Abstract
Purpose: PECAM-1 (CD31) is a glycoprotein expressed on endothelial and bone marrow precursor cells. It plays important roles in angiogenesis, maintenance and integration of the cytoskeleton and direction of leukocytes to the site of inflammation. We aimed to clone the cDNA coding for human CD31 from KG1a for further subcloning and expression in NIH-3T3 mouse cell line.
Methods: CD31 cDNA was cloned from KG1a cell line after total RNA extraction and cDNA synthesis. Pfu DNA polymerase-amplified specific band was ligated to pGEMT-easy vector and sub-cloned in pCMV6-Neo expression vector. After transfection of NIH-3T3 cells using 3 μg of recombinant construct and 6 μl of JetPEI transfection reagent, stable expression was obtained by selection of cells by G418 antibiotic and confirmed by surface flow cytometry.
Results: 2235 bp specific band was aligned completely to human CD31 reference sequence in NCBI database. Transient and stable expression of human CD31 on transfected NIH-3T3 mouse fibroblast cells was achieved (23% and 96%, respectively) as shown by flow cytometry.
Conclusion: Due to murine origin of NIH-3T3 cell line, CD31-expressing NIH-3T3 cells could be useful as immunogen in production of diagnostic monoclonal antibodies against human CD31, with no need for purification of recombinant proteins.
Keywords: CD31, Cloning, NIH-3T3, Angiogenesis, Antibody
Introduction
Human platelet endothelial cell adhesion molecule-1 PECAM-1 or CD31 gene consists of 16 exons and located on chromosome 17 in the region 17q23. It codes for a 130 kDa transmembrane glycoprotein belongs to immunoglobulin (Ig) superfamily. Cytoplasmic region contains two immunoreceptor tyrosine-based inhibitory motifs (ITIM) and up on phosphorylation of tyrosine residues can lead to initiation of signaling pathways. CD31 is expressed on various cells including monocytes, polymorphonuclears (PMNs), platelets, and some subsets of T lymphocytes. It also presents on endothelial cells and functions in extravasation of leukocytes, angiogenesis and activation of integrins.1-5 So, the role of CD31 in inflammation and especially in nervous system has been also considered.6,7 Because expression of CD38 correlates with poor prognosis in B-cell chronic lymphocytic leukemia (B-CLL),8 clinical implication of its natural ligand, CD31,9 has also been investigated10 and showed that expression of CD31 further defines a sub-group of disease11 and low expression of this marker is an adverse prognostic factor.12
Anti-CD31 mouse monoclonal antibodies (MAbs) and their derivatives chimeric and humanized MAbs, therefore could be useful tools in diagnosis, research and therapy of diseases. Production of MAbs by hybridoma technology was first introduced by George Kohler and Cesar Milestain.13 Up to now, huge number of investigators have employed hybridoma technology, but with some modifications including different strategies for immunization of mice. Of them, some groups have stably expressed the gene coding for protein of interest in mouse fibroblast cell line, NIH-3T3,14 and have used the cells as immunogen.15-17 Because of murine origin of NIH-3T3 cell line, the only immunogen part of stably transfected cells is ectopically expressed protein. Using this strategy, all problems encountered in purification of recombinant proteins in eukaryotic systems are bypassed, and intact protein with complete conformational structure is used as immunogen. In addition, transfection of cDNA coding for a specific protein in NIH-3T3 cell line has been performed for purposes other than immunization of mice, e.g. the signaling potential or functional properties of the molecule.18-21
Here, we reported cloning of human CD31 cDNA and stable expression on NIH-3T3 mouse fibroblast cell line for upcoming experiments to produce monoclonal antibodies against CD31.
Materials and Methods
Cells and bacteria
KG1a and NIH-3T3 cell lines were purchased from National Cell Bank of Iran (NCBI, Tehran, Iran) and cultivated in RPMI 1640 cell culture medium (Gibco, Darmstadt, Germany) supplemented by 20% Fetal Bovine Serum (FBS) (Gibco, Darmstadt, Germany), 100 μg/ml Penicillin and 100 IU/ml Streptomycin (Gibco, Darmstadt, Germany) under humidified and 5% CO2 conditions. E.Coli strain DH5α was purchased from Promega Inc. (WI, USA) and cultured in Luria Bertani medium.
