Abstract
Foodborne pathogens are a leading cause of mild-to-severe gastrointestinal illnesses worldwide. Escherichia coli pathotypes have been known to cause gastrointestinal illnesses in children less than 5 years old in Colombia. However, insufficient information is available on the prevalence of E. coli contamination of food products and the kind of E. coli food product reservoirs. The two objectives of this study were designed to address this issue. The first objective was to ascertain coliform, E. coli, and pathogenic E. coli contamination of food products readily available for human consumption in Cartagena, Colombia. The second objective was to evaluate the relationship between pathogenic E. coli isolated from food products and those isolated from cases of diarrhea in children. Food product samples consisting of pasteurized milk, unpasteurized fruit juice, ground beef, cheese, and vegetables were obtained at four retail stores. The food samples were cultured in liquid media and tested for the presence of coliforms and E. coli. E. coli isolates were tested by polymerase chain reaction for the presence of pathogenic E. coli. Coliforms, E. coli, and E. coli intestinal pathotypes contamination were detected in 88.4%, 53%, and 2.1% of food product samples, respectively. Ground beef and cheese were the only food samples contaminated with E. coli intestinal pathotypes including enteropathogenic (EPEC), Shiga toxin–producing (STEC), and enterotoxigenic E. coli (ETEC). Closed multilocus sequencing typing relationships between diarrheagenic E. coli isolates from food products and from individuals with diarrhea suggest that food products readily available at public markets in Cartagena can transmit ETEC and possibly EPEC and STEC. We demonstrated that a high proportion of food products for human consumption available at public markets in Cartagena are contaminated with coliforms, E. coli, and E. coli intestinal pathogens. Furthermore, food products containing E. coli intestinal pathogens may be involved in the transmission of foodborne illnesses among children in Cartagena, Colombia.
Introduction
Diarrheagenic Escherichia coli are well-known foodborne pathogens worldwide. They are classified into six distinct pathogenic categories based on their mechanisms of pathogenesis: enterotoxigenic (ETEC), Shiga toxin–producing/enterohemorrhagic (STEC/EHEC), enteropathogenic (EPEC), enteroinvasive (EIEC), enteroaggregative (EAEC), and diffusely adherent (DAEC) E. coli (Kaper et al., 2004). Shiga toxin–producing E. coli (STEC) O157:H7 strains are highly virulent foodborne pathogens associated with outbreaks of bloody diarrhea, and hemolytic uremic syndrome (HUS) in the United States, Europe, and other countries around the world (Tarr et al., 2005). O157 STEC infections have been estimated to cause 73,000 illnesses in 2005 in the United States, resulting in more than 2000 hospitalizations and 60 deaths (Gould et al., 2013a).
Although clinical studies have indicated the importance of E. coli pathotypes in childhood diarrhea in Colombia, studies on food product contamination by E. coli pathogens in Colombia are limited (Gómez-Duarte et al., 2010; Rúgeles et al., 2010). A few studies report that meats and vegetables at retail were contaminated with two pathogenic E. coli strains, STEC and EAEC (Mattar and Vásquez, 1998; Rúgeles et al., 2010). However, no study has evaluated the genetic relatedness between diarrheagenic E. coli derived from food products and those isolated from children with diarrhea.
A recent case–control study reported a high prevalence of E. coli diseases in Cartagena among children less than 5 year old (Gomez-Duarte et al., 2013). It led us to ask whether foodborne and/or waterborne pathogens are playing a role in the epidemiology of E. coli intestinal illnesses. The objective of this study was to identify the most frequently contaminated food products for human consumption and the most frequent E. coli intestinal pathogens. More importantly, we evaluated whether diarrheagenic E. coli from food products share similarities with previously reported diarrheagenic E. coli from children with diarrhea.
Materials and Methods
Food product collection
Food products were purchased at four main retail stores (stores 1–4) from 2012 to 2013. The stores were located in four different neighborhoods within the city of Cartagena, Colombia. Nineteen samples corresponding to five consumable products (cheese, ground beef, vegetables, pasteurized milk, and unpasteurized juice) were collected at each store for a total of 380 samples. Vegetables collected were carrots, spinach, celery, and/or lettuce. Vegetables collected were fresh, and displayed in open air. None of the vegetables used were prepacked, precooked, or frozen. Solid food samples were collected in aseptic plastic bags. Pasteurized milk was collected in the original container. Unpasteurized juice samples were collected in sterile bottles. All samples were transported at 8°C to the microbiology laboratory at the University of Cartagena for further processing.
Processing of food products
Solid and liquid food product samples were evaluated for bacterial growth, presence of coliforms, and E. coli. Ten grams of solid food product samples were added to 90 mL peptone buffered water (EMD Millipore, Billerica, MA) and homogenized in a sterile blender for 1–2 min at low speed. Liquid samples were not mixed with peptone buffered water. Two hundred milliliters of lactose broth (EMD Millipore) was inoculated with 1 mL of homogenized solid food or liquid food sample. The suspension was incubated at 37°C for 24 h with shaking. Samples with no bacterial growth on lactose broth were discarded. Samples positive for bacterial growth with no gas production were presumed to contain Gram-positive organisms and were also discarded. Food samples positive for bacterial growth with gas production in lactose broth were assumed to contain Gram-negative organisms. Lactose fermenter bacterial suspensions were subsequently cultured on Fluorocult medium (Fluorocult® LMX broth; Thomas Scientific, Swedesboro, NJ) to determine total coliforms and E. coli. One-hundred milliliters of Fluorocult broth was inoculated with 1 mL of lactose broth bacterial suspension (gas-producer) and incubated for 24 h at 37°C. Fluorocult medium bacterial suspensions that emitted fluorescence under ultraviolet light were considered positive for E. coli. An aliquot of the suspension was plated on McConkey agar for isolation of individual colonies. Individual bacterial isolates were confirmed as E. coli by the presence of lactose-fermenting colonies on McConkey agar, metallic green colonies on eosin methylene blue (EMB) agar, positive testing for indol, β-glucuronidase, methyl red, and negative testing for citrate. Up to two E. coli isolates per food sample were stored in Luria broth (LB) supplemented with 20% glycerol at −80°C for further testing.
