Significance
About 40% of East Asians and over 500 million people worldwide carry a specific polymorphism, ALDH2*2, and exhibit “Asian flush” after alcohol drinking. We generated a mouse strain with this engineered polymorphism and demonstrated its resemblance to human carriers in terms of defective alcohol metabolism. With this model, we show that murine ALDH2*2 increases ALDH2 protein turnover and promotes chemical-induced liver tumor development. Importantly, ALDH2 is unstable in ALDH2*2 human liver samples and is significantly down-regulated in human liver tumors. Data from our mouse and clinical studies suggest that ALDH2 is a liver tumor suppressor and the ALDH2*2 polymorphism is a risk factor for liver cancer.
Keywords: ALDH2*2 polymorphism, Asian flush, alcohol metabolism, mouse model, liver cancer
Abstract
Mitochondrial aldehyde dehydrogenase 2 (ALDH2) in the liver removes toxic aldehydes including acetaldehyde, an intermediate of ethanol metabolism. Nearly 40% of East Asians inherit an inactive ALDH2*2 variant, which has a lysine-for-glutamate substitution at position 487 (E487K), and show a characteristic alcohol flush reaction after drinking and a higher risk for gastrointestinal cancers. Here we report the characterization of knockin mice in which the ALDH2(E487K) mutation is inserted into the endogenous murine Aldh2 locus. These mutants recapitulate essentially all human phenotypes including impaired clearance of acetaldehyde, increased sensitivity to acute or chronic alcohol-induced toxicity, and reduced ALDH2 expression due to a dominant-negative effect of the mutation. When treated with a chemical carcinogen, these mutants exhibit increased DNA damage response in hepatocytes, pronounced liver injury, and accelerated development of hepatocellular carcinoma (HCC). Importantly, ALDH2 protein levels are also significantly lower in patient HCC than in peritumor or normal liver tissues. Our results reveal that ALDH2 functions as a tumor suppressor by maintaining genomic stability in the liver, and the common human ALDH2 variant would present a significant risk factor for hepatocarcinogenesis. Our study suggests that the ALDH2*2 allele–alcohol interaction may be an even greater human public health hazard than previously appreciated.
Mitochondrial aldehyde dehydrogenase 2 (ALDH2) is essential for alcohol detoxification. It is the second enzyme in the major oxidative pathway of alcohol metabolism, removing acetaldehyde (ACE), a toxic intermediate product from ethanol metabolism (1). More than 500 million people worldwide, mostly in East Asia, have a G-to-A point mutation in their ALDH2 gene (2, 3). This mutation results in a glutamic acid-to-lysine substitution at residue 487 (E487K) of the human ALDH2 protein (designated ALDH2*2). ALDH2*2 has significantly reduced ability to metabolize ACE (4, 5). Importantly, its activity is partially dominant-negative over that of the wild-type ALDH2*1, due to the structural alterations introduced by the mutation to the ALDH2 homotetramer complex (6). As a result, individuals with a heterozygous ALDH2*2/2*1 genotype have less than half the wild-type activity, and ALDH2*2/2*2 homozygotes have very low residual activity (7). Accumulated ACE can cause an alcohol flush reaction, commonly found in Asians with this variant after alcohol consumption (also called “Asian glow”).
ACE binds to cellular proteins and DNA, leading to DNA damage and organ injury (8). Specifically, endogenous aldehydes are detrimental to hematopoietic stem cells that are defective in Fanconi anemia DNA repair (9, 10). As a result, Fanconi anemia patients with the ALDH2*2 allele exhibit accelerated disease progression (11). ALDH2*2 can also increase the risk for gastrointestinal cancers, such as gastric carcinoma (12), esophageal cancer (13), and colon cancer (14). Despite the liver being the major organ of ethanol detoxification, the relationship between ALDH2*2 and the risk for liver cancer remains unclear (15, 16).
ALDH2 is highly conserved in humans and mice (17, 18), and several mouse models with modified ALDH2 activities have been developed (19). The closest model to the human ALDH2*2 polymorphism is the Aldh2 knockout (KO) mouse, which expresses no protein or enzymatic activity (20). Although Aldh2 KO mice are useful for investigating the impact of complete lack of ALDH2 activity (21), they fail to faithfully reproduce the structural and biochemical properties of ALDH2*2 in human physiology and pathology (21–23). In particular, ALDH2*2 is expressed with reduced but not loss of enzymatic activity and increased protein turnover (24).
To provide better mechanistic links between the ALDH2(E487K) mutation and human disease, we generated an ALDH2*2 knockin (KI) mouse. We observed impaired clearance of ACE from hepatocytes in these mutants after acute or chronic alcohol challenges. The ALDH2(E487K) mutation reduced total liver ALDH2 protein levels via a dominant-negative effect on protein stability, as has been documented for human tissues (24, 25). We also revealed a surprising role for ALDH2 as a liver tumor suppressor, raising the concern that this common human polymorphism may expose over 500 million carriers to greater risk of liver cancer.
Results
Generation of Aldh2E487K KI Mice.
The Aldh2E487K KI mutation was generated through homologous recombination in mouse embryonic stem (ES) cells via introduction of a G-to-A substitution in exon 12 of the endogenous Aldh2 locus, producing a missense mutation orthologous to the human ALDH2(E487K) mutation (Fig. 1A). The KI allele was confirmed by PCR detection of the remaining loxP site (Fig. 1B) after Cre-mediated deletion of the neomycin-resistance cassette and by direct sequencing of either genomic DNA (Fig. 1C) or cDNA amplified from transcribed ALDH2 mRNA. Aldh2E487K KI mice generated from these ES cells were born at the expected Mendelian ratio, had a normal life span, and were healthy and fertile.
Fig. 1.
Generation of Aldh2E487K KI mice. (A) Schematic drawings of the murine Aldh2 allele and targeting construct, Aldh2E487K KI allele generated by homologous recombination in ES cells, and final Aldh2E487K KI allele after Cre-mediated removal of the neomycin-resistance cassette. The dotted lines indicate regions of homology and asterisks indicate the altered exon. (B) PCR genotyping of WT, Aldh2E487K/+, and Aldh2E487K/E487K mice (“M” indicates molecular weight marker). (C) Confirmation of the E487K mutation by direct sequencing of PCR-amplified genomic DNA; *indicates the position of the nucleotide alteration producing the E487K mutation.
Reduced Enzymatic Activity in Aldh2E487K KI Hepatocytes.