Flow cytometry
Evaluation of surface expression of CD31 molecule on KG1a, as a source for cloning of human CD31, was performed by indirect staining of KG1a cells. 5×105 cells were harvested and washed by PBS 1× containing 0.1% NaN3. Mouse monoclonal anti-human CD31 antibody (Biolegend, London, UK) was added on cells in final concentration of 5 μg/ml. In parallel, cells were stained with isotype control antibody (Biolegend, London, UK), as negative control. After 1 hour incubation at 4°C, cells were washed two times and then FITC-conjugated sheep anti-mouse immunoglobulin (Avicenna Research Institute, Tehran, Iran) was added in 1/50 dilution. Cells were incubated in a dark place for 1 hour at 4°C and after two times washing, they were scanned in flow cytometer (BD FACSCalibur flow cytometer).
Total RNA extraction and cDNA synthesis
5×106KG1a cells were harvested and washed two times by RPMI 1640 culture medium. After final centrifugation, supernatant was completely discarded and the pellet was thoroughly resuspended. Cells were lysed by 1 ml RNX-plus solution (CinnaGen, Tehran, Iran) and total RNA was extracted according to manufacturer´s recommendations. Briefly, after adding 200 μl chloroform (Merck, Darmstadt, Germany) and incubation for 5 minutes on ice, the solution was centrifuged at 12000 rpm for 15 minutes at 4°C and colorless aqueous phase was transferred to other tube carefully. Isopropanol (Merck, Darmstadt, Germany) in equal volume was added and after mixing and incubation for 15 minutes on ice, the solution was centrifuged and the precipitated RNA was seen. RNA was washed in 75% ethanol (Merck, Darmstadt, Germany) and resolved in RNase-free double distilled water. Quantity and quality of RNA was evaluated by UV spectrophotometry and agarose gel electrophoresis, respectively. Five μg RNA was employed for synthesis of first strand cDNA using MMuLV reverse transcriptase (RT) enzyme (Thermo Fisher Scientific, Inc., MA, USA) and random hexamer (N6) primer (Thermo Fisher Scientific, Inc., MA, USA). Amplification of beta actin as a house keeping gene in polymerase chain reaction (PCR), as discussed later, was performed for confirming synthesis of cDNA.
Primer design and Polymerase chain reaction
To amplify human CD31 cDNA, specific primers were designed so that forward and reverse primers contained KpnI and HindIII restriction sites, respectively. Selection of restriction enzymes was on the basis of multiple cloning sites (MCS) of expression vector will used for expression of protein in eukaryotic system. For this purpose, reference sequence for human CD31 mRNA was obtained and the sequence was imported in NEBCutter web-based software (version 2.0) to find if the sequence has restriction sites for KpnI and HindIII restriction enzymes. The sequence of forward and reverse primers were selected from the beginning and ending part of reference sequence, respectively and were analyzed in OligoCalc web-based software to check for self-complementarity and hairpin formation of primers. Also, melting temperature (Tm) of both primers was calculated for upcoming experiments and adjusted against each other. To facilitate efficient translation in eukaryotic expression system during subsequent experiments, Kozak consensus sequence (GCCACC) was considered after the sequence for KpnI restriction site (at the beginning of forward primer) and upstream of the start codon. As presence of G nucleotide at position +4 is considered as strong consensus,22 i.e it maximizes the expression level of gene of interest, nucleotide +4 was replace by G nucleotide. To terminate the translation, on the other hand, stop codon was considered at the end of sequence for CD31 cDNA and before the sequence for HindIII restriction site. Optimized PCR conditions were obtained by performing reactions in different concentrations of MgCl2 and a spectrum of annealing temperatures. Twenty-five μl reaction mixture contained 2.5 μl 10× PCR buffer, 1.5 μl 10 mM dNTPs (Thermo Fisher Scientific, Inc., MA, USA), 1μl each primer (10 pmol/μl), 0.2 μl Taq DNA polymerase (10 U/μl) (CinnaGene, Tehran, Iran) and 1 μl cDNA. Each PCR reaction was underwent initial denaturation at 95°C for 5 minutes followed by 39 cycles of denaturation (95°C for 30 seconds), annealing (different temperatures for 30 seconds) and extension (72°C for 45 seconds), and final extension at 72°C for 10 minutes. For obtaining PCR product with high fidelity, amplification of CD31 cDNA was performed using Pfu DNA polymerase (Thermo Fisher Scientific, Inc., MA, USA) in previously optimized PCR conditions, except of extension time for 2.5 minutes. A-tailing was done at 72°C for 7 minutes using Taq DNA polymerase. PCR products were subjected to agarose gel electrophoresis, visualized by ethidium bromide at final concentration 5 μg/ml and documented in UVP Gel Documentation System (UVP, CA, USA).
