Significance
Although transfer RNAs (tRNAs) are best known as adapter molecules essential for translation, recent biochemical and computational evidence has led to a previously unexpected conceptual consensus that tRNAs are not always end products but can further serve as a source of small functional RNAs. Here we report that a novel type of tRNA-derived small RNA, termed SHOT-RNAs, are specifically and abundantly expressed in sex hormone-dependent breast and prostate cancers. SHOT-RNAs are produced from aminoacylated mature tRNAs by angiogenin-mediated cleavage of the anticodon loop, which is promoted by sex hormones and their receptors. We identified the complete repertoire of SHOT-RNAs, and also found their functional significance in cell proliferation. These results have unveiled a novel tRNA-engaged pathway in tumorigenesis.
Keywords: tRNA, tRNA half, sex hormone, breast cancer, prostate cancer
Abstract
Sex hormones and their receptors play critical roles in the development and progression of the breast and prostate cancers. Here we report that a novel type of transfer RNA (tRNA)-derived small RNA, termed Sex HOrmone-dependent TRNA-derived RNAs (SHOT-RNAs), are specifically and abundantly expressed in estrogen receptor (ER)-positive breast cancer and androgen receptor (AR)-positive prostate cancer cell lines. SHOT-RNAs are not abundantly present in ER− breast cancer, AR− prostate cancer, or other examined cancer cell lines from other tissues. ER-dependent accumulation of SHOT-RNAs is not limited to a cell culture system, but it also occurs in luminal-type breast cancer patient tissues. SHOT-RNAs are produced from aminoacylated mature tRNAs by angiogenin-mediated anticodon cleavage, which is promoted by sex hormones and their receptors. Resultant 5′- and 3′-SHOT-RNAs, corresponding to 5′- and 3′-tRNA halves, bear a cyclic phosphate (cP) and an amino acid at the 3′-end, respectively. By devising a “cP-RNA-seq” method that is able to exclusively amplify and sequence cP-containing RNAs, we identified the complete repertoire of 5′-SHOT-RNAs. Furthermore, 5′-SHOT-RNA, but not 3′-SHOT-RNA, has significant functional involvement in cell proliferation. These results have unveiled a novel tRNA-engaged pathway in tumorigenesis of hormone-dependent cancers and implicate SHOT-RNAs as potential candidates for biomarkers and therapeutic targets.
Transfer RNAs (tRNAs) are universally expressed in all three domains of life, and play a central role in protein synthesis as an adapter molecule translating codon triplet sequences into amino acids. Mature tRNAs are 70- to 90-nucleotides (nt) noncoding RNA molecules forming a cloverleaf secondary structure that further folds into an L-shaped tertiary structure. The human nuclear genome encodes over 500 tRNA genes (1), along with numerous genes of tRNA-lookalikes resembling nuclear and mitochondrial tRNAs (2). The multiple encoded sites and high stability place tRNAs among the most abundant RNA molecules in the cellular transcriptome. The abundance of tRNAs varies among different cell and tissue types, and altered tRNA abundance and heterogeneity have been implicated in gene-expression regulation and diseases (2–5).
Although tRNAs are best known as adapter molecules essential for translation, recent experimental and computational evidence has led to a previously unexpected conceptual consensus that tRNAs are not always end products but can further serve as a source of small functional RNAs. In many organisms, small RNAs are produced from mature tRNAs or their precursor transcripts not as random degradation products, but as functional molecules involved in many biological processes beyond translation (6–8). According to the proposed nomenclature (7), tRNA-derived small RNAs identified to date can be classified into two groups: tRNA halves and tRNA-derived fragments. tRNA halves are composed of 30- to 35-nt fragments derived from either the 5′- (5′-tRNA half) or 3′-part (3′-tRNA half) of mature tRNAs. Shorter than tRNA halves, tRNA-derived fragments range from 13 to 24 nt in length and are derived from 5′- or 3′-parts of mature tRNAs, or 5′-leader or 3′-trailer sequences of precursor tRNAs (pre-tRNAs).
The expressions of eukaryotic tRNA halves were shown to be triggered by a variety of stress stimuli, such as oxidative stress, heat/cold shock, and UV irradiation (9). Therefore, tRNA halves are also known as tRNA-derived stress-induced RNAs (tiRNAs) (10), although they are also detected under nonstressed conditions (8, 11). In mammalian cells, the angiogenin (ANG), a member of the RNase A superfamily, was found to be the enzyme that cleaves the anticodon loops of mature tRNAs to produce the tiRNAs (10, 12). Ribonuclease/angiogenin inhibitor 1 (RNH1), an ANG inhibitor, was shown to be a negative regulatory factor for ANG cleavage (10). As other regulatory factors, DNA methyltransferase 2 (Dnmt2) and NOP2/Sun RNA methyltransferase 2 (NSun2) modify many tRNAs to generate the 5-methylcytidine (m5C) modification, which protects tRNAs from stress-induced cleavage (13, 14).
Importantly, stress stimuli accumulate the tRNA halves not as nonfunctional degradation by-products but as functional molecules. Stress-induced 5′-tRNA halves promote the formation of stress granules (15) and also inhibit global translation by displacing translational initiation factor complexes from mRNAs (10, 16). Y-box-binding protein 1 (YB-1), a multifunctional DNA/RNA-binding protein, associates with 5′-tRNA halves and mediate translational inhibition and stress granule formation (16). A unique G-quadruplex structure of a 5′-tRNA half is crucial for translation inhibition by binding to YB-1 (17). Supporting important roles of tRNA halves in pathogenesis, the accumulation of tRNA halves triggers cellular stress responses and apoptosis in NSun2-mutated fibroblasts of patients from neurodevelopmental disorders or Nsun2-deficient mice (14).
Although an increasing number of reports have revealed that tRNA-derived small RNAs are involved in various biological processes beyond translation, information regarding their expression profiles is fragmented and the molecular basis behind their biogenesis and function remain elusive. Here we report a novel type of tRNA halves that are specifically and abundantly expressed in sex hormone-dependent breast and prostate cancers; these are produced by ANG-cleavage of fully aminoacylated mature tRNAs, which are dependent on the presence of sex hormones and their receptors. The complete repertoire of the tRNA halves was determined by developing a method that is able to exclusively amplify and sequence tRNA halves. Furthermore, the tRNA halves have significant functional involvement in cell proliferation, strongly suggesting that our study has unveiled a novel pathway that engages tRNA halves in the development and growth of sex hormone-dependent cancers.
Results
tRNA Halves Are Constitutively Expressed in BmN4 Cells.
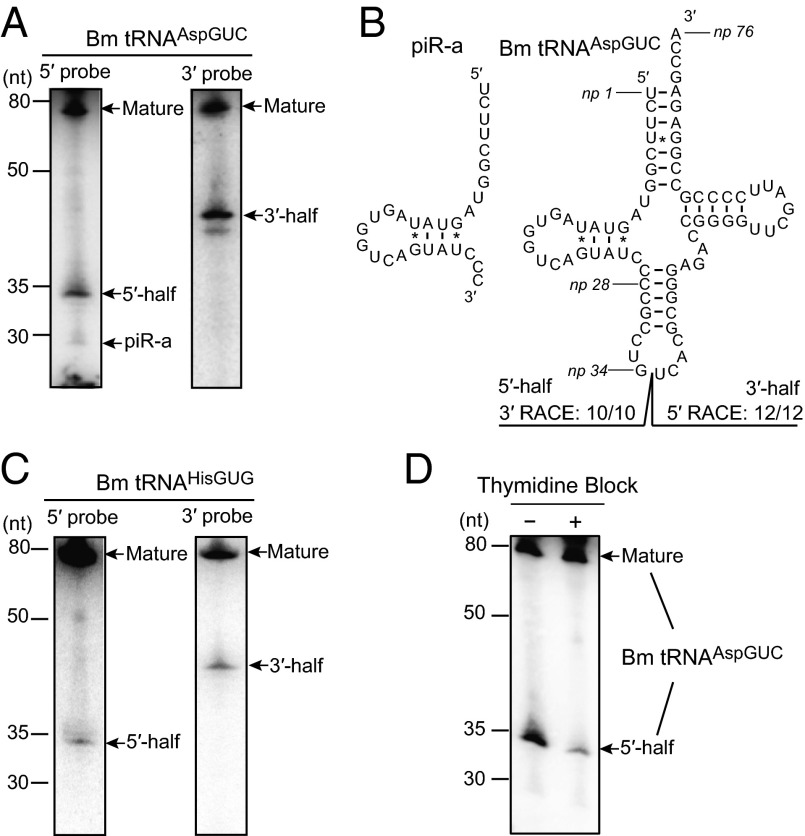
In the course of investigating PIWI-interacting RNAs (piRNAs), a germ-line–specific class of small regulatory RNAs (18), we serendipitously detected the expression of tRNA halves in BmN4 cells. BmN4 cells are Bombyx mori ovary-derived cultured germ cells that endogenously express piRNAs and their bound PIWI proteins; therefore, they present a unique model system for piRNA research (19, 20). When we examined the expression of piRNA-a, an abundant piRNA expressed in BmN4 cells (20) (DNA Databank of Japan no. DRA002562), a Northern blot probe complementary to the piRNA yielded bands of ∼75 and 35 nt that were much more abundant than the ∼28-nt piRNA band (Fig. 1A). Subsequent RACE analyses identified the ∼35-nt band as a 5′-half of a Bombyx cytoplasmic tRNAAspGUC ranging from the 5′-end to anticodon first nucleotide [nucleotide position (np) 1–34 according to the nucleotide numbering system of tRNAs (21)], whereas piRNA-a was found to be derived from np 1–28 of the tRNAAspGUC (Fig. 1B). In addition to the presence of the 5′-tRNAAspGUC half, expression of a 3′-half comprising np 35–76 of the tRNAAspGUC was also detected and identified by Northern blot and RACE analyses (Fig. 1 A and B).
