Abstract
MicroRNAs (miRNAs) are a novel class of endogenous, small, noncoding RNAs that regulate gene expression via degradation, translational inhibition, or translational activation of their target mRNAs. Functionally, an individual miRNA is important as a transcription factor because it is able to regulate the expression of its multiple target genes. As a group, miRNAs are able to directly regulate at least 30% of genes in a cell. In addition, other genes may also be regulated indirectly by miRNAs. It is therefore not surprising that miRNAs could be the pivotal regulators in normal development, physiology, and pathology. Recent studies have identified that miRNAs are highly expressed in vasculature and their expression is dysregulated in diseased vessels. miRNAs are found to be critical modulators for vascular cell functions such as cell differentiation, contraction, migration, proliferation, and apoptosis. Accordingly, miRNAs are involved in the vascular dysfunction, ischemic angiogenesis, re-endothelialization, and vascular neointimal lesion formation under diverse vascular diseases. miRNAs may serve as novel therapeutic targets for vascular diseases such as impaired angiogenesis or re-endothelialization, restenosis, atherosclerosis, hypertension, and diabetic vascular complication. This review article summarizes the research progress regarding the roles of miRNAs in vascular diseases.
Keywords: MicroRNAs, vascular disease, angiogenesis, neointima, restenosis, atherosclerosis
Introduction
MicroRNAs (miRNAs) have emerged as a novel class of endogenous, small, non-coding RNAs that can pair with sites in 3' untranslated regions in mRNAs of protein-coding genes to regulate their expression [1]. The first miRNA, lin-4, was discovered in Caenorhabditis elegans in 1993 [2, 3]. However, the presence of miRNAs in vertebrates had not been confirmed until 2001 [4]. Currently, more than 700 miRNAs have been cloned and sequenced in human [5], and the estimated number of miRNA genes is as high as 1000 in the human genome [6]. More importantly, one miRNA is able to regulate the expression of multiple genes because it can bind to its mRNA targets as either an imperfect or a perfect complement. Thus, a miRNA can be functionally as important as a transcription factor [7]. As a group, miRNAs may directly regulate at least 30% of the genes in a cell [8]. It is therefore not surprising that miRNAs are involved in the regulation of all major cellular functions [9].
It is well established that vascular diseases such as hypertension, atherosclerosis, coronary artery disease, restenosis after angioplasty or transplantation, and diabetic vascular complication are among the leading causes of morbidity and mortality in developed countries. In addition, angiogenesis and re-endothelialization are also common vascular consequences in many diseases including cancer [10], atherosclerosis [11] and ischemic heart disease [12]. Differentiation, contraction, migration, proliferation, and apoptosis of vascular smooth muscle cells (VSMCs) and/or endothelial cells (ECs) are critical cellular evens responsible for the development of angiogenesis and vascular disease. In addition, other cellular events such leukocyte infiltration and activation in the vascular walls are also involved in vascular disease. Recent studies have demonstrated that miRNAs are highly expressed in vascular walls and their expression is dysegulated in diseased vessels [13–15]. miRNAs are found to play important roles in vascular diseases via regulating key vascular cellular events through their target genes [13–18]. The current review article is for summarizing the research progress regarding the roles of miRNAs in vascular pathology.
miRNAs in restenosis and atherosclerosis
Recent studies have revealed that miRNAs are aberrantly expressed in stenotic and atherosclerotic arteries, in which neointimal lesion was formed [13, 19–21]. Using mircroarray analysis, Zhang and colleagues identified that many of the detected miRNAs are dysregulated in rat vascular walls with neointimal growth induced by balloon-catheter angioplasty [13]. The aberrant expression of miRNA was not limited to rat model. In mouse arteries with neointimal growth after ligation injury, both miR-143 and miR-145 was downregulated as reported by Srivastava and colleagues [19], and our group [20]. Moreover, in atherosclerotic human and mouse arteries without mechanical injuries, some of miRNAs were also deregulated as demonstrated in recent reports by Condorelli and colleagues [21], as well as our group [20]. Table 1 shows the miRNAs that are highly expressed in vascular walls and are aberrantly expressed in stenotic arteries induced by angioplasty or atherosclerosis.