TA-cloning of CD31 cDNA
After agarose gel electrophoresis, specific band with correct size was extracted using GeneJet Gel Extraction Kit (Thermo Fisher Scientific, Inc., MA, USA) and then was ligated into pGEMT-easy vector (Thermo Fisher Scientific, Inc., MA, USA) using 3 units T4 DNA lygase (Thermo Fisher Scientific, Inc., MA, USA). After overnight incubation at 4°C, ligation mixture was transformed by heat shock method into DH5α competent bacteria, previously prepared by 0.1M CaCl2 solution. After refreshing bacteria by adding fresh LB broth and incubation for 1 hour at 37°C and shaking in 150 rpm, they were transferred on LB agar medium containing 100 μg/ml Ampicillin (Dana, Tabriz, Iran ), 40 µl X-Gal (20 μg/ml) (Thermo Fisher Scientific, Inc., MA, USA) and 40 µl IPTG (0.1mM) (Thermo Fisher Scientific, Inc., MA, USA). Plate was incubated overnight in 37°C and white colonies were evaluated by colony-PCR in conditions similar to conditions employed for amplification of CD31 cDNA. One positive colony was selected according to specific band with correct size and cultured in LB broth medium overnight at 37°C and shaking in 250 rpm. Miniprep preparation was performed by Gene JET Plasmid miniprep kit (Thermo Fisher Scientific, Inc., MA, USA) according to manufacturer´s recommendations. For initial confirmation of presence of a gene insert with correct size and proper restriction sites i.e. for KpnI and HindIII restriction enzymes (Thermo Fisher Scientific, Inc., MA, USA), double digestion was performed using 10 units of each enzyme overnight at 37°C. Digestion product along with undigested construct was run in agarose gel to visualize excised band. Consequently, selected construct was subjected for sequencing by T7 promoter and SP6 universal primers, to confirm accurate nucleotide sequence of insert. Chromatogram was analyzed first for good peaks for all nucleotides, and then entire sequence was aligned in NCBI database for checking proper sequence of inserted gene. On the other hand, flanking region of inserted gene was checked for vector-specific sequences to insure proper cloning vector and avoid cross-contamination by other commonly used vectors.
Preparation of pCMV6-Neo/CD31 recombinant construct
Double digested CD31 cDNA was sub-cloned in pCMV6-Neo digested in the same way. Ligation was done as previously described and competent DH5α bacteria were transformed by ligation mixture. Screening of colonies was performed on LB-Agar ampicillin plate and positive colonies were identified by colony-PCR reaction. Miniprep for one positive colony was prepared and direct sequencing using of V1.5 and XL39 primers was done for confirmation. To obtain high quality and quantity of finalized recombinant construct without any contamination by bacterial endotoxin, Maxiprep was prepared by EndoFree Plasmid Maxi Purification Kit (Thermo Fisher Scientific, Inc., MA, USA) according to manufacturer´s recommendations.
Eukaryotic expression of human CD31 in mouse fibroblast cell line
105 cells were seed in 6-well cell culture plate and cultured in the presence of different concentrations (300 to 800 μg/ml) of G418 (Thermo Fisher Scientific, Inc., MA, USA) and were monitored for one week. 105 NIH-3T3 cells were cultured overnight in 6-well cell culture plate and transfected by the mixture of 3 μg pCMV6-Neo/CD31 recombinant construct and 6 μl JetPEI transfection reagent (Polyplus-transfection Inc. NY, USA) according to manufacturer´s recommendations. In parallel, cells were transfected by expression vector devoid of CD31 cDNA (mock transfection). 48 hours after transfection, transient expression of CD31 antigen was examined in flow cytometry methods, as described earlier. To obtain stable expression, cells were cultured in complete cell culture medium containing G418. Starting concentration was determined by above-mentioned experiment on NIH-3T3 cells. Growing of transfected cells and dying of untransfected cells were monitored and live cells were subjected to gradually increasing concentrations of G418 up to 1500μg/ml during two months.
