SRF, as mentioned above. Since miR-133 has been recently implicated in the regulation of brown adipose differentiation by directly targeting PRDM16, a master transcription factor for brown adipogenesis [53], and there are common Myf5 positive progenitor cells for brown fat and skeletal muscle during embryonic development, miR-133 likely participates in regulating the adipogenic/myogenic fate determination in such progenitor cells [54]. In fact, miR-133 was recently found to control the brown adipose fate determination of satellite cells [55]. This is particularly intriguing and warrants future investigation into miR-133's functions in myogenesis and adipogenesis to elucidate how this single miRNA can regulate the cell fate determination and switching.
During terminal muscle differentiation, numerous cell signals can affect the activity of myogenic transcription factors, thus fine-tuning and adjusting the outcome of myogenic differentiation [6, 8]. Therefore, miRNAs that regulate these signaling pathways will also profoundly affect the outcome of myogenic differentiation. For example, myostatin, a member of the transforming growth factor β (TGFβ) family, has been implicated in the negative regulation of muscle growth and regeneration [56]. Recently, miR-27a/b was shown to promote myogenic differentiation by targeting the 3′ UTR of myostatin, in addition to inhibiting Pax3 levels during embryonic myogenesis [57, 58]. Inhibiting miR-27a/b by antagomir led to increased myostatin levels and decreased myoblast proliferation and satellite cell activation, whereas miR-27a/b overexpression can reduce myostatin levels and induce skeletal muscle hypertrophy [57, 58]. Likewise, TGF-βs and BMPs both inhibit myogenesis through the activation of Smads [56]. Many miRNAs are found to regulate Smad levels in myocytes, thus affecting the robustness of myogenesis. For example, miR-24 can promote myogenic differentiation by targeting and inhibiting Smad 3 [59], while miR-26a represses Smad 1 and Smad 4 expressions [49]. miR-675-3p is another recently identified miRNA that promotes muscle differentiation by targeting Smads 1 and 5; interestingly, it is encoded in the exon of a long noncoding RNA, H19, which also plays a critical role in myogenesis [60].
As the final step of myogenesis, newly formed multinuclear myotubes will undergo further maturation processes that eventually result in functional myofibers with specialized structures such as neuromuscular junctions (NMJs). Interestingly, miRNAs have also been implicated in the maturation of muscle fibers. For example, miR-206 is enriched in neuromuscular junctions (NMJs), where it plays important roles in promoting the formation of NMJ endplates [61]. This function of miR-206 is mediated, at least in part, by its target, HDAC4, which inhibits the expression and release of FGFBP1 [61]. Since increased FGF pathway activity is required for NMJ reformation during muscle fiber maturation, removing its repressor HDAC4 locally at NMJs by miR-206 is critical for forming functional skeletal muscle [61]. Consistent with its NMJ-specific role, miR-206 deregulation has been associated with the adult motor neuron disease Amyotrophic Lateral Sclerosis (ALS), also known as Lou Gehrig's disease and myotonic dystrophy type 1 (DM1) [61, 62]. Lastly, miR-206 has been found to regulate various other muscle-specific genes also important for the maturation of skeletal muscle, including Connexin 43 (CX43), Follistatin-like 1 (Fstl1), and Utrophin (Utrn) [41, 63, 64].
In summary, as myogenesis progresses from myogenic precursors or satellite cells to functional myofiber formation, miRNAs participate in virtually every step of the way, tightly integrate into the myogenic regulatory network. As the miRNA field enters a more mature stage, more and more muscle-specific mechanisms of miRNA regulation will certainly be discovered.
2.3. miRNAs in Skeletal Muscle Physiology (Fiber Type and Hypertrophy/Atrophy)
All mammals contain a variety of skeletal muscle subtypes differing in their origin and physiological properties [8]. One of the physiological characteristics that reflect muscle heterogeneity is the difference in distinct fiber types between various muscle groups [65]. Conventionally, muscle fibers have been classified as type I (slow) and type II (fast) fibers, where type II fibers consist of three subtypes, IIa, IIb, and IIX. The main difference among these fiber types is in the composition of myosin heavy chain (MyHC) isoforms [65]. Separate genes located at different genomic loci encode these MyHC isoforms in humans [65]. Thus, fiber type determination was traditionally thought to be regulated predominantly at the transcription level [66]. However, this view has been recently challenged by the identification of a group of miRNAs called MyomiRs. These intronic miRNAs, miR-208a, miR-208b, and miR-499, are embedded in three muscle-specific MyHC genes (Myh6, Myh7, and Myh7b, resp.) [67, 68]. Among these three miRNAs, miR-208a is specifically expressed only in cardiac muscle, whereas miR-208b and miR-499 are also expressed in type I (slow) muscle fibers [67].
These MyomiRs play important roles in regulating muscle myosin isoforms, thus regulating muscle fiber type. The two MyomiRs miR-208b and miR-499 have redundant functions in controlling muscle fiber type by repressing fast myofiber genes while activating slow muscle specific genes [67, 68]. Overexpression of miR-499 in skeletal muscle completely converts the fast myofibers of the soleus muscle into slow fibers [67]. Conversely, double knockout of miR-499 and miR-208b in mouse leads to a dramatic loss of type I fibers in the soleus muscle [67]. The effects of MyomiRs on muscle fiber identities are largely mediated by targeting transcriptional repressors of slow muscle fiber genes, such as Sox6, Purβ, and Sp3 [67]. Interestingly, knockout of Sox6 in skeletal muscle in mice indeed results in a fast-to-slow myofiber conversion [69–71]. It is noteworthy that MyomiRs are all embedded in the introns of various muscle myosin heavy chain genes, thus sharing the same expression levels as their host genes [72]. On the other hand, the expression of these MyHC genes is in turn regulated by the transcription factors targeted by these MyomiRs. Therefore, it appears that nature has utilized an endogenous feedback mechanism between MyomiRs and myosin heavy chains for fine-tuning muscle fiber type determination in order to modulate muscle physiology and performance.
Skeletal muscle not only has a remarkable regenerative capacity but also is a very adaptive tissue. Exercise improves muscle function, enhances muscle energy metabolism, and leads to muscle hypertrophy, which is characterized by increasing muscle mass [73, 74]. On the contrary, immobilization, lack of exercise, or other diseases, such as cancer, often induce muscle atrophy, which is defined as loss of muscle mass and reduced metabolic activity. Besides changes in the expression and activity of many muscle enzymes and proteins, the expression of many miRNAs is also altered during muscle hypertrophy or atrophy [75–77]. For example, miR-23a and miR-696 are increased in muscle hypertrophy after endurance exercise and conversely decreased in muscle atrophy induced by muscle immobilization [77–79]. Both of these miRNAs can negatively regulate peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC-1α), a key regulator of metabolism and mitochondria biosynthesis in skeletal muscles [78, 79]. In addition, miR-23a can regulate Atrogin and MuRF1, two E3 ubiquitin ligases important for degradation of muscle sarcomeric proteins during muscle atrophy [80]. Another miRNA participating in muscle atrophy regulation is miR-486, which inactivates atrophy signaling in skeletal muscle through combinatory inhibition of its targets PTEN and FoxO1 [81].