ALDH2 activity was assessed in primary hepatocytes isolated from Aldh2E487K KI mice (Fig. 2A). Hepatocytes from mice carrying one Aldh2E487K allele retained only 44% of ALDH2 activity compared with wild-type mice. In contrast, Aldh2E487K/E487K homozygous hepatocytes produced hardly detectable ALDH2 activity. Because ALDH2 catalyzes oxidation of ACE to acetic acid in alcohol metabolism, the cellular kinetics of ACE clearance was quantitated by liquid chromatography mass spectrometry in cultured hepatocytes with isotopically labeled [13C]ACE (Fig. 2 B and C). Primary hepatocytes of heterozygous Aldh2E487K mice cleared [13C]ACE more slowly than those of wild-type mice, whereas homozygous Aldh2E487K hepatocytes failed to clear [13C]ACE altogether. These results indicate that the ALDH2(E487K) mutant is a loss-of-function enzyme defective in metabolizing its substrate in the liver.
Fig. 2.

Significantly reduced ALDH2 enzymatic activity in Aldh2E487K KI mouse hepatocytes. Primary hepatocytes from WT and Aldh2E487K KI mice were analyzed for ALDH2 activity in vitro (mean ± SEM; **P < 0.01; n = 6) (A) and for ACE clearance over a short (B) or long (C) time course (mean ± SEM; n = 3).
Increased Ethanol Intoxication in Aldh2E487K KI Mice.
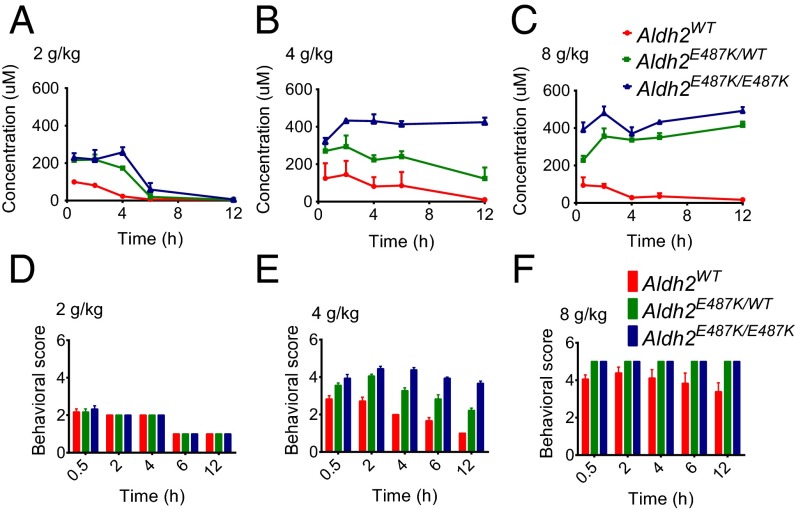
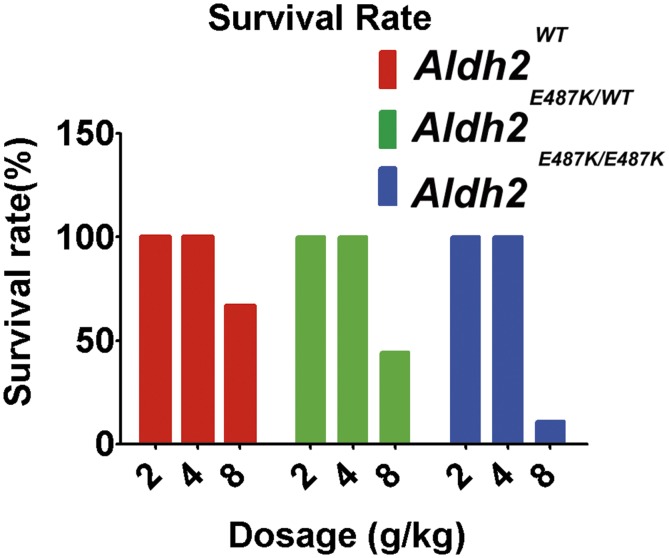
To investigate the effects of the Aldh2E487K mutation on alcohol metabolism, we acutely challenged wild-type and KI mice with ethanol (2, 4, or 8 g/kg) and monitored serum ACE levels, behavioral abnormality, and survival. When exposed to low-dose ethanol (2 g/kg), heterozygous and homozygous Aldh2E487K mice exhibited similarly slow ACE clearance but no behavioral abnormality compared with wild-type mice (Fig. 3 A and D). After 4 g/kg ethanol, homozygous Aldh2E487K mice failed to clear serum ACE (Fig. 3B), and both groups of Aldh2E487K mice showed genotype-dependent mild to severe ataxia (Fig. 3E). High-dose ethanol (8 g/kg) abolished ACE clearance and caused severe behavioral abnormalities including loss of righting reflex in both heterozygous and homozygous Aldh2E487K mice (Fig. 3 C and F). High-dose ethanol (8 g/kg) was lethal in all three groups, with the homozygous mice most sensitive (Fig. S1), consistent with their impaired ACE clearance.
Fig. 3.
Defective alcohol metabolism in Aldh2E487K KI mice after acute ethanol challenge. Serum ACE concentrations from WT and Aldh2E487K KI mice acutely challenged with ethanol at 2 g/kg (A), 4 g/kg (B), or 8 g/kg (C) over 12 h (mean ± SEM; n = 3). Behavioral scores of mice subjected to 2 g/kg (D), 4 g/kg (E), or 8 g/kg (F) ethanol over the same time course (mean ± SEM; n = 6).
Fig. S1.
Survival rates after acute ethanol challenge. WT and Aldh2E487K KI mice were acutely challenged with ethanol at 2, 4, or 8 g/kg (mean ± SEM; n = 6 each).
To study the chronic effects of ethanol consumption, we treated wild-type and Aldh2E487K mice with 4 g/kg ethanol (every day) for 6 wk. This dose was chosen to induce remarkable differences in serum ACE clearance and behavioral abnormality without acute lethal effect. Some lethality was observed in both heterozygous and homozygous Aldh2E487K mice, beginning at 2 wk (Fig. 4A). Moreover, all KI mice lost more body weight than wild-type controls during ethanol treatment (Fig. 4B). Whole-blood smears revealed significantly reduced numbers of white cells in the KI mice relative to wild-type controls (Fig. 4C and Fig. S2A), indicating possible injury of the hematopoietic system caused by chronic ethanol treatment. Furthermore, we observed hepatocyte apoptosis by histology analysis and cleaved caspase 3 staining on liver sections of Aldh2E487K mice (Fig. 5D). However, no inflammatory or necrotic changes were identified in other organs of Aldh2E487K mice, including esophagus and stomach (Fig. S2B). Taken together, these data suggest that mice with even one copy of the ALDH2 variant are more susceptible to ethanol intoxication and organ injury.
Fig. 4.