Results
Surface Expression of CD31
Flow cytometric analysis of surface expression of human CD31 on KG1a myeloid cell line, as a source for amplification of CD31 cDNA, showed strong fluorescent detected in 99 % of cells by flow cytometer. In contrast, surface staining of cells by isotype control, as negative control, showed very weak signal (1%).
Amplifying CD31 cDNA
Total RNA was purified from KG1a cells and cDNA was synthesized using 5 μg of total RNA. Processes were confirmed by agarose gel electrophoresis and beta actin gene amplification, respectively. Designed forward and reverse primers were 5´-GGTACCGCCACCATGGAG CCGAGGTGGGCCCAAG-3´ and 5´-AAGCTT CTAA GTTCCATCAAGGGAGCCTTCC-3´. They showed no self-complementarity and hairpin formation. On the other hand, the reference sequence had no restriction sites for selected restriction enzymes i.e. KpnI and HindIII. Using Taq DNA polymerase, conditions for amplification of CD31 cDNA were optimized. In 1 mM of MgCl2 and annealing temperature of 62°C, specific band with 2235 bp length (2217 bp for CD31 cDNA and 18 bp for two restriction sites and Kozak consensus sequence ) was obtained and the reaction was repeated using Pfu DNA polymerase, but using 2.5 minutes extension time at 72°C. After A-tailing by Taq DNA polymerase, PCR product was run in agarose gel electrophoresis (Figure 1A) and specific band was then extracted.
Figure 1 .
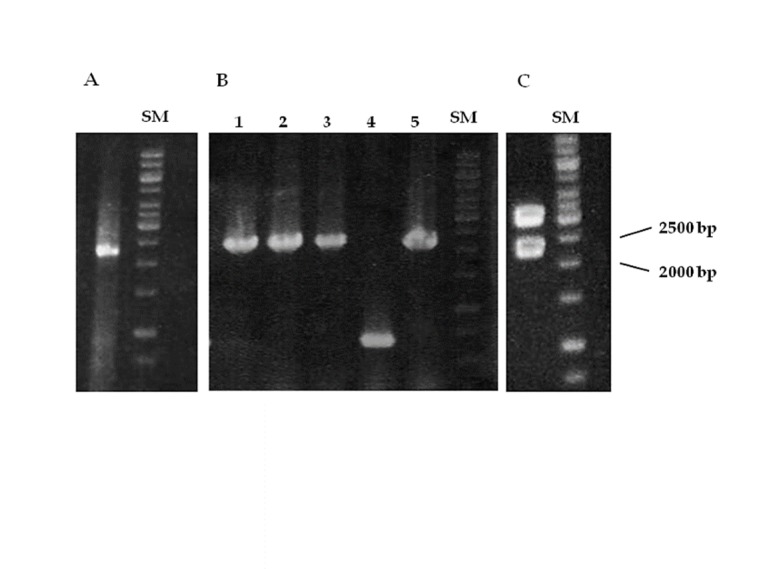
Cloning and subcloning of CD31 cDNA. (A) Amplification of specific band for human CD31 cDNA using Pfu DNA polymerase, (B) Colony–PCR reaction on five white colonies (1-5) after blue/white selection. (C) Excision of 2235 bp band for human CD31 cDNA after double digestion of the construct using KpnI and HindIII restriction enzymes. SM: DNA size marker (bp)
TA-cloning of CD31 cDNA
Extracted PCR product and pGEMT-easy TA-cloning vector were mixed in ratio 5:1 and incubated overnight. Transformation of competent bacteria was performed using heat shock method and after refreshing of bacteria, they spread on LB agar containing ampicillin, X-gal and IPTG. After overnight incubation in 37°C, five white colonies were selected for colony-PCR reaction. Using the same PCR conditions for amplifying CD31 cDNA, 4 colonies showed strong single bands comparable to the size for CD31 cDNA (Figure 1B). One of the colonies was cultured overnight in LB broth medium containing Ampicillin. Preparation of Miniprep was performed, and quality and quantity were measured by agarose gel electrophoresis and UV spectrophotometry, respectively. According to cloning strategy, the construct was subjected to double digestion by KpnI and HindIII restriction endonucleases and successful excision of inserted CD31 cDNA was seen (Figure 1C). Initial analysis of two-sided sequencing of the construct showed good quality peaks and alignment of the inserted sequence in NCBI data base showed complete matching of cloned CD31 cDNA to reference sequence for CD31 mRNA isoform 1 (Figure 2).
