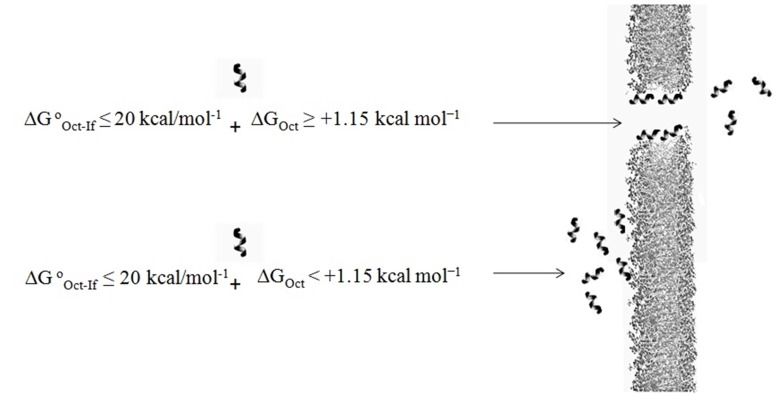Abstract
Purpose: Cell-penetrating peptides (CPPs) are used for delivering drugs and other macromolecular cargo into living cells. In this paper, we investigated the relationship between the structural/physicochemical properties of four new synthetic peptides containing arginine-tryptophan in terms of their cell membrane penetration efficiency.
Methods: The peptides were prepared using solid phase synthesis procedure using FMOC protected amino acids. Fluorescence-activated cell sorting and fluorescence imaging were used to evaluate uptake efficiency. Prediction of the peptide secondary structure and estimation of physicochemical properties was performed using the GOR V method and MPEx 3.2 software (Wimley-White scale, helical wheel projection and total hydrophobic moment).
Results: Our data showed that the uptake efficiency of peptides with two tryptophans at the C- and N-terminus were significantly higher (about 4-fold) than that of peptides containing three tryptophans at both ends. The distribution of arginine at both ends also increased the uptake efficiency 2.52- and 7.18-fold, compared with arginine distribution at the middle of peptides.
Conclusion: According to the obtained results the value of transfer free energies of peptides from the aqueous phase to membrane bilayer could be a good predictor for the cellular uptake efficiency of CPPs.
Keywords: Cell-penetrating peptides (CPPs), Wimley-White scale, GOR (Garnier-Osguthorpe-Robson) V method, Model amphipathic peptide (MAP)
Introduction
Transfer of cargos across cellular membranes is a major issue in cell biology and medicine. During the past few decades, significant progress has been made in focusing on the manufacture of new carriers for cargo delivery into cells. Cell-penetrating peptides or protein transduction domains (PTDs)1 are new carriers that have been widely used to promote the uptake of various macromolecules. CPPs have been shown to efficiently improve transportation of PMO (phosphorodiamidate morpholinos),2 PNA (peptide nucleic acids),3 DNA,4 siRNA (short interfering RNA),5 drug,6 and nanoparticles7 across biological membranes. Recently several classes of CPPs have been introduced. Model amphipathic peptides (MAP) are one of the most common classes of CPPs that have been used in scientific research.8 Amphipathic peptides contain hydrophobic and cationic residues in the backbone, this class of peptides have the ability to enter cells and can have antimicrobial properties.9
A group of amphipathic peptides containing tryptophan and arginine residue have been widely used as CPPfor delivery of various drug and biological compounds.10-12 Tryptophan has an aromatic side chain13 and can participate in hydrophobic interactions to the lipid hydrocarbon chains of the cell membrane.14 Arginine is a hydrophilic and basic amino acid;15 the guanidino groups of arginine tend to bind with phosphate in the cell membrane lipids.16 In this study, four amphipathic peptides containing tryptophan and arginine (RRWWWWRR, RRRWWWWRRR, WWRRRRWW and WWWRRRRWWW) were designed and synthesized in order to explore the relationships between the structural/physicochemical properties and penetration into the MCF-7 cells.