Fig. 1.
tRNA halves were highly expressed in BmN4 cells. (A) BmN4 total RNA was subjected to Northern blot using a probe targeting the 5′- (5′-probe) or 3′-part (3′-probe) of Bombyx cytoplasmic tRNAAspGUC. Detected mature tRNA, piRNA-a, and 5′- and 3′-tRNA halves are indicated by arrows. (B) The cloverleaf secondary structure of Bombyx cytoplasmic tRNAAspGUC-V1 (SI Appendix, Fig. S1A) is shown. Nucleotide positions (np) are indicated according to the nucleotide numbering system of tRNAs (21). Sequences of the tRNAAspGUC halves were identified by RACE analyses. All 3′-RACE products (10 of 10) cloned from 5′-tRNA halves had their 3′-terminal positions at np 34, whereas all 5′-RACE products (12 of 12) cloned from 3′-tRNA halves had their 5′-terminal positions at np 35. piRNA-a was found to be derived from np 1–28 of tRNAAspGUC. (C) BmN4 total RNA was subjected to Northern blot targeting Bombyx cytoplasmic tRNAHisGUG. (D) BmN4 cells were subjected to thymidine block treatment and total RNA from the cells was subjected to Northern blot using a 5′-probe for tRNAAspGUC.
Exploration of the expression of tRNA halves from other randomly chosen Bombyx cytoplasmic tRNAs led to the detection of both the 5′- and 3′-halves derived from tRNAHisGUG (Fig. 1C), but failed to show clear signals for tRNA halves from tRNAGlyUCC, tRNAGlyGCC, and tRNASerCGA. These results suggested that only specific tRNA species produce abundant tRNA halves in BmN4 cells. The 3′-terminal position of the 5′-tRNAHisGUG half was identified to be anticodon first position (np 34) by RACE (SI Appendix, Fig. S2). RACE analysis failed to amplify the 3′-tRNAHisGUG half, which could be attributed to the presence of a posttranscriptional modification. tRNAHisGUG contains a guanine residue at np 37, which is often modified to 1-methyl-guanosine (m1G). The m1G modification inhibits reverse-transcription (22), which would result in the unsuccessful amplification of the 3′-tRNAHisGUG half by RACE. Interestingly, reduced levels of the 5′-tRNAAspGUC expression were observed in BmN4 cells whose cell cycle was arrested by double thymidine block (Fig. 1D), implying that the expressions of tRNA halves in BmN4 cells are correlated with cell proliferation.
tRNA Halves Are Constitutively Expressed in Breast Cancer Cells, but Not in Noncancerous Breast Epithelial Cells.
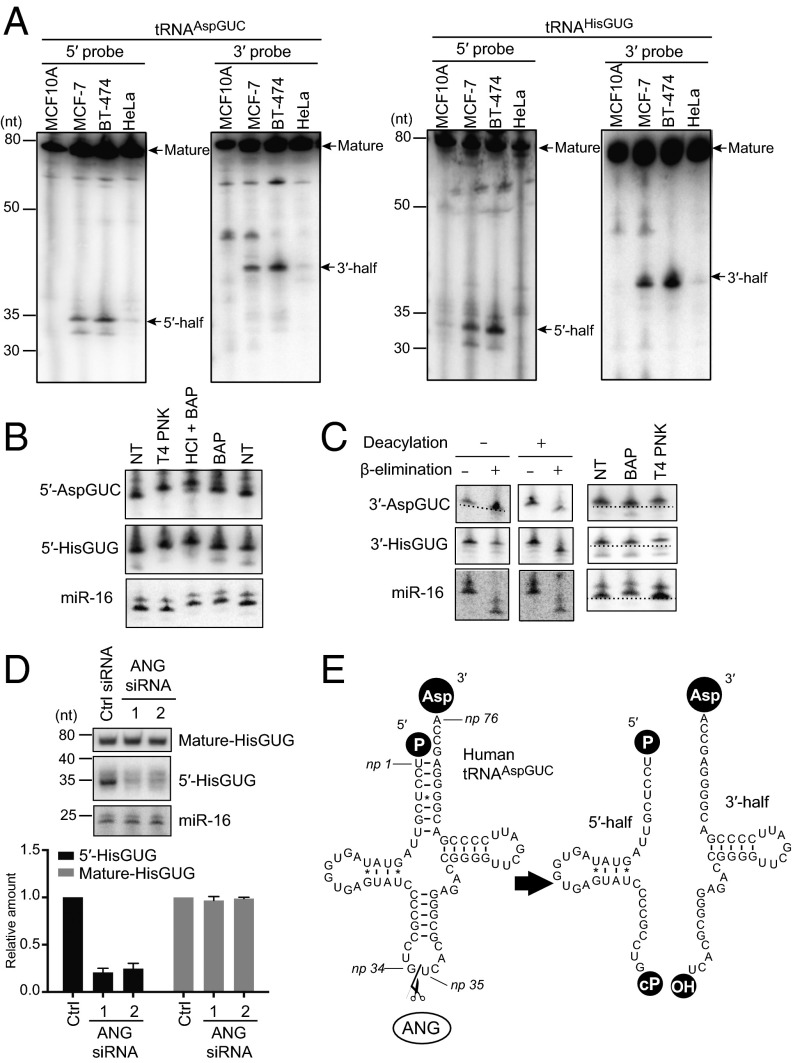
The observed potential connection between tRNA halves and cell proliferation prompted us to analyze the involvement of tRNA halves in tumorigenesis. Northern blots revealed that both 5′- and 3′-tRNA halves derived from human cytoplasmic tRNAAspGUC and tRNAHisGUG were abundantly expressed in MCF-7 and BT-474 human breast cancer cells (Fig. 2A), but only merely present in HeLa cells and absent in noncancerous breast epithelial MCF10A cells. RACE analysis using BT-474 total RNA showed that the 5′-tRNAAspGUC half and 5′-tRNAHisGUG half were commonly derived from np 1–34 of respective mature tRNAs, whereas a majority of the 3′-tRNAAspGUC halves were derived from np 35–76 (Fig. 2E and SI Appendix, Fig. S4). We were not able to amplify the 3′-tRNAHisGUG half, again most likely because of the np 37 m1G modification. The examined cells were cultured in medium optimized for the respective cell lines under appropriate conditions (e.g., appropriate cell confluence), and only specific cell lines showed the expression of tRNA halves. These results imply that the observed expression of tRNA halves was a constitutive rather than a stress-induced phenomenon as with tiRNA expression.
Fig. 2.
tRNA halves were abundantly present in human breast cancer cells. (A) Total RNA from the indicated cells was subjected to Northern blot using a 5′- or 3′-probe targeting human cytoplasmic tRNAAspGUC and tRNAHisGUG. Detected mature tRNAs and tRNA halves are indicated by arrows. (B) Terminal structure of 5′-tRNA halves in BT-474 cells were analyzed enzymatically. Total RNA was treated with T4 PNK, BAP, or acid followed by BAP treatment (HCl+BAP). NT designates nontreated sample used as a negative control. The treated total RNA was subjected to Northern blots targeting the 5′-tRNAAspGUC half, 5′-tRNAHisGUG half, and microRNA-16 (miR-16). miR-16 was investigated as a control RNA containing 5′-phosphate and 3′-hydroxyl terminal structures, which are thought to react with BAP but not with T4 PNK. Indeed, BAP treatment up-shifted the miR-16 band, whereas T4 PNK did not. (C) To analyze the terminal structure of 3′-tRNA halves, BT-474 total RNA was subjected to deacylation treatment, sodium periodate oxidation followed by β-elimination, BAP treatment, or T4 PNK treatment. The 3′-tRNAAspGUC half, 3′-tRNAHisGUG half, and miR-16 in the treated total RNA were analyzed by Northern blots. Because miR-16 contains 5′-phosphate and 3′-hydroxyl ends, β-elimination shortened miR-16 regardless of deacylation treatment. BAP treatment, but not T4 PNK treatment, up-shifted the miR-16 band by removing its 5′-phosphate. The PAGE for 3′-AspGUC detection in the deacylation minus lane leaned (entire picture of the PAGE is shown in SI Appendix, Fig. S5), and a dotted line indicates the extent of the leaning. (D) BT-474 cells were transfected with control siRNA or the two different siRNAs targeting ANG. (Upper) Total RNA was extracted and subjected to Northern blots for detection of mature tRNAHisGUG, 5′-tRNAHisGUG, and miR-16 (negative control). (Lower) The Northern blot bands were quantified and shown as relative abundance; amounts in control cells were set as 1. (E) Schematic description of the production of tRNA halves in breast cancer cells.
The 5′-tRNA Halves Contain a Phosphate at the 5′-End and a 2′,3′-Cyclic Phosphate at the 3′-End.
To analyze terminal structure of the 5′-tRNA halves, total RNA from BT-474 cells was treated with T4 polynucleotide kinase (T4 PNK) or bovine alkaline phosphatase (BAP), and then the mobility of the bands of 5′-tRNA halves in Northern blots was examined. T4 PNK treatment removes both a phosphate and a 2′,3′-cyclic phosphate from the 3′-end of RNAs, whereas BAP removes a phosphate from both the 5′- and 3′-ends of RNAs. Although BAP treatment alone cannot remove a cyclic phosphate from the 3′-end of RNAs, a combination treatment of acid and BAP can remove a cyclic phosphate, because the acid converts the cyclic phosphate to a phosphate. As shown in Fig. 2B, the bands of the 5′-tRNA halves were shifted up by BAP treatment, suggesting the presence of a phosphate in the 5′-tRNA halves. T4 PNK treatment similarly shifted the bands up, and further up-shifts were observed by the acid plus BAP treatments, indicating the additional presence of a 2′,3′-cyclic phosphate in the 5′-tRNA halves. These results indicate that 5′-tRNA halves contain a phosphate at the 5′-end and a 2′,3′-cyclic phosphate at the 3′-end.
