Abstract
The three species Neisseria meningitidis, Neisseria gonorrheae, and Neisseria lactamica are often regarded as highly recombining bacteria. N. meningitidis has been considered a paradigmatic case of the “semiclonal model” or of “epidemic clonality,” demonstrating occasional bouts of clonal propagation in an otherwise recombining species. In this model, occasional clonality generates linkage disequilibrium in the short term. In the long run, however, the effects of clonality are countered by recombination. We show that many data are at odds with this proposal and that N. meningitidis fits the criteria that we have proposed for predominant clonal evolution (PCE). We point out that (i) the proposed way to distinguish epidemic clonality from PCE may be faulty and (ii) the evidence of deep phylogenies by microarrays and whole-genome sequencing is at odds with the predictions of the semiclonal model. Last, we revisit the species status of N. meningitidis, N. gonorrheae, and N. lactamica in the light of the PCE model.
Keywords: deep phylogeny, linkage disequilibrium, near-clade, molecular epidemiology, predominant clonal evolution
The clonality/sexuality controversy in microbiology has gone on for more than 35 y in bacteria (1, 2) as well as in parasitic protozoa (3–6). Early on, several instances of bacterial and parasitic species were removed from the clonal paradigm and instead were considered as “highly recombining” (5). The controversy now can be reconsidered in the light of major results obtained with modern technologies, such as whole-genome sequencing (WGS), single nucleotide polymorphism analysis (SNP), microarrays, and megacomputing, which have made possible considerable advances in evolutionary biology, population genetics, and molecular epidemiology. We reconsider the issue of the population structure of Neisseria meningitidis in the light of these advances.
N. meningitidis and the “Epidemic Clonality” and “Semiclonal” Models
The molecular epidemiology and population genetics of N. meningitidis, a major agent of meningitis, has received much attention. Thousands of strains have been characterized by various markers, including multilocus enzyme electrophoresis (MLEE), multilocus sequence analysis (MLST), random primed polymorphic DNA (RAPD), and WGS and SNP analysis (reviewed in ref. 7). Pioneering MLEE studies showed that, although this pathogen has a drastically restricted ecological niche, infecting only humans, it exhibits considerable genetic diversity, suggesting that N. meningitidis undergoes extensive genetic recombination. However, the natural populations of this species consistently exhibit a strong linkage disequilibrium (LD), i.e., nonrandom association between genotypes at different loci (8). To reconcile these two observations, which at first appear mutually inconsistent, Maynard Smith et al. (5) proposed the epidemic clonality model, which states that the species under study undergoes occasional bouts of clonal propagation in an otherwise recombining population structure. “Epidemic” clones, which are greatly favored by both positive and purging selection, are repeatedly represented in natural populations as repeated multilocus genotypes (MLGs), which bias LD tests, and obscure the general pattern of predominant recombination. Therefore, to distinguish epidemic clonality from “true” clonality, it has been proposed that the repeated MLGs be counted only once (5). The disappearance of LD with such a counting method would be evidence that epidemic clonality is the cause of the LD. The semiclonal model (9) is similar: It states that recombination is obscured by epidemic clonality in the short term but deletes the effects of clonality in the long term. The population of N. meningitidis would appear as a highly diverse and freely recombining mixture of different genotypes, on which the epidemic clones are superimposed, as predicted by the epidemic model (10).
The Predominant Clonal Evolution Model
We have proposed that “predominant clonal evolution” (PCE) be defined as “strongly restrained genetic recombination” (11). This definition is used widely (5, 11), including by many authors studying N. meningitidis (12–14), although it is not accepted by all scientists working on pathogen population genetics (11). Restrained recombination generates both a strong (i.e, statistically significant) LD and discrete, stable genetic subdivisions that can be clouded by occasional recombination/hybridization (near-clades). In contrast to the epidemic clonality/semiclonal model, PCE in the long run overcomes the effects of recombination, and the near-clades, which are distinct from cryptic, biological species (5), tend to separate from each other more and more. Within near-clades, PCE occurs and generates LD and lesser near-clades (the “Russian doll” pattern) (15). Near-clades and Russian doll patterns are evidenced by (i) surveys over the whole ecogeographical range of the species, retrospective studies, and the analysis of ancient collections, which make it possible to ascertain their strong stability in space and time, and (ii) flexible phylogenetic analyses based on the “congruence principle” (16), that is, as more reliable data are taken into account, the phylogenetic signal increases. This evidence shows that the species under study has passed a “clonality threshold.” beyond which the effects of recombination are increasingly overcome and divergence among the near-clades becomes irreversible. Data based on the congruence principle can include (i) adding a higher number of gene fragments in MLST studies (phylogenies based on individual genes may show some discrepancies, but the phylogenetic signal increases when more genes are considered); (ii) comparisons of trees obtained from different software or phylogenetic trees obtained with nonphylogenetic methods to see whether these different approaches give convergent results; (iii) comparisons of data obtained from different genetic markers; and (iv) the use of approaches of great resolution power (WGS and SNPs) rather than methods with lower resolution (MLST). (It should be noted that the information obtained from WGS is several orders of magnitude greater than that obtained with MLST.)