Materials and Methods
Materials
Fmoc–Trp (Boc) -OH, Rink amid AM and Fmoc-Arg (Pbf) -OH were purchased from AAPPTec (Louisville, KY, USA). N,N-Diisopropylethylamine (DIPEA) was obtained from Fluka (Buchs, Switzerland). O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) and triisopropylsilane (TIS) purchased from Sigma–Aldrich (St. Louis, MO, USA). 1,2-ethanedithiol (EDT), Trifluoroacetic acid (TFA) and other reagents were purchased from Merck (Darmstadt, Germany).
Peptide synthesis
The synthesis of peptides was carried out manually with Rink amide resin using the solid-phase synthesis method. O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) and N,N-diisopropylethylamine (DIPEA) in dimethylformamide (DMF) were used as coupling and activating reagents, respectively. Fmoc protecting group was removed from the resin and amino acids by addition of a solution of 20% (v/v) Piperidine/DMF. The Kaiser test was performed to monitor deprotection of peptides in solid phase peptide synthesis. A cleavage cocktail (TFA/H2O/TIS/EDT: 94 /2.5 /2.5 /1) was used for side-chain deprotection and the final cleavage of the peptide from the Rink amide resin. Crude peptides were precipitated by cold diethyl ether under vigorous stirring. The peptides containing the cleavage cocktail and Et2O were centrifuged at 4000 rpm for three minutes, followed by decantation in order to obtain the solid peptide precipitate.17,18
FITC Labeling of Peptides
A solution of 1.1 equivalent of FITC in pyridine/DMF/DCM (12:7:5) was prepared and incubated with the deprotected peptide for 12 h under gentle mixing conditions.19 The completion of the reaction was confirmed by the Kaiser test. The obtained FITC-peptide was cleaved from resin with the cleavage cocktail and was precipitated by diethyl ether.
Cell Culture
The human breast cancer cell line MCF-7 was purchased from the Pasteur Institute (Tehran, Iran). The cells were grown on 25 cm2 cell culture flasks with RPMI-1640 medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), sodium pyruvate (1 mM), penicillinG100 U/ml and streptomycin 100 μg/ml (AppliChem, Darmstadt, Germany) in a humidified atmosphere of 5% CO2, 95% air at 37 °C.
Cytotoxicity analysis
The Mcf-7 cells were seeded in 200 µL of medium per well in 96-well plates 24 h prior to the experiment. The old medium was replaced by different concentrations of peptides synthesized in serum containing medium and plates incubated for 24–72 h at 37°C in a humidified atmosphere of 5% CO2. Cell viability was then determined by measuring the color intensity of the formazan solution at 570nm using an ELISA reader (Bioteck Instruments, Wmooski, VT, USA).
The percentage of inhibition of proliferation was calculated as follows:
Fluorescent microscopy
The human breast cancer cell line MCF-7 was cultured in 6-well plates (200×103 cells/well) for 24 h in an incubator at 37°C in a 5% CO2 environment. The Cells were treated with 15 μM concentration of R2W4R2- FITC, W2R4W2- FITC, W3R4W3-FITC and R3W4R3-FITC. After 1 h incubation at 37 °C, the medium containing the peptide was removed. The cells were washed with PBS and examined with an emission filter of 530 nm BP (green emission) using an Olympus IX81 fluorescence microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
Flow cytometry assay
Flow cytometry was used to determine the percentage of labelled cells and compared with control cells. MCF-7 cells were treated with 5 μM, 15 μM and 25 μM concentration of peptides in separate experiments for 1 h at 37°C and the cells were washed twice with phosphate buffered saline. However, because flow cytometry does not discriminate between cell membrane-bound and internalized FITC-peptides, the cells were treated with trypsin/EDTA (0.25% trypsin and 0.02% EDTA for 5 min) to minimize any contribution of peptides attached to the outer surface of the cell membrane.20
Mean fluorescence ratios were determined for 10,000 cells sampled after the addition of peptides. Fluorescence ratios were calculated in arbitrary units set to a value of 100 for MCF-7 cells. The results were expressed as a percentage of gated populations and data were analysed using control cells. Flow cytometry was carried out using FACSCalibur (Becton Dickinson, San Jose, CA, USA) applying an argon laser. Excitation occurred at 488 nm for FITC.