The 3′-tRNA Halves Contain a Hydroxyl Group at the 5′-End and an Amino Acid at the 3′-End.
To analyze the terminal structures of the 3′-tRNA halves, BT-474 total RNA was subjected to deacylation treatment for removal of amino acids from aminoacylated tRNAs, followed by sodium periodate oxidation and β-elimination, which reacts with 3′-hydroxyl end and removes the 3′-terminal nucleotide. As shown in Fig. 2C, β-eliminated 3′-tRNA halves migrated faster than unreacted RNAs only after the RNAs were subjected to deacylation treatment. In other words, without deacylation treatment, β-elimination was unable to react with the 3′-end of the 3′-tRNA halves because of the presence of an amino acid. These results strongly suggest that 3′-tRNA halves are fully aminoacylated at the 3′-end. For the 5′-end of the 3′-tRNA halves, BAP treatment did not influence band positions, whereas T4 PNK treatment with ATP was able to phosphorylate the 5′-end, resulting in up-shifting of the bands (Fig. 2C). These results indicate the absence of a phosphate at the 5′-end. Taken together, 3′-tRNA halves contain a hydroxyl group at the 5′-end and an amino acid at the 3′-end.
ANG Produces the tRNA Halves in Breast Cancer Cells.
In mammalian cells, ANG cleaves the anticodon loops of tRNAs upon stress stimuli to produce tiRNAs (10, 12). To determine whether ANG is required for the production of tRNA halves in breast cancer, we performed RNAi knockdown of ANG expression in BT-474 cells. Both of the two different siRNAs designed to target the ANG gene were effective to decrease ANG mRNA levels to around 40% compared with control siRNAs (SI Appendix, Fig. S6A). Strikingly, transfection with each of the ANG-targeting siRNAs commonly resulted in the reduction of tRNA halves from tRNAAspGUC (SI Appendix, Fig. S6B) and tRNAHisGUG (Fig. 2D), whereas the levels of corresponding mature tRNAs and control miR-16 did not change. These results strongly suggest that the ANG catalyzes the anticodon cleavage of mature tRNAs to produce tRNA halves in breast cancer cells (Fig. 2E).
Quantification of Sex Hormone-Dependent TRNA-Derived RNAs by TaqMan Quantitative RT-PCR.
Because there was a clear difference in the abundance of tRNA halves between the breast cancer cell lines and HeLa (Fig. 2A), we reasoned that the expression profiles of tRNA halves differ among cancer cell lines. To perform wide screening of cancer cell lines for the tRNA half expressions, there was a need to develop a convenient and accurate method for quantification of tRNA halves. Although a PCR-based detection method is more efficient and sensitive than Northern blot, standard quantitative RT-PCR (qRT-PCR) amplification of the interior sequences of the tRNA halves is not able to distinguish signals among tRNA halves, mature tRNAs, and pre-tRNAs. To circumvent this issue, we developed a TaqMan qRT-PCR-based method that is able to exclusively quantify 5′- or 3′-tRNA halves (Fig. 3 A and B). Total RNA was first treated with T4 PNK and ATP to dephosphorylate the 3′-end of the 5′-tRNA halves, and at the same time to phosphorylate the 5′-end of the 3′-tRNA halves. Subsequently, a 3′- or 5′-RNA adapter was ligated to the 5′- or 3′-tRNA halves, respectively, followed by quantification of the ligation products by TaqMan qRT-PCR. The method successfully amplified and quantified the 5′-tRNAAspGUC half, 5′-tRNAHisGUG half (Fig. 3A), and 3′-tRNAAspGUC half (Fig. 3B), whereas no signal was detected from attempted amplification of the 3′-tRNAHisGUG half, which was most likely because np 37 m1G hindered reverse transcription. Because the TaqMan probes were designed to target the boundary of the tRNA halves and RNA adapters, only adapter-ligated tRNA halves, but not mature tRNAs and pre-tRNAs, were specifically quantified by this method. Indeed, a method without an RNA ligation step failed to produce detectable signals, and a method without T4 PNK treatment severely reduced the signals (Fig. 3 A and B). The abundance of tRNA halves was much greater in BT-474 than in HeLa cells, which is consistent with the Northern blot results (Fig. 2A); this confirmed the credibility of the method.
Fig. 3.
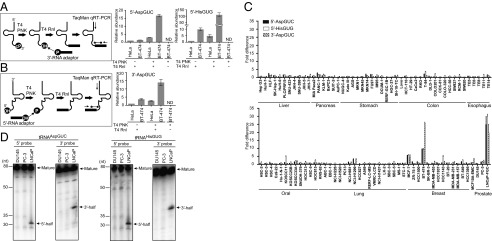
Screening of cancer cell lines for the expressions of tRNA halves. (A, Left) A schematic representation of 5′-tRNA half quantification. (Right) The 5′-tRNAAspGUC half and 5′-tRNAHisGUG half in BT-474 and HeLa total RNA were quantified using a method with or without T4 PNK or T4 RNA ligase (T4 Rnl) treatment. (B, Left) A schematic representation of 3′-tRNA half quantification. (Right) The 3′-tRNAAspGUC half was quantified by using the method described for BT-474 and HeLa total RNA. (C) Expression levels of the 5′-tRNAAspGUC half, 5′-tRNAHisGUG, and 3′-tRNAAspGUC half in the indicated cancer cell lines were examined. Expression levels in BT-20 cells were set as 1, and fold-changes are indicated. Averages of three independent experiments with SD values are shown. (D) Total RNA from the indicated prostate cancer cell lines was subjected to Northern blot for detection of tRNA halves derived from tRNAAspGUC and tRNAHisGUG. Detected mature tRNAs and tRNA halves are indicated by arrows.
Abundant Accumulation of tRNA Halves in Specific Breast and Prostate Cancer Cell Lines.
With this novel method, the expression of the three tRNA halves was profiled in 94 cell lines from liver, pancreas, stomach, colon, esophagus, oral, lung, breast, and prostate cancers, as well as in noncancerous breast epithelial MCF10A cells (Fig. 3C). Each cell line exhibited different expression patterns of the tRNA halves, suggesting that production of the tRNA halves was specifically regulated. The three cell lines, MCF-7 and BT-474 breast cancer cells and LNCaP-FGC prostate cancer cells, showed prominent expression of all three examined tRNA halves. None of the other examined cancer cell lines exhibited abundant expression levels of all three tRNA halves. As clearly detected in BT-474 and MCF-7 cells (Fig. 2A), both 5′- and 3′-tRNA halves from tRNAAspGUC and tRNAHisGUG were abundant in LNCaP-FGC cells by Northern blot, whereas the other two examined prostate cancer cells, DU145 and PC-3, did not show a detectable band for the tRNA halves (Fig. 3D). These results confirm the effectiveness of our screening method, and the particularly abundant expression of tRNA halves in specific breast and prostate cancer cell lines.
Expression of tRNA Halves Is Dependent upon Sex Hormones and Their Receptors.
What are the molecular factors regulating the expression levels of the tRNA halves? Notably, the expression levels of mature tRNAAspGUC and tRNAHisGUG, as determined by Northern blots (Figs. 2A and 3D), were not particularly high in the MCF-7, BT-474, or LNCaP-FGC cell lines compared with those in other cancer cell lines. Thus, the abundant expression of tRNA halves are not simply because of the enhanced transcription or stability of mature tRNAs.
One of the main characteristics that distinguish breast and prostate cancers from the other cancers screened in Fig. 3C is the crucial involvement of sex hormones and their receptors in caner development and progression. High-level exposures of estrogen are a major risk factor for breast cancer and ∼70–75% of breast cancers express estrogen receptor-α (ERα), which contributes to estrogen-dependent tumor growth (23). Among four breast cancer subtypes, such ER+ breast cancers are classified into luminal type A or B, whereas the other two subtypes comprise the human epidermal growth factor receptor 2 (HER2)-positive type, which is ER− but expresses HER2, and the triple-negative type, which is negative for ER, progesterone receptor, and HER2 (24). Similar to the involvement of estrogen and ERα in breast cancer, androgens [mainly testosterone and 5-α-dihydrotestosterone (DHT)] and the androgen receptor (AR) play crucial roles in the tumorigenesis and progression of prostate cancer (25). Interestingly, MCF-7 and BT-474 cells, which abundantly express tRNA halves, are both ER+ luminal-type breast cancer cell lines (26), whereas all examined ER− breast cancer cell lines (HER2+ type: SK-BR-3 and MDA-MB-453; and triple-negative type: HCC1937, HCC1143, BT-20, MDA-MB-231, MDA-MB-157, BT-549, and HCC1395) showed low levels of tRNA halves (Fig. 3C). Similarly, among the three examined prostate cancer cell lines, AR+ LNCaP-FGC cells express abundant tRNA halves, whereas AR− DU145 and PC-3 cells commonly had low levels of tRNA halves (Fig. 3 C and D). These correlations allowed us to hypothesize that sex hormone sensitivity and hormone receptor expression could be a driving force to trigger and regulate the production of tRNA halves in breast and prostate cancers.
