Abstract
Glycosylation is an important plant defense mechanism and conjugates of Fusarium mycotoxins often co-occur with their parent compounds in cereal-based food and feed. In case of deoxynivalenol (DON), deoxynivalenol-3-O-β-d-glucoside (D3G) is the most important masked mycotoxin. The toxicological significance of D3G is not yet fully understood so that it is crucial to obtain this compound in pure and sufficient quantities for toxicological risk assessment and for use as an analytical standard. The aim of this study was the biochemical characterization of a DON-inactivating UDP-glucosyltransferase from rice (OsUGT79) and to investigate its suitability for preparative D3G synthesis. Apparent Michaelis constants (Km) of recombinant OsUGT79 were 0.23 mM DON and 2.2 mM UDP-glucose. Substrate inhibition occurred at DON concentrations above 2 mM (Ki = 24 mM DON), and UDP strongly inhibited the enzyme. Cu2+ and Zn2+ (1 mM) inhibited the enzyme completely. Sucrose synthase AtSUS1 was employed to regenerate UDP-glucose during the glucosylation reaction. With this approach, optimal conversion rates can be obtained at limited concentrations of the costly co-factor UDP-glucose. D3G can now be synthesized in sufficient quantity and purity. Similar strategies may be of interest to produce β-glucosides of other toxins.
Keywords: masked mycotoxin, glycosylation, sucrose synthase, UDP-glucose recycling, Fusarium
1. Introduction
Deoxynivalenol (DON) is the main trichothecene toxin produced by the Fusarium species and a relevant virulence factor in Fusarium head blight disease (FHB) of cereal crops. Trichothecene toxins inhibit eukaryotic protein synthesis and elicit a wide range of pathophysiological effects in humans and animals. Examples include immuno-suppression, apoptotic cell death, and aberrant activation of proinflammatory gene expression [1,2]. The effects of DON on the gastrointestinal system, the immune system, and the brain have recently been reviewed [3]. For consumer protection, maximum tolerated levels of DON in grain and food commodities have been enacted in the European Union [4,5], which should prevent a toxin intake higher than the provisional maximum tolerable daily intake (PMTDI) of 1 μg/kg bodyweight for DON and its acetylated derivatives [6]. It was recently shown that at doses close to the PMTDI, DON already exerts a significant influence on intestinal physiology [7,8]. Furthermore, the estimated intake may exceed the PMTDI in years with high Fusarium incidence [9] and due to individual dietary preferences [10]. By monitoring the excretion of DON and DON-derivatives in urine, a recent study showed that even when consuming a regular diet, one-third of the participants exceeded the PMTDI of 1 μg/kg bodyweight in Austria [11].
The total exposure to DON (based on DON measured in food commodities) is most likely underestimated due to the occurrence of derivatives originating from plant detoxification systems. Historically, such compounds have been termed “masked mycotoxins”, which implies that they are not routinely detected in standard analytical procedures and may be reactivated to the parental toxins during food processing or digestion [12]. This term has been re-defined recently to be used for plant metabolites of mycotoxins solely [13]. Of particular importance are phase II detoxification metabolites of plants. This route involves conjugation to glucose, malonic acid, or glutathione to form hydrophilic molecules which are stored in vacuoles or transported to the apoplast [14,15]. The actual toxicity of such conjugates for humans and animals is mainly unknown and there is a potential risk that the parental toxins are released through hydrolysis during food processing and in the digestive tract [12,16].
Glycosylation is the major route of phase II detoxification and plants possess a respectable arsenal of UDP-glycosyltransferases (UGT). For example, about 100–180 putative glycosyltransferase family 1 (GT1, [17]) genes have been identified in the genomes of the model plants Arabidopsis thaliana and Brachypodium distachyon [18,19]. Glucosylation effectively reduces the acute toxicity of DON, as demonstrated in vitro by reduced inhibition of wheat ribosomes by DON-3-O-β-d-glucoside (D3G) [20]. The importance of glycosylation in response to Fusarium infections was also indicated by induction of UGT genes in wheat [21,22,23,24]. Increased DON resistance and formation of D3G upon infection with DON has been observed in wheat lines harboring the quantitative trait locus gene Fhb1 [25,26]. However, whether glucosylation of DON is directly responsible for the Fusarium resistance of wheat as conferred by Fhb1 has been disputed [27]. Recent evidence clearly shows that overexpression of UGT genes with the ability to detoxify DON leads to increased Fusarium resistance in Brachypodium [28] and in wheat overexpressing the barley glucosyltransferase HvUGT13248 [29].
D3G is currently present in a wide range of cereal commodities with concentrations typically in the range of up to 20% relative to DON, but higher levels have been reported as well [12,30,31,32]. Efforts to increase Fusarium resistance by breeding or through biotechnological approaches may increase the molar ratio of D3G/DON. Yet, little information still exists on bioavailability, metabolism, and long-term toxicity of D3G. It was shown that the bioavailability of D3G is probably low compared to that of DON, as human Caco-2 cells do not absorb D3G [33]. Although D3G is highly resistant to acidic hydrolysis [34], it can be enzymatically hydrolyzed by intestinal bacteria [35] and the released DON may be (partially) absorbed in the distal part of the gut. It was shown that D3G is indeed effectively hydrolyzed in the digestive tracts of humans and animals [36,37,38,39]. Evidence from a pig feeding study indicated that about half of the orally administered D3G is taken up into the bloodstream as DON and is further metabolized [38]. However, a large portion of DON generated by D3G hydrolysis is not resorbed but removed via feces. The European Food Safety Authority (EFSA) Panel on Contaminants in the Food Chain (CONTAM) concluded that with the currently available information, it should pragmatically be assumed that masked or, generally, all modified forms of Fusarium toxins possess the same toxicity as the parent compounds [40]. Evidently, D3G is a dietary risk factor and accurate estimation of its toxicological significance is necessary. Toxicological risk assessment can be achieved through feeding trials, but such strategies depend on large amounts of D3G that are currently unavailable.