DNA amplification by polymerase chain reaction (PCR)
Multilocus sequence typing (MLST)
E. coli clinical isolates were analyzed by MLST as described online (http://mlst.warwick.ac.uk/mlst/). Internal fragments from seven housekeeping genes were amplified by PCR and DNA sequenced as described before (Wirth et al., 2006). Sequence editing was conducted with DNADynamo software (Blue Tractor Software Ltd., North Wales, UK). Seven genes sequences for each strain were concatenated to generate a 3423-bp aligned DNA sequence, and concatamers were aligned using ClustalW software (available online at: http://www.phylogeny.fr/version2_cgi/simple_phylogeny.cgi) (Dereeper et al., 2008, 2010). Phylogenetic trees from E. coli MLST concatamers were conducted by the bootstrapping procedure. E. coli control MLSTs included STEC, EPEC, and ETEC strains reported in the http://mlst.warwick.ac.uk/mlst/ database and previously isolated from human cases of diarrhea. Control E. coli ancestral phylogenetic groups sequences were also obtained from the same database. MLST-based clonal groups were defined as a group of more than one E. coli strain MLST sequence that do not seem to share ancestral origin with other E. coli and that have at least two strains with identical MLST DNA sequence.
DNA amplification by PCR assays were performed as described earlier with slight modifications (Gómez-Duarte et al., 2010). In brief, individual bacterial isolates were cultured overnight on 2 mL LB at 37°C with shaking. Bacterial cultures were centrifuged, pellet resuspended in Tris-EDTA buffer (10 mM Tris, pH 8.0, 0.1 mM EDTA in distilled water), and boiled for 5 min at 95°C. Bacterial suspensions were centrifuged and the crude genomic DNA within the supernatant was transferred to a fresh tube and used as DNA template for PCR reactions. Multiplex PCR (mPCR) for detection of E. coli pathotypes was performed as described earlier (Gannon et al., 1997; Gómez-Duarte et al., 2010) with the following modifications. Two reactions were run for each sample using two different sets of primers. Each 25-μL reaction mixture contained 23 μL of Platinum Blue PCR SuperMix (Invitrogen, Carlsbad, CA), 1 μL of primer mix (Mix1 or Mix2) (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/fpd), and 1 μL of template genomic DNA. Primer Mix1 (58°C annealing temperature) was prepared for amplification of eae/bfpA, stx1/stx 2, and aggR for detection of EPEC, STEC, and EAEC pathotypes, respectively. Primer Mix2 (56°C annealing temperature) was prepared for amplification of ST/LTb, virF/ipaH, and daaE for detection of ETEC, EIEC, and DAEC, respectively. Both PCR mixes were denatured at 94°C for 5 min, followed by 40 cycles of 92°C for 30-s denaturation, annealing for 30 s (at 56°C for PCR mix1 and 58°C for 30 s for PCR mix2), and extension for 30 s at 72°C. PCR products were separated on 3% (wt/vol) agarose gel in Tris acetate EDTA buffer (pH 8.2), stained with ethidium bromide (10 μg/mL), visualized, and recorded under ultraviolet light. Diarrheagenic E. coli reference strains were used as positive controls: EPEC E2348/69 (eae+, bfpA+), STEC O157:H7 2060-004 (eae, stx1, stx2), EIEC EC-12 (ipaH, virF), DAEC C1845 (daaE), and EAEC JM221 (aggR) (Gómez-Duarte et al., 2009). The E. coli DH5 α strain (negative for all virulence genes) was used as a negative control. Each pathotype identified by mPCR was confirmed by single PCR amplification of specific gene targets. This mPCR assay has been validated in prior studies, and differential amplification of DNA did not affect assay sensitivity or specificity (Gómez-Duarte et al., 2009, 2010).
Serotyping
O and H typing of diarrheagenic E. coli isolates were performed at Penn State E. coli Reference Center (Pennsylvania State University, University Park, PA). O serotyping was conducted using antisera generated against E. coli serogroups designated O1-O187 with the exceptions of O31, O47, O67, O72, O94, and O122, as these are not yet designated. H typing was performed by PCR–restriction fragment length polymorphism of fliC flagellar gene responsible for H types.
Antimicrobial susceptibility testing
BBL™ Prompt™ Inoculation System (Becton Dickinson & Co., Franklin Lakes, NJ) was used to prepare standardized bacterial suspensions for the Bauer-Kirby disc-diffusion antimicrobial susceptibility test. In brief, bacterial isolates were grown overnight on LB agar plates at 37°C. Five individual colonies were harvested to create a bacterial suspension following the manufacturer recommendations. Muller-Hinton agar plates were inoculated with the BBL Prompt Innoculation system bacterial suspension. Antimicrobial susceptibility to 12 different antibiotics was tested using BD BBL™ Sensi-Disc™ Susceptibility Test Discs methods (Becton, Dickinson and Company). Strain activity was tested against cefazolin (CZ), ceftriaxone (CTX); ampicillin (A), amoxicillin/clavulanic acid (AC); ceftazidime (CZ), cefuroxime (CX), cefepime (CP), ciprofloxacin (CIP), gentamicin (GM), meropenem (MEM), trimethoprim/sulfamethoxazole (STX), and piperacillin/tazobactam (TZP). The zones of bacterial growth inhibition were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (Qin et al., 2008). The E. coli ATCC 29522 strain was used as the negative control (susceptible to all antibiotics). Klebsiella pneumoniae ATCC 700603D-5 strain was used as the positive control (resistant to all β-lactam antibiotics).