Table 1.
Aberrantly expressed miRNAs in stenotic arteries induced by angioplasty or atherosclerosis.
| Upregulated miRNAs | Downregulated miRNAs |
|---|---|
| miR-21 | miR-125a |
| miR-146 | miR-125b |
| miR-214 | miR-133a |
| miR-221 | miR-143 |
| miR-222 | miR-145 |
| let-31 | miR-347 |
| let-352 | miR-365 |
| Let-7 | |
The molecular mechanisms of the aberrant expression of miRNAs in diseased vessels are currently unclear. Our unpublished data suggested that two key steps of miRNA biogenesis were affected in the vascular walls under disease conditions: miRNA transcription from miRNA genes to pri-miRNAs, and the mature process from pri-miRNAs to pre-miRNAs, although the mechanisms are still unclear. However, the mature process from pre-miRNAs to mature miRNAs was not affected.
It is well established that VSMC dedifferentiation, migration, proliferation, and apoptosis are critical cellular events responsible for the development of a number of proliferative vascular diseases such as restenosis and atherosclerosis. Indeed, VSMCs are the major cells within neointimal lesions in these vascular diseases. The biological roles of miRNAs in VSMCs have been identified by recent studies [13, 14, 20, 21]. We demonstrated that miR-21, a miRNA that is upregulated in vascular walls with neointima, was a critical regulator for VSMC proliferation and apoptosis [13]. In cultured cells, upregulation of miR-21 via pre-miR-21 resulted in the increased VSMC proliferation, but decreased VSMC apoptosis. In contrast, VSMC proliferation was decreased, but apoptosis was increased via miR-21 inhibition through its inhibitor. In addition, we identified that the target genes of miR-21 involved in miR-21-mediated cellular effects on VSMCs were phosphatase and tensin homology deleted from chromosome 10 (PTEN) [13] and programmed cell death 4 (PDCD4) [20]. In our study targeting miR-221/222, we identified that miR-221/222 were critical pro-proliferative miRNAs in cultured VSMCs via their target genes, p27(Kip1) and p57(Kip2) [14]. The proliferative effect of miR-221/222 on VSMCs was further confirmed by another independent research group, in which Davis et al demonstrated that miR-221 increased the proliferation and migration of VSMCs through its target gene, p27(Kip1) [21].
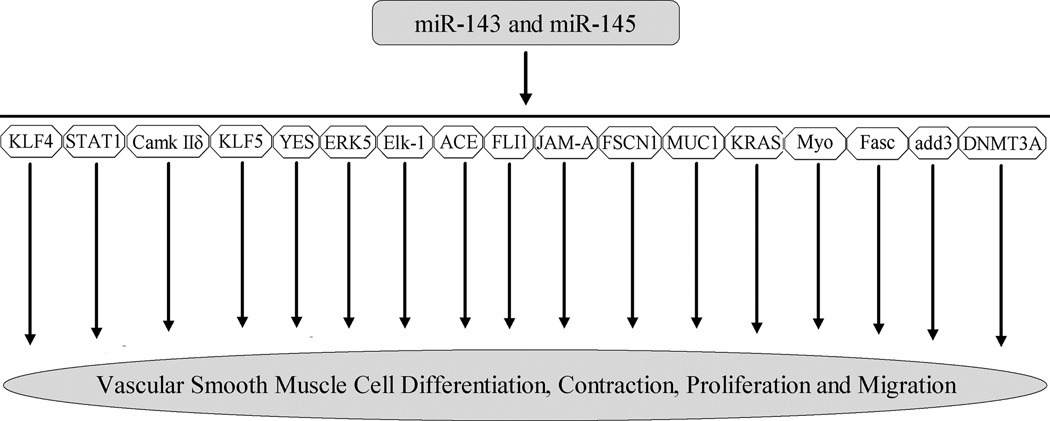
More recently, the roles of miR-145 and miR-143 in VSMC biology have received special attention, because they are abundant miRNAs in normal vascular walls and their expression is downregulated in diseased arteries [13, 15, 19–23]. We identified that miR-145 was selectively expressed in VSMCs of arterial walls and its expressed was significantly downregulated in dedifferentiated, proliferative VSMCs [15]. Both in cultured cells in vitro and in balloon-injured rat carotid arteries in vivo, we demonstrated that miR-145 was a novel biomarker and a critical modulator for VSMC phenotype [15]. Indeed, the expression of miR-145 was consistent with that of VSMC differentiation maker genes, SM α–actin, calponin, and SM-MHC. Moreover, the expression of VSMC differentiation maker genes was significantly upregulated by overexpression of miR-145, but was downregulated by miR-145 inhibition. Furthermore, VSMC proliferation was inhibited by miR-145 overexpression via pre-miR145. To determine the potential gene targets of