Abstract
Purpose: Microbial assay is used to determine the potency of antibiotics and vitamins. In spite of its advantages like simplicity and easiness, and to reveal the slight changes in the molecules, the microbial assay suffers from significant limitations; these methods are of lower specificity, accuracy and sensitivity. The objective of the present study is to evaluate the efficacy of real time-PCR technique in comparison with turbidimetric method for microbial assay of amikacin.
Methods: Microbial determination of amikacin by turbidimetric method was performed according to USP. Also amikacin concentrations were determined by microbial assay using taq-man quantitative PCR method. Standard curves in different concentration for both methods were plotted and method validation parameters of linearity, precision and accuracy were calculated using statistical procedures.
Results: The RT-PCR method was linear in the wider concentration range (5.12 – 38.08 for RT-PCR versus 8.00 – 30.47 for turbidimetric method) with a better correlation coefficient (0.976 for RT-PCR versus 0.958 for turbidimetric method). RT-PCR method with LOQ of 5.12 ng/ml was more sensitive than turbidimetric method with LOQ of 8.00 ng/ml and the former could detect and quantify low concentrations of amikacin. The results of accuracy and precision evaluation showed that the RT-PCR method was accurate and precise in all of the tested concentration.
Conclusion: The RT-PCR method described here provided an accurate and precise technique for measurement of amikacin potency and it can be a candidate for microbial determination of the antibiotics with the same test organism.
Keywords: Microbial assay, Real time-PCR, Turbidimetric method, Amikacin
Introduction
Bioassay is referred to as “a procedure for determining the concentration, purity, and/or biological activity of a substance by measuring its effect on a biological system compared to a standard system”. Vitamins, hormones, plant growth factors, antibiotics or enzymes are substances that can be assayed using bioassay.1 The biological systems used for measuring these effects ranged from sub-cellular components and certain types of microorganisms to groups of animals. Bioassays are most frequently used when there are a number of complicated steps between the system exposure to the substance and the behavior observed, or when the substance is a complex mixture of materials and it is not clear what the active components are.2
One of the well known bioassays is microbial assay which is used to determine the potency of growth-inhibiting substances-especially antibiotics- and growth-promoting substances- like amino acids and vitamins of B group. There are two main procedures for microbial assay including agar diffusion assay and turbidimetric method (also known as tube assay). The basis of both procedures is to measure the microbial growth inhibition or promotion in exposure to the substance to be assayed. In the agar diffusion assay, it is performed based on the sizes of the zones formed as a result of microbial growth inhibition or promotion around the substance reservoir on a nutrient agar medium. On the other hand, the turbidimetric method is based on the photometrical changes in turbidity of the microbial culture as a result of the inhibition or promotion of growth of microbial culture in response to the substance.3 The turbidimetric method has been officially applied for determination of different antibiotics like amikacin as stated in pharmacopeias.4,5
The microbial assay comprises some advantages over instrumental analytical procedures. Firstly, it is economical and easy to perform with no need for specialized equipment or toxic solvents. Moreover, the microbial assay can reveal slight changes which are not demonstrable by using most of conventional chemical methods. However like other bioassays, the microbial assay suffers from significant limitations; these methods are of lower specificity, accuracy and sensitivity.3,6,7
Some efforts to combat the shortcomings of microbial assay have been reported so far,8 for example Marengo and coworkers used a special test organism for microbiological assay of amikacin to improve the specificity of the method. They employed a strain of Providencia stuartii which is resistant to multiple antibiotics except amikacin. According to their study, application of the resistant strain to other antibiotics, allowed the utilization of this assay method in the presence of other concurrently administered antibiotics without the need for their inactivation in the assay procedure and consequently promote the specificity of the method.9
In another effort, Lourenço et al developed a turbidimetric method accomplished by kinetic-reading microplate system instead of spectroscopy. Their proposed method requires significantly lower amount of materials like solutions, culture media, test tubes and consequently physical space. They claimed that the proposed method merged the benefits of both microbiological and physico-chemical assays.10
Real-time quantitative PCR is a rapid method in which quantifies DNA in a very sensitive and reliable manner and thus has the potential for accurate enumeration of microorganisms. Real-time PCR allows accurate detection and quantification of specific nucleic acids in a mixture even if the starting amount of nucleic acids is at a very low concentration. This is accomplished by monitoring the amplification of a target sequence in real-time using fluorescent technology. How quickly the amplified target reaches a threshold detection level correlates with the amount of starting material present. In addition, the potential for PCR contamination in the laboratory is reduced because amplified products can be analyzed and disposed without opening the reaction tubes.11-13 This method has been successfully applied to microbial quantification of certain bacterial populations and total bacteria in diverse cases. It has been proven that real-time PCR was able to detect cells over a linear range of 10 to over than 108 cells.14
Considering the significant advantages of real-time PCR technique over conventional turbidimetric method in quantifying bacterial load, here we aimed to develop a microbial assay method for amikacin, using real-time PCR technique and to validate the linearity, limits of detection and quantification, accuracy and precision of developed method.