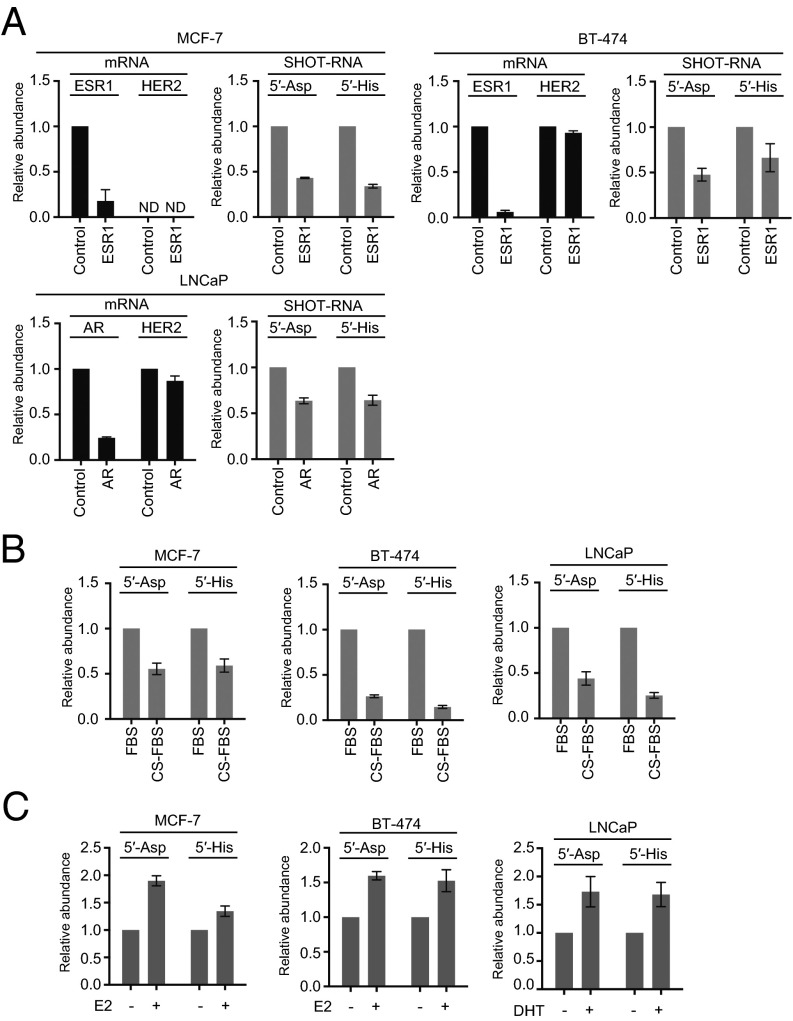
To test our hypothesis, we performed RNAi knockdown of ERα expression in MCF-7 and BT-474 breast cancer cells and of AR expression in LNCaP-FGC prostate cancer cells. siRNAs targeting ERα or AR successfully decreased levels of the target mRNA below 25%, but there was no effect on HER2 mRNA (Fig. 4A). Strikingly, in the all three cell lines, ER or AR knockdowns commonly reduced the amounts of 5′-tRNAAspGUC half and 5′-tRNAHisGUG half (Fig. 4A). To further confirm our hypothesis, we cultured the three cell lines with hormone-free medium as an alternative method to create hormone-insensitive states. As shown in Fig. 4B, the amounts of tRNA halves were decreased in all three cell lines cultured in hormone-free medium. Moreover, we added the corresponding sex hormones, estradiol, and DHT, to the medium culturing MCF-7 and BT-474 breast cancer cells and LNCaP-FGC prostate cancer cells, respectively, and observed that the addition of the hormones commonly increased the levels of tRNA halves in all three cell lines (Fig. 4C). These results clearly indicate the dependency of tRNA half expressions on sex hormones and their receptors. Therefore, we termed this novel type of tRNA halves as Sex HOrmone-dependent TRNA-derived RNAs (SHOT-RNAs). Hereafter, we will use 5′- or 3′-SHOT-RNAamino acid/anticodon sequence to denote SHOT-RNA derived from 5′- or 3′-halves of the tRNA.
Fig. 4.
Dependency of tRNA halves expression on sex hormones and their receptors. (A) MCF-7 and BT-474 cells were transfected with control siRNA or siRNA targeting the ESR1 gene encoding ERα, whereas LNCaP-FGC cells were treated with control siRNA or siRNA targeting the AR. After 72 h of transfection, expression levels of ESR1, AR, and HER2 (negative control) mRNAs and of the 5′-tRNAAspGUC half and 5′-tRNAHisGUG half were quantified. Expression levels of control siRNA-treated cells were set as 1. Each dataset represents the average of three independent experiments with bars showing the SD. (B) The indicated cells were cultured in medium containing normal FBS or hormone-free CS-FBS. After culturing for 120 h, the 5′-tRNAAspGUC half and 5′-tRNAHisGUG half were quantified. (C) The indicated cells were cultured in medium containing 17-β estradiol (E2) or DHT. After culturing for 48 h, the 5′-tRNAAspGUC half and 5′-tRNAHisGUG half were quantified.
Both ERα and AR act as transcription factors to regulate the transcription of many target genes upon binding of corresponding hormones (27, 28). However, mRNA expression levels of both ANG and RNH1 in LNCaP-FGC cells were not changed when cultured in hormone-free medium (SI Appendix, Fig. S7A), indicating that SHOT-RNA expression is not triggered by the direct regulation of ANG and RNH1 transcription by hormone receptors. In addition, we observed that RNH1 protein levels were unchanged after culturing in hormone-free medium (SI Appendix, Fig. S7B). These results suggest that SHOT-RNA generation may not be regulated by RNH1, in contrast to the case of tiRNAs, where the quantitative changes in RNH1 regulate expression (29).
Development of cP-RNA-seq Method to Specifically Identify Cyclic Phosphate-Containing RNA Species.
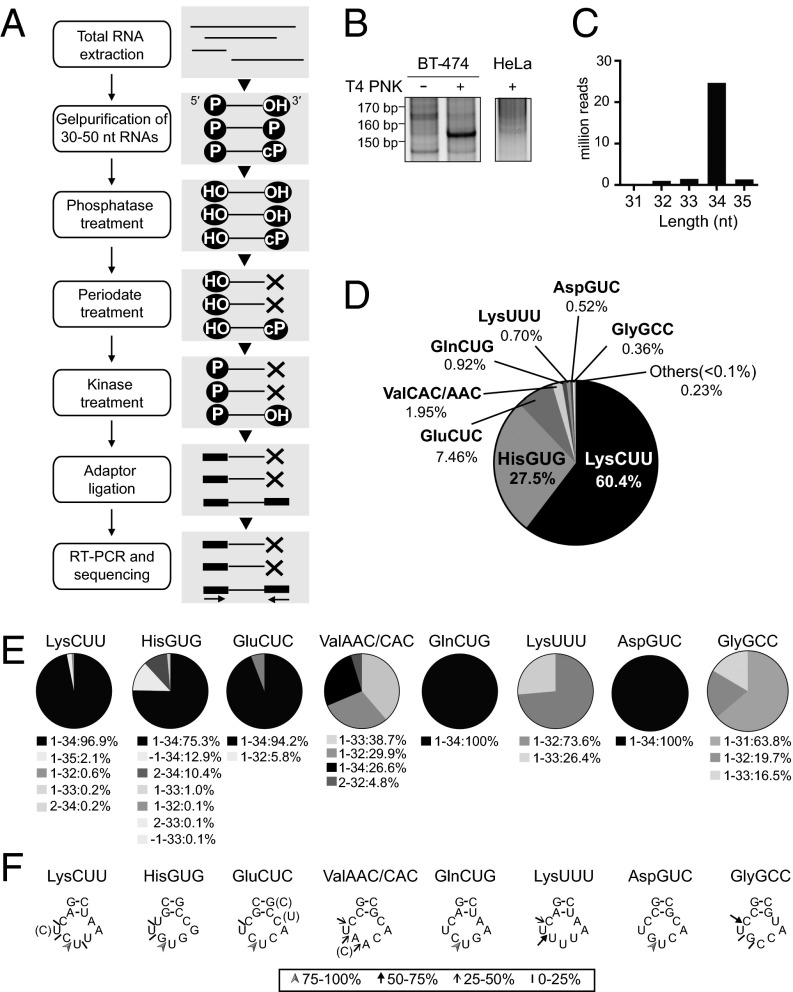
Next, we aimed to clarify the tRNA species that generate SHOT-RNAs. 5′- and 3′-SHOT-RNAs contain the 3′-terminal modifications, a 2′,3′-cyclic phosphate (cP) and an amino acid (Fig. 2 B and C), which would inhibit the adapter ligation step included in RNA sequencing methods. Therefore, RNA sequencing methods without procedures to remove such 3′-modifications cannot accurately capture SHOT-RNA expressions. To identify a comprehensive repertoire of 5′-SHOT-RNAs, we developed a method termed “cP-RNA-seq” that is able to exclusively amplify and sequence RNAs containing a 3′-terminal cyclic phosphate (Fig. 5A). In the cP-RNA-seq targeting 5′-SHOT-RNAs, 30- to 50-nt RNAs were first gel-purified from total RNA. Then, phosphatase treatment was used to remove a phosphate from both the 5′- and 3′-ends of the gel-purified RNAs, although the state of cyclic phosphate was not changed. Eventually, the RNA pools mainly contained two RNA subgroups, one with both 5′- and 3′-hydroxyl ends, and a second with 5′-hydroxyl and 3′-cyclic phosphate ends. Subsequent periodate treatment cleaved the cis-diol group of the 3′-hydroxyl end of the former group to generate 2′,3′-dialdehydes that no longer serve as a substrate for adapter ligation. The latter group survived periodate treatment because of the presence of a 3′-cyclic-phosphate. Therefore, after removal of cyclic phosphate by kinase treatment, the latter group became the only RNA group that bears a 3′-hydroxyl end for 3′-adapter ligation. Subsequent 5′-adapter ligation, RT-PCR amplification, and next-generation sequencing of the cDNAs thus exclusively identified the sequences of 3′-cyclic phosphate-containing RNAs.
Fig. 5.