miR-145 in VSMCs, both bioinformatic and experimental approaches were applied. We identified that kruppel-like factor 5 (KLF5) was one of the critical target genes responsible for miR-145-mediated cellular effects on VSMCs [15]. The role of miR-145 in VSMC biology was further confirmed by Cordes et al [19]. These investigators demonstrated KLF4 and Calmodulin kinase IIdelta (CamkIIδ) are two target genes of miR-145 in VSMCs [19]. In addition, they also found that myocardin may be a direct target gene of miR-145 in VSMCs. miR-143 is another important miRNA in VSMC biology (19, 21–23). It was found that miR-143 was an important regulator for proliferation of VSMC by targeting Elk-1 [19] and ACE [22]. The roles of miR-143 and miR-145 in VSMC biology were further verified by miR-143/145 knockout approach (21–23). Braun and colleagues demonstrated that VSMCs from miR-143/145–deficient mice were locked in the synthetic state (dedifferentiated phenotype) [22]. The incomplete differentiation of VSMCs in miR-143/145–deficient mice was also demonstrated by Condorelli’s group (21). Disarray of actin stress fibers of VSMCs in miR-143/145–deficient mice was identified by Olson and colleagues [23]. The biological roles of miR-143/145 in VSMCs and their identified targets were summarized in Figure 1.
Figure 1. The biological functions of miR-143/145 and their identified target gens.
Kruppel-like factor 4 (KLF4), kruppel-like factor 5 (KLF5), myocardin (Myo), Calmodulin kinase II delta (CamkIIδ), Ets-like protein 1(Elk-1), angiotensin I-converting enzyme (ACE), Junctional Adhesion Molecule A (JAM-A), fascin (Fasc), Friend leukemia virus integration 1 gene (FLI1), Fascin homolog 1 (FSCN1), Yamaguchi sarcoma (YES), activator of transcription 1 (STAT1), mucin 1 (MUC1), versican, adducin3 (add3), DNA methyltranferase 3A (DNMT3A), V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), extracellular signal-regulated kinase-5 (ERK5).
The effects of miRNAs on vascular neointimal growth, the known pathological feature of restenosis and atherosclerosis, have recently been intensively explored [13–15, 22–25]. We demonstrated that miR-21, miR-221/222, and miR-145 were dysregulated in balloon-injured rat carotid arteries [13–15]. Inhibition of the upreguated miR-21 [13], and miR-221/222 [14] inhibited vascular neointimal growth. In addition, the neointimal formation in carotid arteries was also significantly reduced by restoration of miR-145 after angioplasty [15]. The negative effect of miR-145 on vascular neointimal growth was also demonstrated in another study using the same rat model [21]. More recently, the role of miR-143/145 in neointimal formation was further confirmed using gene knockout approach, in which Braun and colleagues found that spontaneous huge neointimal lesion formation was displayed in arteries from miR-143/145 knockout mice without any additional injuries [22]. However, Xin et al reported that neointimal formation after carotid artery ligation injury was decreased in these miR-143/145 knockout mice [23]. More recently, Weber and colleagues demonstrated that delivery of miR-126 by apoptotic body limited atherosclerosis, promoted the incorporation of Sca-1+ progenitor cells, and conferred features of plaque stability in mouse models of atherosclerosis via CXCL12-dependent mechanisms [25].