Physiological muscle atrophy (sarcopenia) also occurs with aging. Several recent studies profiling miRNA expression in skeletal muscle of young and old mice have revealed that several miRNAs, including miR-206, miR-698, miR-744-5p, and miR-468, are increased, whereas others, such as miR-29, miR-434, miR-455, miR-382, miR-181a, and miR-221, are reduced in skeletal muscle cells of old animals [82–84]. While the exact targets and molecular mechanisms of most of these differentially expressed miRNAs are still elusive, it is likely that they play important roles in skeletal muscle by coordinating with regulatory proteins of muscle atrophy.
2.4. miRNAs in Skeletal Muscle Disease Pathophysiology
Skeletal muscle diseases are a large group of heterogeneous muscle disorders that are characterized, at least in part, by muscle wasting. Most studies on miRNA function in muscle diseases have been focused on identifying dysregulated miRNAs using genome-wide analyses, such as microarrays or next-generation RNA deep sequencing. For example, work from Kunkel's lab has profiled miRNA expression in 10 major muscular disorders in humans and identified 185 miRNAs that are significantly up- or downregulated in pathological conditions. Of these, miR-146b, miR-155, miR-214, miR-221, and miR-222 are of special interest, as their expression is altered in almost all of the muscle diseases studied [85]. With miRNA profiling data becoming increasingly available, more and more miRNAs are implicated in the physiopathology of various muscle diseases. Here we provide a brief discussion on some of the most well-studied miRNAs in muscle pathophysiology.
The most common skeletal muscle disorder is muscular dystrophy, a large group of over 30 inherited muscle diseases characterized by progressive muscle wasting accompanied by repeated muscle degeneration and regeneration [86, 87]. The most common and severe muscular dystrophy is Duchenne muscular dystrophy (DMD), which is caused by mutations in the X-linked dystrophin gene [88]. Loss of dystrophin at sarcolemma in skeletal muscle results in fragility of myofibers, leading to muscle degeneration, chronic inflammatory responses, and fibrotic and fatty tissue deposition. Currently, there is no effective therapy for DMD. In the skeletal muscle of DMD patients, muscle tissues undergo profound cellular and molecular changes. Several microarray studies have been conducted using muscle tissues from either DMD patient or the mdx mouse, an animal model for human DMD [85]. Many MyomiRs and muscle-enriched miRNAs, such as miR-1, miR-133, and miR-206, are all increased in the serum of DMD patients and/or in muscle tissues of mdx mice [89–96]. This is consistent with their known role in myogenesis, since there is constant muscle degeneration and regeneration in dystrophic muscle, which requires the upregulation of these miRNAs to direct appropriate myogenic differentiation. Although there are still some inconsistences between various microarray studies, several miRNAs have been consistently identified to be dysregulated and to play a role in the pathophysiology of dystrophic muscles.
One of such miRNAs is miR-486, whose expression is downregulated in the muscle of DMD patients and mdx mice [85]. Using miR-486 transgenic mice, Alexander and colleagues demonstrate that overexpression of miR-486 alleviates the dystrophic phenotype of mdx mice, likely by regulating dedicator of cytokinesis 3 (DOCK3), platelet-derived growth factor receptor β (PDGF-β), and the PTEN/AKT pathway, thus affecting the cell cycle and muscle regeneration in mdx muscle [97]. Another downregulated miRNA in dystrophic muscles is miR-29, which positively regulates myogenic differentiation and reduces fibrosis. Consistent with its function, overexpression of miR-29 in mdx mice improves muscle regeneration, accompanied by reduced fibrosis [98]. In contrast, miR-199-5a is increased in mdx muscle [85]. miR-199-5a regulates the levels of several Wnt signaling pathway proteins, including Frizzled 4 (FZD4), Jagged1 (JAG1), and WNT2 [99]. All of these signaling proteins collectively regulate myoblast proliferation and differentiation; therefore, blocking this signaling pathway through excessive miR-199-5a in dystrophic muscle would likely affect myogenesis and muscle regeneration [99]. Of particular interest, miR-31 is also increased in dystrophic muscles. Besides enlisting a special mechanism to keep satellite cells in a quiescent state [39], miR-31 also directly targets the 3′ UTR of dystrophin transcripts [100]. Thus, miR-31 plays a dual role in dystrophic muscle: preventing satellite cell activation on one hand and promoting degradation of dystrophin mRNA on the other.
Studies using mice lacking both miR-133a-1 and miR-133a-2 (miR-133 dKO) reveal an important role of miR-133a in muscle pathophysiology [101]. In adult skeletal muscle of miR-133 dKO mice, a higher proportion of central nuclei was observed in type II (fast) muscle fibers, reminiscent of human central nuclear myopathy (CNM), a group of congenital myopathies characterized by abnormally centrally localized nuclei and muscle weakness [101]. Mutations of the dynamin 2 gene in human have been associated with CNMs in a dominant negative fashion, and increasing the level of dynamin 2 also leads to CNMs [102, 103]. Interestingly, miR-133a targets the 3′ UTR of dynamin 2 to repress its protein level. Therefore, the central nuclear myopathy observed in miR-133a dKO mice is at least partially due to the elevated protein level of dynamin 2 [101].
Rhabdomyosarcomas (RMS) are the only muscle cancer in humans, and they are the most common soft tissue sarcomas in children [104, 105]. One of the defining characteristics of RMS is the overexpression of myogenic differentiation markers, such as MyoD and Desmin [104, 105]. It has been reported that miR-1 and miR-206 levels are repressed in RMS [106], likely resulting in the inhibition of terminal differentiation of myogenic progenitor cells. Conversely, overexpression of miR-206 in RMS cells promotes their terminal differentiation and blocks tumor growth, thus highlighting the important role of miR-206 in RMS pathogenesis. One miR-206 target, the MET tyrosine-kinase receptor (MET) oncogene, may underlie such functions of miR-206 in RMS [107, 108]. Nevertheless, further studies are needed to definitively establish the role of miR-206 in RMS.
Over the past few years, miRNAs have been implicated in many aspects of muscle diseases. Owing to its molecular nature, small, abundant, and stable, miRNA can be easily detected. Indeed, several studies have attempted to use miRNAs as biomarkers in diagnosing specific forms of muscle diseases [62, 89, 90, 109]. Remarkably, several miRNAs, including MyomiRs and muscle-enriched miRNAs, have been detected in the serum of either DMD patients or mdx mice [62, 90, 91, 109]. The ability to detect such miRNAs in the serum of patients will provide valuable biomarkers and open up opportunities for developing miRNA-based diagnostic assays. Most importantly, given their important roles in skeletal muscle, miRNAs hold great promise for developing effective therapeutic interventions for a wide range of muscle diseases.