The aim of this study was to investigate an enzymatic strategy to produce D3G. Several plant UGTs capable of glucosylating DON have been identified in our group previously [18,20,41]. Of these candidates, a rice UGT variant (OsUGT79) could be successfully expressed as an active protein in high yield in Escherichia coli. This paper reports the kinetic properties of the recombinant enzyme and its application in the production of D3G. A strategy to limit production costs by UDP-glucose recycling during conversion [42,43] was also successfully employed.
2. Results and Discussion
2.1. Expression and Purification of OsUGT79
Expression of functional OsUGT79 in E. coli was attempted with two fusion-protein variants. The first employed a standard expression system (pET21a) with a C-terminal His6-tag (UGT-cHis6) encoding a protein with a calculated molecular mass of 52 kDa. The second variant was comprised of an N-terminal His6-tag and a maltose binding protein (MalE) linked to the C-terminal OsUGT79 via the Tobacco Etch Virus (TEV) protease recognition site (vector pKLD116, [44]). This protein (nHis6-MalE-UGT) has a predicted molecular mass of 95 kDa.
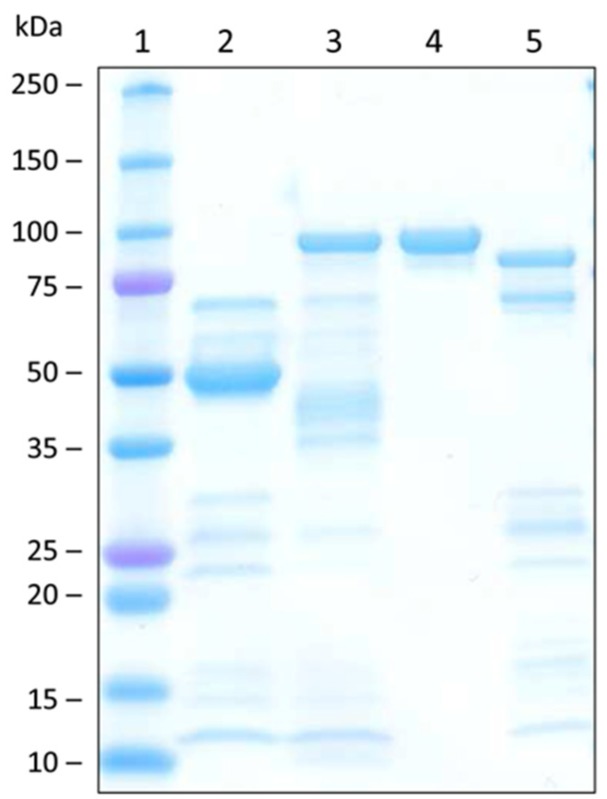
Protein expression levels were estimated from the total protein (Bradford assay) obtained after one-step purification by immobilized metal ion affinity chromatography (IMAC). First experiments with E. coli BL21 yielded low expression levels of <7 mg protein per liter broth with both constructs. Expression could be improved with E. coli Rosetta: 30 mg of protein per liter (4.4 mg per g of wet biomass) were obtained with nHis6-MalE-UGT and 85 mg per liter (5.7 mg per g of wet biomass) with UGT-cHis6. This implies an almost three-fold better yield of the construct without the maltose binding protein. However, activity measurements (1 mM DON/10 mM UDP-glucose) suggested that with a specific activity of 0.15 μmol min−1 mg−1 (referring to D3G formation), IMAC-purified nHis6-MalE-UGT is much more active than UGT-cHis6 with only 0.015 μmol min−1 mg−1. Despite the lower expression levels, the solubility enhancing maltose binding protein MalE appears to improve the yield of active protein. Therefore, further experiments were performed only with nHis6-MalE-UGT. The IMAC fraction of this protein was further purified by size exclusion chromatography (SEC). The specific activity (1 mM DON/10 mM UDP-glucose) of the purest fraction (Figure 1, lane 4) was 0.28 μmol min−1 mg−1.
Figure 1.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis of the purified rice UDP-glucosyltransferase OsUGT79 and sucrose synthase AtSUS1. Lane 1: Precision Plus Protein Standard (Bio-Rad); lane 2: immobilized metal ion affinity chromatography (IMAC)-purified OsUGT79 (UGT-cHis6, 52 kDa); lane 3: IMAC-purified OsUGT79 (nHis6-MalE-UGT, 95 kDa); lane 4: nHis6-MalE-UGT after size exclusion chromatography; lane 5: IMAC-purified AtSUS1 (93 kDa).
2.2. Kinetic Characteristics of OsUGT79
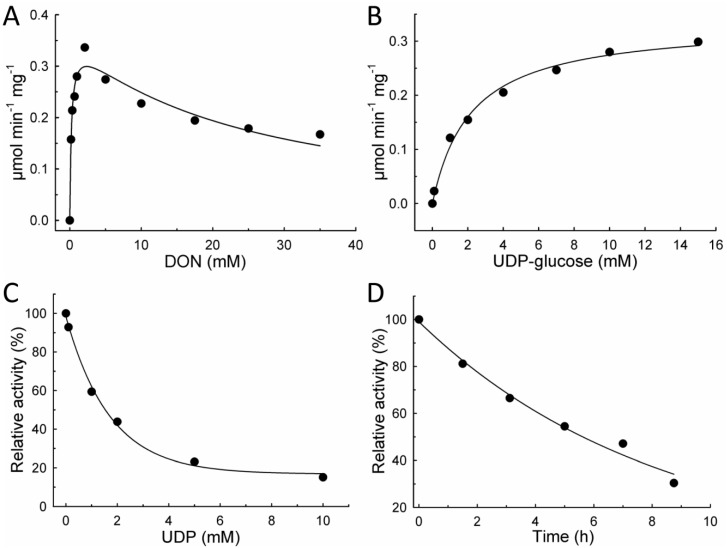
Kinetic characterization of OsUGT79 was performed with SEC-purified enzyme. Increasing the concentration of DON at a constant UDP-glucose concentration of 10 mM revealed substrate [S] inhibition by DON (Figure 2A). Maximum reaction velocity (0.34 μmol min−1 mg−1) was observed at 2 mM DON. Regression analysis using the kinetic model of Haldane (Equation (1)) yielded an apparent Michaelis constant (Km) of 0.23 ± 0.06 mM DON and a theoretical Vmax of 0.36 ± 0.02 μmol min−1 mg−1, corresponding to a kcat of 0.57 s−1. Substrate inhibition by DON was characterized by an estimated inhibitory constant (Ki) of 24 ± 5 mM. Response to variable UDP-glucose concentrations (1 mM DON) can be described with the typical Michaelis-Menten model (Equation (2), Figure 2B) with a Km of 2.2 ± 0.3 mM UDP-glucose. Inclusion of UDP in enzyme assays (1 mM DON/10 mM UDP-glucose) indicated that UDP is an effective inhibitor of OsUGT79 (Figure 2C). Using the model of exponential decay (Equation (3); v(I) reaction velocity as function of inhibitor concentration [I], vmin velocity at maximum inhibitor concentration ([I]→∞), v0 velocity at [I] = 0, λ decay constant), 50% activity reduction was estimated at 1.5 mM UDP. The stability of the enzyme was tested by incubation at reaction conditions (100 mM Tris, 37 °C). Samples were taken in regular intervals and assayed for activity with DON (Figure 2D). This revealed a half-life of 5–6 h at 37 °C (Equation (4); v(t) reaction velocity at incubation time t, v0 initial velocity at t = 0).