Last, unlike the semiclonal/epidemic clonality model, the PCE model proposes that restrained recombination is explained mainly by the built-in biological properties of the organism under consideration rather than by natural selection (although selection obviously impacts the pathogen’s population structure). PCE comes about as the result of evolutionary strategies used by pathogens to avoid the “recombination load” [i.e., the disruption of favorable MLGs (17)]. The PCE concept includes not only mitotic reproduction (“strict clonality”) (11) but also selfing/inbreeding (18), strong homogamy, several kinds of parthenogenesis (19), and “unisex strategies” (20).
Some Problems with the Semiclonal/Epidemic Clonality Model
Can Natural Selection Be the Main Process Accounting for Apparent Clonality?
The maintenance of strong LD and of widespread, worldwide distributed clones and near-clades by natural selection (by host immune defense, among other possible causes) in an otherwise freely recombining species would imply that most possible MLGs are eliminated in every generation. Such widespread elimination could be the case for one or a few loci. However, the number of ad hoc selection explanations increases geometrically with the number of loci considered and would amount to a considerable genetic load (21, 22). This enormous genetic load is why the maintenance of multilocus associations by natural selection, for example by immune selection (23), is questionable (24). It is worth noting that the population structure of Leishmania infantum, a parasitic protozoan, does not change in immunocompromised patients compared with immunocompetent ones (25). Another problem with the selection hypothesis is that it hardly accounts for the consistent maintenance of the same genetic clones and near-clades throughout the world, in highly diversified ecological and host conditions.
Bias in the Epidemic Clonality Approach.
Separating epidemic clonality from true clonality by counting each repeated MLG only once relies on the working hypothesis that such repeated MLGs can be equated with epidemic clones (i.e., clones of recent origin). Such an approach is highly dependent on the level of resolution and molecular clock of the marker used. In the approach proposed in ref. 5, the MLGs are electrophoretic types (ETs), i.e, MLEE MLGs. MLEE has a rather slow molecular clock. Therefore, the most recent common ancestor (MRCA) of each MLG could be very ancient. In the parasite L. infantum, the widespread MLEE MLG MON 1 (MON stands for the nomenclature established by the laboratory of parasitology in Montpellier, France) reveals high microsatellite diversity within it (26). Each microsatellite MLG probably corresponds better to a more recent clone than MON 1, which could have a very ancient MRCA. The same situation obtains with N. meningitidis: The antigen sequence–typing MLGs (finetypes) show a much greater diversity than the ETs (14). Therefore, individual finetypes or MLGS characterized by any other marker of high-resolution power should correspond much better than the ETs to recent clones and should be used for the analysis proposed in ref. 5. It might well be the case that ETs and “sequence types” (STs, i.e., MLST MLGs) might not correspond to recent clones and might have an ancient MRCA. In such cases, the approach proposed in ref. 5 would be strongly biased, with the drawback of considerably lowering sample sizes and hence the power of the statistical tests of LD, leading to a risk of statistical type II error (22). It has been argued that, because this bacterium is highly recombining, STs in N. meningitidis must have a very recent origin (10). This could be the case. However, the recombination rate in N. meningitidis is the very question under concern.
N. meningitidis Data: Which Model Fits Better?
As we have proposed (11), the population genetics data of N. meningitidis fit the PCE model very well. This observation does not mean that this bacterium does not undergo frequent recombination or that the strength of PCE is the same as in other species such as Staphylococcus aureus (10). However, the data support the view that N. meningitidis is beyond the clonality threshold.
Some results are consistent with both semiclonality/epidemic clonality and PCE, although they fit the latter better. Other data definitely are at odds with the former.
LD and Widespread MLGs.