Peptide secondary structure
Structure prediction of synthesized peptides was performed to determine their secondary structure. We applied the GOR V algorithm in order to explain the different levels of uptake by MCF-7 cells. This method has high accuracy (Q3=73.5%) with a wide range of amino acid residues for predicting the secondary structures of peptides.21 It is based on the use of information theory and Bayesian statistics for predicting the secondary structure of peptides and proteins,22 by analysis and comparison of an input sequence with 513 proteins, to calculate the helix, sheet and coil probabilities at each residue position.23 The GOR V server uses the FASTA sequence format and calculates the probability helix (α-helix), extended (β-sheet) and coil structures. The probabilities are then normalized to 0 and 1 using the following equation:23,24
PH+PE+PC=1
The GOR V server is available online at http://gor.bb.iastate.edu (the Plant Sciences Institute at Iowa State University).
Helical wheel projection and thermodynamic parameters
Helical wheel projection illustrates the correct angular displacement from one amino acid to another amino acid in a cylindrical plot of the α-helix structure.25 The Wimley-White is one of the most common scales that can be used for free energy calculation of peptides. This value is provided by the free-energy contributions of the side chains and backbone of peptides.26 The free energy of peptides may be used to ascertain the feasibility of peptide transport across the cell membrane. Helical wheel projection, estimation of the transfer free energy and the total hydrophobic moment of peptides were performed by Membrane Protein Explorer (MPEx 3.2), free software provided by the Stephen White Laboratory at the University of California (Irvine, CA).
Results and Discussion
Cytotoxicity in cell culture
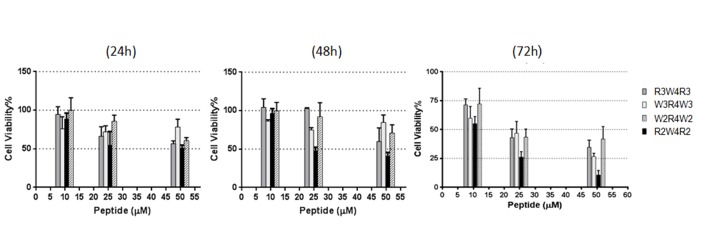
The cell viability of the human breast cancer cell line MCF-7 are summarized in Figure 1. Compared to control, the cells underwent reduced proliferation following treatment with R2W4R2. All of the peptides had cytotoxic effects at concentrations of 25µM and 50 µM, following a 72- hour incubation period. The cell toxicity was dose and time dependent.
Figure 1.
Cytotoxic effects of R3W4R3, R2W4R2, W3R4W3, W2R4W2. The peptides were incubated with 104 cells in 5, 25, and 50 μM concentrations for 24 h, 48h, and 72h. Each value is the mean ± SD of three separate determinations
Cellular Uptake and labeling
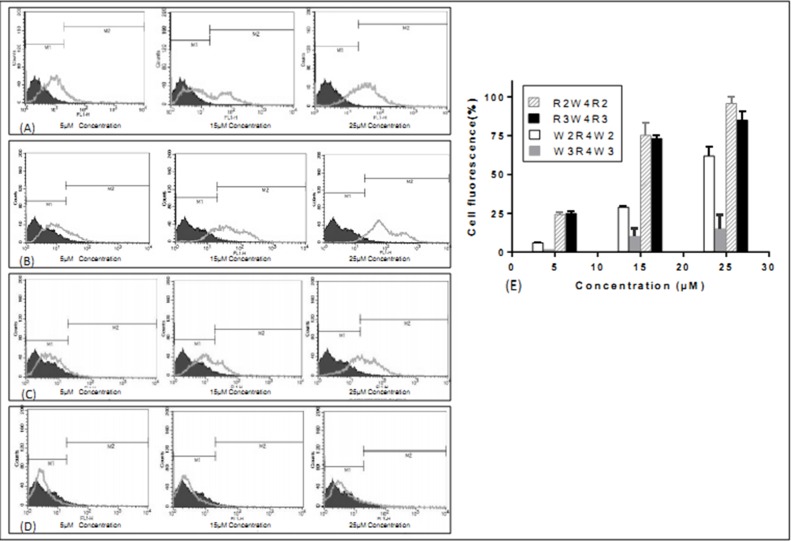
Increasing the concentration of all peptides resulted in an increased population of FITC-labelled cells (Figure 2). This effect was relatively linear in the concentration range of 5, 15 and 25μM of the peptides. The major difference was observed when comparing the two peptides containing four and six tryptophans at both ends. Accumulation of both R2W4R2 and R3W4R3 in MCF-7 was suitable. However the mean cellular uptake of W2R4W2 and W3R4W3 in 15μM concentrations of peptides was 2.52 and 7.18-fold less than that of R3W4R3 respectively. According to the results, there was no significant difference between penetration of R2W4R2 and R3W4R3 sequences. It therefore appears that the location of tryptophan and arginine residues have a dramatic effect on cellular uptake. It seems that the number of tryptophan residues in W2R4W2 and W3R4W3 sequences affects the cellular uptake (Figure 2 and 3).