| (1) |
| (2) |
| (3) |
| (4) |
Figure 2.
Kinetic characterization of recombinant OsUGT79 (nHis6-MalE-UGT) at 37 °C, 100 mM Tris pH 7. (A) 10 mM UDP-glucose, varying deoxynivalenol (DON) concentrations; (B) 1 mM DON, varying UDP-glucose concentrations; (C) Inhibition by UDP (1 mM DON); (D) Stability at 37 °C in 100 mM Tris pH 7.
Divalent cations can have an influence on the activity of glycosyltransferases [45]. Structural studies of a GT-B fold UGT showed that Ca2+, Mn2+, and Mg2+ can interact with the β-phosphate group of UDP and thus possibly play a role in facilitating product release [46,47]. Addition of ethylenediaminetetraacetic acid (EDTA) in the assay caused only low reduction of activity (Table 1), implying that OsUGT79 does not strongly depend on metal ions. Ca2+, Mg2+, and Fe2+ caused a moderate increase of activity (Table 1). Strong inhibition of a UGT by Cu2+, Mn2+, and Zn2+ (1 mM) was previously reported [48]. Here, complete inhibition of OsUGT79 was observed by Cu2+ and Zn2+, but not by Mn2+ (all 1 mM), which increased activity to about 140%.
Table 1.
Influence of ethylenediaminetetraacetic acid (EDTA) and several metal ions (1 mM each) on the activity of OsUGT79. ND, deoxynivalenol-3-O-β-d-glucoside not detectable.
| Compound | Activity (%) |
|---|---|
| Control | 100 |
| EDTA (1 mM) | 98 |
| EDTA (5 mM) | 95 |
| CaCl2 | 108 |
| CuSO4 | ND |
| MgSO4 | 117 |
| MnCl2 | 143 |
| MnSO4 | 139 |
| ZnSO4 | ND |
| FeSO4 | 114 |
2.3. UDP-Glucose Recycling and DON Production
A prerequisite for the preparative production of D3G is a complete conversion, with residual DON concentrations below 1%. In order to achieve this within a reasonable time frame, a considerable excess of UDP-glucose would be required to maintain reaction velocity, especially to compensate for feedback inhibition by UDP accumulating during the reaction.
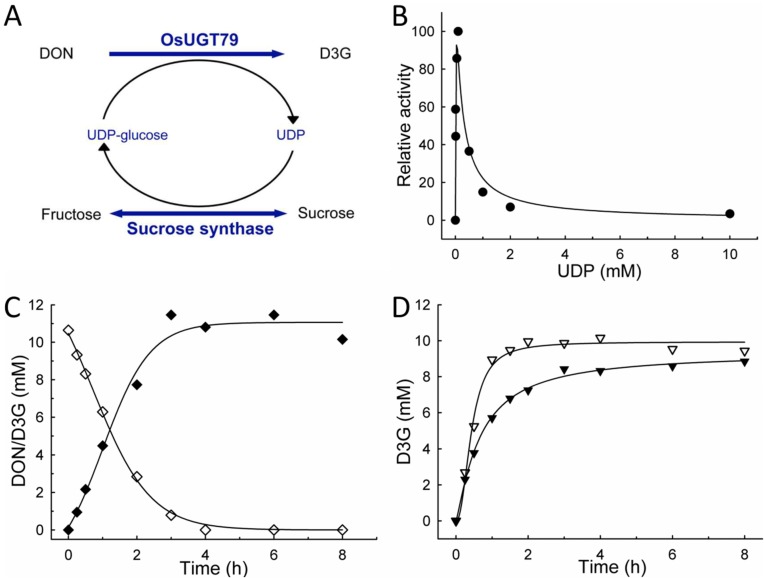
Based on previous studies [42,43], the potential of a sucrose synthase from Arabidopsis thaliana (AtSUS1) was investigated in order to recycle UDP-glucose during DON glucosylation, and the reaction scheme is illustrated in Figure 3A. AtSUS1 catalyzes the reversible formation of UDP-glucose from sucrose and UDP with Km values of 53 and 1.2 mM, respectively [43]. In the reversed sucrose synthesizing reaction, Km values of 25 mM fructose and 50 μM UDP-glucose were reported [49].
Figure 3.
In situ UDP-glucose recycling with OsUGT79 and sucrose synthase AtSUS1 from Arabidopsis thaliana. (A) Reaction scheme; (B) Initial reaction velocities (1 mM deoxynivalenol (DON), 100 mM sucrose) resulting from different UDP concentrations, 0.2 mg mL−1 of each protein in assay; (C) Glucosylation of 10 mM DON with 0.1 mM UDP, 1 mg mL−1 of each protein in assay. All assays performed at 37 °C, 100 mM Tris pH 7, DON (◊), DON-3-O-β-d-glucoside (D3G; ♦); (D) Glucosylation of 10 mM DON with 10 mM UDP-Glucose at 37 °C, 100 mM Tris pH 7, comparison of reaction with (▽) and without (▼) recycling of UDP-glucose, OsUGT79/AtSUS1 each 1 mg mL−1 in assay.