3. lncRNAs, Novel Regulators of Muscle Biology
3.1. lncRNAs, Definition and Classification
Although lncRNAs were only recently discovered, it has become increasingly clear that this class of noncoding RNAs regulates a variety of biological responses and that they do so via multiple different mechanisms [134]. So far, more than 10,000 lncRNAs have been identified in humans, and the number is likely to continue growing [135]. Present in both the nucleus and cytosol, lncRNAs are probably the most functionally diverse RNAs; they can act at nearly all steps of gene expression within the cell. This is largely due to the intrinsic nature of RNA molecules and their ability to form complex secondary structures, which enable them to bind to a diverse set of molecules, such as DNA, RNA, and proteins. Most lncRNAs are transcribed by RNA polymerase II and share mRNA-like features such as the 5′ cap, polyA tail, and splicing sites. In general, the transcription level of lncRNAs is lower compared to that of mRNAs, and transcription is cell context-dependent [136].
Based on their genomic locations and contexts, lncRNAs can be classified into several types: (1) intergenic lncRNAs (lincRNAs); (2) intronic lncRNAs; (3) sense-overlapping lncRNAs, and (4) antisense lncRNAs [3, 137]. Intergenic lncRNAs are long noncoding RNAs located between annotated protein-coding genes, generally in close proximity to neighboring protein-coding genes. In contrast, both sense and antisense lncRNAs at least partially overlap with exon sequences of annotated protein-coding genes and only differ from each other by their transcriptional directions. As the name suggests, an intronic lncRNA is an lncRNA that overlaps with the intronic region of a coding gene, and its transcription can be in either the sense or antisense direction.
Functionally, lncRNAs can also be divided into several subcategories according to their mode of action. (1) Many nucleus-enriched lncRNAs can exert their functions at the transcriptional level, either through cotranscriptional interactions between the nascent lncRNAs and transcriptional complexes or by the recruitment of such complexes like the chromatin modification enzymes, to transcription sites in cis or in trans. The in cis nature of an lncRNA refers to its ability to act on a neighboring gene on the same allele from which itself is transcribed; thus, this type of lncRNA commonly forms a feedback loop for regulation of itself and its neighboring genes. Still, many lncRNAs are found to also function in a trans mode to target gene loci distant from where the lncRNAs are transcribed. (2) Many lncRNAs can act as decoys for certain molecules, such as transcriptional/splicing factors in the nucleus and miRNAs or RNA degradation complexes in cytoplasm, to regulate the expression of targets of those biological pathways. (3) Furthermore, lncRNAs can also serve as a scaffold for forming complex molecular machineries or nuclear subdomains, profoundly affecting the expression level of many genes under various biological contexts. Clearly, additional modes of action for lncRNAs will likely be proposed as more functional lncRNAs are discovered. Nevertheless, example lncRNAs for all of the above-mentioned mechanisms have been identified in skeletal muscle development or pathophysiology, supporting the notion that lncRNAs are a class of evolutionary conserved molecules that affect a wide spread of biological processes (Figure 2).
Figure 2.
Functional mechanism of lncRNAs in skeletal muscle. (A) lncRNAs (in green) are able to positively or negatively regulate transcription at their own loci and at neighboring genes in cis by modulating transcriptional machinery or recruiting chromatin modification complexes. (B) Similarly, lncRNAs can also regulate a wide range of muscle gene expression in trans by recruiting chromatin modification enzymes and transcription machinery to their promoters. (C) lncRNAs can also regulate muscle gene mRNA splicing events by enlisting various splicing factors. (D) lncRNAs can serve as scaffolding for nuclear domains and/or long range chromosomal looping. (E) In cytosol, lncRNAs can serve as a miRNA sponge by competitively sequestering certain miRNA, thus inhibiting their effects on gene expression. (F) Recently, both SINE containing lncRNAs and H19 have been found to regulate mRNA decay by distinct mechanisms, therefore affecting muscle mRNA stability and regulating myogenesis.
Despite the rich information that we have learned about the transcription network regulating muscle development in the last few decades, we are still at the beginning of an “era of noncoding RNA.” Emerging literatures regarding several newly identified lncRNA functions in skeletal muscle have demonstrated that lncRNAs indeed are another integral part of the regulatory network of muscle biology. Clearly, lncRNAs have emerged as important novel regulators of skeletal muscle biology and diseases.
3.2. lncRNAs in Transcriptional Regulation of Skeletal Muscle Genes
Recently, a large number of lncRNAs were identified as being transcribed from enhancer regions of genes. These chromatin loci are usually occupied by transcriptional coactivator p300/CBP and RNA polymerase II and marked with histone H3K4 monomethyl (H3K4me1) and H3K27 acetyl (H3K27ac) modifications [138, 139]. These enhancer RNAs can regulate chromosome structure or transcription machinery through either a cis- or trans-mediated mechanism [140–147]. In two enhancer regions of the MyoD gene, the Distal Regulatory Regions (DRR) and the Core Enhancer (CE), two enhancer-associated lncRNAs, DRRRNA and CERNA, were recently identified through a series of ChIP-seq experiments [148]. It has been proposed that CERNA facilitates the occupancy of RNA Pol II in cis by increasing chromatin accessibility, stimulating the expression of MyoD, while DRRRNA functions in trans to promote the expression of myogenin, a key member of the myogenic transcription factor family [148]. More recently, Mueller and colleagues also identified an lncRNA transcribed upstream of MyoD, named MUNC (MyoD upstream noncoding RNA), and demonstrated that one of the spliced isoforms of MUNC is indeed DRRRNA. Consistent with the previous study, experimentally decreasing MUNC expression blocked myoblast differentiation, further highlighting the role of these enhancer-associated lncRNAs during myogenesis [149]. Interestingly, these enhancer RNAs (eRNAs) seem to provide a positive feedback mechanism to reinforce the myogenic differentiation commitment upon satellite cell activation. Additional studies, in particular genetic studies in animal models, will be required to further establish the function of these lncRNAs in vivo. It will also be interesting to determine whether the expression and function of these lncRNAs are associated with any muscle diseases.
Similarly, a ChIP-seq study of Yin Yang 1 (YY1), an important component of the PcG complex that negatively regulates myogenesis, identified a number of lncRNAs regulated by YY1 (YY1-associated muscle lncRNAs or Yams) [150]. Individual Yams show distinct expression patterns during myogenesis and affect muscle differentiation differently; while Yam-2 and Yam-3 can promote muscle differentiation, Yam-1 and Yam-4 are negative regulators of myogenesis. Among the Yams, Yam-1, a single exon lncRNA, is the best studied. Like YY1, Yam-1 is downregulated during myoblast differentiation and during muscle development. Knockdown of Yam-1 can overcome YY1-mediated inhibitory effects on myogenesis, suggesting that Yam-1 is an important mediator of YY1 during skeletal muscle differentiation [150]. Interestingly, Yam-1 displays a cis effect on the expression of neighboring genes, one of which encodes miR-715, which targets and represses Wnt7b in skeletal muscle [150]. Thus it is also likely that some of the Yam-1 myogenic inhibitory effects can be attributed to in cis activation of miR-715, which in turn downregulates Wnt7b.
Another example of an in cis regulatory lncRNA is Glt2/Meg3, an lncRNA originating from the complex, imprinted Dlk1-Dio3 region of the human genome [151, 152]. Containing protein-coding RNAs, lncRNAs, miRNAs, and snoRNAs, the imprinted Dlk1-Dio3 locus has a complicated transcriptional regulation mechanism [151, 152]. The Glt2/Meg3 lncRNA is found to bind and recruit Polycomb Repressive Complex 2 (PRC2) to repress the transcription of Dlk1 and Glt2-as, another lncRNA encoded by the Dlk1-Dio3 region [152]. Knockout of Glt2/Meg3 results in perinatal lethality with defects in skeletal muscle development, highlighting its critical role in myogenesis [153]. It remains to be determined whether Glt2/Meg3 can also function in trans to regulate the expression and function of additional genes in skeletal muscle.