Statistical analysis
Fisher exact test and one-way analysis of variance were used to determine statistical significances of the differences among food product samples or among retail stores. All statistical calculations were done using IBM SPSS Statistics for Windows, Version 22. For all calculations, the p value for statistical significance was set at <0.01.
Results
Identification of total coliforms and E. coli isolates from food products
Three hundred eighty food samples collected from four different retail stores in Cartagena, Colombia were assayed to detect total coliforms and E. coli contamination (Supplementary Tables S2 and S3). Coliform was found in 334 of the 380 samples. Cheese, ground beef, unpasteurized juice and vegetables were the most frequently contaminated products, with 297 (97.7%) samples positive out of 304 (Supplementary Table S2). In contrast, pasteurized milk was the least frequently contaminated food product, with only 37 (48.7%) samples positive out of a total of 76. The lower frequency of pasteurized milk contamination was statistically significant (p<0.01) when compared to the remaining products. The numbers of coliforms-contaminated products per retail store ranged from 80 to 89 of 95 samples each. The difference in numbers was not statistically significant, suggesting a similar frequency of coliform contamination across retail stores in Cartagena.
Food products contaminated with E. coli were detected in 201 (52.8%) of 380 samples (Table 1). Cheese and ground beef were the most frequently contaminated samples at 96% and 100%, respectively (Supplementary Table S3). Pasteurized milk, unpasteurized juice, and vegetable samples were less frequently contaminated. Pasteurized milk was the least frequently contaminated product, affecting only 6.6% of the samples. The difference in the frequencies of E. coli contamination between pasteurized milk and ground beef or cheese was statistically significant (p<0.01). Similarly, the difference in E. coli contamination between unpasteurized juice or vegetables when compared to either ground beef or cheese was statistically significant (p<0.01). As with the coliform, the frequencies of E. coli–contaminated products across the retail stores were similar to each other, and the differences were not statistically significant.
Table 1.
Identification of Escherichia coli from Food Products
| Positive E. coli | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative E. coli | Pathotypes | No pathotypes | Total E. coli | Total | ||||||
| Sample | No. | % | No. | % | No. | % | No. | % | No. | % |
| Cheese | 3 | 4 | 1 | 1 | 72 | 95 | 73 | 96 | 76 | 100 |
| Pasteurized milk | 71 | 93 | 0 | 0 | 5 | 7 | 5 | 7 | 76 | 100 |
| Unpasteurized juice | 55 | 72 | 0 | 0 | 21 | 28 | 21 | 28 | 76 | 100 |
| Ground beef | 0 | 0 | 7 | 9 | 69 | 91 | 76 | 100 | 76 | 100 |
| Vegetable | 50 | 66 | 0 | 0 | 26 | 34 | 26 | 34 | 76 | 100 |
| Total | 179 | 8 | 193 | 201 | 380 | |||||
Diarrheagenic E. coli isolates present in ground beef and cheese samples
Diarrheagenic E. coli, defined as those isolates carrying virulence genes unique to EPEC, STEC, and ETEC, were identified in 8 (2.0%) of the 380 food samples (Table 1). The frequency of contamination of samples with diarrheagenic E. coli in all the samples that were contaminated with E. coli was 4.0%. Diarrheagenic E. coli was found in 9.2% of the ground beef samples. From eight diarrheagenic E. coli strains identified, seven were detected in ground beef and one in a cheese sample. Four ground beef samples were contaminated with STEC non-O157, three samples with ETEC, and one sample with EPEC. The diarrheagenic E. coli–contaminated cheese sample was positive for EPEC (Table 2).
Table 2.
Genotype and Phenotype of Diarrheagenic Escherichia coli Food Product Isolates
| No. | Name | Pathotype | Virulence genes | Serotype O:H | MLST seq. type | Antibiotic resistance | Food product |
|---|---|---|---|---|---|---|---|
| 1 | CCtF019 | ETEC | lt | 128:16 | 1717 | A, AC | Ground beef |
| 2 | CCtF047 | STEC | stx-2 | 8:19 | 4495a | A, AC | Ground beef |
| 3 | CCtF054 | STEC | stx-2 | 8:19 | 1431 | A, AC | Ground beef |
| 4 | CCtF171.2 | ETEC | lt | 25:12 | 607 | A | Ground beef |
| 5 | CCTF197 | ETEC | st | -:10 | 10 | — | Ground beef |
| 6 | CCtF203.1 | STEC | stx-2 | 8:28 | 4496a | A, AC | Ground beef |
| 7 | CCtF203.2 | STEC | stx-2 | 8:28 | 4496a | A, AC | Ground beef |
| 8 | CCtF317 | EPEC | eae | 126:+ | 58 | A, AC, T/S | Cheese |
New sequence types submitted to the E. coli multilocus sequence typing (MLST) database website: http://mlst.warwick.ac.uk/mlst/.
A, ampicillin; AC, amoxicillin/clavulanate; ETEC, enterotoxigenic E. coli; STEC, Shiga toxin–producing E. coli; T/S, trimethoprim/sulfamethoxazole.