Figure 2.
Cellular uptake of FITC-labeled peptides in live MCF-7 cells after incubation for 1 h at 37 °C. The uptake values of intensity are seen for R2W4R2 (A) and R3W4R3 (B), followed by, in order of decreasing intensity, W2R4W2 (C), W3R4W3 (D). (E) A comparison of the cellular uptake of peptides in 5, 15, and 25μM concentrations. The results are expressed as a percentage of gated populations. Each value is the mean ± SD of two separate determinations.
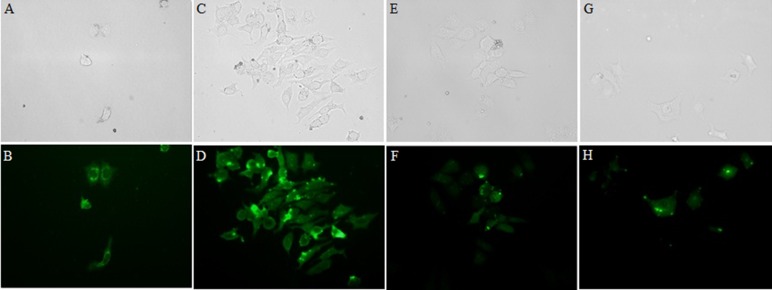
Figure 3.
Fluorescence microscopy, visualization of FITC- labeled,W3R4W3(G,H), R3W4R3(C,D), R2W4R2(A,B), W2R4W2(E,F) in MCF-7 cells. Bright field (top) and fluorescence (bottom) microscopy of MCF-7 cells. Live cells were treated with 15 µM of peptides for 1h at 37°C.
Secondary structure prediction
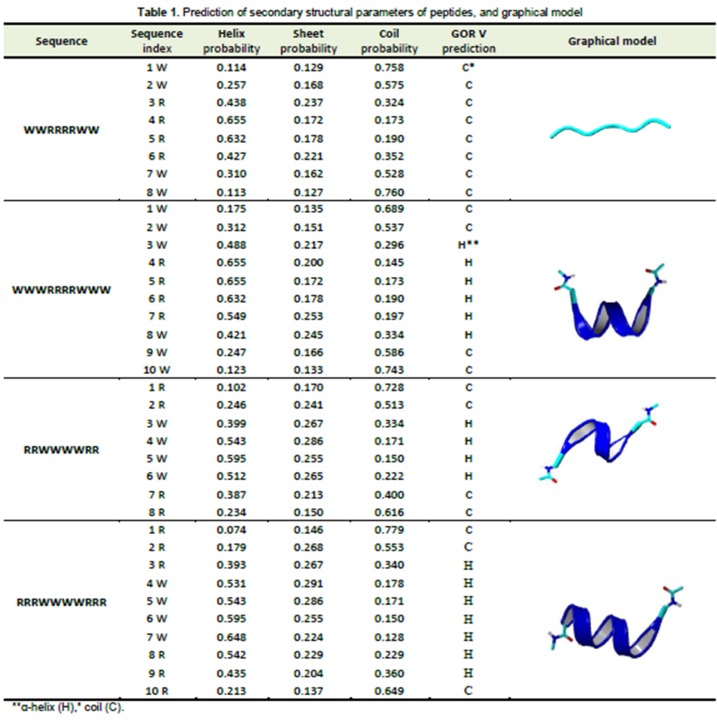
The results of prediction of secondary structure and the graphical model are shown in Table 1. According to the results, for W2R4W2 sequence, coil structure is dominant with no α-helix structure. However, in other sequences, a high probability of α-helix formation was indicated. The amphipathic α-helical is important for the effective transportation of peptides across bilayer lipid membranes. It has been suggested that this structure leads to pore formation followed by peptide insertion into the cell membrane.27 No relevance between the α-helix structure and cellular uptake of W2R4W2 and W3R4W3 sequences was observed.