Materials and Methods
Materials
Staphylococcus aureus ATCC 29737 (National Cell Bank, Pasteur Institute, Iran), antibiotic agar no. 1 and 3 media (Merck, Germany), Amikacin powder (Exir Co., Iran), proteinase K, Ins TAcloneTM PCR Cloning Kit, ethidium bromide, DNA taq polymerase, MgCl2, dNTPs (Fermentas, EU), QIA quick gel extraction kit (Qiagen, USA). All the other chemicals and reagents were purchased from Merck, Germany.
Bacterial strain, culture condition and inoculum preparation
Staphylococcus aureus ATCC 29737 was selected as test organism for amikacin bioassay according to United States Pharmacopeia and it was cultured in antibiotic agar no. 1 medium at 35-37 °C, for 24 hours. The grown colonies were collected using sterile saline and sterile glass beads to prepare the bacterial suspension. Then appropriate amount of the bacterial suspension was added to 400 ml of antibiotic agar no. 3 medium to prepare the inoculum.5
Preparation of amikacin standards
Amikacin standards were prepared according to USP as follow: The stock solution of amikacin was prepared by dissolving appropriate amount of amikacin in 10 ml of sterile water to obtain the concentration of 1 mg/ml. Using this stock solution and sterile water as diluent, ten different standard solutions were prepared by serial dilution method in the ratio of 1: 1.25. The median dose was equal to the 10 µg/ml5
Turbidimetric method
For turbidimetric assay one ml of each standard solution as well as one ml water as positive control were added to each of three prepared test tubes in the test tube rack. Then 9 ml of inoculum was added to each tube in the rack and the completed rack immediately was placed in an incubator at 37 °C, for 4 hours. Each rack also included one blank tube containing one ml of water as diluent and 9 ml of sterile antibiotic agar no. 3. After incubation period, firstly 5 ml of contents of each tube was transferred into another test tube, and then immediately centrifuged for 1 hour at 11,000 rpm. The pellets was frozen and maintained at -20 °C until real time-PCR assay. 0.5 ml of diluted formaldehyde was added to the rest of tubes content. Absorbance of these tubes was read in Cecil spectrophotometer (Cambridge, England) fitted with 580 nm filter.5
DNA extraction
Genomic DNA was extracted from the pellets by SDS-Proteinase K method15 as bellow:
The pellet was dissolved in 475 µl of Tris-EDTA buffer. 60 µl of 10% sodium dodecyl sulphate (SDS) and 7 µl of proteinase K was added to the above suspension, and the mixture incubated for 1 hour at 37°C, then 100 µl 5M NaCl solution was added and mixed well. 80 µl of pre-warmed CTAB/NaCl solution was added, mixed well and the mixture incubated at 65ºC for 10 minutes. After cooling, 700 µl of chloroform-Isoamylalcohol (24: 1) was added, vortexed well and centrifuged for 10 minutes at 11,000g. The aqueous phase was transferred into a new micro tube, to which cold isopropanol was added with the ratio of 0.6 volume of aqueous phase. The mixture was mixed three times and then incubated at -20°C for 30 minutes and finally centrifuged for 15 min at 12000g. The DNA pellet was washed with ethanol and centrifuged for 10 minutes at 12, 000g (the step repeated twice). Then the supernatant was discarded carefully and DNA pellet was dissolved in 100 µl of TE buffer after complete drying.
