Inherited retinal dystrophies (IRD) are a heterogeneous group of blinding diseases that affect more than 200,000 Americans and millions worldwide (1). Identification of genes that are responsible for IRD when defective is of great importance to the basic understanding as well as development of efficient gene diagnosis and treatment. Thus far, 272 genetic loci are linked to various forms of IRD; among these, 232 protein-coding genes have been identified to cause IRD when defective (RetNet, https://sph.uth.edu/Retnet, last updated April 29, 2015). microRNAs (miRNAs) are endogenous, small, noncoding, regulatory RNAs (2). Since their discovery in 1993 (3, 4), miRNAs have proven to play important roles in the fine-tuning of gene expression at posttranscriptional levels (2). In spite of their importance in gene-expression regulation, few mutations of individual miRNAs have been found to cause Mendelian inherited diseases in humans, raising doubts about their importance in normal development and functions, and whether mutations in a miRNA can have significant functional impact to cause Mendelian inherited diseases, including IRD. Recently, a point mutation in the seed sequence of miR-184 was reported to cause familial keratoconus with cataract (5) and an anterior-segment dysplasia syndrome with endothelial dystrophy, iris hypoplasia, congenital cataract, and stromal thinning (6), suggesting that miR-184 is required for anterior-segment development and function, and proving that mutations in an individual miRNA can cause inherited ocular disease in humans. In mice, Lumayag et al. demonstrated that inactivation of the miR-183/96/182 cluster gene causes syndromic IRD with multisensory defects (7), suggesting that defects in miRNA genes can cause IRD in mammals. However, until now, no mutations in miRNAs have been found to cause IRD in humans. In PNAS, Conte et al. report a dominant mutation in miR-204 that causes IRD associated with ocular coloboma in a large five-generation family (8). This paper is the first report to show that a point mutation in an individual miRNA results in significant functional consequence and causes IRD in humans, and the third miRNA in which mutations cause Mendelian inherited disorders in humans, after miR-96, which causes nonsyndromic hearing loss (9, 10), and miR-184 (5, 6). Conte et al’s report (8) provides a new molecular mechanism of IRD and underscores the importance of miRNA in IRD.
miR-204 resides in intron 8 of the transient receptor potential (TRP) channel gene, TRPM3, on human chromosome (chr)9q21.12. Through a classic parametric linkage analysis, Conte et al. zoomed in on four regions segregating with the phenotypes, including 9q21 (8). Through exon sequencing, extensive bioinformatic studies to eliminate nondisease-causing variants using multiple publically available databases, including the Exon Variant Server of the National Heart, Lung and Blood Institute Exome Sequencing Project, dbSNP, and the 1000 genomes project, and segregation studies, Conte et al. found that a point mutation in miR-204, n.37C > T, cosegregates with the disease but is not present in all control databases and 21 sporadic cases of retinal dystrophy with microphthalmia anophthalmia and coloboma (MAC), 457 patients with isolated MAC, and 672 patients with autosomal dominant IRD, suggesting that n.37C > T is the disease-causing mutation in this family (8). Publically available control databases have become invaluable sources for evaluating the nature of a mutation and excluding nondisease-causing genomic variants. The Conte et al. report exemplifies an effective approach for identification of disease-causing mutations in a postgenomic era.
In the family studied by Conte et al. (8), all affected individuals had progressive retinal dystrophy, vision loss, and coloboma; multiple affected individuals underwent bilateral cataract surgery at early ages, suggesting lens defects; and the proband also showed iridolenticular adhesions and scattered retinal pigmented epithelia (RPE) mottling. Mutation in miR-204 as the cause of the disease is consistent with the observation that miR-204 is preferentially expressed in multiple ocular tissues, including the retina, RPE, ciliary body, and lens (11–13). In addition, previous in vitro and in vivo studies in medaka fish suggested its important roles in the development and function of the retina (13, 14), lens (13–15), and RPE (12). Knockdown of miR-204 in fish resulted in microphthalmia, abnormal lens formation, altered dorso-ventral (D-V) patterning of the retina, and coloboma (13).
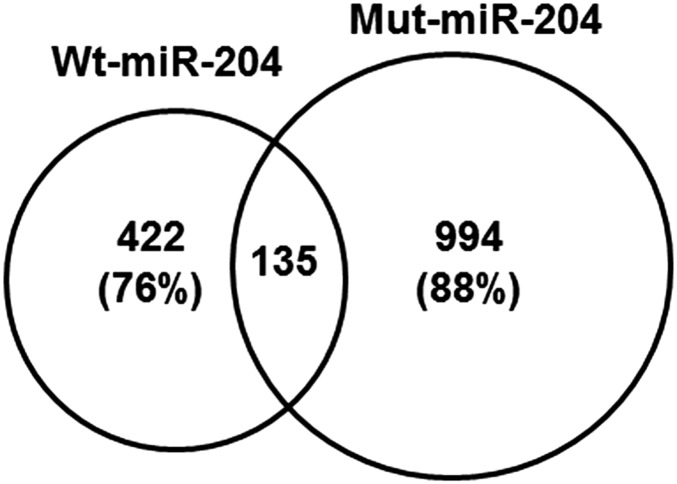
Mutations in miRNAs either disrupt their biogenesis and, therefore, expression levels of the mature miRNAs, and/or change their downstream target genes (5, 6, 9, 10). The seed sequence of a miRNA is the major determinant of its downstream target genes and, therefore, functions of the miRNA (16). The mutation identified in this family, n.37C > T, is located at the fourth residue of the seed sequence of miR-204. Conte et al. demonstrated that this mutation has no significant impact on the secondary structure of pre-miR-204 and biogenesis of miR-204 (8). However, target prediction analysis revealed a loss of 442 predicted targets of wild-type miR-204 (wt-miR-204) and a gain of 994 new predicted target genes of the mutant miR-204 (mut-miR-204) (Fig. 1) (8), in which genes involved in eye development, synaptic function, cell adhesion, axon guidance, D-V axis formation, and cellular response to stress are significantly enriched.