Increased sensitivity of Aldh2E487K KI mice to damage induced by chronic ethanol challenge. (A and B) Survival (A) and body weight (B) of WT and Aldh2E487K KI mice treated with 4 g/kg ethanol for 6 wk (mean ± SEM; n = 9). (C) White blood cell counts from whole-blood smears of chronically treated mice (mean ± SEM; *P < 0.05, **P < 0.01; n = 3). (D) Histology and cleaved caspase 3 staining of treated mouse liver sections (400×).
Fig. S2.
Chronic ethanol challenge in Aldh2E487K knockin mice. (A) Representative images of white blood cells stained from a whole-blood smear. (B) H&E staining of esophagus and stomach in Aldh2E487K knockin mice (n = 3).
Fig. 5.
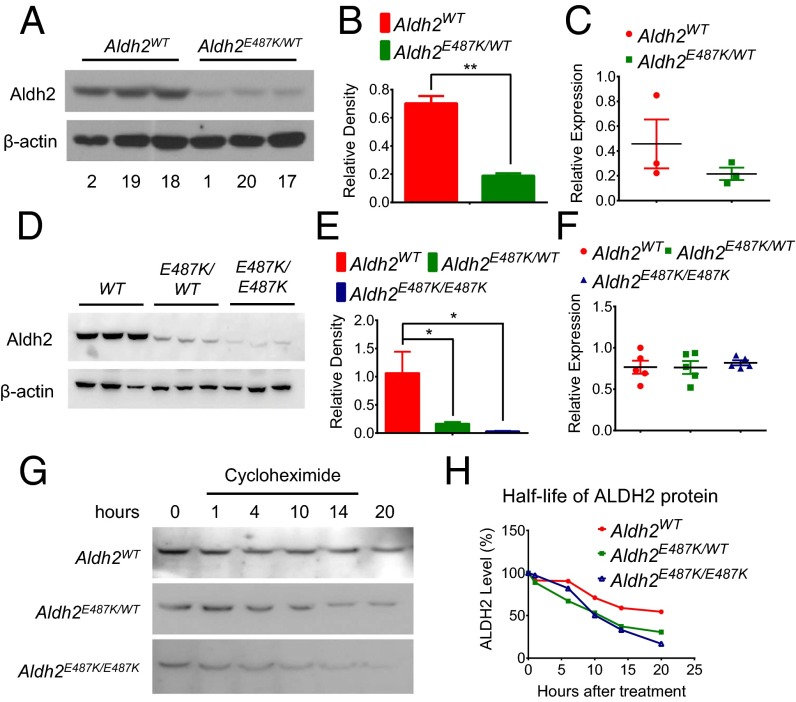
Reduced levels and stability of ALDH2 protein in liver of ALDH2*2 humans and Aldh2E487K KI mice. (A and B) Representative Western blot (A) and quantification (B) relative to β-actin of human liver ALDH2 with the indicated genotypes. (C) Measurement of ALDH2 mRNA by real-time quantitative PCR in these human samples. (D and E) Representative Western blot (D) and quantification (E) normalized to β-actin of ALDH2 in heterozygous and homozygous Aldh2E487K KI mouse liver. (F) Measurement of ALDH2 mRNA by real-time quantitative PCR in these mouse samples. (G and H) Western blot (G) and quantification (H) of ALDH2 in primary hepatocytes from Aldh2E487K KI mice after cycloheximide treatment to determine the half-life of ALDH2 proteins (mean ± SEM; *P < 0.01; **P < 0.001; n = 3).
Increased ALDH2 Protein Turnover in Aldh2E487K KI Mice.
It has been observed in prior studies that ALDH2 protein levels are very low in ALDH2*2 liver (24, 26). The ALDH2(E487K) variant increases overall ALDH2 protein turnover through a dominant but unknown mechanism (24). We examined the levels of ALDH2 protein in lysates of 20 human liver surgical specimens (Fig. S3). Consistent with previous reports, humans with single-allele ALDH2E487K mutations expressed only about 27% of liver ALDH2 protein relative to wild-type individuals (Fig. 5 A and B). Likewise, one allele of Aldh2E487K in mice caused an 85% decrease in liver ALDH2 protein, whereas two mutant alleles led to a nearly complete loss of ALDH2 protein (Fig. 5 D and E). ALDH2 mRNA levels were not different in humans or mice (Fig. 5 C and F), suggesting a posttranscriptional regulation. Cycloheximide chasing experiments on purified mouse hepatocytes indicate that total ALDH2 protein is less stable in heterozygous or homozygous KI cells compared with wild type (Fig. 5 G and H), revealing a reduced stability of the ALDH2(E487K) protein itself and a dominant effect on wild-type ALDH2 protein.
Fig. S3.

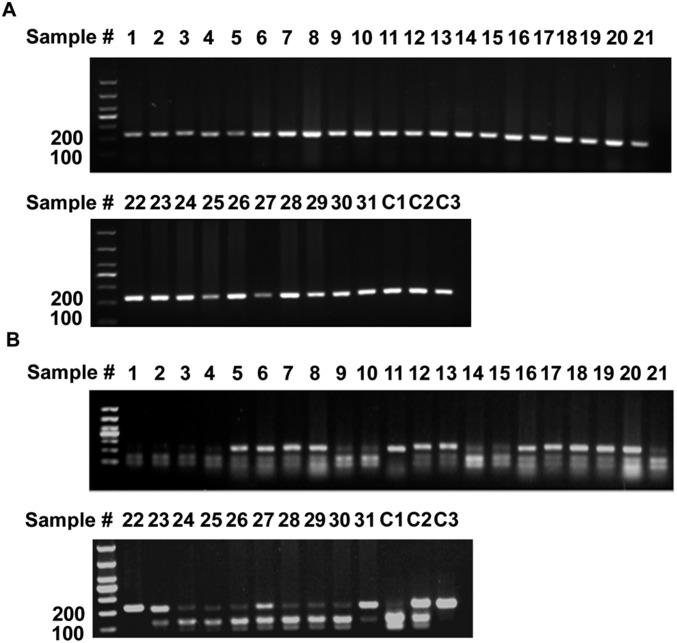
PCR genotyping of the ALDH2 polymorphism in human liver tissues. (A) PCR fragments spanning the polymorphism amplified from genomic DNA of 20 human liver tissues. (B) Digested PCR fragments from A to identify the presence of the mutation. C1, C2, and C3 are positive controls for ALDH2*1/2*1, ALDH2*1/2*2, and ALDH2*2/2*2 genotypes, respectively. Patients 1, 4, 5, 9, 12, 15, 17, and 20 are heterozygotes.
Reduced ALDH2 Levels in Human HCC Samples.
Given the defects in clearing ACE and detoxifying ethanol by ALDH2E487K liver, we reasoned that ALDH2*2 humans might be more prone to develop liver cancer. We analyzed the genetic polymorphism of ALDH2E487K mutations in HCC samples and observed a 40% mutation rate (17 of 43) comprising 5% homozygous and 35% heterozygous mutations (Fig. S4). The mutation rate of ALDH2E487K in HCC samples was close to the 40% mutation rate found in normal livers (Fig. S3), suggesting that within our limited and random tumor samples no obvious correlation exists between ALDH2E487K mutation and HCC incidence.