Cloning of nuc a DNA target into plasmid vector pTZ57R
The nuc a gene of the extracted DNA from S. aureus was amplified using below primers: (sense) 5'-ACACCTGAAACAAAGCATCC, and (Antisense): 5'-ATACGCTAAGCCACGTCCA. The reaction was performed by eppendorf PCR termocycler (Hamburg, Germany) at a final volume of 20 µL contained 2 µl of buffer, 0.4 µl of MgCl2, 0.4 µl of dNTPs, 0.36 µl of PCR primers, 0.4 µl of DNA taq polymerase, and 1µl of DNA template. The reactions were run for 35 cycles with denaturation at 94°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 50s. An initial 5-min denaturing step at 95°C was used. The products were analyzed by 1% agarose gel electrophoresis containing ethidium bromide. The gels were visualized under UV transillumination (Vilber Lourmat, France).
The PCR product was extracted from the gel by QIA quick gel extraction kit and inserted into the pTZ57R vector using Ins TAcloneTM PCR Cloning Kit according to the manufacturer’s instruction. The cloned targets, designated as pTZ57R-nuc a, were amplified by transformation to the competent E. coli DH5α followed by its extraction using alkaline lysis method. The double stranded DNA approximation was performed using the UV/Vis spectrophotometer (Cambridge, England). Then, the corresponding plasmid copy number (PCN) was calculated using below Eq.1.16
Real-time PCR primers and Taq Man probes
In this study, we designed a primer set from the sequence of the nuc a gene for real-time PCR detection of S. aureus. Sequence representing S. aureus was downloaded from the Gen Bank nucleotide database. A region of the nuc a gene was selected for the design of real-time PCR primers and a fluorogenic 5'-nuclease assay probe (Taq-Man probe) using Beacon designer software (PREMIER Biosoft USA). Sequences of S. aureus primers and dual labeled Taq Man probe are as follows:
StaNuc1 (sense: 5'-TGAAACAAAGCATCCTAAA);
StaNuc 2 (antisense: 5'- TTCTTTGCATTTTCTACCA);
StaNucPR (sense: 5'-FAM- ACTTGCTTCAGGACCATATTTCTCT -BHQ1).
BLAST searches17 predicted that the S. aureus primer/probe sets would be specific for their respective targets and would not be expected to cross react with any other microbial sequences.
Real-time PCR amplification and detection
Firstly, a standard curve was plotted with Ct values obtained from amplification of known quantities of pTZ57R-nuc a. The standard curves were used to translate Ct values to the absolute number of DNA molecules. A ten-fold serial dilution series of the pTZ57R-nuc a, ranging from 1 × 102 to 1 × 107 copies/ml, was used to construct the standard curves. Threshold cycle (Ct) values in each dilution were measured in duplicate and were plotted against the logarithm of their initial template copy numbers. Each standard curve was generated by a linear regression of the plotted points. From the slope of each standard curve, PCR amplification efficiency (E) was calculated according to the Eq. 2:
PCR runs were accepted when the standard curve correlation co-efficient was ≥ 0.99. The contamination in each run was determined with the control reaction that did not contain DNA.
In the next step, the DNA of frozen samples from turbidimetric method was extracted, amplified and quantified in the instrument RotorGen 6000 (Corbett, Australia) using the PCR primers and probe. The reactions were run for 35 cycles, with denaturation at 94°C for 45s, annealing at 62°C for 45s, and extension at 72°C for 45s. An initial 5-min denaturing step at 95°C was used.
PCRs were performed at a final volume of 20 µL contained 2 µl of buffer, 0.4 µl of MgCl2, 0.4 µl of dNTPs, 0.36 µl of PCR primers, 0.4 µl of DNA taq polymerase, 0.24 µl probe and 1µl of DNA template. At the end of each reaction, the critical threshold cycle (Ct) is defined automatically by RotorGen software, as the cycle at which the fluorescence becomes detectable above background levels and is inversely proportional to the logarithm of the initial number of template molecules.
Method validation
Both methods were validated in terms of linearity, accuracy, and precision, limit of detection (LOD) and Limit of quantification (LOQ).