Leukocyte infiltration and activation are also important cellular events in both restenosis and atherosclerosis. The biological roles of miRNAs in leukocyte infiltration and activation in restenosis and atherosclerosis have not yet been characterized; however, significant differential expression of miRNAs was observed in human peripheral blood monocytes treated with oxidized LDL [26]. Inhibition of endogenous miR-125a-5p levels in THP-1 cells significantly increased the secretion of inflammatory cytokines (TGF-b, TNF-a, IL-2, and IL-6) and increased the expression of macrophage scavenger receptors (LOX-1 and CD68), which resulted in increased lipid uptake [26]. These data suggest that miR-125a-5p is anti-atherogenic in the macrophage. The roles of miRNAs in endothelial cell repair (re-endothelialization) are described later.
miRNAs in angiogenesis and re-endothelialization
Angiogenesis is an important mechanism for the maintenance of adequate blood supply to the tissues both in normal development and in the process of diseases such as ischemic heart disease, cancer, and atherosclerosis [10–12]. The first evidence showing miRNAs in the regulation of angiogenesis came from Dicer knockout mice [17]. Dicer is a critical enzyme for miRNA synthesis [17]. These Dicer deficient mice died early during development due to the thinning of vascular walls and severe disorganization of the network of blood vessels [17]. Similarly, knockdown of Dicer in both human and animal vascular ECs via RNAi approach significantly reduced EC migration, capillary sprouting, and tube formation [18]. Dicer knockout and knockdown accompanied the expression changes of a number of angiogenesis-related genes including vascular endothelial growth factor and its receptor, tek-TIE-2, Tie-1, endothelial nitric oxide synthase, and interleukin-8 [17–18].
The above phenotypic change in angiogenesis observed in these Dicer deficient animals and ECs led an explosion in studying the biological roles of individual miRNAs in EC biology and angiogenesis. In this respect, Rainaldi and colleagues identified that miR-221 and miR-222 had significant inhibitory effects on EC migration, tube formation and wound healing in cultured ECs [16]. These investigators further identified that miR-221/miR-222-meditaed effects on ECs occurred, at least in part, through their target gene, c-Kit. More recently, a series of studies were conducted by different research groups in this new research area to identify the angiogenesis-related miRNAs [17, 18, 27–37]. Based on the results, these miRNAs can be divided into two groups: pro-angiogenic miRNAs or anti-angiogenic miRNAs. Pro-angiogenic miRNAs include miR-126 [27, 28], miR-17–92 [29], Let-7 [17], miR-130a [30], miR-210 [31], miR-378 [32], and miR-296 [33]. In contrast, the current known anti-angiogenic miRNAs include miR-221/222 [16, 34], miR-328 [35], miR-92a [36], and miR-214 [37]. It should be noted that the effects of these miRNAs on EC biology and angiogenesis are not only identified in cultured ECs in vitro, but also in ischemia-induced angiogenesis in vivo using limb ischemia and myocardial infarction models [28, 36].
Another endothelial cell growth or recovery related event in athersocelrosis and angioplasty is re-endothelialization. Loss of endothelial integrity induced by damaged or apoptotic cells is a major event is in atherosclerosis. Circulating endothelial progenitor cells have been demonstrated to play an integral role in endothelial integrity due to their ability to reinforce the endothelium with new healthy endothelial cells to replace damaged or apoptotic cells [38, 39]. In a recent study, individuals with atherosclerosis showed significantly higher expression of miR-221 and miR-222 in endothelial progenitor cells (EPC) compared with individuals without atherosclerosis [34]. Furthermore, miR-221/222 levels were observed to be inversely related to EPC levels, as the individuals with atherosclerosis had significantly less EPC numbers. Atorvastatin, a drug that has previously been shown to increase circulating EPC numbers, was shown to decrease miR-221 and miR-222 expression in EPCs [34]. The result suggests that miRNAs may be involved in re-endothelialization during the pathogenesis of atherosclerosis. It is well-known that re-endothelialization is a key cellular events in the vascular walls after angioplasty. Our unpublished studies have revealed that multiple miRNAs are involved in the process of re-endothelialization. For example, we found miR-221 and miR-22 were two inhibitory miRNAs for the EC recovery after angioplasty.