In order to establish whether this strategy is applicable for D3G production, initial experiments were performed in the absence of UDP-glucose, but with 100 mM sucrose and different initial UDP concentrations. Both proteins (OsUGT79 and AtSUS) were applied at concentrations of 0.2 mg mL−1 (IMAC purification stage, Figure 1). The results (Figure 3B) show that D3G formation rates strongly depend on initial UDP concentrations. In agreement with the strong inhibition of OsUGT79 by UDP, maximum conversion rates were found at low UDP concentrations of 0.1 mM. Regression analysis (Equation (1)) implied that the reaction can be described analogous to an enzyme subject to substrate inhibition. An apparent Km of 0.014 mM and a Ki of 0.18 mM UDP were estimated. Therefore, maximum conversion rates occurred at virtual UDP-glucose concentrations (<0.1 mM) that are at least 20-fold below the Km (2 mM). This would imply that using UDP as sole substrate is of low practical value due to kinetic limitations. Nevertheless, with 0.1 mM UDP, 10 mM DON were rapidly converted to D3G (Figure 3C), with residual DON (molar ratio DON/D3G) below 1% after 6 h.
In the case that an aglycon is not stable in aqueous conditions, it is essential to complete the reaction as fast as possible. Rapid conversion is also of interest in view of the limited half-life of OsUGT79 (5–6 h, Section 2.1) at reaction conditions. Therefore, recycling of UDP-glucose is also of interest to maintain high donor concentrations and to avoid accumulation of UDP during the reaction. To demonstrate this, glucosylation of 10 mM DON with equimolar initial UDP-glucose concentration was compared with and without UDP-glucose recycling. The results (Figure 3D) confirm that inclusion of AtSUS1 is effective to maintain optimal reaction conditions. With the aid of donor recycling, glucosylation was almost completed within 2 h. In absence of AtSUS1, the reaction velocity declined rapidly.
2.4. Preparative Production and Purification of D3G
A typical batch for larger scale D3G production contained 50–100 mg DON with concentrations of 10 mM. Reaction conditions were as described above with 1 mg mL−1 OsUGT79 (IMAC purification stage). UDP-glucose concentrations were 2 mM (concentration at Km) when AtSUS1 was included. Batches without donor recycling contained UDP-glucose in 1.5 molar excess over DON. The reactions were usually carried out overnight.
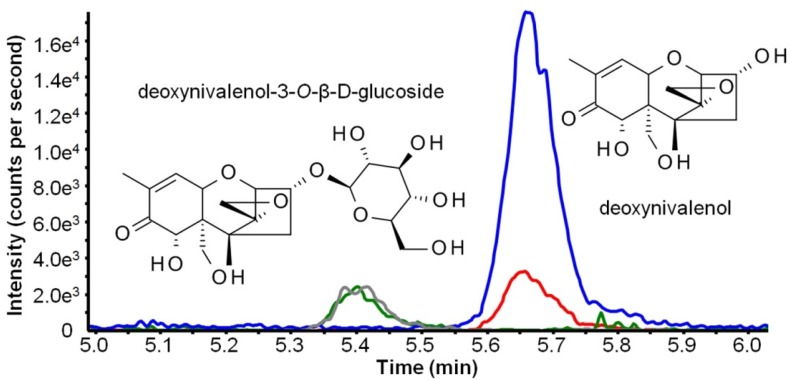
High performance liquid chromatographic-mass spectrometric (HPLC-MS/MS) measurements were performed prior to purification to confirm successful and complete conversion of DON to D3G. Both selected reaction monitoring (SRM) transitions of D3G were present at a retention time of 4.97 min and DON was not found in the final product (<0.05%). A chromatogram of a standard containing 100 μg/L of both compounds is shown in Figure 4.
Figure 4.
Structure formula and extracted ion chromatogram of a standard containing 100 μg L−1 deoxynivalenol (DON) and DON-3-O-β-d-glucoside (D3G). Green (m/z 517.3 > m/z 59.1) and gray (m/z 517.3 > m/z 427.1) lines show selected reaction monitoring (SRM) traces for D3G. Blue (m/z 355.1 > m/z 59.2) and red (m/z 355.1 > m/z 265.2) lines show SRM traces for DON.
Purification was carried out on a preparative high performance liquid chromatography system and the obtained yield of D3G in a crystalline form after evaporation and lyophilization was 53 mg for a batch containing 50 mg DON. A subsample of the crystalline D3G was dissolved in MeOH and its purity was determined as follows: The overall purity was determined by HPLC-ultraviolet (UV) determination at 200 nm and was above 98% based on a 500 mg L−1 solution, which is in agreement with the used DON. The absence of DON was confirmed by LC-MS measurements and verified to be below the limit of detection (corresponding to <0.05% overall). Furthermore, enhanced product ion (MS/MS) scans were performed at three collision energies (10, 20, 30 eV). Comparison with MS/MS measurements of a commercially available certified D3G standard under the same conditions proved the compounds to be identical.
3. Experimental Section
3.1. Materials
Uridine 5′-diphosphoglucose disodium salt hydrate (UDP-glucose) from Saccharomyces cerevisiae (cat. no. U4625) and Uridine 5′-diphosphate disodium salt hydrate (UDP, cat. no. 94330) were purchased from Sigma-Aldrich (Vienna, Austria). E. coli BL21 Star™ (DE3) was from Invitrogen (Carlsbad, CA, USA), E. coli Rosetta™ (DE3) from Novagen (Madison, WI, USA). DON (purity >98%) was purified at the IFA-Tulln following a published procedure [50].
3.2. Cloning of OsUGT79 and AtSUS1
The OsUGT79 gene (GenBank accession NM_001058779) was amplified from plasmid pWS57 [18] using the forward primer 5′-ATGGGCTCTATGTCCACTCCTGC-3′ and the reverse primer 5′-ATTGGAATACTTTGCTGCAAACTC-3′. Using the Quikchange method [51,52], the resulting product was introduced into pET21a (Novagen, Madison, WI, USA) and plasmid pKLD116, a pET31b derivative containing His6-tagged maltose binding protein (MalE), followed by a TEV protease cleavage site [44].