Table 1.
Prediction of secondary structural parameters of peptides, and graphical model
Helical wheel projection and total hydrophobic moment
It has been suggested that the cellular uptake efficiency of peptide is dependent on the regular arrangement of hydrophobic and hydrophilic amino acid residues in one side of the helical wheel projection.11,28 The helical wheel projection for R2W4R2, R3W4R3 and W3R4W3 sequences are shown in (Figure 4). According to this projection, these peptides do not show any regular structure. Therefore, it cannot be used to explain differences in cellular uptake of the peptides.
Figure 4.
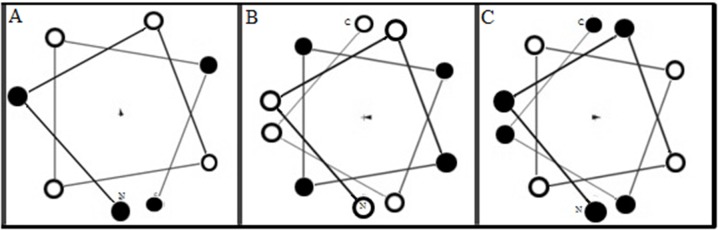
Illustration of sequences in helical wheel projection of W3R4W3 (B), R2W4R2 (A), and R3W4R3 (C) generated using the program MPEx 3.2. Black symbols represent basics and hydrophilic residues (Arg), white, aromatics and hydrophobic residues (Trp).
The hydrophobic moment was performed to explain the high permeability of some irregular hydrophobic and hydrophilic residues in the α-helix structure, such as the amphiphilic peptide melittin.29 The results of the total hydrophobic moment (μH) are shown in Table 2. Despite the fact that these peptides had the same value for the total hydrophobic moment, W3R4W3 had much lower cellular uptake activity than the other peptides studied in this study.
Table 2. Hydrophobicity scales and the total hydrophobic moment .
| Sequence | NH2-R2W4R2-CONH2 | NH2-R3W4R3-CONH2 | NH2-W3R4W3-CONH2 | NH2-W2R4W2-CONH2 |
| Total. Hyd. Moment (μH)* | 3.26 | 3.26 | 3.49 | 3.49 |
| Wimley-White Scale(Oct-If)** (kcal mol-1) | 9.51 | 11.51 | 9.03 | 9.51 |
| Wimley-White Scale (Oct)*** (kcal mol-1) | 3.18 | 6.8 | -1 | 3.18 |
*Total. Hyd. Moment= the total hydrophobic moment
**Wimley–White Scale (Oct-If) ==difference between the Wimley–White whole-residue octanol hydrophobicity scale and interfacial hydrophobicity scale.
***Wimley–White Scale (Oct) = Wimley–White whole-residue octanol hydrophobicity scale
Wimley-White scales
It is believed that difference between octanol hydrophobicity scale and interfacial hydrophobicity scale can be an effective parameter to predict behavior of peptides to penetrate the cell membrane. According to the research conducted in this field, peptides with ∆G°Oct-w≤20 kcal/mol-1 would be good candidates for penetrating the cell membrane.30 Based on the values obtained from the W.W scale (Table 2) and flow cytometry data, ∆G°Oct-w does not appear to be a suitable parameter for predicting cell penetrating peptides, since ∆G°Oct-w of all peptides were less than 20 kcal/mol-1.