Besides regulating chromatin accessibility, some lncRNAs can also directly regulate specific transcription factors to modulate gene expression during myogenesis. Steroid receptor RNA activator (SRA) is such an lncRNA. Enriched in myofibers, it can selectively enhance transcriptional activation through steroid receptors [154, 155]. As a matter of fact, SRA lncRNA was the first lncRNA found to regulate myogenic differentiation [154, 156]. During myogenic differentiation, SRA lncRNA forms a functional complex with p68/p72, a member of the DEAD-box family of RNA helicases, and MyoD to facilitate chromatin remodeling and the formation of the transcription initiation complex at a subset of muscle-specific genes [154, 156]. Remarkably, the SRA transcripts also undergo alternative splicing and give rise to an mRNA encoding the protein SRAP, an RNA binding protein that specifically binds to SRA lncRNA and antagonizes its interaction with MyoD and p68/p72 [156, 157]. Interestingly, the ratio of the protein-coding and noncoding isoforms of the SRA transcripts changes during muscle differentiation. Thus, SRA lncRNA and protein form a reciprocal negative feedback loop that fine-tunes MyoD activity during myogenic differentiation.
3.3. lncRNAs as Molecular Decoys to Regulate Muscle Differentiation
lncRNA H19, the first lncRNA identified in mammals, originates from the imprinted H19/IGF2 region [158]. Transcribed from the maternal allele, the H19 lncRNA is highly expressed in embryos as well as in adult skeletal muscle, suggesting its conserved function in regulating myogenesis [60, 159, 160]. MyoD directly activates H19 lncRNA transcription by interacting with a mesodermal enhancer of the H19/Igf2 locus. H19 in turn downregulates IGF2 expression in trans by recruiting the PRC2 repressor complex during muscle differentiation [161–163]. In addition, the H19 lncRNA contains several binding sites for the let-7 family of miRNAs, thus functioning as a molecular sponge/decoy for the major miRNAs of the let-7 family, which play critical roles in a wide range of physiological and pathological processes, including myogenesis [164]. Indeed, overexpression of let-7 rescues the premature myogenic differentiation phenotype caused by H19 knockdown, confirming that H19 affects myogenesis at least in part by antagonizing the function of the let-7 family of miRNAs. Consistent with this view, two of the well-documented targets of let-7, HMGA2 and IGF2, have also been found to be downstream effectors of the H19 lncRNA during muscle differentiation [164]. Intriguingly, two conserved miRNAs (miR-675-3p and miR-675-5p) that play important roles during myogenic differentiation were recently found to reside in the first exon of the H19 lncRNA, thus suggesting that H19 lncRNA also serves as a primary miRNA transcript [60]. It would be interesting to further study how H19 achieves both functions of miRNA sponge and miRNA precursor during myogenesis and discover which function is more critical or important in vivo.
Another excellent example of lncRNAs acting as molecular decoys is linc-MD1, the first muscle-specific lncRNA identified by Gabellini and colleagues. Linc-MD1 is encoded by a genomic locus that overlaps with the bicistronic miR-206 and miR-133b transcript-coding region [165]. Linc-MD1 is required for appropriate muscle differentiation, at least in part because it regulates the levels of Myocyte Enhancer Factor 2C (MEF2C) and Mastermind-like protein 1 (MAML1), via the mechanism of sponging endogenous miR-133 and miR-135 in cytoplasm [165]. Similar to H19, linc-MD1 can also serve as a pre-miRNA transcript, as it encodes miR-133b [165]. Interestingly, HuR, an RNA binding protein that stabilizes the mRNAs of several myogenic factors during muscle differentiation, is a target of miR-133. HuR facilitates the formation of the linc-MD1-miRNA complex and its accumulation in cytoplasm, thus promoting linc-MD1's miRNA sponge activity [166]. Therefore, there is a delicate and complex regulatory circuit between linc-MD1, miR-133, and HuR, which is critical for appropriate muscle differentiation. Lastly, the substantial downregulation of linc-MD1 in primary myoblasts of patients with DMD suggests that it is likely involved in the pathogenesis of this muscle disorder [165].
Amazingly, about one-third of lncRNAs are found to contain at least one short interspersed element (SINE) sequence, suggesting that they may actively participate in the Staufen 1- and Staufen 2- (STAU1- and STAU2-) mediated mRNA decay (SMD) process [167, 168]. When a SINE within the 3′ UTR of a protein-coding RNA forms intermolecular base pairing with a partially complementary SINE of one or more lncRNAs, the resulting double-stranded RNA (dsRNA) can be recognized by Staufen 1 and Staufen 2 and degraded through SMD [169, 170]. Several mRNAs that encode proteins which play important roles during myogenesis have SINE sequences in their 3′ UTRs (two recently reported examples are Cdc6 and Traf6) and thus can be bound by SINE-containing lncRNAs and targeted for degradation through SMD, inhibiting their translation and thus their functions during myogenesis [170].
3.4. lncRNAs as Subcellular Domain Scaffold to Regulate Muscle Differentiation
The lncRNA Malat1 is abundantly expressed in various types of cells and regulates their proliferation and metastasis [171, 172]. Malat1 is predominantly located in nuclear speckles and regulates both gene transcription and pre-mRNA splicing [173, 174]. Through interaction with Cbx4, Malat1 regulates gene expression by modulating chromatin translocation among nuclear domains [174], whereas it regulates alternative splicing of certain pre-mRNAs through interaction with the SR family of splicing factors [173]. During muscle differentiation, Malat1 is upregulated, and it has been identified as a downstream target of myostatin, a well-known negative regulator of myogenesis [175]. Knockdown of Malat1 suppresses myoblast proliferation and differentiation [176]. However, the exact mechanism underlying this observation remains elusive and needs further investigation.
3.5. lncRNAs and Duchenne Muscular Dystrophy (DMD)
Dystrophin is the causative gene of DMD; however, the molecular mechanisms and cellular events underlying DMD pathophysiology are still not fully understood. A recent study using tiling arrays has identified about 14 new lncRNAs that originate from the dystrophin gene, which consists of 79 exons, making it the largest human gene. Some of these lncRNAs appear to target the promoters of the dystrophin gene in trans to repress the expression of some dystrophin isoforms [177]. However, it remains to be determined how these lncRNAs contribute to the pathophysiology of the dystrophic muscle. In addition, in DMD patients with mental retardation, a specific chromosomal inversion event generates an lncRNA, KUCG1, which may account for the clinical mental retardation symptoms [178]. Furthermore, as mentioned above, the level of linc-MD1 is greatly reduced in the muscle of DMD patients, and linc-MD1 overexpression can rescue the defective myogenic differentiation and restore the normal expression of Maml1, Mef2c, Myog and Mhc [165]. Therefore, enhancing linc-MD1 levels in DMD patients promises to be a potential therapeutic strategy.