The sucrose synthase gene (AtSUS1) from Arabidopsis thaliana (TAIR accession AT5G20830.1; GenBank accession BAH19538.1) was amplified from A. thaliana cDNA using the forward primer 5′-CATATGGCAAACGCTGAACGTATG-3′ and the reverse primer 5′-GCGGCCGCGTCATCTTGTGCAAGAGG-3′. The restriction sites NdeI and NotI were used for cloning into pET21a in frame with the C-terminal His-tag. AtSUS1 was expressed with E. coli BL21 Star.
3.3. Protein Expression and Purification
Protein expression was carried out in terrific broth with 100 mg L−1 ampicillin (E. coli BL21 (DE3)), additionally supplemented with 35 mg L−1 chloramphenicol for E. coli Rosetta. Isopropyl-β-D-1-thiogalactopyranoside (IPTG, 0.5 mM final concentration) was added when the optical density (OD600) reached 0.5. The flasks were further incubated for 16 h at 25 °C and 100 rpm. After that period, the biomass was harvested by centrifugation (4000 g, 15 min) and resuspended in 25 mM Tris pH 7.5 + 500 mM NaCl/25 mM imidazole, the binding buffer for the first purification step. The cells were disrupted in a French Pressure Cell Press (Aminco, Silver Spring, MD, USA), in three passes at 1200 psi (ca. 8270 kPa). The cell extract was cleared by centrifugation at 70,000 g. Protein purification was performed by IMAC on Ni2+-charged HisTrap Crude FF columns, 5 mL (GE Healthcare, Vienna, Austria). Target protein was bound to the column in the above specified binding buffer. Protein was eluted with the same buffer containing 500 mM imidazole. After IMAC, the buffer was changed to 50 mM potassium phosphate pH 7 + 150 mM NaCl by gel filtration with Sephadex G25 (GE Healthcare). OsUGT79 was stored as such or further purified by size exclusion chromatography on Superose 12 (GE Healthcare) with 50 mM potassium phosphate pH 7 + 150 mM NaCl. Column dimensions were 2 cm2 area, 90 cm bed height, flow 0.5 cm min−1. Purified OsUGT79 (IMAC purification stage) was stored at −80 °C. AtSUS1 almost completely lost activity by freezing/thawing and had to be prepared fresh prior to use.
Protein concentrations were determined with the Bio-Rad (Vienna, Austria) protein assay based on the dye-binding method of Bradford. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) including Coomassie blue staining was performed with the Mini-PROTEAN system with precast gels (4%–20%) from Bio-Rad. The molecular mass marker used was High Precision Dual Color (10–250 kDa range, Bio-Rad).
3.4. Glycosylation Assays
Unless mentioned otherwise, enzyme assays were performed in 100 mM Tris, pH 7 at 37 °C, 10 min reaction time, OsUGT79 was added to concentrations of 0.1–1 mg mL−1. Assays for UDP-glucose recycling with AtSUS1 additionally contained 100 mM sucrose. The assays were stopped by transferring 20 μL of sample to 180 μL methanol. After centrifugation (20,000 g, 5 min) to remove precipitated protein, the samples were further diluted in H2O to an expected concentration range of 1 mg L−1 DON/D3G. The concentrations of DON and D3G were determined by HPLC-MS/MS (see Section 3.5). The activity units (μmol min−1 mg−1) displayed refer to the formation of D3G per mg of protein. All assays were performed in duplicate. Data analysis (i.e., for kinetic enzyme characterization) was performed with SigmaPlot 11.0 (Systat Software, San Jose, CA, USA) using Equations (1)–(4).
3.5. DON and D3G Determination by HPLC-MS/MS
Concentrations of DON and D3G were determined on a 1290 HPLC system coupled to a 4000 QTrap LC-MS/MS System (AB Sciex, Foster City, CA, USA). Briefly, chromatographic separation was achieved on a Gemini C18 column (150 × 4.6 mm, 5 μm, Phenomenex, Aschaffenburg, Germany) at 25 °C with a flow rate of 0.8 mL min−1. The following water-methanol gradient (eluent A: 80:20, v:v; eluent B: 3:97, v:v; both containing 5 mM ammonium acetate) was used: initial conditions at 0% B were held for 1 min, followed by a linear increase to 50% B within 5 min. Afterwards, 100% B were reached within 0.1 min. Following a holding time of 2 min at 100% B, a fast switch to the initial conditions was performed and column equilibration was achieved until the end of the run (10 min). The mass spectrometer was operated in negative electrospray ionization mode. The following source settings were used: temperature 550 °C, ion spray voltage −4 kV, curtain gas 30 psi (207 kPa of 99.5% nitrogen), source gas one and two both 50 psi (345 kPa of zero grade air), collision gas (nitrogen) set to high. For quantitation two SRM transitions per compound were acquired with a dwell time of 25 ms. The acetate adducts (m/z 355.1 for DON, m/z 517.3 for D3G) were chosen as precursors and the declustering potential (DP) was −40 V for DON, −50 V for D3G. The product ions were for DON m/z 59.2 (collision energy (CE) of −40 V) and m/z 265.2 (CE −22 V), for D3G m/z 427.1 (CE −30 V) and m/z 59.1 (CE −85 V).
3.6. Preparative High-Performance Liquid Chromatography
Purification of D3G was carried out using an 1100 series preparative HPLC system equipped with an automatic fraction collector and a multiple wavelength detector (MWD) (all Agilent Technologies, Waldbronn, Germany). A Gemini NX column (150 × 21.2 mm, 5 µm, Phenomenex, Aschaffenburg, Germany) and gradient elution (eluent A: water, eluent B: methanol) was used for the separation of D3G from residual glucose and other impurities. The initial conditions of 20% B were maintained for 1 min, followed by a linear increase to 60% B within 4 min and to 100% B within 0.1 min. Following a hold time of 1 min at 100%, the initial conditions were achieved with a fast switch to 25% B and the column was equilibrated prior to the next injection. The flow rate was 16 mL min−1 and the injection volume was set to 900 μL, in general. The fractions were collected from 4 to 6 min with the maximum peak duration of 0.5 min using threshold working mode. The collected fractions were pooled, the organic phase was evaporated on a rotary evaporator at 30 °C, and the rest of water phase was removed by lyophilization. The obtained crystals of D3G were weighed in a glass vial on a micro balance and stored at −20 °C.