The linearity of the methods was investigated using the analysis of 10 serially diluted samples of amikacin in the concentration range of 6.4 to 38.08 ng/ml. Then correlation between logarithm (log) of amikacin concentration and absorbance in turbidimetric method or between log of concentration and log of bacterial number in RT-PCR method, were obtained by linear regression analysis. Since our method is based on the antibacterial activity of amikacin, LOQ was calculated according to the minimum concentration of amikacin which produced a visually detectable antibacterial effect against S. aureus.18 The obtained LOQ values were confirmed according to the accuracy and precision of the method at these levels.
The intra- and inter-day precisions and the accuracy of the methods were determined using 3 concentrations at the lower, the middle and the upper level of the of calibration curves. For intra-day precision, relative standard deviation of four independent assays carried out on the same day was reported. Inter-day precision was evaluated by three groups and each group containing four independent assays carried out on the same day. For accuracy determination, samples with a known concentration were analyzed in triplicate, and then the experimentally derived concentrations were calculated from the responses and the calibration equation. Then the accuracy at each concentration was reported as a percentage of the experimentally derived concentration to the nominal concentration.18,19
The agreement between two methods was evaluated by computing the mean square error and the Bland-Altman method.20
Results
Standard curve construction using pTZ57R-nuc a
The nuc a gene of the extracted DNA from S. aureus was amplified using the designed primers listed in method section and the products were analyzed by agarose gel electrophoresis. As can be seen in Figure 1, the presences of a DNA fragment of 153 bps confirmed the amplification of nuc a gene of S. aureus. The nuc a encoding DNA was cloned into pTZ57R vector using Ins TAcloneTM PCR Cloning Kit, and the recombinant vector, designated as pTZ57R-nuc a, was further amplified by transformation to the competent E. coli DH5α. The concentration of the extracted pTZ57R-nuc a was approximated using the UV/Vis spectrophotometer. To make a standard curve translating the Ct values to the absolute number of DNA molecules a ten-fold serial dilution series of the pTZ57R-nuc a, ranging from 1 × 102 to 1 × 107 copies/ml, were quantified in real time PCR using the primers and probes mentioned in method section. Furthermore, the DNA of the frozen samples from turbidimetric method was simultaneously amplified and quantified using the Real Time PCR method.
Figure 1 .
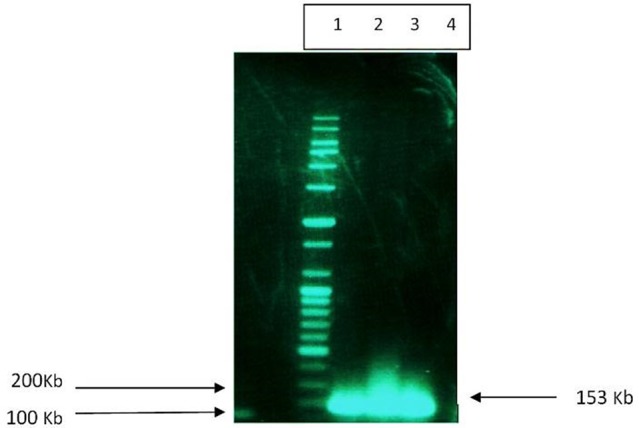
Agarose gel electrophoresis of PCR products for amplification of nuc a gene of S. aureus. Lane 1: 100bs DNA ladder; Lane 2-4: samples amplified with primers for nuc a gene of S. aureus.
Validation of the turbidimetric and RT- PCR method
Linear relationship between bacterial absorbance at λ max = 580 nm and concentration (in log scale) of the amikacin in turbidimetric method are shown in Figure 2. Also Figure 3 depicted the linear relationship between cell number and concentration of amikacin (in log scale) in RT-PCR method.
Figure 2 .