Figure 2 .
Alignment of amplified human CD31 cDNA with reference sequence in NCBI database. Comparing the amplified sequence with reference sequence for human CD31 showed complete alignment. Only 5´ and 3´ ends of sequence have been briefly shown.
Construction of recombinant pCMV6-Neo/CD31 construction
pGEMT-easy/CD31 recombinant cloning vector was digested by KpnI and HindIII restriction enzymes. Excised CD31 cDNA with 2235 bp band was ligated into double digested pCMV6-Neo expression vector by the same enzymes and pCMV6-Neo/CD31 construction was transformed into competent DH5α bacteria. After transformation, 8 colonies were obtained from cultured DH5α bacteria in LB agar/ampicillin medium. After performing colony-PCR reaction, 3 colonies showed successful insertion and one of the colonies was selected for preparation of Miniprep, direct sequencing and preparation of Maxiprep for subsequent experiments.
Transfection of NIH-3T3 cells and expression of human CD31 protein
NIH-3T3 mouse fibroblastic cell line was transfected by pCMV6-Neo/CD31 recombinant expression vector using JetPEI transfection reagent and transient expression of CD31 was confirmed by surface flow cytometry 48 hours after transfection (Figure 3C). After that, pCMV6-Neo/CD31- and mock-transfected and also untransfected NIH-3T3 cells were subjected to growth in complete cell culture medium containing 400 μg/ml G418, minimal dose for dying NIH-3T3 cells. Untransfected cells died and transfected cells were continued to be grown in G418 up to 1500 μg/ml during two months. Cells were finally analyzed by surface flow cytometry and significant positive reactivity of pCMV6-Neo/CD31- transfected cells (98%) was found (Figure 3D). On the other hand, untransfected and mock-transfected NIH-3T3 cells showed no positive signal in flow cytometry (5% and 4%, respectively) (Figure 3 A and B).
Figure 3 .
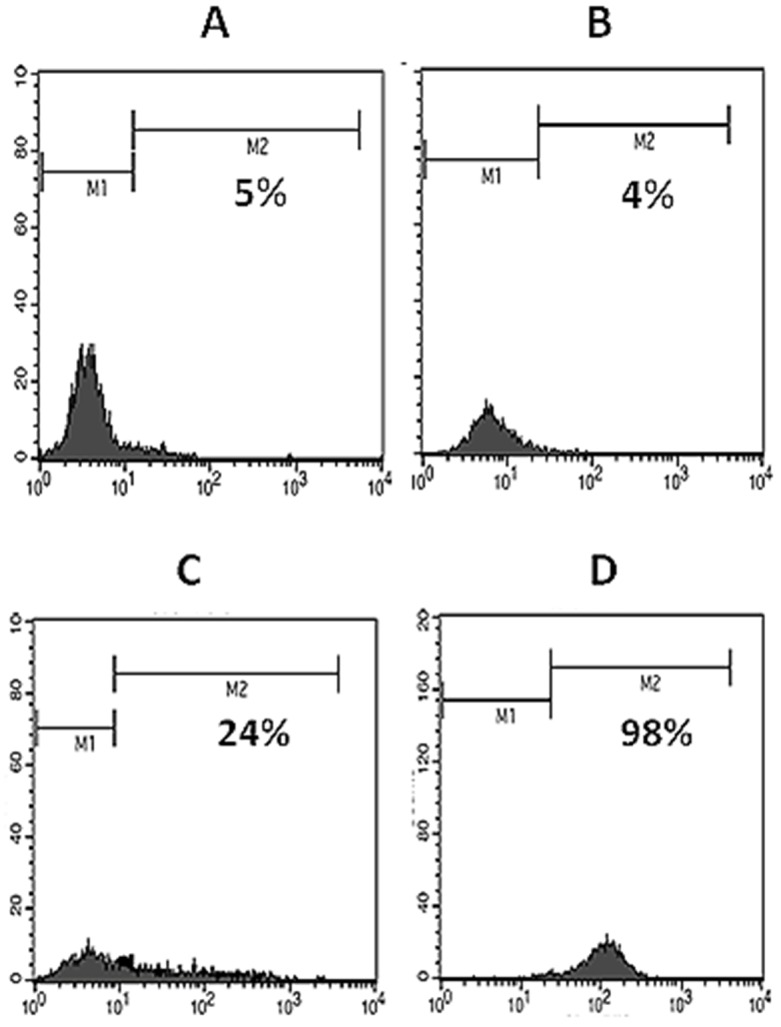
Flow cytometric analysis of expression of human CD31. Using specific monoclonal antibody, expression of human CD31 on NIH-3T3 (A), mock-transfected NIH-3T3 (B) and pCMV6-Neo/CD31-transfected NIH-3T3 cells (C, for transient and D, for stable expression) were analyzed.
Discussion
Human PECAM-1 or CD31 is a transmembrane glycoprotein with variety of functions in angiogenesis, trans-endothelial migration of leukocytes, and also in inflammation.3,6,7,11,23 More interests have been paid to CD31 after its introduction as ligand for CD38 and therefore its implication in some hematologic malignancies especially B-CLL.9-11 Specific monoclonal antibodies (MAbs) against CD31 could be therefore valuble tools in research, diagnostic and therapeutic areas. We aimed in this study to clone full-length cDNA coding for human CD31 and ectopically express in mouse fibroblast cell line, NIH-3T3. This artificial cell could be useful as immunogen for production of CD31-specific MAbs.