3.7. D3G Purity Analysis by HPLC-UV
For the purity measurement of the crystalline D3G, an 1100 Series HPLC system (Agilent Technologies, Waldbronn, Germany) including an 1100 Series HPLC MWD and a Gemini C18 column (150 × 4.6 mm, 5 μm, Phenomenex, Aschaffenburg, Germany) was used. Pure water (A) and acetonitrile (B) were used as eluents and the following gradient was applied: 0–1 min 5% B; 1–26 min linear increase to 100% B, 26–29 min 100%, 29.1–32 min 5% B. During the whole run the UV absorption was measured at the wavelengths 200 nm, 210 nm, and 220 nm, but only the 200 nm signal was used for the purity assessment. The final product of D3G was weighed with a microbalance and dissolved in methanol to obtain a concentration of ca. 500 mg L−1. Of this solution 10 μL were injected into the system and the peak area of D3G was divided by the sum of all peak areas and multiplied with 100 to obtain the overall purity in percent.
4. Conclusions
This paper reports the kinetic characteristics of a DON-conjugating UDP-glucosyltransferase from rice (OsUGT79), which can be used for preparative synthesis of D3G. The fact that this enzyme can be functionally expressed with E. coli is a great advantage. Previously, engineered yeast strains expressing plant UGTs were employed to produce β-glucosides of zearalenone and its derivatives in the mg range [53,54]. The availability of a functional cell-free catalyst simplifies the production process drastically and allows catalysis to be performed in small volumes and, most importantly, facilitates product clean-up. Recycling of UDP-glucose is an effective strategy to optimize conversion kinetics and is of economic interest to reduce the required amounts of the costly co-factor UDP-glucose. Therefore, it is now possible to provide sufficient amounts of D3G for toxicological risk assessment by a relatively simple and rapid procedure. Similar approaches may be of interest to produce β-glucosides of other mycotoxins that are so far not commercially available.
Acknowledgments
This work was supported by the Vienna Science and Technology Fund (WWTF LS12-012) and the Austrian Science Fund (FWF) (SFB F3708 and F3706). Furthermore, the financial support by the Austrian Federal Ministry of Science, Research and Economy, the National Foundation of Research, Technology and Development, and the Lower Austrian Government is acknowledged. We thank Imer Maloku (Bsc) for the support in the purification of DON and Jeanette Moulinier-Anzola for providing A. thaliana cDNA.
Author Contributions
H.M. and G.A. conceived and designed the experiments; H.M., A.M., E.V., J.K., M.L., and S.N. performed the experiments; I.R., F.B., and G.A supervised the experimental work. H.M., A.M., and E.V. wrote the paper and all authors amended and corrected the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bhat R., Rai R.V., Karim A.A. Mycotoxins in food and feed: Present status and future concerns. Compr. Rev. Food Sci. Food Saf. 2010;9:57–81. doi: 10.1111/j.1541-4337.2009.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Wu F., Groopman J.D., Pestka J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014;5:351–372. doi: 10.1146/annurev-food-030713-092431. [DOI] [PubMed] [Google Scholar]
- 3.Maresca M. From the gut to the brain: Journey and pathophysiological effects of the food-associated trichothecene mycotoxin deoxynivalenol. Toxins. 2013;5:784–820. doi: 10.3390/toxins5040784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Commission Commision Regulation (EC) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off. J. Eur. Union. 2006;L364:5–24. [Google Scholar]
- 5.European Commision Commission Regulation (EC) No 1126/2006 of 28 September 2008 amending Regulation (EC) No 1881/2006 setting maximum levels for certain contaminants in foodstuffs as regards Fusarium toxins in maize and maize products. Off. J. Eur. Union. 2007;L255:14–17. [Google Scholar]
- 6.Joint FAO/WHO Expert Committee on Food Additives (JECFA) Evaluation of certain contaminants in food. [(accessed on 17 July 2015)];WHO Tech. Rep. Ser. 2011 959 Available online: http://whqlibdoc.who.int/trs/WHO_TRS_959_eng.pdf. [Google Scholar]
- 7.Graziani F., Pujol A., Nicoletti C., Pinton P., Armand L., di Pasquale E., Oswald I.P., Perrier J., Maresca M. The Food-Associated Ribotoxin Deoxynivalenol Modulates Inducible NO Synthase in Human Intestinal Cell Model. Toxicol. Sci. 2015;145:372–382. doi: 10.1093/toxsci/kfv058. [DOI] [PubMed] [Google Scholar]
- 8.Pinton P., Graziani F., Pujol A., Nicoletti C., Paris O., Ernouf P., di Pasquale E., Perrier J., Oswald I.P., Maresca M. Deoxynivalenol inhibits the expression by goblet cells of intestinal mucins through a PKR and MAP kinase dependent repression of the resistin-like molecule β. Mol. Nutr. Food Res. 2015;59:1076–1087. doi: 10.1002/mnfr.201500005. [DOI] [PubMed] [Google Scholar]
- 9.Gratz S.W., Richardson A.J., Duncan G., Holtrop G. Annual variation of dietary deoxynivalenol exposure during years of different Fusarium prevalence: A pilot biomonitoring study. Food Addit. Contam. Part A. 2014;31:1579–1585. doi: 10.1080/19440049.2014.937772. [DOI] [PubMed] [Google Scholar]