3.6. Facioscapulohumeral Muscular Dystrophy (FSHD), an lncRNA Link
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal-dominant hereditary muscle disease that causes progressive weakness and loss of skeletal muscles [179]. As the third most common muscular dystrophy, it affects 1 in 14,000 people [179]. The genetic region involved in FSHD does not encode any protein but rather contains a 3.3 kb macrosatellite D4Z4 repeat [180]. While normal people have anywhere from 11 to more than 100 copies of D4Z4 repeats in the subtelomeric region of chromosome 4q35, FSHD patients tend to have less than ten repeats [181–183]. A recently identified chromatin-associated lncRNA, DBE-T, is selectively produced in FSHD patients and is associated with the derepression of genes in the 4q35 locus [184]. Mechanistically, DBE-T directly interacts with and recruits the Trithorax group protein Ash1L, a histone methyltransferase, to the FSHD locus. Locally increasing Ash1L results in histone H3K36 dimethylation, which in turn relaxes the local chromatin, resulting in the activation of DBE-T as well as neighboring genes at the FSHD loci [184]. Therefore, a feedforward mechanism involving the lncRNA DBE-T and Ash1L in FSHD patients promotes the derepression of chromosomal 4q35 region and thus contributes to the pathogenesis of FSHD.
4. Perspectives
The discovery of noncoding RNAs (miRNAs and lncRNAs) has dramatically expanded our understanding of how gene expression is regulated. miRNAs, which regulate gene expression by targeting protein-coding genes, add a posttranscriptional regulatory mechanism in gene regulation. With the increasing number of publications regarding miRNA function in skeletal muscles, it has become clear that miRNAs are an integral part of myogenic regulatory networks. Although much progress has been made in identifying and verifying targets of specific miRNAs in skeletal muscle cells and in elucidating the functional mechanism of many miRNAs in skeletal muscle biology, the relationship between miRNAs and various muscle disorders has yet to be fully understood. In addition, how to harness these small RNAs to develop effective and economic diagnostic and therapeutic tools is still a question to be addressed. Finally, the function of miRNAs in skeletal muscle metabolism appears to be one of the understudied areas of miRNA biology. Skeletal muscle is a metabolically active tissue, and it would be of great interest to define whether miRNAs are also an integral part of the metabolic regulatory network in muscle. Undoubtedly, future work about the molecular mechanisms of miRNAs in skeletal muscle diseases and their therapeutic applications will emerge and contribute to a comprehensive understanding of miRNA biology in skeletal muscles.
Unlike miRNAs, lncRNAs are identified only recently and we are still at the “infant stage” of studying this novel class of noncoding RNAs in skeletal muscle. Overall, as for miRNAs, there is emerging evidence that lncRNAs are important regulators of muscle biology. It is likely that there are still many muscle-related lncRNAs to be discovered and that they will act via multiple distinct mechanisms, some of which may be unprecedented. Future studies will also need to examine whether aberrant lncRNAs are linked with various muscle diseases and what their roles are in pathogenesis. In addition, given the RNA nature of lncRNAs and miRNAs, it is intriguing to speculate that lncRNAs and miRNAs can coordinately regulate the expression of certain mRNAs. To some extent, the fact that lncRNAs can reduce the levels of miRNAs by functioning as their sponges supports this hypothesis. Furthermore, in the light of the recently identified muscle-specific micropeptide myoregulin, encoded by a putative noncoding RNA [185], we cannot rule out the possibility that some “noncoding” lncRNAs might actually encode physiological active micropeptides. In conclusion, we are at an exciting time in noncoding RNA biology and in the study of their functions in skeletal muscle biology and muscle diseases. We are confident that more and more noncoding RNA-based diagnostic and therapeutic applications will emerge in the future.
Acknowledgments
The authors are supported by Muscular Dystrophy Association (186548 to J. Liu, 294854 to D.-Z. Wang), NIH (HL085635, HL116919 to D.-Z. Wang), and The National Natural Science Foundation of China (81272005 to Z.-L. Deng).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lander E. S., Linton L. M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Rinn J. L., Chang H. Y. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chargé S. B. P., Rudnicki M. A. Cellular and molecular regulation of muscle regeneration. Physiological Reviews. 2004;84(1):209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 5.Dhawan J., Rando T. A. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends in Cell Biology. 2005;15(12):666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Kuang S., Rudnicki M. A. The emerging biology of satellite cells and their therapeutic potential. Trends in Molecular Medicine. 2008;14(2):82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature Reviews Molecular Cell Biology. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 8.Bentzinger C. F., Wang Y. X., Rudnicki M. A. Building muscle: molecular regulation of myogenesis. Cold Spring Harbor Perspectives in Biology. 2012;4(2) doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berardi E., Annibali D., Cassano M., Crippa S., Sampaolesi M. Molecular and cell-based therapies for muscle degenerations: a road under construction. Frontiers in Physiology. 2014;5, article 119 doi: 10.3389/fphys.2014.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callis T. E., Deng Z., Chen J.-F., Wang D.-Z. Muscling through the microRNA world. Experimental Biology and Medicine. 2008;233(2):131–138. doi: 10.3181/0709-mr-237. [DOI] [PubMed] [Google Scholar]
- 11.Sharma M., Juvvuna P. K., Kukreti H., McFarlane C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Frontiers in Physiology. 2014;5, article 239 doi: 10.3389/fphys.2014.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neguembor M. V., Jothi M., Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skeletal Muscle. 2014;4(1, article 8) doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X., Hagedorn C. H., Cullen B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory R. I., Yan K.-P., Amuthan G., et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 15.Denli A. M., Tops B. B. J., Plasterk R. H. A., Ketting R. F., Hannon G. J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y., Ahn C., Han J., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 17.Chendrimada T. P., Gregory R. I., Kumaraswamy E., et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipowicz W., Bhattacharyya S. N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Reviews Genetics. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 19.Espinoza-Lewis R. A., Wang D. Z. MicroRNAs in Heart Development. Current Topics in developmental biology. 2012;100:279–317. doi: 10.1016/b978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 21.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N., Olson E. N. MicroRNA regulatory networks in cardiovascular development. Developmental Cell. 2010;18(4):510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small E. M., Olson E. N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein E., Kim S. Y., Carmell M. A., et al. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 25.O'Rourke J. R., Georges S. A., Seay H. R., et al. Essential role for Dicer during skeletal muscle development. Developmental Biology. 2007;311(2):359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J.-F., Mandel E. M., Thomson J. M., et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim L. P., Lau N. C., Garrett-Engele P., et al. Microarray analysis shows that some microRNAs downregulate large numbers of-target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 29.Lagha M., Sato T., Bajard L., et al. Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:307–315. doi: 10.1101/sqb.2008.73.006. [DOI] [PubMed] [Google Scholar]