Potential involvement of miRNAs in other vascular diseases
Differential expression of some miRNAs in the aorta of spontaneously hypertensive rats (SHR) has been identified recently [40]. Xu and colleagues determined the expression of miR-1, miR-133a, miR-155 and miR-208 in these hypertensive aortas [40]. Among the detected miRNAs, the expression of miR-155 was significantly lower in aortas of 16-week-old SHR than that of age-matched control Wistar-Kyoto (WKY) rats. In addition, miR-155 level was negatively correlated to blood pressure. Braun and colleagues identified that angiotensin-converting enzyme (ACE), a critical enzyme in hypertension, is a target gene of miR-143 and miR-145 [22]. More excitedly, miR-miR-143 and 145 were found to play a critical role in keeping the contractive functions of VSMCs and vessels [22]. Accordingly, decreased blood pressure was found in these miR-143/145–deficient mice due to the decreased contractile abilities of the vessels [22, 23]. The biological roles of other miRNAs in hypertension should be determined in future studies.
Despite current optimal treatment, vascular complication has still emerged as a leading cause of morbidity and mortality in diabetic patients in the USA and worldwide. It is well known that multiple genes are responsible for diabetes-mediated vascular diseases. miRNAs are important regulators for gene expression, it is therefore reasonable to hypothesize that miRNAs may play important roles in diabetic vascular complications. Our unpublished microarray data suggested that multiple miRNAs were aberrantly expressed in both type I and type II diabetic vessels. Moreover, these aberrantly miRNAs were also found in cultured vascular cells stimulated with high glucose. Among these aberrantly expressed miRNAs, miR-143 and miR-145 were found to be downregulated, whereas miR-21 was found to be upregulated in diabetic vessels. We have confirmed that these 3 miRNAs are related to VSMC growth and vascular neointimal lesion formation. In a recent study, Li et al identified that miR-221 was able to regulate high glucose-induced endothelial dysfunction [41].
Clinical applications of miRNAs in vascular disease
Tissue- and cell-specific expression is one important characteristic of miRNA expression [42]. Indeed, one miRNA may be highly expressed in one tissue or one cell but has no or low expression in other tissues or other cells. For example, miR-1 is reported to be a muscle or heart specific miRNA [42], whereas miR-145 is a vascular smooth muscle cell specific miRNA as described in our recent article [15]. Interestingly, recent studies have revealed that miRNAs are able to be released into circulating blood from the injured cells and tissues induced by ischemic damage. In contrast to our original thought, the cell-free miRNAs are relatively stable due to binding with other materials such as exosomes in circulating blood [43]. Thus, circulating miRNAs could be used as novel diagnostic biomarkers in patients with occlusive vascular diseases. For example, recent studies from us and other groups have demonstrated that circulating miRNAs such as miR-1 and miR-208 could be used biomarkers for coronary artery diseases in human [44–48].
Several in vivo animal studies have revealed the promising therapeutic results in vascular disease [13–15, 22–25]. These pre-clinical investigations indicate that miRNAs may represent new therapeutic targets for diverse vascular diseases. It is well-established that multiple miRNAs are aberrantly expressed in vascular diseases, some being upregulated and others downregulated. Thus, two major miRNA-based therapeutic strategies are restoring the expression of miRNAs reduced in diseases and, conversely, inhibiting overexpressed miRNAs. Although downregulation of miRNAs in vascular walls is relatively easy, the upregulation of some miRNAs in the diseased vascular walls is still a challenge due to the impairment of the mature process of miRNAs under disease conditions. We should realize that we are still at the early stages in miRNA-based therapy.