Another parameter used to predict the penetration efficiency of peptides is the transfer free energy values of peptide from water to membrane. The amount of energy required for transferring a random-coil and α-helix insertion into the hydrophobic core of the cell membrane is +1.15 kcal mol-1 (∆GOct ≥ +1.15 kcal mol-1).31 According to the data in Table 2, R2W4R2, R3W4R3 and W2R4W2 have enough energy for penetration from water to membrane. The data also shows that W3R4W3 with lowest cellular uptake has not sufficient free energy value. It seems that uptake efficiency and transfer free energy values correlate with each other and this parameter may be used to explain differences in uptake of the peptides into the MCF-7 cells (Figure 5).
Figure 5.
Effect of thermodynamic parameters for the transfer of peptide from water to the intracellular space.
Conclusion
In this study, four amphipathic peptides containing tryptophan and arginine amino acid residues were synthesized to assess the importance of different parameters in their cell uptake. The flow cytometry results showed different cellular uptake efficiency. Various thermodynamic and structural parameters that might influence peptide transport through the cell membrane were investigated. The secondary structure and the helical wheel projection failed to describe the differences in cellular uptake of the peptides. On the other hand it seems that the uptake efficiency correlates well with the transfer free energy values and this parameter may be used to describe the different uptake efficiency of the cell penetrating peptides into MCF-7 cells.
Acknowledgments
The financial support from the "Drug Applied Research Center" and "Research Center for Pharmaceutical Nanotechnology" of Tabriz University of Medical Sciences is greatly acknowledged. This paper is based on an MSc thesis submitted by A. Shirani in Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences (Grant No. 5/79/1709).
Ethical Issues
Not applicable.
Conflict of Interest
The authors report no conflicts of interest.
References
- 1.Vives E, Schmidt J, Pelegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta. 2008;1786(2):126–38. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Yin H, Moulton HM, Betts C, Merritt T, Seow Y, Ashraf S. et al. Functional rescue of dystrophin-deficient mdx mice by a chimeric peptide-PMO. Mol Ther. 2010;18(10):1822–9. doi: 10.1038/mt.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siwkowski AM, Malik L, Esau CC, Maier MA, Wancewicz EV, Albertshofer K. et al. Identification and functional validation of PNAs that inhibit murine CD40 expression by redirection of splicing. Nucleic Acids Res. 2004;32(9):2695–706. doi: 10.1093/nar/gkh584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottschalk S, Sparrow JT, Hauer J, Mims MP, Leland FE, Woo SL. et al. A novel DNA-peptide complex for efficient gene transfer and expression in mammalian cells. Gene Ther. 1996;3(5):448–57. [PubMed] [Google Scholar]
- 5.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558(1-3):63–8. doi: 10.1016/s0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Carneado J, Kogan MJ, Pujals S, Giralt E. Amphipathic peptides and drug delivery. Biopolymers. 2004;76(2):196–203. doi: 10.1002/bip.10585. [DOI] [PubMed] [Google Scholar]
- 7.Sawant R, Torchilin V. Intracellular delivery of nanoparticles with CPPs. Methods Mol Biol. 2011;683:431–51. doi: 10.1007/978-1-60761-919-2_31. [DOI] [PubMed] [Google Scholar]
- 8.Kilk K, Mahlapuu R, Soomets U, Langel U. Analysis of in vitro toxicity of five cell-penetrating peptides by metabolic profiling. Toxicology. 2009;265(3):87–95. doi: 10.1016/j.tox.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita H, Demizu Y, Shoda T, Sato Y, Oba M, Tanaka M. et al. Amphipathic short helix-stabilized peptides with cell-membrane penetrating ability. Bioorg Med Chem. 2014;22(8):2403–8. doi: 10.1016/j.bmc.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37(42):14713–8. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 11.Rydberg HA, Matson M, Amand HL, Esbjorner EK, Norden B. Effects of tryptophan content and backbone spacing on the uptake efficiency of cell-penetrating peptides. Biochemistry. 2012;51(27):5531–9. doi: 10.1021/bi300454k. [DOI] [PubMed] [Google Scholar]