- 10.Šarkanj B., Warth B., Uhlig S., Abia W.A., Sulyok M., Klapec T., Krska R., Banjari I. Urinary analysis reveals high deoxynivalenol exposure in pregnant women from Croatia. Food Chem. Toxicol. 2013;62:231–237. doi: 10.1016/j.fct.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Warth B., Sulyok M., Fruhmann P., Berthiller F., Schuhmacher R., Hametner C., Adam G., Fröhlich J., Krska R. Assessment of human deoxynivalenol exposure using an LC-MS/MS based biomarker method. Toxicol. Lett. 2012;211:85–90. doi: 10.1016/j.toxlet.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Berthiller F., Crews C., Dall’Asta C., Saeger S.D., Haesaert G., Karlovsky P., Oswald I.P., Seefelder W., Speijers G., Stroka J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013;57:165–186. doi: 10.1002/mnfr.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rychlik M., Humpf H.-U., Marko D., Dänicke S., Mally A., Berthiller F., Klaffke H., Lorenz N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014;30:197–205. doi: 10.1007/s12550-014-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman J.O.D., Blake-Kalff M.M.A., Davies T.G.E. Detoxification of xenobiotics by plants: Chemical modification and vacuolar compartmentation. Trends Plant Sci. 1997;2:144–151. doi: 10.1016/S1360-1385(97)01019-4. [DOI] [Google Scholar]
- 15.Bowles D., Lim E.-K., Poppenberger B., Vaistij F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- 16.Broekaert N., Devreese M., de Baere S., de Backer P., Croubels S. Modified Fusarium mycotoxins unmasked: From occurrence in cereals to animal and human excretion. Food Chem. Toxicol. 2015;80:17–31. doi: 10.1016/j.fct.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Coutinho P.M., Deleury E., Davies G.J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 18.Schweiger W., Pasquet J.C., Nussbaumer T., Paris M.P.K., Wiesenberger G., Macadré C., Ametz C., Berthiller F., Lemmens M., Saindrenan P., et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol. Plant-Microbe Interact. 2013;26:781–792. doi: 10.1094/MPMI-08-12-0205-R. [DOI] [PubMed] [Google Scholar]
- 19.Bowles D., Isayenkova J., Lim E.K., Poppenberger B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005;8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Poppenberger B., Berthiller F., Lucyshyn D., Sieberer T., Schuhmacher R., Krska R., Kuchler K., Glössl J., Luschnig C., Adam G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003;278:47905–47914. doi: 10.1074/jbc.M307552200. [DOI] [PubMed] [Google Scholar]
- 21.Desmond O.J., Manners J.M., Schenk P.M., Maclean D.J., Kazan K. Gene expression analysis of the wheat response to infection by Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2008;73:40–47. doi: 10.1016/j.pmpp.2008.12.001. [DOI] [Google Scholar]
- 22.Hill-Ambroz K., Webb C.A., Matthews A.R., Li W., Gill B.S., Fellers J.P. Expression analysis and physical mapping of a cDNA library of Fusarium head blight infected wheat spikes. Crop Sci. 2006;46:S15–S26. doi: 10.2135/cropsci2005.07.0206tpg. [DOI] [Google Scholar]
- 23.Ma L., Shang Y., Cao A., Qi Z., Xing L., Chen P., Liu D., Wang X. Molecular cloning and characterization of an up-regulated UDP-glucosyltransferase gene induced by DON from Triticum aestivum L. cv. Wangshuibai. Mol. Biol. Rep. 2010;37:785–795. doi: 10.1007/s11033-009-9606-3. [DOI] [PubMed] [Google Scholar]
- 24.Steiner B., Kurz H., Lemmens M., Buerstmayr H. Differential gene expression of related wheat lines with contrasting levels of head blight resistance after Fusarium graminearum inoculation. Theor. Appl. Genet. 2009;118:753–764. doi: 10.1007/s00122-008-0935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemmens M., Scholz U., Berthiller F., Dall’Asta C., Koutnik A., Schuhmacher R., Adam G., Buerstmayr H., Mesterházy Á., Krska R., et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant-Microbe Interact. 2005;18:1318–1324. doi: 10.1094/MPMI-18-1318. [DOI] [PubMed] [Google Scholar]
- 26.Horevaj P., Gale L.R., Milus E.A. Resistance in winter wheat lines to initial infection and spread within spikes by deoxynivalenol and nivalenol chemotypes of Fusarium graminearum. Plant Dis. 2011;95:31–37. doi: 10.1094/PDIS-03-10-0167. [DOI] [PubMed] [Google Scholar]
- 27.Gunnaiah R., Kushalappa A.C., Duggavathi R., Fox S., Somers D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE. 2012;7:e40695. doi: 10.1371/journal.pone.0040695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasquet J.-C. Doctoral Thesis. Ecole doctorale Sciences du Végétal; Orsay, France: Nov, 2014. Détoxication des Mycotoxines par les Plantes : Analyse de l’Interaction entre Brachypodium distachyon et Fusarium graminearum. [Google Scholar]
- 29.Li X., Shin S., Heinen S., Dill-Macky R., Berthiller F., Clemente T., McCormick S., Muehlbauer G.J. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol. Plant-Microbe Interact. 2015 doi: 10.1094/MPMI-03-15-0062-R. submitted. [DOI] [PubMed] [Google Scholar]
- 30.Berthiller F., Dall’asta C., Corradini R., Marchelli R., Sulyok M., Krska R., Adam G., Schuhmacher R. Occurrence of deoxynivalenol and its 3-β-d-glucoside in wheat and maize. Food Addit. Contam. Part A. 2009;26:507–511. doi: 10.1080/02652030802555668. [DOI] [PubMed] [Google Scholar]
- 31.Desmarchelier A., Seefelder W. Survey of deoxynivalenol and deoxynivalenol-3-glucoside in cereal-based products by liquid chromatography electrospray ionization tandem mass spectrometry. World Mycotoxin J. 2011;4:29–35. doi: 10.3920/WMJ2010.1236. [DOI] [Google Scholar]
- 32.Kostelanska M., Hajslova J., Zachariasova M., Malachova A., Kalachova K., Poustka J., Fiala J., Scott P.M., Berthiller F., Krska R. Occurrence of deoxynivalenol and its major conjugate, deoxynivalenol-3-glucoside, in beer and some brewing intermediates. J. Agric. Food Chem. 2009;57:3187–3194. doi: 10.1021/jf803749u. [DOI] [PubMed] [Google Scholar]