Fig. S4.
PCR genotyping of the ALDH2 polymorphism in human liver tumors. (A) PCR fragments spanning the polymorphism amplified from genomic DNA of 31 human HCC samples. (B) Digested PCR fragments from A to identify the presence of the mutation. C1, C2, and C3 are positive controls for ALDH2*1/2*1, ALDH2*1/2*2, and ALDH2*2/2*2 genotypes, respectively.
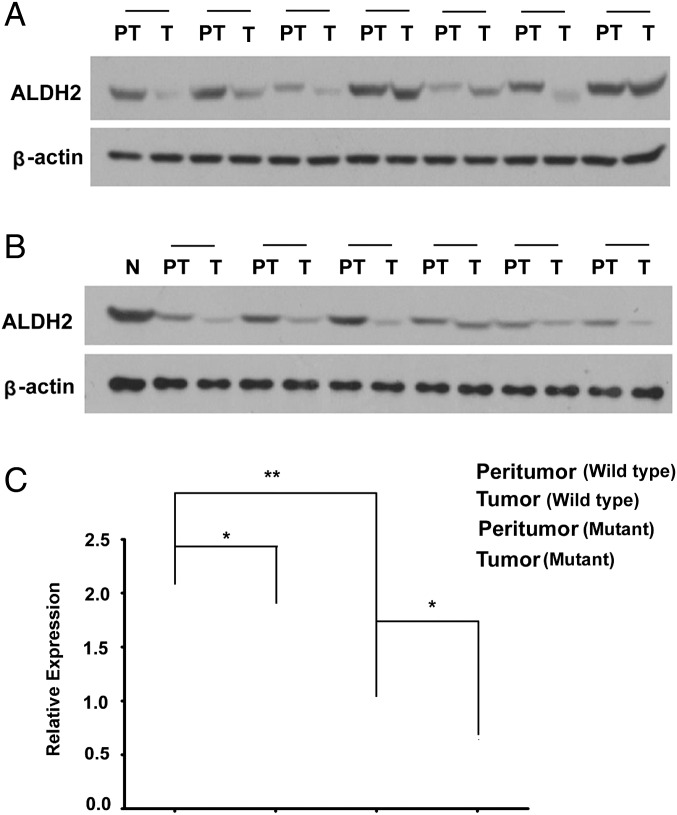
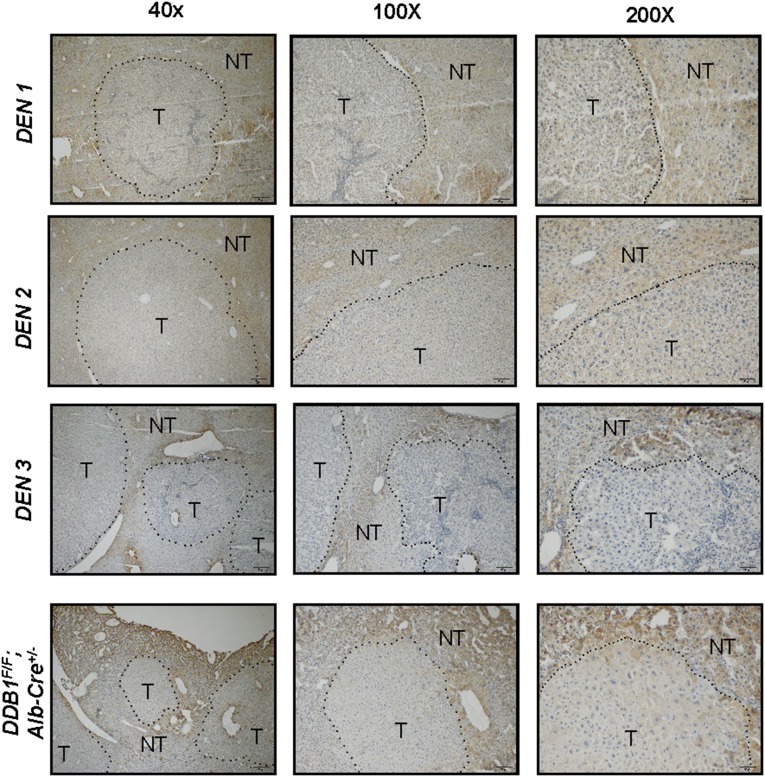
However, surprisingly, ALDH2 protein levels were significantly reduced in almost all HCC samples we examined compared with matching peritumor liver tissues by Western blot analysis (Fig. 6). This decrease was found in six out of nine wild-type and six out of seven ALDH2*2 mutant patients compared with their matching peritumor tissues (Fig. 6). In addition, immunostaining revealed that ALDH2 expression was also reduced in all mouse hepatomas from either a chemical-induced model (27) or a spontaneous genetic model (28) compared with immediately neighboring nontumor liver tissues (Fig. S5). These data suggest that ALDH2 might play a tumor suppressor role in the liver, independent of the ALDH2 polymorphism.
Fig. 6.
Reduced ALDH2 expression in human liver tumor samples independent of the ALDH2 polymorphism. (A and B) Western blot of ALDH2 in liver tumor (T) and peritumor (PT) tissues from (A) ALDH2*1/2*1 patients or (B) ALDH2*2/2*1 patients. N, normal WT liver. (C) Quantification of ALDH2 levels normalized to β-actin (mean ± SEM; *P < 0.05, **P < 0.01).
Fig. S5.
Immunohistochemical staining for ALDH2 in hepatoma sections from chemical (DEN)-induced and DDB1F/F;Alb-Cre+/− genetic liver cancer models. Dashed lines outline tumor (T) and nontumor (NT) areas; 40×, 100×, and 200× amplified sections are shown. Three DEN mice are shown.
Increased Hepatocarcinogenesis in Aldh2E487K KI Mice.
Because ALDH2E487K KI mice express significantly reduced ALDH2 protein in the liver (Fig. 5), we investigated their susceptibility to liver tumor development. We induced liver tumors in wild-type and Aldh2E487K KI mice by injecting diethylnitrosamine (DEN) at postnatal day (P)14 and examined the livers after 8 mo (Fig. 7A). Mice of all three groups developed liver tumors (Fig. 7B); however, Aldh2E487K mutant mice developed a greater number and a larger volume of liver tumors than wild-type controls (Fig. 7 C and D). Serum alanine transaminase (ALT) activity was also higher in Aldh2E487K/WT mice and further elevated in Aldh2E487K/E487K mice (Fig. 7E), indicating more severe liver injury. Finally, liver histology revealed more focal hepatic lesions in Aldh2E487K KI mutants (Fig. 7F, Upper), and immunostaining for phosphorylated histone H2AX demonstrated a substantially higher degree of DNA damage in the mutants (Fig. 7F, Lower). We conclude that ALDH2E487K KI mice are more susceptible to carcinogen-induced liver tumor development.