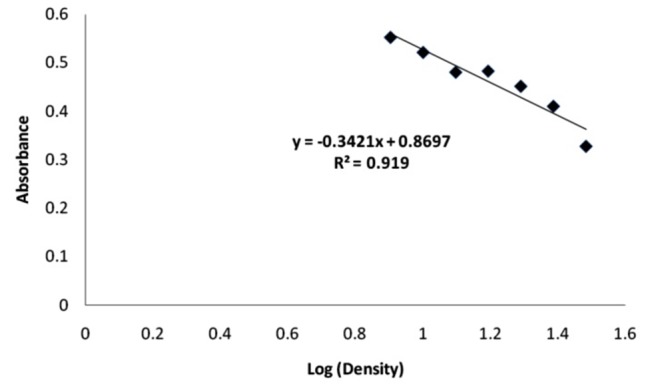
Linear relationship between bacterial absorbance at λ max = 580 nm and concentration (in log scale) of the amikacin in turbidometric method.
Figure 3 .
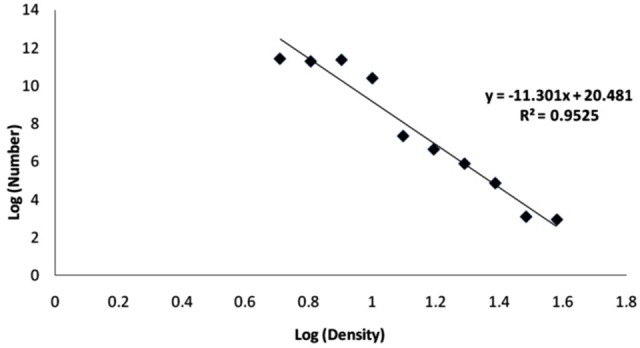
Linear relationship between cell number and concentration of amikacin (in log scale) in rt-PCR method. (rt-PCR conditions: 35 cycles, denaturation at 94°C for 45 s, annealing at 62°C for 45 s, extension at 72°C for 45s, final volume = 20 µL contained 2 µl of buffer, 0.4 µl of MgCl2, 0.4 µl of dNTPs, 0.36 µl of PCR primers, 0.4 µl of DNA taq polymerase, probe and 1µl of DNA template.)
According to the linearity parameters reported in Table 1, the RT-PCR method was linear in the wider concentration range (5.12 – 38.08 for RT-PCR versus 8.00 – 30.47 for turbidimetric method) with a better correlation coefficient (0.976 for RT-PCR versus 0.958 for turbidimetric method). The LOQ values for amikacin against S. aureus based on visual method were to be 5.12 and 8.00 ng/ml for RT-PCR and turbidimetric respectively. The values were located in the linear range with accuracy of 80-120% and precision less than 20%.
Table 1. Linearity parameters of the methods.
| Method | Linearity range (ng/ml) | Y-intercept ± SD* | Slope ± SD | Point number | Correlation coefficient (r) | LOQ (ng/ml) |
| Turbidometry | 8.00 – 30.47 | 0.869 ± 0.02 | -0.342 ± 0.03 | 7 | 0.958 | 8.00 |
| Rt-PCR | 5.12 – 38.08 | 20.48 ± 0.16 | 11.30 ± 0.16 | 10 | 0.976 | 5.12 |
*SD: Standard deviation
Comparing the LOQ values of two methods indicated that RT-PCR method was more sensitive and it could detect lower concentrations of amikacin. Our method could not detect the concentrations lower than the LOQ values because that these levels fall under the minimum inhibitory concentrations of amikacin against S. aureus. Therefore the LOD values were considered equal to LOQ values.
The results of accuracy and precision evaluation were shown in Table 2. According to the ICH and FDA guidelines [18-19], the RT-PCR method was accurate and precise in all of the tested concentration, as an accuracy of between 85 and115% and precision of less than 15%; therefore it could be concluded that the proposed method was accurate and precise in whole concentration range. On the other hand turbidimetric method was precise at all of the tested concentration, but it was accurate only in middle and low concentrations. According to the guidelines, the method was valid because two out of three concentrations met the acceptable criteria. Therefore, it can be said that, although the methods was valid (because only one of concentrations were out of range), the obtained results at high concentration of amikacin were not reliable.