Identification of 5′-SHOT-RNAs by cP-RNA-seq. (A) A flowchart of the cP-RNA-seq procedure. (B) Total RNA extracted from BT-474 and HeLa cells was applied to cP-RNA-seq method. The method amplified ∼153-bp cDNA products (5′-adapter, 55 bp; 3′-adapter, 63 bp; and therefore inserted sequences, ∼35 bp) from BT-474 cells. (C) Read-length distribution of the tRNA-mapped reads. (D) Pie charts showing the percentage of reads derived from respective cytoplasmic tRNA species. (E) Pie charts showing the mature tRNA regions from which the sequenced 5′-SHOT-RNAs were derived. (F) Based on the 3′-terminal sequences of 5′-SHOT-RNAs, the ANG cleavage sites in the anticodon loops were predicted. Arrowheads, thick arrows, thin arrows, and short strokes designates the 3′-terminal positions of each 5′-SHOT-RNA occupying 75–100%, 50–75%, 25–50%, and 10–25% of the reads, respectively.
Identification of a Comprehensive Repertoire of SHOT-RNAs.
To capture 5′-SHOT-RNAs, we applied the cP-RNA-seq method to total RNA isolated from BT-474 breast cancer cells. The method successfully amplified ∼153-bp bands (considering adapters’ lengths, inserted RNAs were estimated to be ∼35 nt) (Fig. 5B) whose dependency on T4 PNK treatment suggests that as expected the amplified bands were derived from cyclic-phosphate–containing RNAs. Consistent with the low levels of 5′-tRNA halves in HeLa cells detected by Northern blots (Fig. 2A), this method failed to yield clear bands from HeLa total RNA (Fig. 5B). Illumina sequencing of the amplified bands from BT-474 cells yielded ∼33 million reads that aligned to the human genome, of which around 28 million reads (∼85%) were mapped to tRNAs. Among the tRNA mapped reads, we focused the RNA species with >10,000 reads that in aggregate accounted for >98.9% of the reads. Of these, almost all sequences were unanimously derived from 5′-halves of mature tRNAs with lengths of 32–35 nt (Fig. 5 C and D), which validated the use of the cP-RNA-seq method to capture the repertoire of 5′-SHOT-RNAs.
Only eight cytoplasmic tRNA species were identified to be a source of 5′-SHOT-RNAs with >0.1% reads of the 5′-tRNA half sequences identified by cP-RNA-seq (Fig. 5D). 5′-SHOT-RNALysCUU and 5′-SHOT-RNAHisGUG were particularly enriched, which comprised 60.4% and 27.5% of the total reads, respectively. As with Northern blot (Fig. 2) and qRT-PCR analyses (Figs. 3 and 4), 5′-SHOT-RNAAspGUC was also detected by cP-RNA-seq. The majority of the 5′-SHOT-RNAs were derived from a region that started at np 1 (5′-end) and ended at np 32–34 in the anticodon loop of the mature tRNAs (Fig. 5E). The sequence results of 5′-SHOT-RNAAspGUC and 5′-SHOT-RNAHisGUG were consistent with RACE results (SI Appendix, Fig. S4B). The identified 3′-terminal position of the 5′-SHOT-RNAs exhibited focal patterns of ANG cleavage (Fig. 5F). As the RNase A superfamily has been reported to share substrate preferences for a pyrimidine (30, 31), major sites for ANG cleavage within the anticodon loop were located between cytidine and uridine or guanosine and uridine residues, implying that ANG tends to cleave the 5′-side of uridines in the anticodon loop.
The 5′-SHOT-RNAs Enhance Cell Proliferation.
Because the expression of tRNA halves was correlated to cell proliferation in BmN4 cells (Fig. 1C), we hypothesized that SHOT-RNAs are not just nonfunctionally accumulated but required as functional molecules for cell proliferation in hormone-dependent cancers. To test this hypothesis, we investigated the impact of SHOT-RNA absence on cell proliferation by performing siRNA knockdown of SHOT-RNAs. Because SHOT-RNAs share identical sequences with mature tRNAs or pre-tRNAs, there was a possibility that siRNAs targeting SHOT-RNAs affect mature tRNAs or pre-tRNAs as well as SHOT-RNAs, leading to confusing results. However, Lee et al. (32) reported the successful siRNA knockdown of 3′-trailer-derived tRNA fragments without affecting pre-tRNA levels. Moreover, Garcia-Silva et al. (33) succeeded in selectively staining tRNA fragments without obtaining signals from mature tRNAs by avoiding the denaturation step in their modified FISH method. Considering these reports and the fact that mature aminoacylated tRNAs form rigid L-shaped tertiary structures and tightly bind to an elongation factor in the cytoplasm, we assumed that siRNAs targeting SHOT-RNAs could preferably function toward SHOT-RNAs without greatly affecting mature tRNAs. Indeed, as described below, all of our siRNA transfections targeting SHOT-RNAs did not affect mature tRNA levels but specifically reduced the levels of targeted SHOT-RNAs.
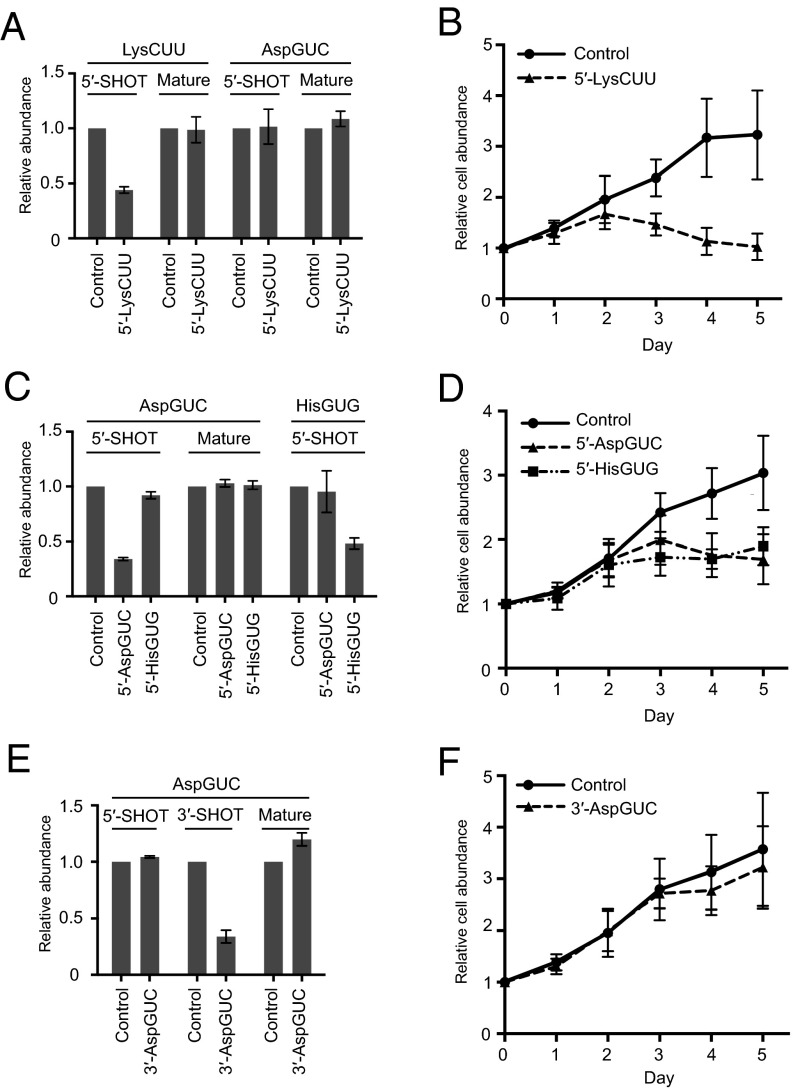
We first transfected siRNA targeting 5′-SHOT-RNALysCUU, the most abundantly sequenced 5′-SHOT-RNA, into LNCaP-FGC prostate cancer cells. Using both qRT-PCR (Fig. 6A) and Northern blot (SI Appendix, Fig. S8), we confirmed that the siRNA transfection specifically reduced the levels of 5′-SHOT-RNALysCUU. Upon 5′-SHOT-RNALysCUU reduction, strikingly, the cell growth rate was decreased compared with control siRNA-transfected cells (Fig. 6B). Because the levels of mature tRNA were not changed by siRNA transfection (Fig. 6A and SI Appendix, Fig. S8), inhibition of cell proliferation appeared to be solely attributable to the change in SHOT-RNA levels. 5′-SHOT-RNALysCUU depletion strongly impaired cell proliferation from 3 days after siRNA transfection (days 3, 4, and 5). Compared with the day of transfection (day 0), the amounts of cells transfected with control siRNA grew by ×3.17 ± 0.717 on day 4, whereas the number of 5′-SHOT-RNALysCUU–silenced cells was almost at the same level (×1.13 ± 0.253) on day 4.
Fig. 6.
Knockdown of 5′-SHOT-RNA inhibited cell proliferation. (A, C, and E) LNCaP-FGC cells were transfected with control siRNA or siRNA targeting the indicated SHOT-RNAs. After 72 h of transfection, expression levels of SHOT-RNAs were quantified by the method described in Fig. 3. Mature tRNA levels were quantified by FL-PCR (49). Expression levels from control siRNA-treated cells were set as 1. Each dataset represents the average of three independent experiments with bars showing the SD. (B, D, and F) The relative abundance of LNCaP-FGC cells at 1, 2, 3, 4, and 5 d after transfection of the indicated siRNAs were examined using AlamarBlue assay. Cell abundance on the day of transfection was set at 1. Each dataset represents the average of three independent experiments with bars showing the SD.
To explore the universality of the effect of SHOT-RNA reduction on cell growth, we further performed siRNA knockdown of 5′-SHOT-RNAAspGUC and 5′-SHOT-RNAHisGUG in LNCaP-FGC cells. As with the case of 5′-SHOT-RNALysCUU knockdown, respective siRNAs commonly reduced the levels of the targeted SHOT-RNA, whereas nontargeted SHOT-RNA and mature tRNA were not affected by the siRNA (Fig. 6C), validating siRNA knockdown as an effective method to reduce specific SHOT-RNAs without affecting the corresponding mature tRNAs. The depletion of each of 5′-SHOT-RNAAspGUC and 5′-SHOT-RNAHisGUG commonly impaired cell growth, resulting in increased cell numbers by only ×1.76 ± 0.320 and ×1.77 ± 0.145 cells on day 4 compared with day 0, whereas the numbers of control cells increased by ×2.71 ± 0.373 (Fig. 6D). These results indicate that 5′-SHOT-RNAAspGUC and 5′-SHOT-RNAHisGUG, as well as 5′-SHOT-RNALysCUU, are required for cell proliferation.