- 30.Williams A. H., Liu N., van Rooij E., Olson E. N. MicroRNA control of muscle development and disease. Current Opinion in Cell Biology. 2009;21(3):461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniou A., Mastroyiannopoulos N. P., Uney J. B., Phylactou L. A. MiR-186 inhibits muscle cell differentiation through myogenin regulation. The Journal of Biological Chemistry. 2014;289(7):3923–3935. doi: 10.1074/jbc.m113.507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naguibneva I., Ameyar-Zazoua M., Polesskaya A., et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nature Cell Biology. 2006;8(3):278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 33.Chen J.-F., Tao Y., Li J., et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. The Journal of Cell Biology. 2010;190(5):867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goljanek-Whysall K., Sweetman D., Abu-Elmagd M., et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):11936–11941. doi: 10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirai H., Verma M., Watanabe S., Tastad C., Asakura Y., Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. Journal of Cell Biology. 2010;191(2):347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N., Williams A. H., Kim Y., et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(52):20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crist C. G., Montarras D., Pallafacchina G., et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dey B. K., Gagan J., Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Molecular and Cellular Biology. 2011;31(1):203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crist C. G., Montarras D., Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11(1):118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Huebscher U., Kuenzle C. C., Spadari S. Variation of DNA polymerases-alpha, -beta. and -gamma during perinatal tissue growth and differentiation. Nucleic Acids Research. 1977;4(8):2917–2929. doi: 10.1093/nar/4.8.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. Journal of Cell Biology. 2006;174(5):677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu N., Bezprozvannaya S., Williams A. H., et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes and Development. 2008;22(23):3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato T., Yamamoto T., Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nature Communications. 2014;5(article 4597) doi: 10.1038/ncomms5597. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar S., Dey B. K., Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Molecular Biology of the Cell. 2010;21(13):2138–2149. doi: 10.1091/mbc.e10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes and Development. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee T. I., Jenner R. G., Boyer L. A., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Molecular Cell. 2009;36(1):61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung F. W., Tellam R. L. MicroRNA-26a targets the histone methyltransferase enhancer of Zeste homolog 2 during myogenesis. The Journal of Biological Chemistry. 2008;283(15):9836–9843. doi: 10.1074/jbc.m709614200. [DOI] [PubMed] [Google Scholar]
- 49.Dey B. K., Gagan J., Yan Z., Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes and Development. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. The Journal of Biological Chemistry. 2012;287(30):25255–25265. doi: 10.1074/jbc.m112.357053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H., Garzon R., Sun H., et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei W., He H.-B., Zhang W.-Y., et al. MiR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death and Disease. 2013;4(6, article e668) doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seale P., Bjork B., Yang W., et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trajkovski M., Ahmed K., Esau C. C., Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nature Cell Biology. 2012;14(12):1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- 55.Yin H., Pasut A., Soleimani V. D., et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metabolism. 2013;17(2):210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Derynck R., Akhurst R. J. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nature Cell Biology. 2007;9(9):1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 57.Huang Z., Chen X., Yu B., He J., Chen D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochemical and Biophysical Research Communications. 2012;423(2):265–269. doi: 10.1016/j.bbrc.2012.05.106. [DOI] [PubMed] [Google Scholar]
- 58.McFarlane C., Vajjala A., Arigela H., et al. Negative auto-regulation of myostatin expression is mediated by Smad3 and MicroRNA-27. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0087687.e87687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Q., Zhang Y., Yang G., et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Research. 2008;36(8):2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dey B. K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes and Development. 2014;28(5):491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams A. H., Valdez G., Moresi V., et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toivonen J. M., Manzano R., Oliván S., et al. MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089065.e89065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson C., Catoe H., Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Research. 2006;34(20):5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenberg M. I., Georges S. A., Asawachaicharn A., Analau E., Tapscott S. J. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. Journal of Cell Biology. 2006;175(1):77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiaffino S., Reggiani C. Fiber types in mammalian skeletal muscles. Physiological Reviews. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 66.Hughes S. M., Chi M. M.-Y., Lowry O. H., Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. Journal of Cell Biology. 1999;145(3):633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Rooij E., Quiat D., Johnson B. A., et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Developmental Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 69.Quiat D., Voelker K. A., Pei J., et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(25):10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hagiwara N., Ma B., Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Developmental Dynamics. 2005;234(2):301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- 71.Hagiwara N., Yeh M., Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Developmental Dynamics. 2007;236(8):2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- 72.McCarthy J. J., Esser K. A., Peterson C. A., Dupont-Versteegden E. E. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiological Genomics. 2009;39(3):219–226. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreyer H. C., Fujita S., Glynn E. L., Drummond M. J., Volpi E., Rasmussen B. B. Resistance exercise increases leg muscle protein synthesis and mTOR signalling independent of sex. Acta Physiologica. 2010;199(1):71–81. doi: 10.1111/j.1748-1716.2010.02074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayhew D. L., Kim J. S., Cross J. M., Ferrando A. A., Bamman M. M. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. Journal of Applied Physiology. 2009;107(5):1655–1662. doi: 10.1152/japplphysiol.91234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baggish A. L., Hale A., Weiner R. B., et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. The Journal of Physiology. 2011;589(16):3983–3994. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davidsen P. K., Gallagher I. J., Hartman J. W., et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. Journal of Applied Physiology. 2011;110(2):309–317. doi: 10.1152/japplphysiol.00901.2010. [DOI] [PubMed] [Google Scholar]
- 77.Hudson M. B., Woodworth-Hobbs M. E., Zheng B., et al. miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. The American Journal of Physiology—Cell Physiology. 2014;306(6):C551–C558. doi: 10.1152/ajpcell.00266.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aoi W., Naito Y., Mizushima K., et al. The microRNA miR-696 regulates PGC-1α in mouse skeletal muscle in response to physical activity. The American Journal of Physiology—Endocrinology and Metabolism. 2010;298(4):E799–E806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 79.Safdar A., Abadi A., Akhtar M., Hettinga B. P., Tarnopolsky M. A. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57BI/6J male mice. PLoS ONE. 2009;4(5) doi: 10.1371/journal.pone.0005610.e5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wada S., Kato Y., Okutsu M., et al. Translational suppression of atrophic regulators by MicroRNA-23a integrates resistance to skeletal muscle atrophy. The Journal of Biological Chemistry. 2011;286(44):38456–38465. doi: 10.1074/jbc.m111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu J., Li R., Workeneh B., Dong Y., Wang X., Hu Z. Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney International. 2012;82(4):401–411. doi: 10.1038/ki.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J. Y., Park Y. K., Lee K. P., et al. Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging (Albany NY) 2014;6(7):524–544. doi: 10.18632/aging.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Drummond M. J., McCarthy J. J., Fry C. S., Esser K. A., Rasmussen B. B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. The American Journal of Physiology—Endocrinology and Metabolism. 2008;295(6):E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamrick M. W., Herberg S., Arounleut P., et al. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochemical and Biophysical Research Communications. 2010;400(3):379–383. doi: 10.1016/j.bbrc.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eisenberg I., Eran A., Nishino I., et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davies K. E., Nowak K. J. Molecular mechanisms of muscular dystrophies: old and new players. Nature Reviews Molecular Cell Biology. 2006;7(10):762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 87.Rahimov F., Kunkel L. M. The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy. The Journal of Cell Biology. 