Conclusions
miRNAs in vascular disease has emerged as a new research area. The initial exciting results have demonstrated that multiple miRNAs are involved in the development of both human and animal vascular diseases via regulating vascular cell differentiation, contraction, migration, proliferation, and apoptosis through their target genes. miRNAs may represent novel biomarkers and new therapeutic targets for diverse vascular diseases.
Acknowledgements
The author's research was supported by National Institutes of Health Grant HL080133, HL095707, and a grant from American Heart Association 09GRNT2250567.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial interest: No
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Friedman JM, Jones PA. MicroRNAs: Critical mediators of differentiation, development and disease. Swiss Med Wkly. 2008;139:466–472. doi: 10.4414/smw.2009.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33(2):139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 10.De Paepe B. Anti-angiogenic agents and cancer: current insights and future perspectives. Recent Pat Anticancer Drug Discov. 2009;4:180–185. doi: 10.2174/157489209788452821. [DOI] [PubMed] [Google Scholar]
- 11.Di Stefano R, Felice F, Balbarini A. Angiogenesis as risk factor for plaque vulnerability. Curr Pharm Des. 2009;15:1095–1106. doi: 10.2174/138161209787846892. [DOI] [PubMed] [Google Scholar]
- 12.Smart N, Dubé KN, Riley PR. Coronary vessel development and insight towards neovascular therapy. Int J Exp Pathol. 2009;90:262–283. doi: 10.1111/j.1365-2613.2009.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Cheng Y, Zhang S, et al. A necessary role of miR-222 and miR-221 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Liu X, Yang J, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, Tuccoli A, Mariani L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 17.Kuehbacher A, Urbich C, Zeiher AM, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 18.Suárez Y, Fernández-Hernando C, Pober JS, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 19.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C. MicroRNA-145 in vascular smooth muscle cell biology: a new therapeutic target for vascular disease. Cell Cycle. 2009;8:3469–3473. doi: 10.4161/cc.8.21.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elia L, Quintavalle M, Zhang J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pushparaj PN, Aarthi JJ, Kumar SD, et al. RNAi and RNAa - The Yin and Yang of RNAome. Bioinformation. 2008;2:235–237. doi: 10.6026/97320630002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 26.Chen T, Huang Z, Wang L, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 27.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Solingen C, Seghers L, Bijkerk R, et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med. 2009;13:1577–1585. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasanaro P, D'Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–1583. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee DY, Deng Z, Wang CH, et al. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Würdinger T, Tannous BA, Saydam O, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minami Y, Satoh M, Maesawa C, et al. Effect of atorvastatin on microRNA 221 / 222 expression in endothelial progenitor cells obtained from patients with coronary artery disease. Eur J Clin Invest. 2009;39(5):359–367. doi: 10.1111/j.1365-2362.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 35.Wang CH, Lee DY, Deng Z, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3(6):e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 37.Chan LS, Yue PY, Mak NK, et al. Role of MicroRNA-214 in ginsenoside-Rg1-induced angiogenesis. The European Journal of Pharmaceutical Sciences. 2009;38(4):370–377. doi: 10.1016/j.ejps.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Zampetaki A, Kirton JP, Xu Q. Vascular repair by endothelial progenitor cells. Cardiovasc Res. 2008;78:413–421. doi: 10.1093/cvr/cvn081. [DOI] [PubMed] [Google Scholar]
- 39.Asahara T, Murohara T, Sullivan A, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 40.Xu CC, Han WQ, Xiao B, et al. Differential expression of microRNAs in the aorta of spontaneously hypertensive rats. Sheng Li Xue Bao. 2008;60:553–560. [PubMed] [Google Scholar]
- 41.Li Y, Song YH, Li F, et al. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem Biophys Res Commun. 2009;381:81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 43.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol. Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji X, Takahashi R, Hiura Y, et al. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 46.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 47.Adachi T, Nakanishi M, Otsuka Y, et al. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 48.Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]