To examine differences between 5′- and 3′-SHOT-RNA, 3′-SHOT-RNAAspGUC was further silenced by siRNA transfection. As shown in Fig. 6E, siRNA specifically reduced the levels of targeted 3′-SHOT-RNAAspGUC, but not those of 5′-SHOT-RNAAspGUC or mature tRNAAspGUC, indicating that siRNA silencing of targeted SHOT-RNAs does not influence their counterpart SHOT-RNAs or corresponding mature tRNAs. In contrast to the case with 5′-SHOT-RNAs, 3′-SHOT-RNAAspGUC depletion failed to impair cell growth (Fig. 6F). Taken together, these results strongly suggest that SHOT-RNAs are expressed as functional molecules, and different species of 5′-SHOT-RNA universally play roles in cell proliferation, whereas a 3′-SHOT-RNA bears no impact.
Abundant Accumulation of SHOT-RNAs in ER+ Breast Cancer Patient Tissues.
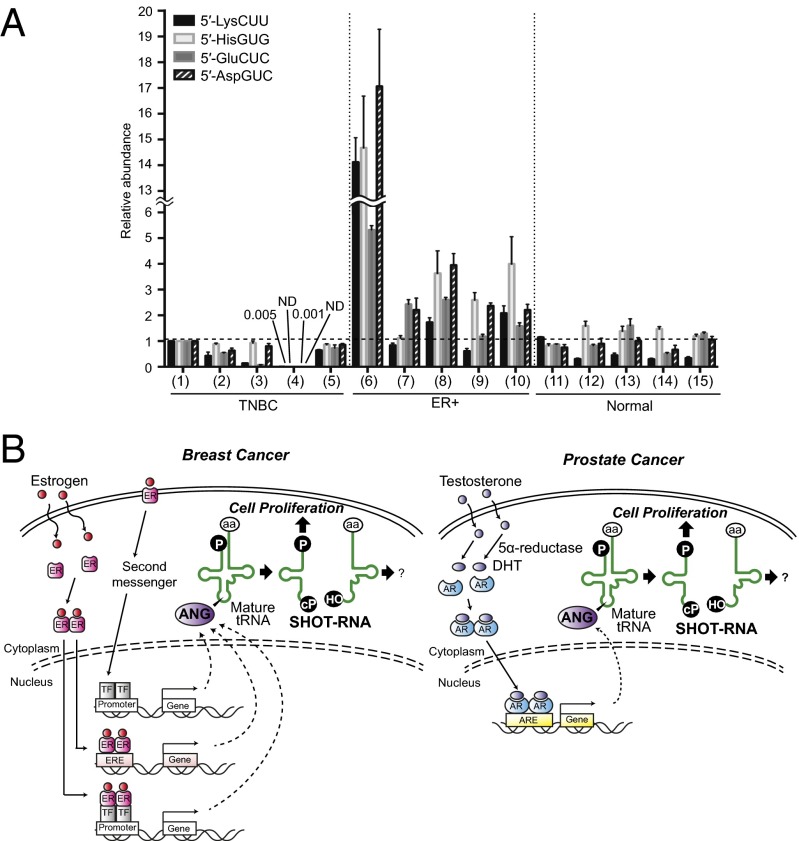
To examine SHOT-RNA expressions in actual patient tissues, total RNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue samples obtained from patients with ER+ luminal-type or triple-negative-type (ER−) breast cancers and from normal breast tissues. Expression of the four 5′-SHOT-RNAs (5′-SHOT-RNALysCUU, 5′-SHOT-RNAHisGUG, 5′-SHOT-RNAAspGUC, and 5′-SHOT-RNAGluCUC) in the total RNAs was then quantified using the TaqMan qRT-PCR (Fig. 7A). Although each tissue sample exhibited different expression patterns, the ER+ patient tissues in particular showed a prominent expression level of SHOT-RNAs, but none of the ER− patient tissues or normal breast tissues exhibited abundant expression levels of SHOT-RNAs. These results confirm the effectiveness and sensitivity of our method to quantify SHOT-RNAs in patient tissues, and indicate that the phenomenon of hormone-dependent SHOT-RNA accumulation is not limited to a cell culture system but it also occurs in actual cancer patient tissues.
Fig. 7.
SHOT-RNA expressions in breast cancer patient tissues. (A) Total RNA was extracted from FFPE tissues obtained from five ER+ breast cancer patients (ER+), five triple-negative breast cancer patients (TNBC), and five normal breast tissues (Normal), and subjected to TaqMan qRT-PCR quantification (shown in Fig. 3) of the indicated SHOT-RNAs. Expression levels of sample 1 (TNBC) were set as 1. Each dataset represents the average of three independent experiments with bars showing the SD. (B) A proposed model for SHOT-RNA involvement in sex hormone-dependent breast and prostate cancers.
Discussion
Here we report the identification of SHOT-RNAs as a novel class of small functional RNAs whose expression is dependent on sex hormones and corresponding receptors. Hormone dependency was demonstrated by: (i) SHOT-RNAs were prominently and constitutively enriched only in ER+ breast cancer and AR+ prostate cancer cell lines, whereas ER− breast cancer, AR− prostate cancer, and all examined cancer cell lines from other tissues expressed low levels of tRNA halves; (ii) SHOT-RNAs were prominently enriched only in tissues from ER+ breast cancer patients but not in those from triple-negative patients and in normal breast tissues; (iii) depletion of sex hormone receptors, ERα and AR, commonly reduced SHOT-RNA levels; (iv) culturing cells in hormone-free medium decreased SHOT-RNA levels; and (v) addition of the sex hormones to the cell culture medium increased SHOT-RNA levels. We propose the model that the sex hormone-signaling pathways activate ANG cleavage of aminoacylated mature tRNAs, and the resultant accumulation of SHOT-RNAs contributes to cell proliferation and thereby may promote tumorigenesis and tumor growth (Fig. 7B).
The finding of SHOT-RNAs was motivated by the serendipitous discovery of tRNA halves in B. mori ovary-derived BmN4 cells. Because BmN4 cells were cultured in phenol red- and FBS-free medium, it appears that the expression of tRNA halves in BmN4 cells is independent of hormones and their receptors. In addition, no Bombyx homolog of human ANG is found in the silkworm genome (SilkBase: silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi). Therefore, the biogenesis mechanisms and their regulation of tRNA halves in BmN4 cells are different from those of SHOT-RNAs in cancers.
Although SHOT-RNAs and tiRNAs share an identical biogenesis factor (10, 12), they are also distinct RNAs. First, SHOT-RNAs are constitutively expressed in specific sex hormone-dependent cancer cells, whereas tiRNA expression is triggered by stress stimuli, which is a widely conserved phenomenon in various cells. Second, it’s likely that RNH1 is not involved in SHOT-RNA production, but the reduced levels of RNH1 contribute to tiRNA accumulation (10, 34). Third, tRNA species that generate SHOT-RNAs appear to be different from those producing tiRNAs. tiRNAs are widely produced from various tRNAs such as tRNAAla, tRNACys, and tRNASer (10, 15, 16, 34), but SHOT-RNAs derived from these tRNAs were not identified in our analyses. Fourth, although 5′- and 3′-SHOT-RNAs were expressed with similar quantities, tiRNAs have been reported to be asymmetrically expressed with a much greater abundance of 5′-tiRNAs compared with their 3′-counterpart (12, 14, 35). Therefore, our findings have revealed a novel tRNA-engaged pathway in sex hormone-dependent cancers.
Sex hormones and their receptors play crucial roles in the genesis and progression of breast and prostate cancers (23, 25). Sustained exposure to estrogen is well-known promoter of breast cancer onset and progression, and at least 70% of breast cancers are classified as ER+ luminal type, in which development and growth are dependent on the estrogen-activated ERα (36). In prostate cancer, androgen plays a crucial role in tumorigenesis by promoting AR-mediated gene-expression regulation (25). The high expression specificity of SHOT-RNAs implies their potential use as a novel biomarker in sex hormone-dependent cancers. Actually, the specific and abundant SHOT-RNA accumulation was observed in ER+ breast cancer patient tissues. Further studies are required to clarify the association of SHOT-RNA expression with various prognostic factors. Examining the presence of SHOT-RNAs in exosomes (37) may also be interesting for the purpose of developing a biomarker for noninvasive testing.
SHOT-RNAs are generated by ANG cleavage of the anticodon loop of aminoacylated cytoplasmic tRNAs, resulting in the accumulation of 5′-SHOT-RNAs containing a phosphate at the 5′-end and a 2′,3′-cyclic phosphate at the 3′-end, and of 3′-SHOT-RNAs containing a 5′-hydroxyl group at the 5′-end and an amino acid at the 3′-end. It is notable that ANG leaves a cyclic phosphate, but bovine pancreatic RNase A, the best-studied ribonuclease, leaves a phosphate by way of an intermediate cyclic phosphate state (38). The presence of a 3′-terminal cyclic phosphate and amino acid in SHOT-RNAs suggest that the SHOT-RNAs will not be accurately captured by normal RNA sequencing methods because the 3′-terminal modifications hinder the adapter ligation steps. To fully sequence SHOT-RNAs by RNA-seq, total RNA should be subjected to deacylation and T4 PNK treatment in advance to remove the 3′-terminal cyclic phosphate and amino acid.