A logical consequence of LD is the spread of ubiquitous, stable MLGs that are overrepresented under panmictic expectations (3, 4, 11, 21, 22, 27). Both LD and widespread MLGs are very frequently observed in N. meningitidis.
Strains of N. meningitidis are classified by serological typing based on antigenic properties of the capsular polysaccharide, which identifies the serogroup (7). Remarkably, N. meningitidis exhibits a strong association (LD) between antigen diversity/capsular serogroups and MLGs evidenced by MLEE/MLST (7, 14, 28–30). This association has been taken as evidence that recombination is frequent in this species, because the core genome and the dispensable genome (which drives antigen diversity) would not have had time to diverge (10). However, this strong LD between genotype and phenotype also can be taken as clear evidence for clonality (11, 27) and is one of the most relevant predictions of the PCE model. LD between genetic markers is strong in all N. meningitidis populations surveyed. It has been verified in 84 N. meningitidis serogroup A strains by MLEE, MLST, and RAPD (31). It is worth noting that (i) MLEE and MLST data are not redundant in this study, because different loci were analyzed for these two markers, and (ii) this LD cannot be attributed to epidemic clonality, even with the criteria presented in ref. 5, because this study considered only strains with different genotypes. There also is a strong parity between MLST diversity on one hand and the data revealed by microarrays (29, 32) and WGS (12) on the other hand. In contrast to the sample surveyed in ref. 31, these data concern strains from diversified serogroups, not only from the serogroup A.
Widespread, Overrepresented MLGs.
Many examples of N. meningitidis MLGs that spread over continents and persist for decades are available (13, 14, 28, 32). For example, ET 19 has been sampled from 1967– 1983 in 14 different countries and several continents, and ET 48 has been recorded from 1969–1989 in eight different countries (30). Earlier studies (8) make it possible to confirm the endurance of N. meningitidis MLGs.
These results, although they perfectly fit the PCE expectations, are in general compatible with semi/epidemic clonality. However, the data presented in ref. 31 are not. Moreover, as noted above, if natural selection is mainly responsible for the spread and persistence of these clonal genotypes in an otherwise quasi-panmictic species (5), it would be surprising that natural selection would favor the same MLGs in such different ecosystems, places, clinical contexts, and host environments. Last, the permanency of some MLGs (30) obviously poses the question of what is an epidemic, “ephemeral” clone.
Near-Clades, Russian Dolls, and Deep Phylogenies.
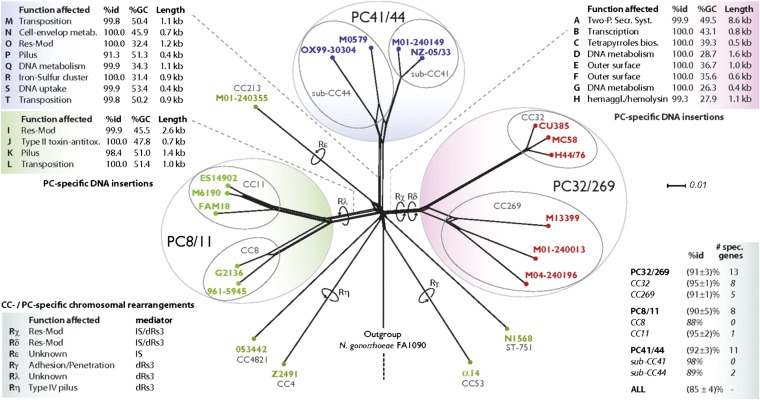
Near-clades correspond to imperfectly separated phylogenetic lines among which occasional recombination/hybridization may occur (11). We have shown that structures that fit the near-clade concept are observed in many pathogen species, including viruses, bacteria, parasitic protozoa, and fungi (11). The durable and widespread genetic clusters (clonal complexes—CCs—evidenced by MLST) recorded in N. meningitidis can be equated to near-clades (11). The term clade, although it is used in the case of N. meningitidis (12, 33), should be dismissed, since recombination is supposedly frequent. N. meningitidis near-clades are corroborated by MLST and MLEE (34). They are linked to pathogenicity and serogroups (7). They can persist for much longer than decades, since the ST1 complex (i.e., MLEE subgroup I/II) has been recorded in Russia and China from 1930–2007, and the ST4 complex (i.e., MLEE subgroup IV) has been observed in Gambia from 1917–1992 (7). Extended MLST with 20 loci corroborates the CCs evidenced with the classical MLST relying on seven loci (35). The CC structure is unexpected from extensive recombination, even in carriage strains (9), despite the classical notion that carriage strains are highly diverse, whereas pathogenic strains are characterized by the spread of hyperinvasive clones (7). The persistence of CC structure is confirmed by long-term MLEE studies dealing with a high number of both carriage and pathogenic strains and several different serotypes. The MLEE subgroups (i.e., near-clades) are not linked to either geographical distance or time. They are associated with antigenic reactivity, by monoclonal antibodies, to conserved pilin epitopes and class 1 outer membrane protein (30). A strong population structure revealing the existence of two major groups has been uncovered by microarrays and has been confirmed by two independent studies dealing with different places, different times, and different strain