- 33.De Nijs M., van den Top H., Portier L., Oegema G., Kramer E., van Egmond H., Hoogenboom L. Digestibility and absorption of deoxynivalenol-3-β-glucoside in in vitro models. World Mycotoxin J. 2012;5:319–324. doi: 10.3920/WMJ2012.1430. [DOI] [Google Scholar]
- 34.Malachova A., Stockova L., Wakker A., Varga E., Krska R., Michlmayr H., Adam G., Berthiller F. Critical Evaluation of indirect methods for the determination of modified deoxynivalenol in cereals. Anal. Bioanal. Chem. 2015 doi: 10.1007/s00216-015-8793-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berthiller F., Krska R., Domig K.J., Kneifel W., Juge N., Schuhmacher R., Adam G. Hydrolytic fate of deoxynivalenol-3-glucoside during digestion. Toxicol. Lett. 2011;206:264–267. doi: 10.1016/j.toxlet.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dall’Erta A., Cirlini M., Dall’Asta M., del Rio D., Galaverna G., Dall’Asta C. Masked mycotoxins are efficiently hydrolyzed by human colonic microbiota releasing their aglycones. Chem. Res. Toxicol. 2013;26:305–312. doi: 10.1021/tx300438c. [DOI] [PubMed] [Google Scholar]
- 37.Nagl V., Schwartz H., Krska R., Moll W.-D., Knasmüller S., Ritzmann M., Adam G., Berthiller F. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in rats. Toxicol. Lett. 2012;213:367–373. doi: 10.1016/j.toxlet.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagl V., Woechtl B., Schwartz-Zimmermann H.E., Hennig-Pauka I., Moll W.-D., Adam G., Berthiller F. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in pigs. Toxicol. Lett. 2014;229:190–197. doi: 10.1016/j.toxlet.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 39.Gratz S.W., Duncan G., Richardson A.J. The human fecal microbiota metabolizes deoxynivalenol and deoxynivalenol-3-glucoside and may be responsible for urinary deepoxy-deoxynivalenol. Appl. Environ. Microbiol. 2013;79:1821–1825. doi: 10.1128/AEM.02987-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.European Food Safety Authority (EFSA) Scientific opinion on the risks for human and animal health related to the presence of modified forms of certain mycotoxins in food and feed. EFSA J. 2014;12:3916. [Google Scholar]
- 41.Schweiger W., Boddu J., Shin S., Poppenberger B., Berthiller F., Lemmens M., Muehlbauer G.J., Adam G. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Mol. Plant-Microbe Interact. 2010;23:977–986. doi: 10.1094/MPMI-23-7-0977. [DOI] [PubMed] [Google Scholar]
- 42.Son M.H., Kim B.-G., Kim D.H., Jin M., Kim K., Ahn J.-H. Production of flavonoid O-glucoside using sucrose synthase and flavonoid O-glucosyltransferase fusion protein. J. Microbiol. Biotechnol. 2009;19:709–712. [PubMed] [Google Scholar]
- 43.Masada S., Kawase Y., Nagatoshi M., Oguchi Y., Terasaka K., Mizukami H. An efficient chemoenzymatic production of small molecule glucosides with in situ UDP-glucose recycling. FEBS Lett. 2007;581:2562–2566. doi: 10.1016/j.febslet.2007.04.074. [DOI] [PubMed] [Google Scholar]
- 44.Rocco C., Dennison K., Klenchin V.A., Rayment I., Escalante-Semerena J. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lairson L., Henrissat B., Davies G., Withers S. Glycosyltransferases: Structures, functions, and mechanisms. Biochemistry. 2008;77:521. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 46.Moréra S., Larivière L., Kurzeck J., Aschke-Sonnenborn U., Freemont P.S., Janin J., Rüger W. High resolution crystal structures of T4 phage β-glucosyltransferase: Induced fit and effect of substrate and metal binding. J. Mol. Biol. 2001;311:569–577. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- 47.Larivière L., Gueguen-Chaignon V., Moréra S. Crystal structures of the T4 phage β-glucosyltransferase and the D100A mutant in complex with UDP-glucose: Glucose binding and identification of the catalytic base for a direct displacement mechanism. J. Mol. Biol. 2003;330:1077–1086. doi: 10.1016/S0022-2836(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 48.Ford C.M., Boss P.K., Høj P.B. Cloning and characterization of vitis vinifera UDP-Glucose: Flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 1998;273:9224–9233. doi: 10.1074/jbc.273.15.9224. [DOI] [PubMed] [Google Scholar]
- 49.Almagro G., Baroja-Fernández E., Muñoz F.J., Bahaji A., Etxeberria E., Li J., Montero M., Hidalgo M., Sesma M.T., Pozueta-Romero J. No evidence for the occurrence of substrate inhibition of Arabidopsis thaliana sucrose synthase-1 (AtSUS1) by fructose and UDP-glucose. Plant Signal. Behav. 2012;7:799–802. doi: 10.4161/psb.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altpeter F., Posselt U. Production of high quantities of 3-acetyldeoxynivalenol and deoxynivalenol. Appl. Microbiol. Biotechnol. 1994;41:384–387. doi: 10.1007/BF01982524. [DOI] [Google Scholar]
- 51.Chen G., Qiu N., Karrer C., Caspers P., Page M. Restriction site-free insertion of PCR products directionally into vectors. BioTechniques. 2000;28:498–500, 504–505. doi: 10.2144/00283st08. [DOI] [PubMed] [Google Scholar]
- 52.Van den Ent F., Löwe J. RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Paris M.P.K., Schweiger W., Hametner C., Stückler R., Muehlbauer G.J., Varga E., Krska R., Berthiller F., Adam G. Zearalenone-16-O-glucoside: A new masked mycotoxin. J. Agric. Food Chem. 2014;62:1181–1189. doi: 10.1021/jf405627d. [DOI] [PubMed] [Google Scholar]
- 54.Krenn P., Berthiller F., Schweiger W., Hametner C., Ludwig R., Adam G., Krska R., Schuhmacher R. Production of zearalenone-4-glucoside, a-zearalenol-4-glucoside and β-zearalenol-4-glucoside. Mycotoxin Res. 2007;23:180–184. doi: 10.1007/BF02946045. [DOI] [PubMed] [Google Scholar]