Fig. 7.
Accelerated liver tumor development in DEN-induced Aldh2E487K KI mice. (A) Outline of the experimental protocol. (B) Representative photographs of livers dissected from WT or KI mice 8 mo after DEN treatment. (C and D) Total number (C) and maximal volumes (D) of liver tumors (>0.5 mm) from WT and KI mice. (E) Serum ALT activity to estimate liver injury. (F) Histology and phospho-H2AX staining (400×) of liver tissue sections (mean ± SEM; *P < 0.05, **P < 0.01; n = 4).
Discussion
Aldh2E487K KI Mice Model Human Deficiency in Alcohol Metabolism.
ALDH2 catalyzes ACE metabolism and plays important roles in the pathogenesis of cardiovascular diseases, diabetes, neurodegenerative diseases, and cancer (19). The ALDH2*2 polymorphism is the most common ALDH2 deficiency in humans (29, 30), and several transgenic mouse models have been developed to investigate the roles of this enzyme (31). These models include Aldh2 knockout (21), ALDH2 overexpression (32) and Aldh2E487K overexpression (33) mice. Here we generated a mouse strain with a mutation in murine ALDH2 corresponding to the E487K substitution in human ALDH2. The same mouse strain was independently generated while this manuscript was under preparation, and was shown to accumulate ACE in the blood upon ethanol challenge and exhibit heightened pain response (34). Here, we provide a comprehensive description of ACE metabolism in these heterozygote and homozygote mutant mice.
First, in addition to increased blood ACE levels, Aldh2E487K KI mice were more susceptible to ethanol toxicity than wild-type mice after acute alcohol challenge, due to the impaired clearance of ACE by their hepatocytes. Second, upon chronic alcohol exposure, the KI mice lost more weight than controls, and homozygotes died early. The number of white blood cells in KI mice was reduced dramatically, indicating possible injury of the hematopoietic system caused by ACE accumulation. Despite the increased apoptotic cells in the liver of the KI mice, no significant injury was observed in the esophagus and stomach, likely because the 6-wk treatment was too short to induce injury. Interestingly, we also observed that all KI mice exhibited hyperpigmentation on the feet (Fig. S6). This cutaneous change is also found in chronic alcoholics, who often demonstrate hyperpigmentation and erythema, particularly on the legs (35, 36).
Fig. S6.
Skin hyperpigmentation in Aldh2E487K KI mice receiving chronic ethanol feeding. (A) Photographs of the enhanced pigmentation of paw skin of Aldh2E487K KI mice treated with ethanol (4 g/kg) for 6 wk. (B) H&E staining of corresponding paw skin.
Dominant-Negative Effect of ALDH2(E487K) on ALDH2 Protein Stability.
It is thought that ALDH2 deficiency in human ALDH2*1/*2 heterozygotes results from a dominant-negative effect of the mutant enzyme on the catalytic activity of the holoenzyme containing the wild-type enzyme (25). However, we observed a significant reduction of ALDH2 protein levels and shortened half-lives of all ALDH2 proteins in the livers of both human and mouse heterozygotes. The protein is barely detectable in mouse homozygotes. No difference of ALDH2 RNA levels was found between mutants and wild types, suggesting a posttranscriptional mechanism regulating the turnover of the mutant ALDH2 protein as well as the wild-type protein.
To investigate the mechanism underlying the instability of ALDH2(E487K) protein, we tested whether the lysine residue introduced in the mutant ALDH2 might make it more susceptible to polyubiquitination and therefore proteasome-mediated degradation. ALDH2(E487K) mutant protein did not exhibit increased ubiquitination in vitro, and treating primary hepatocytes from mutant mice with a proteasome inhibitor did not result in increased ALDH2 protein levels (Fig. S7). Although the precise mechanism awaits further investigation, new routes of pharmaceutical interference might be directed toward increasing ALDH2 protein stability in addition to its enzymatic activity (19, 37).
Fig. S7.
Proteasome-independent mechanism for the instability of ALDH2(E487K) mutant protein. (A) Western blot (WB) analysis of ALDH2 levels in primary hepatocytes isolated from WT or KI mice after treatment with the proteasome inhibitor Mg132. p21 levels were monitored as a control for Mg132 efficiency. “NC” indicates vehicle-only controls. (B) Ubiquitination assays for ALDH2 WT and E487K mutants expressed in HEK293 cells. Flag-ALDH2 was pulled down and probed for HA-UB. Flag-CLC2, positive control. IP, immunoprecipitation; M, E487K mutant; W, WT.
Increased Hepatocarcinogenesis by ALDH2(E487K).
People with the ALDH2*2 polymorphism are reported to be at a greater risk of developing esophageal and other digestive tract cancers (12–14, 38), attributable to ACE-induced DNA damage and genome instability (39, 40). Aldh2E487K KI mice did not exhibit obvious damage in the esophagus or stomach after 2 mo of daily ethanol challenge, judged by histology evaluation. No tumors or dysplastic nodules were found in any organs of these mice, and deletion of one copy of the Trp53 gene, encoding the tumor suppressor p53, in the KI mice did not accelerate the tumorigenesis. Longer chronic exposure to ethanol may be required to evaluate ethanol- and ACE-mediated tumorigenesis in this model.
The relationship between the ALDH2 polymorphism and liver cancer risk remains unclear (15, 16), despite the fact that liver is the major organ for alcohol metabolism and ACE production and that chronic ethanol challenge causes liver damage (Fig. 5B). In this study, we analyzed the ALDH2E487K mutation in human samples and found that the mutation rate in HCC samples (39%) was similar to that in normal livers (40%) and within the range reported for East Asians, suggesting no obvious direct relationship between the ALDH2E487K mutation and HCC incidence. However, we observed lower ALDH2 protein levels in HCC samples compared with matching peritumor liver tissues independent of the ALDH2 polymorphism. To test ALDH2 as a potential HCC tumor suppressor, we induced HCC in Aldh2E487K mutant and wild-type mice with DEN, a chemical carcinogen causing hepatocyte DNA damage. Compared with controls, Aldh2E487K mutant mice developed a greater number and larger volume of HCCs, and the effect was more pronounced in homozygote mutants than in heterozygote mutants. The DNA damage response as indicated by γH2AX staining and liver injury measured by ALT elevation was also greatly increased in the mutants bearing larger HCC loads. How this polymorphism cross-talks with the known HCC-promoting signaling network awaits future investigation (41).