samplings. Both studies comprise carriage as well as pathogenic strains and include several serogroups. The two major groups corroborate the CCs and are linked to serogroups. The first study (32) deals with strains collected from 1962–2002 in several different countries. The second (29) concerns 13 pathogenic strains and 16 carriage strains of six different serogroups, collected from 1983–2002 in Gambia, Germany, the United Kingdom, and the United States. Both studies uncover the same population structure, namely eight “genome groups” distributed into two main groups, with a fine correspondence with CC diversity. In 84 serogroup A strains, two genetic groups (near-clades) are corroborated by MLEE, RAPD, and MLST. These results cannot be explained by a Wahlund effect (separation by either space or time or both) (31). Last, a WGS analysis dealing with 22 strains pertaining to five serogroups collected in five different continents has revealed the existence of three distinct “phylogenetic clades” (i.e., near-clades, each comprising several CCs) (Fig. 1) (12).
Fig. 1.
The three main phylogenetic clades revealed by WGS in N. meningitidis (12).
The results just reviewed show several examples of Russian doll patterns, i.e., PCE features both at the level of the whole species and also within each of the near-clades that subdivide it (15). The two near-clades corroborated by MLEE, RAPD, and MLST in 84 serogroup A strains (31) each exhibit a strong LD evidenced in particular by a high correlation between the genetic distances inferred from the three genetic markers. This condition shows that these two near-clades each undergo PCE and hence cannot be equated with cryptic biological species. In the two microarray studies (29, 32), the two main genetic groups are further distributed into eight lesser genome groups. This sample corresponds to 22 different CCs that correspond to the genome groups. This pattern corresponds to two main near-clades with eight smaller near-clades and 22 even smaller near-clades. Last, the three phylogenetic groups revealed by WGS corroborate the CCs, and each of these phylogenetic groups comprises several CCs (12). As for LD and the existence of widespread genotypes, some of these features are still compatible with the semi/epidemic clonality model, which predicts that some CCs may be widespread and persist for decades. However, (i) the persistence of some near-clades since 1917 and 1930 (7) far exceeds this duration; (ii) again, it would not be expected that the survival of the same CCs would be maintained mainly by natural selection for so long and in so many different environments; (iii) the two near-clades revealed within 83 serogroup A strains (31) do not fit the epidemic clonality model, because a strong LD persists within each of them, although no repeated genotypes are recorded in this sample. More importantly, the two microarray studies (29, 32), which are not limited to the serogroup A and which include both carriage and pathogenic strains, do evidence a strong and persistent population structure. Such a structure based on so many different markers strongly suggests the existence of deep phylogenies. This finding is definitely corroborated by the WGS study (12), which reveals the presence of three phylogenetic clades in a sample comprising several serogroups. The three clades, which are associated with specific restriction modification systems, corroborate the CC classification (12). However, WGS reveals the existence of deep phylogenies that could not be evidenced by the classical MLST approach.
In summary, although some features of the N. meningitidis population are compatible with the semi/epidemic clonality model, others (i.e., persistent LD even when repeated genotypes are absent and the presence of deep phylogenies) definitely imply that, on the whole, the data including both carriage and pathogenic strains and for various serogroups better fit the PCE model. The semi/epidemic clonality model predicts that, in the long term, recombination should erase the effects of clonality, but that prediction is challenged by the existence of deep phylogenies in N. meningitidis. The PCE model states, to the contrary, that, beyond a clonality threshold, PCE will overcome the impact of recombination.
The hypothesis of frequent recombination in N. meningitidis has been inferred mostly from the traditional index calculated by the rate of recombination over the rate of mutation (r/m) (14, 36). It is thought that a population will be panmictic if r ≥ m × 20 (37). However, the reliability of this index is questionable, because it may vary greatly among studies (35). For example, in Streptococcus pneumoniae the r/m has been estimated at 66 by MLST (38) but at 7.2 by WGS (39).