The first and one of the most important steps of hybridoma technology is preparing a good and proper immunogen. It should be as intact as possible for preserving all potential epitopes. On the other hand, using full-length protein with proper post transcriptionally modifications similar to that occurs for native protein, is helpful for obtaining functionally more active and useful MAbs.24 Additionally, immunization of animal with purified protein is done by mixing antigen to complete (CFA) and incomplete Freund´s adjuvant (IFA) usually administered subcutaneously. According to American Association for Laboratory Animal Science (IACUC) Policy on Administering Complete Freund´s Adjuvant,25 this protocol could induce unwanted local inflammation causing skin ulcerations and draining sinuses with granulomas and may lead to improper antibody response to antigen and limited number of antigen-specific clones. All above mentioned reasons force us to produce recombinant proteins in eukaryotic expression systems, in which purification step is tedious and challenging. However, immunization of mouse using stably-transfected murine NIH-3T3 cell line needs neither adjuvant nor purification and the route of injection is intraperitoneal with minimal side-effects for animal. Stable expression of cDNA coding for protein of interest in mouse fibroblast cell line, NIH-3T3, has been used by several investigators as an approach to prepare immunogen for immunization of mouse15-17 and also for other research purposes.18-21
KG1 cell line is a myeloblastic cell line with CD7−CD31+phenotype and was first identified and introduced in 1978. After 35 repetitive passages, a sub-lineage named KG1a (CD7+CD31+) was obtained with absence of some characteristics of its own parent cell line, including response to colony-stimulating factor (CSF) and expression of FCγ receptor.26 According to the study by Turner et al. to explore adhesion molecules on human hematopoietic cell lines,27 KG1a was showed that express high levels of CD31. To insure high expression of CD31 on KG1a cells in our study, they were subjected to indirect immunofluorescent staining, and results showed strong surface expression of CD31. So, KG1a was used in this study as a cellular source for expression of human CD31 gene and amplifying cDNA coding for CD31 protein.