Diarrheagenic E. coli isolates from food-product-shared serotypes and MLST with human diarrheagenic E. coli isolates
To compare the diarrheagenic E. coli isolates from food products and the previously reported diarrheagenic E. coli from children with diarrhea, we performed O:H serotyping and MLST. All E. coli pathotype strains identified from food products were serotyped. A total of 4 O serogroups were identified, including O8, O25, O126, and O128. In one of the strains, no O serogroup was found. A total of 6 H serogroups were identified including H10, H12, H16, H19, H28, and H+. All 4 STEC strains belong to a single O8 serogroup, 2 of them belong to H19 serogroup, and the remaining 2 to H28 serogroup (Table 2).
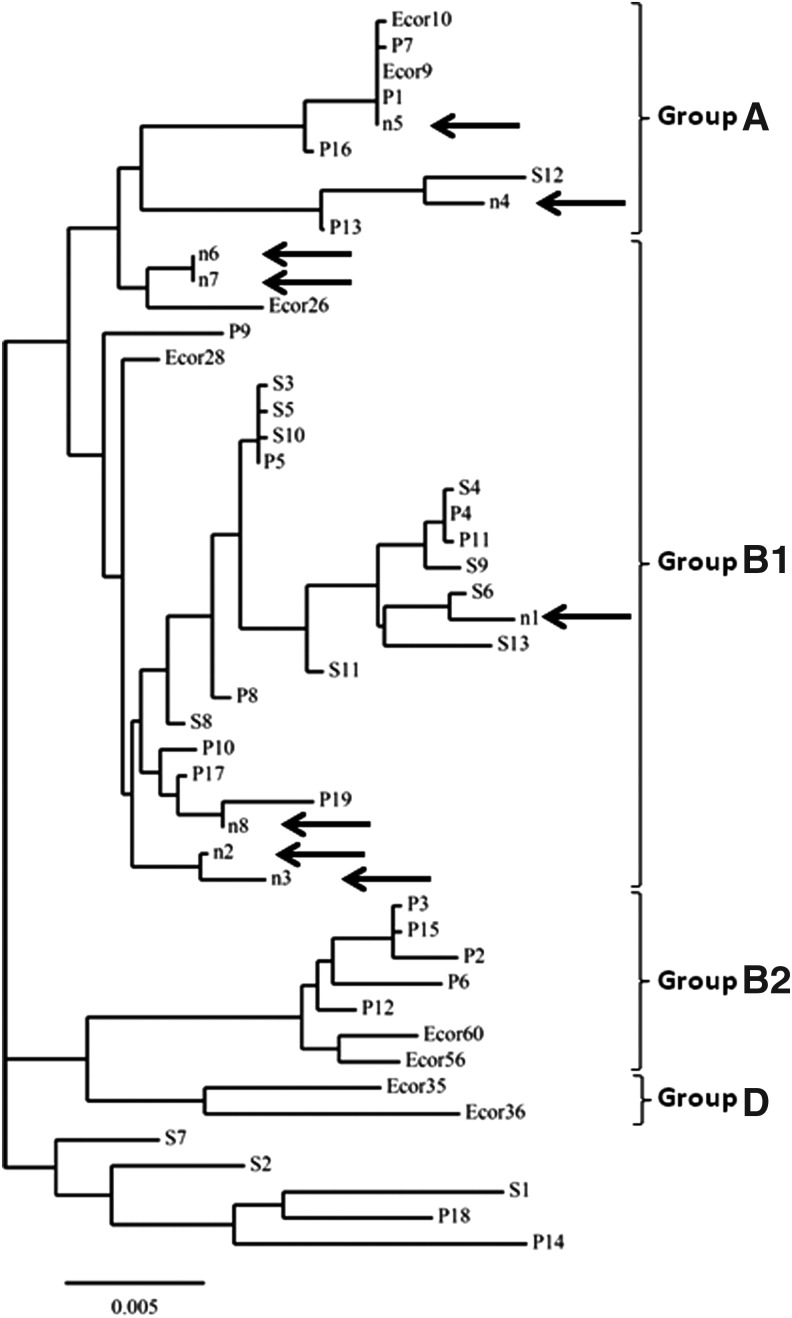
MLST analysis was performed on all diarrheagenic E. coli isolates from food samples to study their genetic diversity. We identified five previously reported sequence types and two novel sequence types among these E. coli isolates (Table 2). The 2 new sequence types ST4495 and ST4496 have been submitted to the MLST database. They were present in three STEC isolates. These strains also expressed O8 serogroup (Table 2). A phylogenetic tree constructed based on the MLST concatamer sequence alignments revealed a close relationship among E. coli pathotypes sharing identical serotype. STEC strains CCtF203.1 and CCtF203.2 share identical O8:H28 serotype and MLST sequence type. Similarly, STEC strains CCtF047 and CCtF054 share an identical O8:H19 serotype and grouped closely in the MLST phylogenetic tree (Fig. 1). All identified E. coli pathotypes from food products clustered with the strains that belong to the ancestral groups B1 and A. No E. coli isolate clustered with the ancestral groups B2 or D.
FIG. 1.