Table 2. Accuracy and precision of the methods .
| Method | Concentration (ng/ml) | Precision (RSD*%) | Accuracy ± SD (%) | |
| Intra-day | Inter-day | |||
| Turbidometry | 8 | 2.82 | 3.37 | 103.92 ± 19.1 |
| 15.6 | 4.03 | 7.29 | 86.01 ± 16.3 | |
| 30.47 | 4.47 | 12.79 | 125.31 ± 8.95 | |
| rt-PCR | 6.4 | 2.22 | 1.39 | 101.45 ± 3.21 |
| 15.6 | 1.19 | 5.59 | 107.03 ± 7.95 | |
| 38.08 | 3.44 | 8.04 | 93.19 ± 4.58 | |
*RSD: Relative standard deviation
Correlation between results obtained from RT-PCR and turbidimetry and evaluating the agreement between two methods
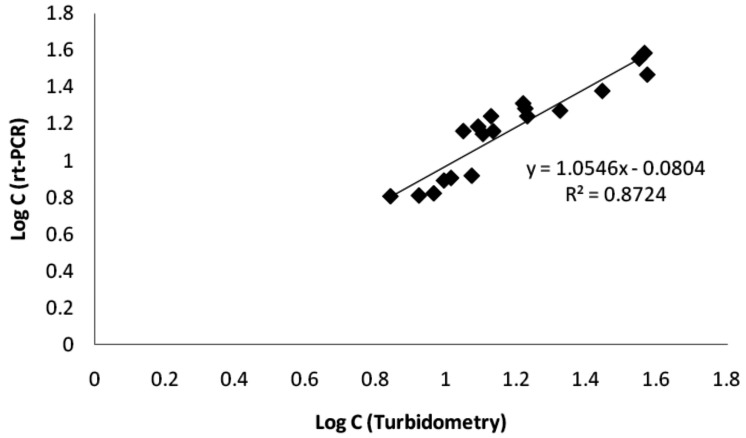
The log of obtained concentrations from RT-PCR method was correlated against those obtained from turbidimetric method with a linear regression method. The concentration range of 8.00 to 30.47 ng/ml was selected for the correlation according to the linearity range of the turbidimetric method. Obtained calibration graph that was shown in Figure 4 demonstrated a linear relationship between them with correlation coefficient of 0.934.
Figure 4 .
Correlation between concentrations obtained using turbidometric and rt-PCR methods in the concentration range of 8.00 to 30.47 µg/ml.
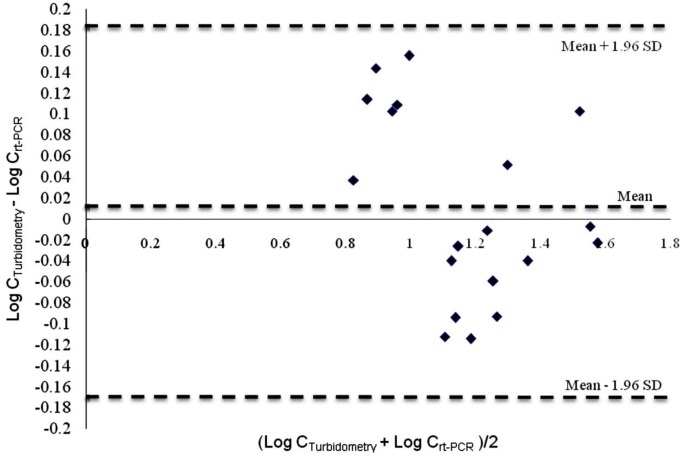
Since the Application of regression method as the only approach to compare between two methods could be misleading; the agreement between methods was analyzed by Bland-Altman method and the Tukey mean-difference plot was drawn by plotting the differences between obtained results of methods against average of these results [20]. As shown in Figure 5, the obtained results from both of the methods were in acceptable range (average difference ± 1.96 SD Differences).
Figure 5 .
Tukey mean-difference plot (Bland-Altman plot) for evaluating the agreement between turbidometry and rt-PCR method in the concentration range of 8.00 to 30.47 µg/ml
According to the regression and Bland-Altman analysis, RT-PCR method could be applied as a reliable alternative for turbidimetry in quantification of amikacin.
Discussion
The turbidimetric method is based on the photometrical changes in turbidity of the microbial culture as a result of the inhibition or promotion of growth of microbial culture in response to the tested substance3. According to the Lambert-Beer law, when light is transmitted through a suspension, its intensity is reduced in proportion to the thickness of the layer of the suspension and to the concentration of the materials suspended.