Fig. 1.
n.37C > T mutation in miR-204 significantly changed its predicted target genes. Venn diagram summarizing the number of overlapping and unique predicted target genes of wt-miR-204 and mut-miR-204.
Through overexpression of wt-miR-204 and mut-miR-204 in a human RPE cell line and gene-expression profiling, Conte et al. showed that n.37C > T mutation resulted in significant changes in expression of target genes of miR-204. Furthermore, overexpression of the mut-miR-204 in fish resulted in phenotypes similar to the ones observed in the affected individuals in the family, including microphthalmia, coloboma, retinal degeneration, and vision loss. The resemblance to fish with mut-miR-204 overexpression is more than to fish with either overexpression (8) or knockdown of wt-miR-204 (13), suggesting that gain-of-function of mut-miR-204 is the major mechanism underlying the defects observed in the affected individuals of the family, although loss-of-function also may contribute to the disease. This conclusion is further supported by the fact that deletion encompassing the TRPM3 gene have not been reported to cause ocular diseases (8). Thus, this is the first example of a miRNA mutation causing an inherited disease in human with gain-of-function as the main mechanism.
Intriguingly, Conte et al. (8) demonstrated that overexpression and knockdown of wt-miR-204 and overexpression of mut-miR-204 in fish resulted in overlapping but different phenotypes. This complexity reflects a characteristic nature of miRNA function—one miRNA can have different functions in different tissues at different development stages. Further studies are needed to unravel detailed mechanisms of the different effects of the wt-miR-204 and mut-miR-204 in different cell types of the retina and other ocular tissues.
Overall, Conte et al. (8) provided convincing evidence that the n.37C > T mutation in miR-204 is responsible for the IRD and iris coloboma in this family, providing the first evidence that mutation in an individual miRNA can cause IRD and other inherited ocular disorders. Although the n.37C > T appears to be a rare mutation, miR-204 should be included in mutation screening for IRD in future studies, because other mutations in miR-204 affecting its biogenesis and target spectra may cause similar or different forms of inherited ocular diseases, as numerous examples have been shown that different mutations in the same protein-coding gene can cause different forms of IRD (1).
The paucity of Mendelian inherited diseases caused by miRNA mutations is attributed, in part, to their small sizes (8). However,
Conte et al. report a dominant mutation in miR-204 that causes IRD associated with ocular coloboma in a large five-generation family.
disease-causing mutations in miRNAs may have been significantly underestimated, as miRNAs are one of noncoding elements of the genome and have been largely neglected in mutation screenings in the past. With the principle proven by the Conte et al. report (8), it is reasonable to propose that mutations in other miRNAs that are enriched in the retina and other ocular tissues could also cause various forms of IRD and other inherited ocular disorders in humans (17).
miRNAs are generally considered as fine-tuners of gene expression, providing quantitative modulation of their downstream genes (2). Mature miRNAs function by base-pairing with target sites in the 3′UTRs of the transcripts of their downstream target genes inducing breakdown and/or translation inhibition of the transcripts. Mutations in target sites of downstream genes could interrupt the miRNA regulatory network and cause or contribute to disease (18, 19). Therefore, mutation screening in the 3′UTRs of protein-coding genes that are known to cause IRD when defective and that carry target sites for miR-204 and other retina-enriched miRNAs is warranted in searching for new disease-causing mutations. More broadly, genetic variants in miRNAs and their target sites in protein-coding genes may quantitatively modulate disease-related genetic networks and contribute to the pathogenesis of multifactorial ocular diseases, including age-related macular degeneration, glaucoma, and diabetic retinopathy.
Footnotes
The author declares no conflict of interest.
See companion article on page E3236 in issue 25 of volume 112.
References
- 1.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84(2):132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 5.Hughes AE, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol Vis Sci. 2012;53(1):348–353. doi: 10.1167/iovs.11-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci USA. 2013;110(6):E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte I, et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci USA. 2015;112(25):E3236–E3245. doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mencía A, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 10.Soldà G, et al. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21(3):577–585. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235(9):2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 12.Wang FE, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24(5):1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte I, et al. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107(35):15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conte I, et al. The combination of transcriptomics and informatics identifies pathways targeted by miR-204 during neurogenesis and axon guidance. Nucleic Acids Res. 2014;42(12):7793–7806. doi: 10.1093/nar/gku498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaham O, et al. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9(3):e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28(2):87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Abelson JF, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 19.Simon D, et al. A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum Mol Genet. 2010;19(10):2015–2027. doi: 10.1093/hmg/ddq083. [DOI] [PubMed] [Google Scholar]