2013;201(4):499–510. doi: 10.1083/jcb.201212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoffman E. P., Brown R. H., Jr., Kunkel L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 89.Matsuzaka Y., Kishi S., Aoki Y., et al. Three novel serum biomarkers, miR-1, miR-133a, and miR-206 for Limb-girdle muscular dystrophy, Facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environmental Health and Preventive Medicine. 2014;19(6):452–458. doi: 10.1007/s12199-014-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeanson-Leh L., Lameth J., Krimi S., et al. Serum profiling identifies novel muscle miRNA and cardiomyopathy-related miRNA biomarkers in golden retriever muscular dystrophy dogs and duchenne muscular dystrophy patients. The American Journal of Pathology. 2014;184(11):2885–2898. doi: 10.1016/j.ajpath.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 91.Mizuno H., Nakamura A., Aoki Y., et al. Identification of muscle-specific MicroRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0018388.e18388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCarthy J. J., Esser K. A., Andrade F. H. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. American Journal of Physiology: Cell Physiology. 2007;293(1):C451–C457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 93.Deng Z., Chen J.-F., Wang D.-Z. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskeletal Disorders. 2011;12, article 115 doi: 10.1186/1471-2474-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cacchiarelli D., Legnini I., Martone J., et al. miRNAs as serum biomarkers for Duchenne muscular dystrophy. The EMBO Molecular Medicine. 2011;3(5):258–265. doi: 10.1002/emmm.201100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zaharieva I. T., Calissano M., Scoto M., et al. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular dystrophy. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0080263.e80263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Endo K., Weng H., Naito Y., et al. Classification of various muscular tissues using miRNA profiling. Biomedical Research. 2013;34(6):289–299. doi: 10.2220/biomedres.34.289. [DOI] [PubMed] [Google Scholar]
- 97.Alexander M. S., Casar J. C., Motohashi N., et al. MicroRNA-486–dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy–associated symptoms. Journal of Clinical Investigation. 2014;124(6):2651–2667. doi: 10.1172/jci73579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L., Zhou L., Jiang P., et al. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Molecular Therapy. 2012;20(6):1222–1233. doi: 10.1038/mt.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alexander M. S., Kawahara G., Motohashi N., et al. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death and Differentiation. 2013;20(9):1194–1208. doi: 10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cacchiarelli D., Incitti T., Martone J., et al. MiR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. The EMBO Reports. 2011;12(2):136–141. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu N., Bezprozvannaya S., Shelton J. M., et al. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. The Journal of Clinical Investigation. 2011;121(8):3258–3268. doi: 10.1172/jci46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bitoun M., Durieux A.-C. C., Prudhon B., et al. Dynamin 2 mutations associated with human diseases impair clathrin-mediated receptor endocytosis. Human Mutation. 2009;30(10):1419–1427. doi: 10.1002/humu.21086. [DOI] [PubMed] [Google Scholar]
- 103.Durieux A.-C., Prudhon B., Guicheney P., Bitoun M. Dynamin 2 and human diseases. Journal of Molecular Medicine. 2010;88(4):339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 104.Merlino G., Helman L. J. Rhabdomyosarcoma—working out the pathways. Oncogene. 1999;18(38):5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 105.Wachtel M., Runge T., Leuschner I., et al. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. Journal of Clinical Oncology. 2006;24(5):816–822. doi: 10.1200/JCO.2005.03.4934. [DOI] [PubMed] [Google Scholar]
- 106.Missiaglia E., Shepherd C. J., Patel S., et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. British Journal of Cancer. 2010;102(12):1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taulli R., Bersani F., Foglizzo V., et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. The Journal of Clinical Investigation. 2009;119(8):2366–2378. doi: 10.1172/jci38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan D., Dong X. D., Chen X., et al. MicroRNA-1/206 targets c-met and inhibits rhabdomyosarcoma development. Journal of Biological Chemistry. 2009;284(43):29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X., Li Y., Zhao L., et al. Circulating muscle-specific miRNAs in Duchenne muscular dystrophy patients. Molecular Therapy—Nucleic Acids. 2014;3(7):p. e177. doi: 10.1038/mtna.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen C., Wang K., Chen J., et al. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. The Journal of Biological Chemistry. 2009;284(8):5362–5369. doi: 10.1074/jbc.m807523200. [DOI] [PubMed] [Google Scholar]
- 111.Liu N., Williams A. H., Maxeiner J. M., et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. Journal of Clinical Investigation. 2012;122(6):2054–2065. doi: 10.1172/jci62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang L., Chen X., Zheng Y., et al. MiR-23a inhibits myogenic differentiation through down regulation of fast myosin heavy chain isoforms. Experimental Cell Research. 2012;318(18):2324–2334. doi: 10.1016/j.yexcr.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 113.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0033766.e33766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Winbanks C. E., Wang B., Beyer C., et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. The Journal of Biological Chemistry. 2011;286(16):13805–13814. doi: 10.1074/jbc.m110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qadir A. S., Woo K. M., Ryoo H.-M., Yi T., Song S. U., Baek J.-H. MiR-124 inhibits myogenic differentiation of mesenchymal stem cells via targeting Dlx5. Journal of Cellular Biochemistry. 2014;115(9):1572–1581. doi: 10.1002/jcb.24821. [DOI] [PubMed] [Google Scholar]
- 116.Ge Y., Sun Y., Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. Journal of Cell Biology. 2011;192(1):69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Motohashi N., Alexander M. S., Shimizu-Motohashi Y., Myers J. A., Kawahara G., Kunkel L. M. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. Journal of Cell Science. 2013;126(12):2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J., Ying Z.-Z., Tang Z.-L., Long L.-Q., Li K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. Journal of Biological Chemistry. 2012;287(25):21093–21101. doi: 10.1074/jbc.M111.330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khanna N., Ge Y., Chen J. MicroRNA-146b promotes myogenic differentiation and modulates multiple gene targets in muscle cells. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0100657.e100657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seok H. Y., Tatsuguchi M., Callis T. E., He A., Pu W. T., Wang D.-Z. miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. The Journal of Biological Chemistry. 2011;286(41):35339–35346. doi: 10.1074/jbc.m111.273276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jia L., Li Y. F., Wu G. F., et al. MiRNA-199a-3p regulates C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal pathway. International Journal of Molecular Sciences. 2014;15(1):296–308. doi: 10.3390/ijms15010296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luo W., Wu H., Ye Y., et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death and Disease. 2014;5(7):p. e1347. doi: 10.1038/cddis.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yan B., Guo J.-T., Zhu C.-D., Zhao L.-H., Zhao J.-L. miR-203b: a novel regulator of MyoD expression in tilapia skeletal muscle. Journal of Experimental Biology. 2013;216(3):447–451. doi: 10.1242/jeb.076315. [DOI] [PubMed] [Google Scholar]
- 124.Feng Y., Cao J.-H., Li X.-Y., Zhao S.-H. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochemistry and Function. 2011;29(5):378–383. doi: 10.1002/cbf.1760. [DOI] [PubMed] [Google Scholar]