Multilocus sequence typing (MLST) phylogenetic tree of Shiga toxin–producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolates from food products. A phylogenetic tree was constructed by assembly and alignment of MLST DNA sequences using the ClustalW program. E. coli strain MLST sequences from ancestral groups A, B1, B2, and D as well as STEC and EPEC sequences, available in the MLST database (http://mlst.warwick.ac.uk/mlst/), were used as controls. STEC controls include the following: S1, STEC 055/89; S2, STEC 493/89; S3, STEC CL/37; S4, STEC DEC11C; S5, STEC 3593/00; S6, STEC 3199/98; S7, STEC 5122/99; S8, STEC 4797/97; S9, STEC E135360; S10, STEC 5244-00-2; S11, STEC 07-07786; S12, STEC 06-EGY30; S13, STEC NIPH-11060424. EPEC controls include: P1, EPEC to HC68; P2, EPEC DEC2A; P3, EPEC 2348/69; P4, EPEC HC10; P5, EPEC HC15; P6, EPEC Trh36; P7, EPEC DEC6A; P8, EPEC DEC12B; P9, EPEC Trh37; P10, EPEC 181; P11, EPEC HC40; P12, EPEC HC36; P13, EPEC HC91; P14, EPEC HC95; P15, EPEC HC59; P16, EPEC HC66; P17, EPEC HC87; P18, EPEC 109; P19, EPEC 219. E. coli ancestral control strain sequences were derived from the ECOR collection. Food product STEC and EPEC isolates are represented by n: n2 CCtF047 (STEC); n3, CCtF054 (STEC); n6, CCtF203.1 (STEC); n7, CCtF2203.2 (STEC); n8, CCtF317 (EPEC).
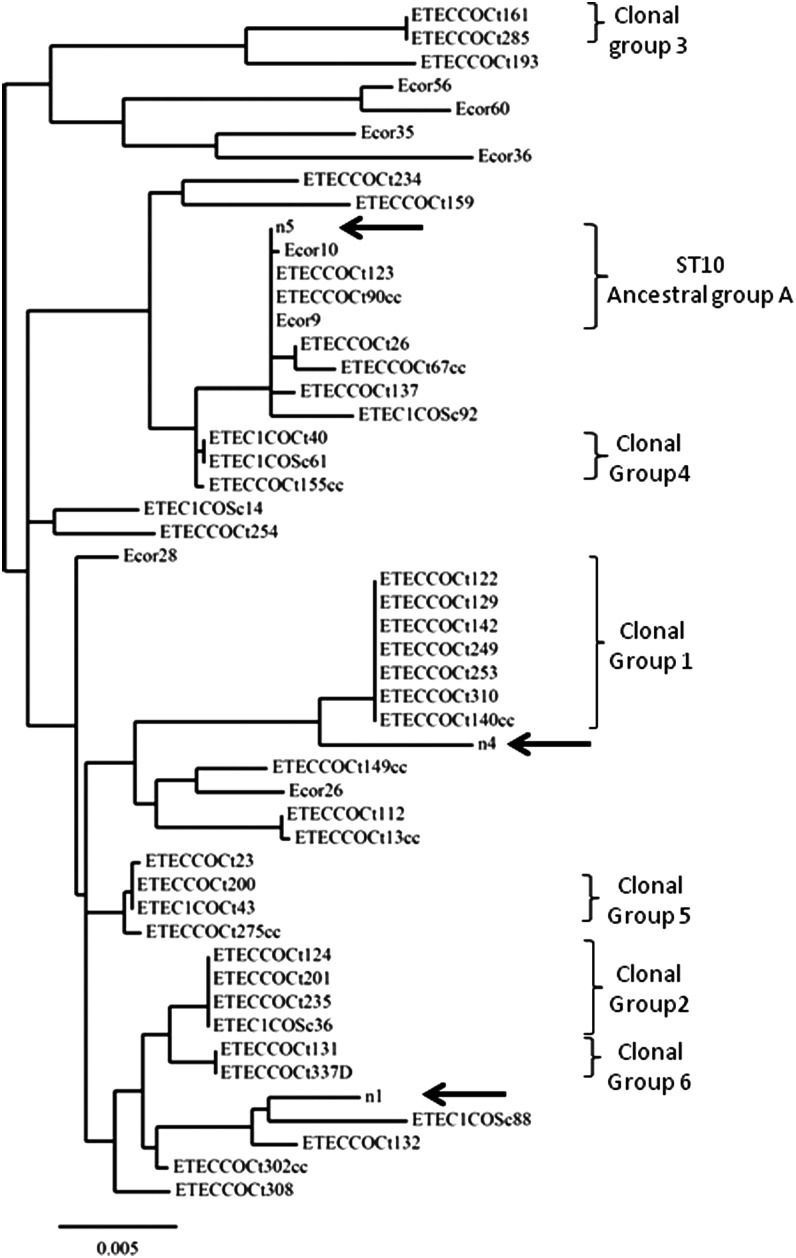
MLST analysis revealed that all ETEC strains from food products grouped with the previously reported Colombian human ETEC strains (Fig. 1). The ETEC CCtF197 isolate had an MLST sequence type identical to the human ETEC clinical isolates COCt123 and COCt90cc and to the ancestral E. coli ECOR9 strain (Fig. 2). These data suggest that the ETEC isolates and the E. coli ECOR9 strain belong to the sequence type ST10 and to the ancestral E. coli group A (Figs. 1 and 2).
FIG. 2.
Multilocus sequence typing (MLST) phylogenetic tree of Escherichia coli pathotypes from food products. The phylogenetic tree was constructed by assembly and alignment of MLST DNA sequences using the ClustalW program. E. coli strain MLST sequences from ancestral groups A, B1, B2, and D as well as enterotoxigenic E. coli (ETEC) sequences, available in the MLST database, were used as controls. E. coli ancestral control strain sequences were derived from the ECOR collection. Food product ETEC isolates are represented by n: n1, CCtF019; n4, CCtF171.2; and n5, CCtF197. MLST-based clonal groups 1–6 correspond to human ETEC clinical isolates previously reported (Guerra et al., 2014).