By devising the cP-RNA-seq method, we succeeded in exclusively amplifying and identifying the complete 5′-SHOT-RNAs repertoire. Only specific tRNAs become a source for SHOT-RNAs. The members of the RNase A superfamily share a substrate preference for a pyrimidine, such that ANG has been reported to cleave the 5′-side of cytidine and uridine (30). In the case of SHOT-RNA production, ANG cleavage mainly occurs between cytidine/guanosine and uridines. However, because many noncleaved cytoplasmic tRNAs also contain cytidine and uridine in their anticodon loops, the sequence preference of ANG may not be the sole reason for the selective production of SHOT-RNAs from specific tRNA sources. It is noteworthy that more than 96% of the SHOT-RNA sequence reads were derived from the tRNAs with corresponding amino acids that were positively or negatively charged, such as lysine, histidine, glutamate, and aspartate. Considering that SHOT-RNAs are produced from fully aminoacylated mature tRNAs, the properties of amino acid species may be involved in ANG recognition of tRNAs. Further studies, such as in vitro ANG cleavage analyses of mature tRNAs, are required to fully elucidate the regulation of ANG substrate recognition.
How are the SHOT-RNA expressions promoted by sex hormones and their receptors? In breast cancer, estrogen-bound ERαs form dimers that activate transcription of target genes by directly binding to estrogen response elements or by interacting with other transcription factors (28) (Fig. 7B). In addition, as a noncanonical pathway, extracellular estrogen binds to the plasma membrane ERs, which initiates signal cascades via second messengers and leads to the activation of other transcription factors. In prostate cancer, testosterone is converted to DHT by 5α-reductase, and the DHT-bound ARs form dimers and bind to androgen response elements, inducing the transcription of target genes (Fig. 7B) (27). These transcriptional activation systems of target genes by sex hormone-signaling pathways may result in the promotion of ANG cleavage, which most likely occurs in the cytoplasm because only mature aminoacylated cytoplasmic tRNAs are targeted by ANG. Although our results showed no quantitative change in ANG mRNA with changing hormone status, ANG protein levels or localization may be affected. Although our results showed no quantitative changes in RNH1 mRNA and protein levels with changing hormone status, other ANG cofactors may be involved with regulation of ANG activity. Alternatively, the altered status of tRNA m5C modification, which can be induced by changes in levels of Dnmt2 and NSun2, may regulate SHOT-RNA expression. Further studies are required to determine how the ligand-activated hormone receptor pathway is connected to enhanced ANG cleavage for SHOT-RNA productions. On the other hand, ER+ luminal A is the most heterogenous subtype in breast cancer (39), and two of the ER+ breast cancer cell lines (ZR-75-1 and HCC1500) did not show abundant SHOT-RNA expression. Our results from breast cancer patients also showed that some ER+ samples did not show abundant levels of all of the examined SHOT-RNAs, implying that the sex hormone pathway is necessary, but not a sole sufficient condition, and other unidentified cellular factors may be involved in the expressed repertoire and quantity of SHOT-RNAs.
Intriguingly, specific knockdowns of three different 5′-SHOT-RNA species, but not a 3′-SHOT-RNA, commonly resulted in severe impairment of cell proliferation, indicating the enhancer roles of SHOT-RNAs in cell proliferation. The important questions remain regarding the mechanism behind the enhancement and why only 5′-SHOT-RNA appeared to have a functional role in cell proliferation. The asymmetrical results of this study show peculiar concordance with those of tiRNA studies reporting that 5′-tiRNAs, but not their 3′-conterparts, play active role in stress granule promotion and translational repression (10, 15, 16). Investigations and comparisons of subcellular localization patterns and interacting proteins of 5′- and 3′-SHOT-RNAs may reveal further insights to answer these questions. Because of their roles in acceleration of cell proliferation, SHOT-RNAs may play important functions in the development and growth of sex hormone-dependent cancers. Although patients with these diseases receive endocrine therapy to suppress hormone receptor activity or hormone exposure to prevent tumor growth (40, 41), many patients encounter de novo or acquired resistance and require more aggressive treatments, such as chemotherapy (42, 43). Further studies may lead to the use of SHOT-RNAs as potential target candidates for future therapeutic applications in sex hormone-dependent breast and prostate cancers.
Materials and Methods
tRNA Sequences.
Sequences of Bombyx and human tRNAs were identified using the tRNAscan-SE program (1, 44). As shown in SI Appendix, Figs. S1, S3, and S9, the tRNA sequences were sorted by mismatches and aligned using DNADynamo software (BlueTractor Software).
Cell Culture.
BmN4 cells were cultured as previously described (20). To arrest the cell cycle, double thymidine block was performed by incubating BmN4 cells with 2.5 mM thymidine for 16 h, then releasing the cells by washing with PBS and incubating for 9 h, and again incubating with 2.5 mM thymidine for 16 h. The MCF-7 cell line was cultured in MEM medium (Life Technologies) containing 10% (vol/vol) FBS, 1 mM sodium pyruvate, 1× nonessential amino acids solution (Life Technologies), and 10 μg/mL insulin (Sigma). HeLa and BT-474 cells were cultured in DMEM (Life Technologies) containing 10% FBS. DU145, PC-3, and LNCaP-FGC cells were cultured in RPMI1640 medium (Life Technologies) containing 10% FBS. Human cancer cell lines used for SHOT-RNA screenings (Fig. 3C) were obtained from the American Type Culture Collection, Japanese Collection of Research Bioresources, Riken Cell Bank, and Korean Cell Line Bank, and cultured in appropriate media and conditions.
RACE Identification of Terminal Sequences of tRNA Halves.
The sequences of adapters and primers for RACE analysis are shown in SI Appendix, Table S1. The fraction of 30- to 50-nt RNAs were gel-purified from the total RNA of BmN4, HeLa, and BT-474 cells. For 5′-RACE to analyze the 5′-terminal sequences of the 3′-tRNAAspGUC half, purified RNAs were treated with T4 PNK (New England Biolabs) and 100 µM ATP to phosphorylate the 5′-end of the 3′-tRNA half. Then, the RNAs were subjected to a ligation reaction with a 5′-RNA adapter using T4 RNA ligase (T4 Rnl; Fisher Scientific). The ligated RNAs were subjected to RT-PCR using the One Step SYBR Ex Taq qRT-PCR Kit (Clontech). For 3′-RACE to analyze the 3′-terminal sequences of 5′-tRNA halves, RNAs were treated with T4 PNK to remove the 3′-terminal cyclic phosphate of the 5′-tRNA halves. Then, the RNAs were subjected to a ligation reaction with a 3′-RNA adapter, followed by RT-PCR. The PCR products were cloned using the StrataClone PCR cloning kit (Agilent Technologies).
RNA Isolation, Enzymatic Treatment, and β-Elimination.
RNA isolation, enzymatic treatment, and β-elimination were performed as previously described (20, 45). In brief, total RNA was isolated using TRIsure reagent (Bioline). To investigate the terminal structures of SHOT-RNAs, total RNA was treated with BAP (Takara), T4 PNK (New England Biolabs, with ATP), or acid (incubation in 10 mM HCl at 4 °C for 3 h). For deacylation treatment, total RNA was incubated in 20 mM Tris⋅HCl (pH 9.0) at 37 °C for 40 min. For β-elimination reaction, total RNA was first incubated with 10 mM NaIO4 at 0 °C for 40 min in the dark. Then, 1 M rhamnose (1/10 volume) was added to quench unreacted NaIO4 and incubated at 0 °C for 30 min. Subsequently, β-elimination was performed by adding an equal volume of 2 M Lys-HCl (pH 8.5) and incubating at 45 °C for 90 min.
Northern Blot.
Northern blot analysis was performed as described previously (20). The sequences of probes are shown in SI Appendix, Table S2. The visualization and quantification were performed with storage phosphor autoradiography using Typhoon-9400 and ImageQuant v5.2 (GE Healthcare).
RNAi Knockdown of ANG.
For RNAi knockdown of ANG, two siRNAs were designed using the siExplorer algorithm (46) and synthesized by Bioland Scientific. The siRNA sequences are shown in SI Appendix, Table S3. ON-TARGETplus Nontargeting siRNA #2 (#D-001810–02; Dharmacon) was used as a negative control. BT-474 cells were transfected with 50 nM of each siRNA using the RNAi/MAX kit (Life Technologies). After 72 h of the transfection, the cells were subjected to a second transfection reaction with 100 nM of siRNAs, followed by another 72-h incubation, and then harvested. The efficiency of the RNAi knockdown was verified by real-time RT-PCR quantification of ANG mRNA using SsoFast EvaGreen Supermix (Bio-Rad), which were normalized by GAPDH expression levels. The sequences of the primers for real-time RT-PCR are shown in SI Appendix, Table S4.
Quantification of SHOT-RNAs by TaqMan qRT-PCR.
The sequences of the adapters and primers for SHOT-RNA quantification by TaqMan qRT-PCR are shown in SI Appendix, Table S5. To dephosphorylate the 3′-end of 5′-SHOT-RNAs, and at the same time to phosphorylate the 5′-end of 3′-SHOT-RNAs, 1 µg of total RNA was treated with T4 PNK and 100 µM ATP. For quantification of 5′-SHOT-RNAs, 100 ng of the treated RNAs were subjected to a ligation reaction (10-µL reaction mixture) with 20 pmol of a 3′-RNA adaptor using T4 Rnl. Subsequently, 1 ng of ligated RNA was subjected to TaqMan qRT-PCR (10-µL reaction mixture) using the One Step PrimeScript RT-PCR Kit (Clontech), 400 nM of a TaqMan probe targeting the boundary of the SHOT-RNA and 3′-RNA adaptor, and 2 pmol each of specific forward and reverse primers. With StepOne Plus Real-time PCR machine (Applied Biosystems), the reaction mixture was incubated at 42 °C for 5 min and then 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The quantified SHOT-RNA levels were normalized to U6 snRNA expression levels quantified by SsoFast EvaGreen Supermix (Bio-Rad). For quantification of 3′-SHOT-RNAAspGUC, T4 PNK-treated RNAs were subjected to a ligation reaction with a 5′-RNA adapter, followed by TaqMan qRT-PCR.