- 125.Tan S.-B., Li J., Chen X., et al. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chemistry & Biology. 2014;21(10):1265–1270. doi: 10.1016/j.chembiol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 126.Cardinalli B., Castellani L., Fasanaro P., et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE. 2009;4(10) doi: 10.1371/journal.pone.0007607.e7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen Y., Melton D. W., Gelfond J. A. L., McManus L. M., Shireman P. K. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiological Genomics. 2012;44(21):1042–1051. doi: 10.1152/physiolgenomics.00052.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gagan J., Dey B. K., Layer R., Yan Z., Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. The Journal of Biological Chemistry. 2011;286(22):19431–19438. doi: 10.1074/jbc.m111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheung T. H., Quach N. L., Charville G. W., et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482(7386):524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Crippa S., Cassano M., Messina G., et al. miR669a and miR669q prevent skeletal muscle differentiation in postnatal cardiac progenitors. Journal of Cell Biology. 2011;193(7):1197–1212. doi: 10.1083/jcb.201011099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Y., Gelfond J., McManus L. M., Shireman P. K. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiological Genomics. 2011;43(10):621–630. doi: 10.1152/physiolgenomics.00136.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dormoy-Raclet V., Cammas A., Celona B., et al. HuR and miR-1192 regulate myogenesis by modulating the translation of HMGB1 mRNA. Nature Communications. 2013;4, article 2388 doi: 10.1038/ncomms3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin C.-Y., Chen J.-S., Loo M.-R., Hsiao C.-C., Chang W.-Y., Tsai H.-J. MicroRNA-3906 regulates fast muscle differentiation through modulating the target gene homer-1b in zebrafish embryos. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0070187.e70187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ulitsky I., Bartel D. P. XLincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Derrien T., Johnson R., Bussotti G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guttman M., Rinn J. L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Batista P. J., Chang H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.de Santa F., Barozzi I., Mietton F., et al. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biology. 2010;8(5) doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim T.-K., Hemberg M., Gray J. M., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Orom U. A., Shiekhattar R. XLong noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154(6):X1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hah N., Murakami S., Nagari A., Danko C. G., Lee Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Research. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lai F., Orom U. A., Cesaroni M., et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lam M. T. Y., Cho H., Lesch H. P., et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li W., Notani D., Ma Q., et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Melo C. A., Drost J., Wijchers P. J., et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 146.Ørom U. A., Derrien T., Beringer M., et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang K. C., Yang Y. W., Liu B., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–126. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mousavi K., Zare H., Dell'Orso S., et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mueller A. C., Cichewicz M. A., Dey B. K., et al. MUNC: a lncRNA that induces the expression of pro-myogenic genes in skeletal myogenesis. Molecular and Cellular Biology. 2014 doi: 10.1128/MCB.01079-14. [DOI] [Google Scholar]
- 150.Lu L., Sun K., Chen X., et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO Journal. 2013;32(19):2575–2588. doi: 10.1038/emboj.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.da Rocha S. T., Edwards C. A., Ito M., Ogata T., Ferguson-Smith A. C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends in Genetics. 2008;24(6):306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 152.Zhao J., Ohsumi T. K., Kung J. T., et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhou Y., Cheunsuchon P., Nakayama Y., et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137(16):2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Caretti G., Schiltz R. L., Dilworth F. J., et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Developmental Cell. 2006;11(4):547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 155.Lanz R. B., McKenna N. J., Onate S. A., et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 156.Hubé F., Velasco G., Rollin J., Furling D., Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Research. 2011;39(2):513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Emberley E., Huang G.-J., Hamedani M. K., et al. Identification of new human coding steroid receptor RNA activator isoforms. Biochemical and Biophysical Research Communications. 2003;301(2):509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 158.Khosla S., Aitchison A., Gregory R., Allen N. D., Feil R. Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Molecular and Cellular Biology. 1999;19(4):2556–2566. doi: 10.1128/mcb.19.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 160.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 161.Forne T., Oswald J., Dean W., et al. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(19):10243–10248. doi: 10.1073/pnas.94.19.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ripoche M. A., Kress C., Poirier F., Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes and Development. 1997;11(12):1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 163.Runge S., Nielsen F. C., Nielsen J., Lykke-Andersen J., Wewer U. M., Christiansen J. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. The Journal of Biological Chemistry. 2000;275(38):29562–29569. doi: 10.1074/jbc.m001156200. [DOI] [PubMed] [Google Scholar]
- 164.Kallen A. N., Zhou X.-B., Xu J., et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Molecular Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cesana M., Cacchiarelli D., Legnini I., et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Molecular Cell. 2014;53(3):506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kapusta A., Kronenberg Z., Lynch V. J., et al. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genetics. 2013;9(4) doi: 10.1371/journal.pgen.1003470.e1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carrieri C., Cimatti L., Biagioli M., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 169.Gong C., Maquat L. E. LncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu eleme. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang J., Gong C., Maquat L. E. Control of myogenesis by rodent SINE-containing lncRNAs. Genes and Development. 2013;27(7):793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gutschner T., Hämmerle M., Eißmann M., et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Research. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tripathi V., Shen Z., Chakraborty A., et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genetics. 2013;9(3) doi: 10.1371/journal.pgen.1003368.e1003368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tripathi V., Ellis J. D., Shen Z., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang L., Lin C., Liu W., et al. NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thomas M., Langley B., Berry C., et al. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. Journal of Biological Chemistry. 2000;275(51):40235–40243. doi: 10.1074/jbc.m004356200. [DOI] [PubMed] [Google Scholar]
- 176.Watts R., Johnsen V. L., Shearer J., Hittel D. S. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. The American Journal of Physiology—Cell Physiology. 2013;304(10):C995–C1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 177.Bovolenta M., Erriquez D., Valli E., et al. The DMD locus harbours multiple long non-coding RNAs which orchestrate and control transcription of muscle dystrophin mRNA isoforms. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0045328.e45328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tran T. H. T., Zhang Z., Yagi M., et al. Molecular characterization of an X(p21.2;q28) chromosomal inversion in a Duchenne muscular dystrophy patient with mental retardation reveals a novel long non-coding gene on Xq28. Journal of Human Genetics. 2013;58(1):33–39. doi: 10.1038/jhg.2012.131. [DOI] [PubMed] [Google Scholar]
- 179.Tonini M. M. O., Passos-Bueno M. R., Cerqueira A., Matioli S. R., Pavanello R., Zatz M. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD) Neuromuscular Disorders. 2004;14(1):33–38. doi: 10.1016/j.nmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 180.Wijmenga C., Sandkuijl L. A., Moerer P., et al. Genetic linkage map of facioscapulohumeral muscular dystrophy and five polymorphic loci on chromosome 4q35-qter. The American Journal of Human Genetics. 1992;51(2):411–415. [PMC free article] [PubMed] [Google Scholar]
- 181.Hewitt J. E., Lyle R., Clark L. N., et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Human Molecular Genetics. 1994;3(8):1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 182.van Deutekom J. C. T., Wijmenga C., van Tienhoven E. A. E., et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Human Molecular Genetics. 1993;2(12):2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 183.Wijmenga C., Hewitt J. E., Sandkuijl L. A., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nature Genetics. 1992;2(1):26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 184.Cabianca D. S., Casa V., Bodega B., et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in fshd muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Anderson D. M., Anderson K. M., Chang C.-L., et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]