The Beer-Lambert law is written as Eq3:
Eq3: A = Ɛ bc
where Ɛ is the wavelength-dependent molar absorptivity coefficient with unit of Lmol-1 cm-1, b is the path length with unit of cm, and c is the analyte concentration with unit of mol L-1.18
It is obvious that the Lambert-Beer law is valid only for concentrations between about 107 and 109 organisms per ml; marked deviations are observed at higher and lower concentrations. In fact, the logarithmic phase in microbial growth phase begins about 1 hour after inoculation and lasts about 3 hours. The deviations from the Lambert-Beer law can be related mostly to the reduced size of the bacteria during the early phase of logarithmic growth.21,22 As a result, incubation times, initial inoculum concentration and the antibiotic concentration are markedly restricted lead to low repeatability of the turbidimetric method.
RT-quantitative PCR is a method based on PCR to amplify and quantify a template DNA molecule in a very sensitive and reliable manner and thus has the potential for accurate enumeration of microorganisms.11-14 There are many different variations of RT-PCR chemistry but three types have been used most frequently; DNA binding dyes like SYBR green; DNA hybridization probes such as FRET (Light cycler) and Taqman probes (Applied Biosystems)11. Sensitive and specific detection is possible with RT-PCR using fluorescent labeled probes. Fluorescent probes are more expensive than intercalating dyes, but offer a more specific way of detecting the accumulation of amplicon. Since these probes use sequence specific oligonucleotides coupled to fluorescent dyes, therefore they allow eliminating false positives due to primer dimmers and non-specific product formation.23,24 Nuc a gene has been previously used to enumerate Staphylococcus aureus.25 Here, we hypothesized that the real-time PCR technique would be a good candidate to replace with turbidimetric method to eliminate the problems of antibiotic assay by microbial method.
The method validation results indicated that, in comparison with the turbidimetric method, RT-PCR was linear in the wider range with better correlation coefficient. Also it was more accurate and precise. The basis of both methods (turbidimetric and RT-PCR method) for assay of amikacin is to measure the microbial growth inhibition in exposure to the antibiotic and the concentrations lower than the MIC value were practically undetectable using these methods. As a result, LOQ was calculated according to the minimum concentration of amikacin which produced a visually detectable antibacterial effect against S. aureus.18 This method for determination of LOQ of microbial assays was previously applied.26,27 According to the results, RT-PCR method was more sensitive than turbidimetric method with less LOQ value. Meanwhile there was a linear correlation with coefficient of 0.985 between obtained results from the turbidimetric and RT-PCR methods, also the results of Bland-Altman analysis showed a good agreement between both methods. Therefore, considering the validation parameters it could be said that, the proposed RT-PCR method could be a precise method for detection and quantification of amikacin. However, it had some drawbacks over turbidimetric method. The first and the most important one is that RT-PCR is an indirect method for detection of amikacin. It is a complex method which requires more expensive reagents and instrument, and also it was very labor intensive.
Conclusion
The RT-PCR method described here provided an accurate and precise technique for measurement of amikacin potency and it can be a candidate for microbial determination of the antibiotics with the test organism of S. aureus with ATCC number of 29737.
Acknowledgments
The authors thank Faculty of Pharmacy and Hematology and Oncology Research Center, Tabriz University of Medical Sciences, Iran. The results described in this paper were part of student thesis.
Ethical Issues
Not applicable.
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Duffus HJ. Glossary for chemists of terms used in toxicology. Pure Appl Chem. 1993;65(9):2003–122. doi: 10.1351/pac199365092003. [DOI] [Google Scholar]
- 2. International Society for Complexity, Information, and Design (ISCID). ISCID Encyclopedia of Science and Philosophy - BETA. Available from: http://www.iscid.org/encyclopedia/Bioassay.
- 3.Hewitt W. Microbiological assay for pharmaceutical analysis: a rational approach. Florida: Interpharm/CRC press; 2004. [Google Scholar]
- 4. The Japanese Pharmacopoeia. General tests, Microbial assay for antibiotics. 15th ed. Available from: http://jpdb.nihs.go.jp/jp14e/14data/General_Test/Microbial_Assay_for_Antibio.pdf.