Low antimicrobial resistance identified among E. coli isolated from food products
Resistance to ampicillin, amoxicillin/clavulanic acid, and trimethoprim/sulfamethoxazole was detected in 7/8 (87.5%), 6/8 (75.0%), and 1/8 (12.5%) diarrheagenic E. coli isolates, respectively. Six of eight diarrheagenic E. coli strains were resistant to more than one antibiotic. One EPEC strain recovered from a cheese sample was resistant to all of the three antibiotics. Twenty-six E. coli food product isolates, 8 diarrheagenic (Table 2) and 18 nondiarrheagenic, were evaluated for antimicrobial susceptibility. Similar to diarrheagenic E. coli, nondiarrheagenic strains were predominantly resistant to ampicillin and amoxicillin/clavulanic acid. They were sensitive to the remaining antibiotics (data not shown).
Discussion
Foodborne illnesses are associated with high morbidity and mortality among humans in both industrialized and developing countries. Bacterial pathogens are the primary etiological agents in foodborne illnesses (Khabbaz et al., 2014). This study reports bacterial contamination in a large proportion of food products available at retail markets in Cartagena, Colombia. Detection of coliforms, E. coli, and diarrheagenic E. coli pathogens among food product samples is an indication of fecal contamination. Furthermore, this level of contamination places the local community, especially children, at a higher risk of foodborne illnesses.
E. coli contamination of ground beef, cheese, and vegetables is an evidence of fecal contamination that may occur at any stage in the food chain, including production, harvesting, storage, packaging, or purchasing. Contamination of vegetables frequently occurs after irrigation using untreated sewage water (Castro-Rosas et al., 2012). Ground beef contamination generally occurs during slaughtering and processing, while cheese contamination may occur during manufacturing and processing (Vernozy-Rozand et al., 2005; Gould et al., 2011). No difference in the total numbers of contaminated food products was identified among different retail stores. Similarly, no difference was observed in the type of contaminated food products detected per store.
Undercooked ground beef, unpasteurized (raw) milk and juice, soft cheese made from raw milk, and raw fruits and vegetables are frequently contaminated with E. coli (Gould et al., 2013a, b). These products are important foodborne pathogen vectors in several South American countries (Pires et al., 2012). The high frequency of pasteurized milk contamination in retail stores in Cartagena, Colombia may indicate postprocessing contamination or ineffective pasteurization technique. It is of great concern that 48.7% of the pasteurized milk samples were contaminated with coliforms and that 6.6% were contaminated with E. coli. This high bacterial contamination level represents a high risk of foodborne illnesses to the local consumers. It also raises the question of whether milk contamination is widespread in Colombia.
Ground beef was frequently contaminated with coliforms and E. coli in this study. Seven (9.2%) of 76 samples were contaminated with pathogenic E. coli, including atypical EPEC, STEC, and ETEC. Cheese samples were also often contaminated with coliforms and E. coli. However, only one of the samples (1.3%) was contaminated with EPEC. Overall, a total of eight (2%) food product samples were contaminated with pathogenic E. coli. Similar E. coli pathotypes have been reported in food products from Mexico with the exception of EAEC, which was not identified in our study (Canizalez-Roman et al., 2013). Ground beef contamination with STEC is a leading cause of dysenteric diarrhea foodborne outbreaks and HUS (Rivas et al., 2006; Werber et al., 2012). Countries in Latin America and Europe, in addition to the United States, have reported ground beef contamination with STEC (Brusa et al., 2012; Soborg et al., 2013; Robbins et al., 2014).
Cattle and pork meat contaminated with O157:H7 STEC was reported in Colombia in 1998 (Mattar and Vásquez, 1998). Subsequent studies indicated that STECs detected in food products are predominantly non-O157 STECs serotypes (Martínez et al., 2007; Rúgeles et al., 2010). O8 was the predominant serogroup among STEC strains isolated from ground beef in this study. While the O8 serogroup has not been associated with cases of human disease in the United States, it has been associated with human illness, including HUS, in Argentina, Germany, and Sweden (Friedrich et al., 2002; Johnson et al., 2006). O8:H19 is one of the most commonly identified serotypes among STECs recovered from beef in Latin American countries, including Mexico and Argentina (Meichtri et al., 2004; Amézquita-López et al., 2012).
Detection of ETEC among ground beef samples in Colombia is surprising, as ETEC is frequently recognized as a waterborne pathogen, and less commonly as a foodborne pathogen (Daniels et al., 2000). However, the identification of ETEC strains in food products is not very unusual as they have been previously reported in Mexico, Brazil, and Burkina Faso (Echeverria et al., 1991; Kagambèga et al., 2012; Gómez-Aldapa et al., 2014; Ayulo et al., 1994). Here we report that ETEC isolates were detected in three individual ground beef samples. One of the ETEC isolates, ETEC CCtF157 strain, was positive for the ST toxin gene and its MLST sequence was identical to 2 previously reported ETEC clinical isolates from Cartagena (Guerra et al., 2014). This isolate was also related to the ECOR9 strain, an ancestral E. coli from clonal group A. The remaining ETEC strains were positive for LT. They clustered with previously reported ETEC strains isolated from children with diarrhea in Colombia with MLST analysis. Phylogenetic typing using MLST suggests that ETEC-contaminated ground beef available at retail may contribute to the transmission and elevated ETEC diarrhea morbidity in children living in Cartagena, Colombia.
We identified an atypical EPEC strain positive for eae and negative for bfpA, from a cheese sample. The EPEC isolate belonged to O126 serogroup that matched to MLST ST58. Although O126 is recognized as one of the classical EPEC O groups (Trabulsi et al., 2002), the sequence type ST58 was not only present among EPEC strains but also among EIEC and some extraintestinal pathogenic E. coli strains (http://mlst.warwick.ac.uk/mlst/). Atypical EPECs have caused large outbreaks of diarrheal disease in both children and adults (Kaper et al., 2004). In industrialized countries, atypical EPEC strains are more frequently isolated from diarrheal cases than typical EPECs expressing the bundle-forming pili.