Our data suggest that the ALDH2*2 population might be more susceptible to HCC if exposed to liver-damaging agents, as is observed in the Aldh2E487K mutant mice exposed to DEN. In support, the ALDH2 polymorphism was reported to be a significant risk factor for HCC development and recurrence in Japanese hepatitis C virus patients (42, 43). The ALDH2 polymorphism may also be a risk factor in other subpopulations of HCC patients when their disease etiology is closely analyzed in association with their ALDH2 genotype. Another explanation for the human and mouse difference in HCC incidence is that human carriers of the ALDH2*2 polymorphism might refrain from drinking alcohol due to “alcohol flush,” and hence are less likely to be subjected to alcohol-induced liver damage and cancer than noncarriers. This population might be more prone to develop liver cancer due to their ALDH2 deficiency if their alcohol consumption were equivalent to that of noncarriers. Therefore, grouping all HCC patients in an association analysis might miss the differential contribution of this mutation to the disease.
Based on our clinical and mouse data, we conclude that ALDH2 may protect liver cells from DNA damage induced by chemical agents such as aldehydes and DEN. Although the ALDH2*2 polymorphism itself does not directly lead to liver cancer, it reduces ALDH2 protein levels and causes enzyme deficiency in the liver, leading to inefficient detoxification of aldehydes and accumulation of cancer-causing mutations.
Methods
Generation of Aldh2E487K Knockin Mice.
The Aldh2E487K knockin targeting vector was assembled using genomic fragments of the mouse Aldh2 gene including a 2.0-kb 5′ arm and a 6.4-kb 3′ arm retrieved from 129S6/SvEvBAC DNA (origin) via an Escherichia coli-based chromosome engineering system (44), a loxP-flanked neomycin-resistance cassette (Neor), and a thymidine kinase gene (TK) as negative selection for genomic integration. The 3′ arm contains murine Aldh2 exon 12 bearing the E487K mutation, which was created by PCR-based mutagenesis. The linearized targeting construct was electroporated into 129S6/SvEv mouse embryonic stem cells. Following homologous recombination in ES cells, a Cre recombinase expression vector was transiently transfected to delete the Neor cassette. Targeted ES cells were identified by PCR and sequencing and injected into mouse blastocysts to produce chimeras. The chimeric mice were bred to C57BL/6J mice to generate heterozygous and homozygous Aldh2E487K mice. Mice were genotyped using PCR primers spanning the position of the residual loxP site (5′-CGGGAATTGAACTTGGTAGCCAG-3′ and 5′-GCGTAAGGCATGCGCCATCAC-3′), generating a 169-bp PCR product in WT mice and a 263-bp PCR product in KI mice. Initial litters were analyzed for the presence of Aldh2E487K by PCR and sequencing using the primers 5′-CCTGAGCCGAATGCTTTTAAG-3′ and 5′-CTCACGCTCCTTACTGGAC-3′. Wild-type littermates of Aldh2E487K mice served as controls for all analyses.
Mice were bred and maintained in a barrier facility under pathogen-free conditions at WuXi AppTec. Mice were housed on a 12-h/12-h light/dark cycle and given food and water ad libitum. Mice were older than 8 wk at the beginning of the experiments and were used in accordance with protocols approved by the Institutional Animal Care and Use Committee.
Patient Samples.
The use of human samples was approved by the local ethics committee (Institutional Review Board of Sir Run Run Shaw Hospital) and included informed consent. HCC tissues were collected from patients undergoing liver resection for HCC, and normal liver tissues were collected from patients undergoing laparoscopic surgery for intrahepatic calculi at the Sir Run Run Shaw Hospital (Zhejiang University). All tissues were stored at −80 °C.
Genetic Polymorphism of ALDH2 in Human Tissues.
Genomic DNA was extracted by a MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa). The human ALDH2 gene was analyzed by PCR using primers spanning exon 12 (5′-TCAAATTACAGGGTCAACTGCTA-3′ and 5′-GCCCCCAACAGACCCCAATC-3′). The 222-bp fragment was digested with Eco57I (Fermentas) and subjected to agarose gel electrophoresis. The wild-type ALDH2 allele yielded two fragments of 132 and 90 bp, and the ALDH2E487K allele yielded a single product of 222 bp (45).
Primary Mouse Hepatocytes.
Hepatocytes were isolated from mice by the modified collagenase method as previously described (46). Mice were anesthetized using 1% pentobarbital sodium, and livers were perfused with buffer solution (10 mM Hepes, 3 mM KCl, 130 mM NaCl, 1 mM NaH2PO4, 10 mM glucose) containing 0.5 mM EGTA (pH 7.4) and then with buffer solution containing 5 mM CaCl2 and 100 U/mL collagenase type IV (Sigma) at a rate of 2 mL/min. Livers were isolated, and cells were dissociated and washed with PBS. Hepatocytes were plated at 1 × 105 viable cells per cm2 on rat tail collagen-coated (BD) tissue-culture dishes in DMEM, 15% FBS (vol/vol), 100 U/mL penicillin, and 100 µg/mL streptomycin and kept in a humidified cell-culture incubator at 37 °C and 5% CO2 until use the following day.
Reverse-Transcription Reaction and Quantitative Real-Time PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen). RNA was reverse-transcribed into cDNA with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNAs were quantified using real-time PCR in a StepOne Plus system according to the standard SYBR Green PCR Kit protocol (Applied Biosystems) in triplicate. β-actin was used to normalize the amount of total mRNA. Relative expression was calculated with the comparative CT method. Primer sequences were as follows: Aldh2: 5′-gctgggctgacaagtaccat-3′ and 5′-ttgatcaagttggccacgta-3′; β-actin: 5′-GACGGCCAGGTCATCACTAT-3′ and 5′-CTTCTGCATCCTGTCAGCAA-3′; ALDH2: 5′-TCAAATTACAGGGTCAACTGCTA-3′ and 5′-GCCCCCAACAGACCCCAATC-3′; β-ACTIN: 5′-CTTAGTTGCGTTACACCCTTTC-3′ and 5′-CACCTTCACCGTTCCAGTTT-3′.
Western Blotting.
Tissues or cell samples were homogenized in RIPA buffer (Sigma) containing protease inhibitor mixture (Roche), incubated on ice for 30 min, and centrifuged for 15 min at 12,000 × g. Equal amounts of proteins were separated by SDS/PAGE, transferred to nitrocellulose membranes (Invitrogen), and incubated overnight at 4 °C with primary antibodies. Immunoblots were visualized with horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology; 1:2,000) using enhanced chemiluminescence (Pierce Biotechnology). Primary antibodies were ALDH2 (goat polyclonal; Santa Cruz Biotechnology; 1:500), p21 (mouse monoclonal; Cell Signaling Technology; 1:1,000), and β-actin (rabbit monoclonal; Cell Signaling Technology; 1:1,000).