RNAi Knockdown of Hormone Receptors and Cell Cultures with Hormone-Free or Hormone-Added Medium.
For RNAi knockdown of the ESR1 gene that encodes ERα, SMARTpool: ON-TARGETplus ESR1 siRNA (#L-003401; Dharmacon) was used, and the ON-TARGETplus Nontargeting Control Pool (#D-001810; Dharmacon) was used as a negative control. For RNAi knockdown of AR, Individual: ON-TARGETplus AR siRNA (#J-003400–06; Dharmacon) was used, and ON-TARGETplus Nontargeting siRNA #2 (#D-001810–02; Dharmacon) was used as a negative control. MCF-7, BT-474, and LNCaP-FGC cells were transfected with 50 nM of each siRNA by a reverse-transfection method using the RNAiMAX kit according to the manufacturer’s instructions, and the cells were harvested 72 h after the transfection. The efficiency of RNAi knockdown was verified by real-time qRT-PCR using the primers shown in SI Appendix, Table S4. The expression levels were normalized to GAPDH levels. As an alternative method to create a hormone-insensitive state, the cells were cultured in phenol red-free medium containing 10% charcoal stripped FBS (CS-FBS; Life Technologies) for 120 h. With respect to hormone addition experiments, MCF7 and BT-474 cells were treated for 48 h with 10 nM and 100 nM of 17β-estradiol (Sigma), respectively, and LNCaP-FGC cells were treated for 48 h with 10 nM of DHT (Sigma). RNH1 protein levels were examined by a Western blot using anti-RNH1 antibody (Proteintech). β-Tubulin was examined as an internal control by using anti–β-tubulin (Developmental Studies Hybridoma Bank).
cP-RNA-Seq to Selectively Amplify and Sequence 5′-SHOT-RNAs.
As shown in Fig. 5A, 35- to 50-nt RNAs were gel-purified from BT-474 total RNA. Then, the purified RNAs were treated by calf intestinal alkaline phosphatase (New England Biolabs). After phenol-chloroform purification, the RNAs were oxidized by incubation in 10 mM NaIO4 at 0 °C for 40 min in the dark, followed by ethanol precipitation. The RNAs were then treated with T4 PNK. After phenol–chloroform purification, directional ligation of adapters, cDNA generation, and PCR amplification were performed using the TruSeq Small RNA Sample Prep kit (Illumina) according to the manufacturer’s protocol. The amplified cDNAs were sequenced using Illumina hiSeq2000 system at the Functional Genomics Core at the University of Pennsylvania.
Bioinformatics Analyses of 5′-SHOT-RNAs.
The analyzed 5′-SHOT-RNA sample contains 50,220,564 raw reads and can be found publically in the Gene Expression Omnibus Database (GEO accession no. SRX1060214). Before mapping we used the cutadapt tool (journal.embnet.org/index.php/embnetjournal/article/pre-tRNAsview/200) to remove 3′-adapter sequence: 5′-TGGAATTCTCGGGTGCCAAGG-3′. We used SHRiMP2 (47) for all read mappings and did not allow for any insertions or deletions when mapping. We used a set of 632 tRNA-reference genes comprised of 610 nuclear tRNAs from the GRCh37 (hg19) human genome assembly [listed in gtRNAdb (1)] and 22 known mitochondrial tRNAs from tRNAdb (48). The 610 entries from gtRNAdb include 508 true tRNAs and 102 psuedo-tRNAs. We purposely excluded selenocysteine tRNA, tRNAs with undetermined anticodon identity, and tRNAs mapping to contigs that are not part of the major chromosome assembly. To help validate the cP-RNA-seq method by confirming that most of the reads landed on tRNAs and to account for the repetitive nature of tRNA sequences, we mapped the reads to the full-genome allowing a 4% mismatch rate and allowing nonunique mappings. Approximately 33 million reads mapped, of which ∼28 million (∼85%) intersected with at least 1 of the 632 tRNA-reference genes.
RNAi Knockdown of SHOT-RNAs.
siRNA targeting each SHOT-RNA was designed using siExplorer (46) and synthesized by Bioland Scientific. The siRNA sequences are shown in SI Appendix, Table S3. The ON-TARGETplus Nontargeting siRNA #2 (#D-001810-02; Dharmacon) was used as a negative control. The LNCaP-FGC cells were transfected with each siRNA (10 nM for 5′-SHOT-RNALysCUU and 3′-SHOT-RNAAspGUC, 20 nM for 5′-SHOT-RNAAspGUC and 5′-SHOT-RNAHisGUG) using the reverse-transfection method with the RNAiMAX kit, according to the manufacturer’s protocol. The efficiency of RNAi knockdown was verified by quantification of SHOT-RNAs 72 h after transfection. The expression levels were normalized to U6 snRNA levels. Mature tRNALysCUU and tRNAAspGUC were quantified by four-leaf clover qRT-PCR (FL-PCR) (49). The sequences of adapters and primers for FL-PCR are shown in SI Appendix, Table S6. Briefly, total RNA was subjected to deacylation treatment, followed by ligation reaction with a DNA/RNA-hybrid stem-loop adapter using T4 RNA ligase 2 (New England Biolabs) that specifically ligates the adapter to mature tRNA. The ligation products were specifically amplified and quantified by TaqMan qRT-PCR.
Cell Proliferation Assay.
LNCaP-FGC cells were transfected with the SHOT-RNA-targeted siRNAs and plated in a 96-well cell culture plate. Cell proliferation and viability were determined with AlamarBlue Cell Viability Reagent (Life Technologies). At 24, 48, 72, 96, and 120 h after transfection, AlamarBlue reagent was added to the medium and the cells were incubated for another 6 h. Absorbance was measured at 570 and 600 nm and relative cell abundance was calculated using the manufacturer’s instructions.
SHOT-RNA Quantification Using Breast Cancer Patient FFPE Samples.
Total RNA was extracted from 1 to 3 10-μm-thick FFPE sections using the RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies) according to the manufacturer’s protocol. A total RNA amount of 100 ng was treated with T4 PNK and 100 µM ATP, following which 50 ng of the treated RNAs was subjected to a ligation reaction (10-µL reaction mixture) with 20 pmol of a 3′-RNA adaptor using T4 Rnl. Subsequently, 500 pg of ligated RNA was subjected to TaqMan qRT-PCR as described above for SHOT-RNA detection using cell lines. 5S rRNA expression levels were quantified by SsoFast EvaGreen Supermix (Bio-Rad) and used for normalization.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for helpful discussions. This study was supported in part by National Institutes of Health Grant GM106047 (to Y.K.); a W. M. Keck Foundation grant (to I.R.); institutional funds (Y.K. and I.R.); a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (26293304) (to I.I.); and a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (to S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. SRX1060214).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510077112/-/DCSupplemental.
References
- 1.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telonis AG, Loher P, Kirino Y, Rigoutsos I. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front Genet. 2014;5:344. doi: 10.3389/fgene.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2(12):e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Ishimura R, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345(6195):455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobala A, Hutvagner G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2(6):853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu M, Honda S, Kirino Y. Transfer RNA as a source of small functional RNA. J Mol Biol Mol Imag. 2014;1(2):1–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138(2):215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schutz K, Hesselberth JR, Fields S. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA. 2010;16(3):621–631. doi: 10.1261/rna.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu H, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco S, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov P, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA. 2014;11(51):18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 19.Kawaoka S, et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15(7):1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda S, et al. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 2013;19(10):1405–1418. doi: 10.1261/rna.040428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26(1):148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7(2):237–247. doi: 10.4161/rna.7.2.11468. [DOI] [PubMed] [Google Scholar]
- 23.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354(3):270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 24.Network CGA. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 2011;10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riaz M, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013;15(2):R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.da Silva HB, et al. Dissecting major signaling pathways throughout the development of prostate cancer. Prostate Cancer. 2013;2013:920612. doi: 10.1155/2013/920612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heldring N, et al. Estrogen receptors: How do they signal and what are their targets. Physiol Rev. 2007;87(3):905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 29.Pizzo E, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126(Pt 18):4308–4319. doi: 10.1242/jcs.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorrentino S. Human extracellular ribonucleases: Multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell Mol Life Sci. 1998;54(8):785–794. doi: 10.1007/s000180050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybak SM, Vallee BL. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry. 1988;27(7):2288–2294. doi: 10.1021/bi00407a007. [DOI] [PubMed] [Google Scholar]
- 32.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Silva MR, et al. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol. 2010;171(2):64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Saikia M, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287(51):42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhahbi JM, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 37.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JE, Venegas FD, Raines RT. Energetics of catalysis by ribonucleases: Fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry. 1994;33(23):7408–7414. doi: 10.1021/bi00189a047. [DOI] [PubMed] [Google Scholar]
- 39.Ciriello G, et al. The molecular diversity of luminal A breast tumors. Breast Cancer Res Treat. 2013;141(3):409–420. doi: 10.1007/s10549-013-2699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole MP, Jones CT, Todd ID. A new anti-oestrogenic agent in late breast cancer. An early clinical appraisal of ICI46474. Br J Cancer. 1971;25(2):270–275. doi: 10.1038/bjc.1971.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huggins C. Endocrine-induced regression of cancers. Cancer Res. 1967;27(11):1925–1930. [PubMed] [Google Scholar]
- 42.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 43.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 44.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14(4):347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 46.Katoh T, Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 2007;35(4):e27. doi: 10.1093/nar/gkl1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: Sensitive yet practical SHort Read Mapping. Bioinformatics. 2011;27(7):1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 48.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honda S, Shigematsu M, Morichika K, Telonis AG, Kirino Y. Four-leaf clover qRT-PCR: A convenient method for selective quantification of mature tRNA. RNA Biol. 2015;12(5):501–508. doi: 10.1080/15476286.2015.1031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.