Antibiotic susceptibility among E. coli strains including diarrheagenic and nondiarrheagenic was similar. Most of the isolates were resistant to ampicillin and to amoxicillin/clavulanic acid. Resistance to trimethoprim/sulfamethoxazole was less frequent. This is in contrast to antibiotic multiresistance patterns reported among diarrheagenic E. coli strains from food product samples in other Latin American countries including Brazil, Mexico, and Peru (Garcia et al., 2011; Canizalez-Roman et al., 2013; Pons et al., 2014).
In summary, this is the first study of diarrheagenic E. coli contamination of the food supply in Cartagena, Colombia. This study highlights the high level of fecal contamination of food products with coliforms and E. coli in public markets and the presence of non-O157 STEC and ETEC isolates among food products, particularly ground beef. MLST analysis of diarrheagenic E. coli isolates revealed striking identity or similarity with diarrheagenic E. coli from children with diarrhea. It suggests that the food products readily available at public markets in Cartagena, Colombia may be involved in the transmission of the diarrheagenic E. coli foodborne illnesses. More food product surveillance studies are needed to evaluate the extent of the food product contamination at retail stores in the Colombian cities to measure the frequency and the types of foodborne pathogens. These studies are important for guiding the health policy changes directed at improving food quality and safety during production, processing, and delivery to the public. These policy changes should ultimately help prevent foodborne illness outbreaks, including those associated with high burden of disease and fatal outcomes.
Supplementary Material
Acknowledgments
We are grateful to Mr. Parimal Samir for manuscript editing and helpful advice. Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Number R01AI095346 to O.G.G-D. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure Statement
No competing financial interests exist.
References
- Amézquita-López BA, Quiñones B, Cooley MB, León-Félix J, Castro-del Campo N, Mandrell RE, Jiménez M, Chaidez C. Genotypic analyses of Shiga toxin–producing Escherichia coli O157 and non-O157 recovered from feces of domestic animals on rural farms in Mexico. PloS ONE 2012;7:e51565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayulo AM, Machado RA, Scussel VM. Enterotoxigenic Escherichia coli and Staphylococcus aureus in fish and seafood from the southern region of Brazil. Int J Food Microbiol 1994;24:171–178 [DOI] [PubMed] [Google Scholar]
- Brusa V, Aliverti V, Aliverti F, Ortega EE, de la Torre JH, Linares LH, Sanz ME, Etcheverría AI, Padola NL, Galli L, et al. . Shiga toxin–producing Escherichia coli in beef retail markets from Argentina. Front Cell Infect Microbiol 2012;2:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canizalez-Roman A, Gonzalez-Nuñez E, Vidal JE, Flores-Villaseñor H, León-Sicairos N. Prevalence and antibiotic resistance profiles of diarrheagenic Escherichia coli strains isolated from food items in northwestern Mexico. Int J Food Microbiol 2013;164:36–45 [DOI] [PubMed] [Google Scholar]
- Castro-Rosas J, Cerna-Cortés JF, Méndez-Reyes E, Lopez-Hernandez D, Gómez-Aldapa CA, Estrada-Garcia T. Presence of faecal coliforms, Escherichia coli and diarrheagenic E. coli pathotypes in ready-to-eat salads, from an area where crops are irrigated with untreated sewage water. Int J Food Microbiol 2012;156:176–180 [DOI] [PubMed] [Google Scholar]
- Daniels NA, Neimann J, Karpati A, Parashar UD, Greene KD, Wells JG, Srivastava A, Tauxe RV, Mintz ED, Quick R. Traveler's diarrhea at sea: Three outbreaks of waterborne enterotoxigenic Escherichia coli on cruise ships. J Infect Dis 2000;181:1491–1495 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, et al. . Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie J-M, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 2010;10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria P, Sethabutr O, Pitarangsi C. Microbiology and diagnosis of infections with Shigella and enteroinvasive Escherichia coli. Rev Infect Dis 1991;13(Suppl 4):S220–S225 [DOI] [PubMed] [Google Scholar]
- Friedrich AW, Bielaszewska M, Zhang W-L, Pulz M, Kuczius T, Ammon A, Karch H. Escherichia coli harboring Shiga toxin 2 gene variants: Frequency and association with clinical symptoms. J Infect Dis 2002;185:74–84 [DOI] [PubMed] [Google Scholar]
- Gannon VP, D'Souza S, Graham T, King RK. Specific identification of Escherichia coli O157:H7 using a multiplex PCR assay. Adv Exp Med Biol 1997;412:81–82 [DOI] [PubMed] [Google Scholar]
- Garcia PG, Silva VL, Diniz CG. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecal microbiota from children with and without acute diarrhea. J Microbiol Seoul Korea 2011;49:46–52 [DOI] [PubMed] [Google Scholar]
- Gómez-Aldapa CA, Rangel-Vargas E, Bautista-De León H, Castro-Rosas J. Presence of non-O157 Shiga toxin–producing Escherichia coli, enterotoxigenic E. coli, enteropathogenic E. coli and Salmonella in fresh beetroot (Beta vulgaris L.) juice from public markets in Mexico. J Sci Food Agric 2014;94:2705–2711 [DOI] [PubMed] [Google Scholar]