- 5. United States Pharmacopeia 30/National Formulary 25. Rockville, MD, USA: United States Pharmacopeial Convention; 2006.
- 6.Gavin JJ. Gavin JJMicrobiological process report: analytical microbiologyIIITurbidimetric methods. Appl Microbiol. 1957;5(4):235–43. doi: 10.1128/am.5.4.235-243.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcotte C, Lacroix C, Kheadr E, Grignon L, Fliss I. A rapid turbidometric microplate bioassay for accurate quantification of lactic acid bacteria bacteriocins. Int J Food Microbiol. 2004;90(3):283–93. doi: 10.1016/s0168-1605(03)00315-5. [DOI] [PubMed] [Google Scholar]
- 8.Pitkin D, Actor P, Holl W, Post A, Weisbach JA. Semiautomated turbidimetric microbiological assay for determination of cefazolin. Antimicrob Agents Chemother. 1974;5(3):223–7. doi: 10.1128/aac.5.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marengo PB, Wilkins J, Overturf GD. Rapid, specific microbiological assay for amikacin (BB-K8) Antimicrob Agents Chemother. 1974;6(4):498–500. doi: 10.1128/aac.6.4.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lourenço FR, Andreoli Pinto TJ. Antibiotic microbial assay using kinetic-reading microplate system. Braz J Pharm Sci. 2011;47(3):573–84. doi: 10.1590/s1984-82502011000300015. [DOI] [Google Scholar]
- 11.Karlen Y, Mcnair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8(1):131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2015-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6(10):986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 14.Ott SJ, Musfeldt M, Ullmann U, Hampe J, Schreiber S. Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. J Clin Microbiol. 2004;42(6):2566–72. doi: 10.1128/jcm.42.6.2566-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW. Molecular cloning, a laboratory manual. 3th ed. New York: Cold spring harbor laboratory press; 2001. [Google Scholar]
- 16.Whelan JA, Russell NB, Whelan MA. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods. 2003;278(1-2):261–9. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18. International Conference on Harmonization. ICH of Technical Requirements for the Registration of Pharmaceuticals for Human Use, Validation of analytical procedures: Methodology, ICH-Q2B, Geneva. 1996.
- 19. Guidelines for industry bioanalytical method validation.http://www.fda.gov/downloads/Drugs/Guidance Compliance Regulatory Information/ Guidances/ ucm 073384.pdf. 2001.
- 20.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327(8476):307–10. doi: 10.1016/s0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 21.Spaun J. Problems in standardization of turbidity determinations on bacterial suspensions. Bull World Health Organ. 1962;26:219–25. [PMC free article] [PubMed] [Google Scholar]
- 22. Todar's Online Textbook of Bacteriology, Kenneth Todar, 2008, available in: http://www.textbookofbacteriology.net/growth_2.html.
- 23.Peters IR, Helps CR, Hall EJ, Day MJ. Real-time RT-PCR: considerations for efficient and sensitive assay design. J Immunol Methods. 2004;286(1-2):203–17. doi: 10.1016/j.jim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira ID, Rosario VE, Cravo PV. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botaro BG, Cortinhas CS, Marco LV, Moreno JF, Silva LF, Benites NR. et al. Detection and enumeration of Staphylococcus aureus from bovine milk samples by real-time polymerase chain reaction. J Dairy Sci. 2013;96(11):6955–64. doi: 10.3168/jds.2013-6559. [DOI] [PubMed] [Google Scholar]
- 26.Zuluaga AF, Agudelo M, Rodriguez CA, Vesga O. Application of microbiological assay to determine pharmaceutical equivalence of generic intravenous antibiotics. BMC Clin Pharmacol. 2009;9(1):1. doi: 10.1186/1472-6904-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelaziz AA, Elbanna TE, Gamaleldeen NM. validated microbiological and HPLC methods for the determination of moxifloxacin in pharmaceutical preparations and human plasma. Braz J Microbiol. 2012;43(4):1291–301. doi: 10.1590/s1517-83822012000400008. [DOI] [PMC free article] [PubMed] [Google Scholar]