Cycloheximide Treatment.
Cycloheximide (20 µg/mL; Sigma) was added to primary mouse hepatocytes to block protein synthesis. Total cell lysates were subjected to Western blotting using an anti-ALDH2 antibody. The relative band intensity was quantified using Quantity software (1D component of ImageQuant LAS 4010; GE Healthcare).
ALDH2 Activity Assay.
The activity of ALDH2 was analyzed using the colorimetric Mitochondrial Aldehyde Dehydrogenase (ALDH2) Activity Assay Kit (Abcam; ab115348) according to the manufacturer’s protocol.
ACE Metabolism in Primary Hepatocytes.
Freshly thawed primary mouse hepatocytes (1 × 106) were plated in Corning T25 flasks and incubated overnight at 37 °C and 5% CO2 in 1:1 DMEM:F12 media with 15% (vol/vol) FBS. Universally labeled [13C]ACE was added to each flask to a concentration of 75 μM, and plug caps were used to prevent ACE loss. A flask containing media and ACE alone (no cells) was used as a control for ACE evaporation. At 0, 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, 4, 5, and 24 h, duplicate 60-µL aliquots of media were frozen on dry ice and stored at −80 °C before liquid chromatography mass spectrometry (LCMS) quantitation.
LCMS Measurement of ACE.
ACE concentrations were determined through derivatization with dinitrophenylhydrazine (DNPH) using butyraldehyde-DNPH (Supelco) as an internal standard. Tissue-culture media or blood (60 μL) was deproteinized by addition of an equal volume of cold acetonitrile (ACN) containing 10 μg/mL butyraldehyde-DNPH and centrifuged for 30 min at 3,500 × g at 4 °C. One hundred microliters of supernatant was derivatized by the addition of 30 μL of 0.75 μg/mL freshly prepared DNPH in ACN and 10 μL of 1 M citric acid (pH 4.0). After 30 min at 25 °C, the reaction was quenched with 120 μL of 0.1% formic acid in water. Samples were analyzed by direct injection onto a Waters ACQUITY UPLC system and Applied Biosystems API 4000 Q MS system. Chromatographic separation was performed using a Kinetex C18 30 × 2.1 mm 100A column at a flow rate of 500 μL/min, using water + 0.1% formic acid as buffer A and ACN + 0.1% formic acid as buffer B. The gradient was 0–0.5 min 95% buffer A, to 3.5 min 5% buffer A, to 4 min at 5% buffer A, return to 95% buffer A at 4.5 min, and reequilibration of the column to 5 min at 95% buffer A. [13C]ACE-DNPH and butyraldehyde-DNPH were monitored using the following multiple reaction monitoring transitions, respectively: [224.960/46, declustering potential (DP) −40, entrance potential (EP) −10, collision energy (CE) −42, collision cell exit potential (CXP) −5] and (250.958/46.0, DP −55, EP −10, CE −52, CXP −5). Quantitation was performed using a [13C]ACE standard curve prepared in parallel, and rate constants of ACE consumption were determined by fitting time/concentration values to a first-order decay model.
Ethanol Challenge.
For acute exposure, mice were administered ethanol (Sigma) in water by gavage once at doses of 2, 4, and 8 g/kg. For chronic exposure, mice were treated with 4 g/kg ethanol daily for 6 wk.
Behavioral Score.
After acute ethanol challenge, mouse behaviors were observed and scored according to the following system: 1, rearing; 2, sedation; 3, mild ataxia (dysfunction of hind limbs); 4, severe ataxia (inability to move, body shaking); 5, loss of righting reflex.
Blood Smear.
EDTA-anticoagulated blood samples were collected and thick/thin smears were prepared. Smears were air-dried, thin smears were fixed in methanol, and thick/thin smears were stained by Giemsa (pH 7.2) (Sigma). Slides were rinsed with water, air-dried, and examined under a microscope. White blood cells were counted in five random high-power (200×) fields and are given as the number of cells per field.
Histology and Immunohistochemistry.
Mouse tissues were fixed overnight at 4 °C in PBS-buffered 4% (wt/vol) paraformaldehyde and embedded in paraffin. Five-micrometer cross-sections were prepared and stained with hematoxylin and eosin. For immunohistochemistry, tissue sections were blocked with blocking buffer and incubated with antibodies against anti-phosphohistone H2AX (Cell Signaling Technology; 1:50).
Tumor Induction and Analysis.
P14 mice were injected intraperitoneally with 25 mg/kg DEN (Sigma) (27) and euthanized after 8 mo. Mouse livers were isolated and separated into individual lobes. Externally visible tumors (>0.5 mm) were counted and volumes were measured by stereomicroscopy.
ALT Activity.
Blood samples anticoagulated by EDTA-K2 were collected and centrifuged at 3,000 × g for 10 min at 4 °C. The activity of serum ALT was analyzed using a Reitman–Frankel ALT Activity Assay Kit (Jiancheng).
Statistical Analysis.
All results are presented as the means ± SEM of at least three independent experiments; each was performed in duplicate or triplicate. Data were analyzed using the two-sided Student’s t test and one-way analysis of variance (ANOVA) with appropriate multiple comparison correction as required, using Prism 5.0 software (GraphPad). Differences were considered significant at P < 0.05 (*P < 0.05, **P < 0.01).
Acknowledgments
We thank all the scientists at Agios Pharmaceuticals, Inc. and WuXi PharmaTech, Inc. for comments and discussion as well as technical assistance. This work was supported in part by funds from the National Natural Science Foundation of China (81090420/H1606 and 81201942) and Zhejiang Provincial Natural Science Foundation of China (LZ14H160002) and Fundamental Research Funds for the Central Universities and Key Construction Program of the National “985” Project.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510757112/-/DCSupplemental.