- 12.Sharma R, Shivpuri S, Anand A, Kulshreshtha A, Ganguli M. Insight into the role of physicochemical parameters in a novel series of amphipathic peptides for efficient DNA delivery. Mol Pharm. 2013;10(7):2588–600. doi: 10.1021/mp400032q. [DOI] [PubMed] [Google Scholar]
- 13.Abidi F, Jacquot S, Lassiter C, Trivier E, Hanauer A, Schwartz CE. Novel mutations in Rsk-2, the gene for Coffin-Lowry syndrome (CLS) Eur J Hum Genet. 1999;7(1):20–6. doi: 10.1038/sj.ejhg.5200231. [DOI] [PubMed] [Google Scholar]
- 14.Killian JA, Von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25(9):429–34. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 15.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv Drug Deliv Rev. 2005;57(4):547–58. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Schug KA, Lindner W. Noncovalent binding between guanidinium and anionic groups: focus on biological- and synthetic-based arginine/guanidinium interactions with phosph[on]ate and sulf[on]ate residues. Chem Rev. 2005;105(1):67–114. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- 17.Amblard M, Fehrentz JA, Martinez J, Subra G. Methods and protocols of modern solid phase Peptide synthesis. Mol Biotechnol. 2006;33(3):239–54. doi: 10.1385/mb:33:3:239. [DOI] [PubMed] [Google Scholar]
- 18.Reichert J, Grasnick D, Afonin S, Buerck J, Wadhwani P, Ulrich AS. A critical evaluation of the conformational requirements of fusogenic peptides in membranes. Eur Biophys J. 2007;36(4-5):405–13. doi: 10.1007/s00249-006-0106-2. [DOI] [PubMed] [Google Scholar]
- 19.Maurya IK, Thota CK, Sharma J, Tupe SG, Chaudhary P, Singh MK. et al. Mechanism of action of novel synthetic dodecapeptides against Candida albicans. Biochim Biophys Acta. 2013;1830(11):5193–203. doi: 10.1016/j.bbagen.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ. et al. Cell-penetrating peptidesA reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278(1):585–90. doi: 10.1074/jbc.m209548200. [DOI] [PubMed] [Google Scholar]
- 21.Kloczkowski A, Ting KL, Jernigan RL, Garnier J. Combining the GOR V algorithm with evolutionary information for protein secondary structure prediction from amino acid sequence. Proteins. 2002;49(2):154–66. doi: 10.1002/prot.10181. [DOI] [PubMed] [Google Scholar]
- 22.Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 23.Cheng H, Sen TZ, Kloczkowski A, Margaritis D, Jernigan RL. Prediction of protein secondary structure by mining structural fragment database. Polymer (Guildf) 2005;46(12):4314–21. doi: 10.1016/j.polymer.2005.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen TZ, Jernigan RL, Garnier J, Kloczkowski A. GOR V server for protein secondary structure prediction. Bioinformatics. 2005;21(11):2787–8. doi: 10.1093/bioinformatics/bti408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vance VB, Huang AH. The major protein from lipid bodies of maizeCharacterization and structure based on cDNA cloning. J Biol Chem. 1987;262(23):11275–9. [PubMed] [Google Scholar]
- 26.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–8. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 27.Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers. 1998;47(6):451–63. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 28.Yandek LE, Pokorny A, Floren A, Knoelke K, Langel U, Almeida PF. Mechanism of the cell-penetrating peptide transportan 10 permeation of lipid bilayers. Biophys J. 2007;92(7):2434–44. doi: 10.1529/biophysj.106.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta. 1999;1462(1-2):71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 30.Almeida PF, Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: from kinetics to thermodynamics. Biochemistry. 2009;48(34):8083–93. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wimley WC, White SH. Designing transmembrane alpha-helices that insert spontaneously. Biochemistry. 2000;39(15):4432–42. doi: 10.1021/bi992746j. [DOI] [PubMed] [Google Scholar]