- Gómez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis 2009;63:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Duarte OG, Arzuza O, Urbina D, Bai J, Guerra J, Montes O, Puello M, Mendoza K, Castro GY. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children's diarrheal stools in two Caribbean-Colombian cities. Foodborne Pathog Dis 2010;7:199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Duarte OG, Romero-Herazo YC, Paez-Canro CZ, Eslava-Schmalbach JH, Arzuza O. Enterotoxigenic Escherichia coli associated with childhood diarrhoea in Colombia, South America. J Infect Dev Ctries 2013;7:372–381 [DOI] [PubMed] [Google Scholar]
- Gould LH, Seys S, Everstine K, Norton D, Ripley D, Reimann D, Dreyfuss M, Chen WS, Selman CA. Recordkeeping practices of beef grinding activities at retail establishments. J Food Prot 2011;74:1022–1024 [DOI] [PubMed] [Google Scholar]
- Gould LH, Walsh KA, Vieira AR, Herman K, Williams IT, Hall AJ, Cole D, Centers for Disease Control and Prevention. Surveillance for foodborne disease outbreaks—United States, 1998–2008. MMWR Morb Mortal Wkly Rep 2013a;62:1–34 [PubMed] [Google Scholar]
- Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, et al. . Increased recognition of non-O157 Shiga toxin–producing Escherichia coli infections in the United States during 2000–2010: Epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 2013b;10:453–460 [DOI] [PubMed] [Google Scholar]
- Guerra JA, Romero-Herazo YC, Arzuza O, Gómez-Duarte OG. Phenotypic and genotypic characterization of enterotoxigenic Escherichia coli clinical isolates from Northern Colombia, South America. BioMed Res Int 2014;2014:236–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin–producing Escherichia coli. Clin Infect Dis 2006;43:1587–1595 [DOI] [PubMed] [Google Scholar]
- Kagambèga A, Barro N, Traoré AS, Siitonen A, Haukka K. Characterization of Salmonella enterica and detection of the virulence genes specific to diarrheagenic Escherichia coli from poultry carcasses in Ouagadougou, Burkina Faso. Foodborne Pathog Dis 2012;9:589–593 [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol 2004;2:123–140 [DOI] [PubMed] [Google Scholar]
- Khabbaz RF, Moseley RR, Steiner RJ, Levitt AM, Bell BP. Challenges of infectious diseases in the USA. Lancet 2014;384:53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez AJ, Bossio CP, Durango AC, Vanegas MC. Characterization of Shiga toxigenic Escherichia coli isolated from foods. J Food Prot 2007;70:2843–2846 [DOI] [PubMed] [Google Scholar]
- Mattar S, Vásquez E. Escherichia coli O157:H7 infection in Colombia. Emerg Infect Dis 1998;4:126–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meichtri L, Miliwebsky E, Gioffré A, Chinen I, Baschkier A, Chillemi G, Guth BEC, Masana MO, Cataldi A, Rodríguez HR, et al. . Shiga toxin–producing Escherichia coli in healthy young beef steers from Argentina: Prevalence and virulence properties. Int J Food Microbiol 2004;96:189–198 [DOI] [PubMed] [Google Scholar]
- Pires SM, Vieira AR, Perez E, Lo Fo Wong D, Hald T. Attributing human foodborne illness to food sources and water in Latin America and the Caribbean using data from outbreak investigations. Int J Food Microbiol 2012;152:129–138 [DOI] [PubMed] [Google Scholar]
- Pons MJ, Mosquito S, Gomes C, Del Valle LJ, Ochoa TJ, Ruiz J. Analysis of quinolone-resistance in commensal and diarrheagenic Escherichia coli isolates from infants in Lima, Peru. Trans R Soc Trop Med Hyg 2014;108:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Zerr DM, Weissman SJ, Englund JA, Denno DM, Klein EJ, Tarr PI, Kwong J, Stapp JR, Tulloch LG, et al. . Prevalence and mechanisms of broad-spectrum beta-lactam resistance in Enterobacteriaceae: A children's hospital experience. Antimicrob Agents Chemother 2008;52:3909–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M, Miliwebsky E, Chinen I, Roldán CD, Balbi L, García B, Fiorilli G, Sosa-Estani S, Kincaid J, Rangel J, et al. . Characterization and epidemiologic subtyping of Shiga toxin–producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog Dis 2006;3:88–96 [DOI] [PubMed] [Google Scholar]
- Robbins A, Anand M, Nicholas DC, Egan JS, Musser KA, Giguere S, Prince H, Beaufait HE, Sears SD, Borda J, et al. . Ground beef recall associated with non-O157 Shiga toxin–producing Escherichia coli, United States. Emerg Infect Dis 2014;20:165–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rúgeles LC, Bai J, Martínez AJ, Vanegas MC, Gómez-Duarte OG. Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int J Food Microbiol 2010;138:282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soborg B, Lassen SG, Müller L, Jensen T, Ethelberg S, Mølbak K, Scheutz F. A verocytotoxin-producing E. coli outbreak with a surprisingly high risk of haemolytic uraemic syndrome, Denmark, September-October 2012. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull 2013;18:20350. [PubMed] [Google Scholar]
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin–producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005;365:1073–1086 [DOI] [PubMed] [Google Scholar]
- Trabulsi LR, Keller R, Tardelli Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis 2002;8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernozy-Rozand C, Montet MP, Berardin M, Bavai C, Beutin L. Isolation and characterization of Shiga toxin–producing Escherichia coli strains from raw milk cheeses in France. Lett Appl Microbiol 2005;41:235–241 [DOI] [PubMed] [Google Scholar]
- Werber D, Krause G, Frank C, Fruth A, Flieger A, Mielke M, Schaade L, Stark K. Outbreaks of virulent diarrheagenic Escherichia coli—Are we in control? BMC Med 2012;10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, et al. . Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol 2006;60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.