Amplification of CD31 cDNA resulted in 2235 bp amplicon, subsequently cloned in pGEMT-easy TA-cloning vector and completely aligned to reference sequence of mRNA for long isoform of human CD31 isoform 1. The inserted sequence in cloning vector was sub-cloned in a eukaryotic expression vector for further transfection in mouse fibroblast cell line, NIH-3T3. For this purpose, pCMV6-Neo as an expression vector with a strong promoter for cytomegalovirus (CMV) was utilized for. pCMV6-Neo contains a resistance gene to G418 antibiotic (neomycin) and it is useable for screening of transfected cells. So transfected NIH-3T3 cells would be resistant and alive against G418 antibiotic.
Conclusion
In this study the full-length cDNA coding for human CD31 was cloned from KG1a myeloid cell line and stably expressed in mouse NIH-3T3 cell line. Stable expression of CD31 molecule in NIH-3T3 resulted in an appropriate immunogen could be helpful in production of monoclonal antibodies used in research, diagnostic and therapeutic areas.
Acknowledgments
We would like to give special thanks to Leila Mohammad-nejad for her technical assistance. This study was supported by a grant from Immunology Research Center, Tabriz University of Medical Sciences.
Ethical Issues
Not applicable.
Conflict of Interest
Authors declare no conflict of interests
References
- 1.Gumina RJ, Kirschbaum NE, Rao PN, Vantuinen P, Newman PJ. The human PECAM1 gene maps to 17q23. Genomics. 1996;34(2):229–32. doi: 10.1006/geno.1996.0272. [DOI] [PubMed] [Google Scholar]
- 2.Newman PJ. Switched at birth: a new family for PECAM-1. J Clin Invest. 1999;103(1):5–9. doi: 10.1172/jci5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson DE. The unfolding tale of PECAM-1. FEBS lett. 2003;540(1-3):7–14. doi: 10.1016/s0014-5793(03)00224-2. [DOI] [PubMed] [Google Scholar]
- 4.Newman PJ, Newman DK. Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler Thromb Vasc Biol. 2003;23(6):953–64. doi: 10.1161/01.atv.0000071347.69358.d9. [DOI] [PubMed] [Google Scholar]
- 5.Engelhardt B, Wolburg H. Mini-review: Transendothelial migration of leukocytes: through the front door or around the side of the house? Eur J Immunol. 2004;34(11):2955–63. doi: 10.1002/eji.200425327. [DOI] [PubMed] [Google Scholar]
- 6.Woodfin A, Voisin MB, Nourshargh S. PECAM-1: a multi-functional molecule in inflammation and vascular biology. Arterioscler Thromb Vasc Biol. 2007;27(12):2514–23. doi: 10.1161/atvbaha.107.151456. [DOI] [PubMed] [Google Scholar]
- 7.Kalinowska A, Losy J. PECAM-1, a key player in neuroinflammation. Eur J Neurol. 2006;13(12):1284–90. doi: 10.1111/j.1468-1331.2006.01640.x. [DOI] [PubMed] [Google Scholar]
- 8.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL. et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. [PubMed] [Google Scholar]
- 9.Deaglio S, Mallone R, Baj G, Arnulfo A, Surico N, Dianzani U. et al. CD38/CD31, a receptor/ligand system ruling adhesion and signaling in human leukocytes. Chem Immunol. 2000;75:99–120. doi: 10.1159/000058765. [DOI] [PubMed] [Google Scholar]
- 10.Chiorazzi N. Implications of new prognostic markers in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:76–87. doi: 10.1182/asheducation-2012.1.76. [DOI] [PubMed] [Google Scholar]
- 11.Ibrahim S, Jilani I, O'brien S, Rogers A, Manshouri T, Giles F. et al. Clinical relevance of the expression of the CD31 ligand for CD38 in patients with B-cell chronic lymphocytic leukemia. Cancer. 2003;97(8):1914–9. doi: 10.1002/cncr.11264. [DOI] [PubMed] [Google Scholar]
- 12.Mainou-Fowler T, Porteous A, Nicolle A, Proctor SJ, Anderson JJ, Summerfield G. CD31 density is a novel risk factor for patients with B-cell chronic lymphocytic leukaemia. Int J Oncol. 2008;33(1):169–74. doi: 10.3892/ijo.33.1.169. [DOI] [PubMed] [Google Scholar]