References
- 1.Yin SJ. Alcohol dehydrogenase: Enzymology and metabolism. Alcohol Alcohol Suppl. 1994;2:113–119. [PubMed] [Google Scholar]
- 2.Matsuo K, et al. Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis. 2001;22(6):913–916. doi: 10.1093/carcin/22.6.913. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152(8):1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81(1):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baan R, et al. WHO International Agency for Research on Cancer Monograph Working Group Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8(4):292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Miller S, et al. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17(2):159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrés J, et al. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994;269(19):13854–13860. [PubMed] [Google Scholar]
- 8.Kayani MA, Parry JM. The in vitro genotoxicity of ethanol and acetaldehyde. Toxicol In Vitro. 2010;24(1):56–60. doi: 10.1016/j.tiv.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Garaycoechea JI, et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489(7417):571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 10.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475(7354):53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 11.Hira A, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuo K, et al. The aldehyde dehydrogenase 2 (ALDH2) Glu504Lys polymorphism interacts with alcohol drinking in the risk of stomach cancer. Carcinogenesis. 2013;34(7):1510–1515. doi: 10.1093/carcin/bgt080. [DOI] [PubMed] [Google Scholar]
- 13.Fang P, et al. Meta-analysis of ALDH2 variants and esophageal cancer in Asians. Asian Pac J Cancer Prev. 2011;12(10):2623–2627. [PubMed] [Google Scholar]
- 14.Guo XF, et al. Meta-analysis of the ADH1B and ALDH2 polymorphisms and the risk of colorectal cancer in East Asians. Intern Med. 2013;52(24):2693–2699. doi: 10.2169/internalmedicine.52.1202. [DOI] [PubMed] [Google Scholar]
- 15.Ding J, et al. Alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 genotypes, alcohol drinking and the risk of primary hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev. 2008;9(1):31–35. [PubMed] [Google Scholar]
- 16.Takeshita T, Yang X, Inoue Y, Sato S, Morimoto K. Relationship between alcohol drinking, ADH2 and ALDH2 genotypes, and risk for hepatocellular carcinoma in Japanese. Cancer Lett. 2000;149(1-2):69–76. doi: 10.1016/s0304-3835(99)00343-2. [DOI] [PubMed] [Google Scholar]
- 17.Chang C, Mann J, Yoshida A. Transgenesis of the aldehyde dehydrogenase-2 (ALDH2) locus in a mouse model and in cultured human cells. Adv Exp Med Biol. 1995;372:131–136. doi: 10.1007/978-1-4615-1965-2_17. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa M, et al. A comparison of covalent binding of ethanol metabolites to DNA according to Aldh2 genotype. Toxicol Lett. 2007;168(2):148–154. doi: 10.1016/j.toxlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: New therapeutic opportunities. Physiol Rev. 2014;94(1):1–34. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitagawa K, et al. Aldehyde dehydrogenase (ALDH) 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett. 2000;476(3):306–311. doi: 10.1016/s0014-5793(00)01710-5. [DOI] [PubMed] [Google Scholar]
- 21.Yu HS, et al. Characteristics of aldehyde dehydrogenase 2 (Aldh2) knockout mice. Toxicol Mech Methods. 2009;19(9):535–540. doi: 10.3109/15376510903401708. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto A, et al. Effects of 5-week ethanol feeding on the liver of aldehyde dehydrogenase 2 knockout mice. Pharmacogenet Genomics. 2008;18(10):847–852. doi: 10.1097/FPC.0b013e328307a0a9. [DOI] [PubMed] [Google Scholar]
- 23.Isse T, Matsuno K, Oyama T, Kitagawa K, Kawamoto T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol Clin Exp Res. 2005;29(11):1959–1964. doi: 10.1097/01.alc.0000187161.07820.21. [DOI] [PubMed] [Google Scholar]
- 24.Xiao Q, Weiner H, Crabb DW. The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion. J Clin Invest. 1996;98(9):2027–2032. doi: 10.1172/JCI119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Q, Weiner H, Johnston T, Crabb DW. The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced HeLa cells. J Clin Invest. 1995;96(5):2180–2186. doi: 10.1172/JCI118272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida A, Wang G, Davé V. Determination of genotypes of human aldehyde dehydrogenase ALDH2 locus. Am J Hum Genet. 1983;35(6):1107–1116. [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121(7):977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Yamaji S, et al. Hepatocyte-specific deletion of DDB1 induces liver regeneration and tumorigenesis. Proc Natl Acad Sci USA. 2010;107(51):22237–22242. doi: 10.1073/pnas.1015793108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi S, et al. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet. 1994;343(8899):741–742. doi: 10.1016/s0140-6736(94)91629-2. [DOI] [PubMed] [Google Scholar]
- 30.Thomasson HR, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48(4):677–681. [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6(3):e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doser TA, et al. Transgenic overexpression of aldehyde dehydrogenase-2 rescues chronic alcohol intake-induced myocardial hypertrophy and contractile dysfunction. Circulation. 2009;119(14):1941–1949. doi: 10.1161/CIRCULATIONAHA.108.823799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Endo J, et al. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ Res. 2009;105(11):1118–1127. doi: 10.1161/CIRCRESAHA.109.206607. [DOI] [PubMed] [Google Scholar]
- 34.Zambelli VO, et al. Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med. 2014;6(251):251ra118. doi: 10.1126/scitranslmed.3009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu SW, Lien MH, Fenske NA. The effects of alcohol and drug abuse on the skin. Clin Dermatol. 2010;28(4):391–399. doi: 10.1016/j.clindermatol.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Rao GS. Cutaneous changes in chronic alcoholics. Indian J Dermatol Venereol Leprol. 2004;70(2):79–81. [PubMed] [Google Scholar]
- 37.Chen CH, et al. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang SJ, et al. Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J Gastroenterol. 2007;13(43):5760–5764. doi: 10.3748/wjg.v13.i43.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda T, Yabushita H, Kanaly RA, Shibutani S, Yokoyama A. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol. 2006;19(10):1374–1378. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 40.Oberbeck N, et al. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol Cell. 2014;55(6):807–817. doi: 10.1016/j.molcel.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng GS. Conflicting roles of molecules in hepatocarcinogenesis: Paradigm or paradox. Cancer Cell. 2012;21(2):150–154. doi: 10.1016/j.ccr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato S, et al. Genetic polymorphisms of aldehyde dehydrogenase 2, cytochrome p450 2E1 for liver cancer risk in HCV antibody-positive Japanese patients and the variations of CYP2E1 mRNA expression levels in the liver due to its polymorphism. Scand J Gastroenterol. 2003;38(8):886–893. doi: 10.1080/00365520310004489. [DOI] [PubMed] [Google Scholar]
- 43.Tomoda T, et al. Genetic risk of hepatocellular carcinoma in patients with hepatitis C virus: A case control study. J Gastroenterol Hepatol. 2012;27(4):797–804. doi: 10.1111/j.1440-1746.2011.06948.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee EC, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 45.Wang RS, Nakajima T, Kawamoto T, Honma T. Effects of aldehyde dehydrogenase-2 genetic polymorphisms on metabolism of structurally different aldehydes in human liver. Drug Metab Dispos. 2002;30(1):69–73. doi: 10.1124/dmd.30.1.69. [DOI] [PubMed] [Google Scholar]
- 46.Gaudy AM, Clementi AH, Campbell JS, Smrcka AV, Mooney RA. Suppressor of cytokine signaling-3 is a glucagon-inducible inhibitor of PKA activity and gluconeogenic gene expression in hepatocytes. J Biol Chem. 2010;285(53):41356–41365. doi: 10.1074/jbc.M110.159111. [DOI] [PMC free article] [PubMed] [Google Scholar]