- 13.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 14.Hollingsworth MA, Rebellato LM, Moore JW, Finn OJ, Metzgar RS. Antigens expressed on NIH 3T3 cells following transformation with DNA from human pancreatic tumor. Cancer Res. 1986;46(5):2482–7. [PubMed] [Google Scholar]
- 15.Kazemi T, Tahmasebi F, Bayat AA, Mohajer N, Khoshnoodi J, Jeddi-Tehrani M. et al. Characterization of novel murine monoclonal antibodies directed against the extracellular domain of human HER2 tyrosine kinase receptor. Hybridoma (Larchmt) 2011;30(4):347–53. doi: 10.1089/hyb.2011.0023. [DOI] [PubMed] [Google Scholar]
- 16.Klapper LN, Vaisman N, Hurwitz E, Pinkas-Kramarski R, Yarden Y, Sela M. A subclass of tumor-inhibitory monoclonal antibodies to ErbB-2/HER2 blocks crosstalk with growth factor receptors. Oncogene. 1997;14(17):2099–109. doi: 10.1038/sj.onc.1201029. [DOI] [PubMed] [Google Scholar]
- 17.Buhring HJ, Sures I, Jallal B, Weiss FU, Busch FW, Ludwig WD. et al. The receptor tyrosine kinase p185HER2 is expressed on a subset of B-lymphoid blasts from patients with acute lymphoblastic leukemia and chronic myelogenous leukemia. Blood. 1995;86(5):1916–23. [PubMed] [Google Scholar]
- 18.Moscatelli D, Quarto N. Transformation of NIH 3T3 cells with basic fibroblast growth factor or the hst/K-fgf oncogene causes downregulation of the fibroblast growth factor receptor: reversal of morphological transformation and restoration of receptor number by suramin. J Cell Biol. 1989;109(5):2519–27. doi: 10.1083/jcb.109.5.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayasu M, Shima H, Aonuma S, Nakagama H, Nagao M, Sugimura T. Deletion of transfected oncogenes from NIH 3T3 transformants by inhibitors of poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 1988;85(23):9066–70. doi: 10.1073/pnas.85.23.9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nigro S, Geido E, Infusini E, Orecchia R, Giaretti W. Transfection of human mutated K-ras in mouse NIH-3T3 cells is associated with increased cloning efficiency and DNA aneuploidization. Int J Cancer. 1996;67(6):871–5. doi: 10.1002/(sici)1097-0215(19960917)67:6<871::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Thorgeirsson UP, Turpeenniemi-Hujanen T, Williams JE, Westin EH, Heilman CA, Talmadge JE. et al. NIH/3T3 cells transfected with human tumor DNA containing activated ras oncogenes express the metastatic phenotype in nude mice. Mol Cell Biol. 1985;5(1):259–62. doi: 10.1128/mcb.5.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108(2):229–41. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilan N, Madri JA. PECAM-1: old friend, new partners. Curr Opin Cell Biol. 2003;15(5):515–24. doi: 10.1016/s0955-0674(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 24.Brown MC, Joaquim TR, Chambers R, Onisk DV, Yin F, Moriango JM. et al. Impact of immunization technology and assay application on antibody performance--a systematic comparative evaluation. PLoS One. 2011;6(12):e28718. doi: 10.1371/journal.pone.0028718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. IACUC Policy on Administering Complete Freund’s Adjuvant (CFA) and other Adjuvants. [28 July 2014]; Available from: http://www.iacuc.emory.edu/documents/367_Complete_Freunds_Adjuvent.pdf.
- 26.Furley AJ, Reeves BR, Mizutani S, Altass LJ, Watt SM, Jacob MC. et al. Divergent molecular phenotypes of KG1 and KG1a myeloid cell lines. Blood. 1986;68(5):1101–7. [PubMed] [Google Scholar]
- 27.Turner ML, Masek LC, Hardy CL, Parker AC, Sweetenham JW. Comparative adhesion of human haemopoietic cell lines to extracellular matrix components, bone marrow stromal and endothelial cultures. Br J Haematol. 1998;100(1):112–22. doi: 10.1046/j.1365-2141.1998.00543.x. [DOI] [PubMed] [Google Scholar]