Is the Population Structure of N. meningitidis Better Explained by Predominant Natural Selection or by Built-In Properties?
The hypothesis that epidemic clonality in N. meningitidis is mainly the result of natural selection is pervasive (7, 13, 14, 28, 34). Natural selection would play the role of an “invisible hand,” able to maintain the same population structure, ubiquitous genotypes, and genetic subdivisions over continents for much longer than decades and in totally different ecogeographical and host environments, but at the cost of eliminating most of the possible genotypes in every generation (21, 22). This hypothesis, which is already debatable when semi/epidemic clonality is concerned, can hardly account for PCE features, in particular the presence of deep phylogenies (12). It should be noted that in the case of N. meningitidis the classical panselectionist hypothesis has been challenged by several authors (12, 35, 40).
We have long advanced the view that PCE in pathogens is not a “passive” feature, explainable by environmental parameters that would filter downstream many, if not most, possible MLGs. Instead, we have argued that recombination is restrained upstream by intrinsic biological properties of pathogens (11, 18). Restrained recombination makes it possible for pathogens to escape the recombinational load (17) and could be governed by sexuality/clonality machinery working as a diallelic system (11). Such double machinery, governing either parthenogenesis or outcrossing has been inferred for the cladoceran Daphnia pulex (41). We have hypothesized that in pathogens such double machinery could be ancestral. It is worth noting that in the parasitic protozoan Giardia intestinalis the meiotic machinery is similar to that of higher eukaryotes and is homologous to bacterial genes used for DNA repair (42). Several authors have proposed that the main function of recombination is DNA repair rather than the generation of new MLGs (43, 44). In N. meningitidis, many data are consistent with the hypothesis that this bacterium possesses built-in mechanisms that favor genetic exchange between closely related or identical genotypes (selfing) and that inhibit outcrossing. In this way, N. meningitidis is able both to repair its DNA and avoid recombinational load. Restriction systems are different in different CCs and phylogenetic clades, which favors selfing and “invisible sex,” and inhibits outcrossing (12, 44). The distribution of DNA uptake sequences in the N. meningitidis genome suggests that the main role of transformation in this bacterium is to favor genome stability rather than to generate new MLGs (12, 13). These two features—restriction systems that are specific to CCs and clades and the specific distribution of DNA uptake sequences—suggest that this bacterium does have built-in mechanisms that favor restrained recombination and do not support the hypothesis (7) that PCE is mainly or exclusively explained by natural selection.
It is probable that natural selection plays an important role in N. meningitidis population structure. However, the data surveyed above show that this bacterium possesses built-in mechanisms that are perfectly able to lead to a PCE pattern, either through mitotic clonality or clonality by selfing.
N. meningitidis, N. gonorrheae, and N. lactamica Species Definition: Contribution of the PCE Model
The PCE model makes it possible to adapt the phylogenetic species concept (45) to pathogens. Because bacterial populations always undergo some recombination, even between phylogenetically distant lines, it is impossible to describe species on a strict clade concept. The near-clades, however, are based on a clearly defined evolutionary concept in which the strict cladistic demands are relaxed.
Two cases can be taken into account. In one case, the near-clade concept provides a convenient background for describing new species, if specialists of the field find it useful [e.g., for the “assemblages” (i.e., near-clades) that subdivide the population structure of G. intestinalis (46)]. Alternatively, already described pathogen species can be equated to near-clades from an evolutionary point of view, as is the case for the three species N. meningitidis, N. gonorrhoeae, and N. lactamica. These species are clearly differentiated by MLST, but some recombination occurs among them (47–49). Therefore they can be seen as a set of Russian doll near-clades. The distinction of the three species is medically relevant, because their ecology and pathogenicity are quite different. Therefore they fit the phenotypic species concept. Their correspondence to near-clades (not true clades) also makes it possible to support their species status in the framework of the phylogenetic species concept (45).
Semantic Clarification
As we have pointed out repeatedly (11, 15, 46), the genetic literature dealing with the pathogen population is littered with an incredible inflation of terms, all of which most probably are synonymous with “near-clades.” In the N. meningitidis literature, the terms “clade” (12, 31), “clonal complex” (7, 10, 50), “cluster” (48, 49), “genome group” (29), “lineage” (28), “semi-discrete lineage” (14), and “subgroup” (34) are equivalent to near-clades from an evolutionary point of view. It would seem desirable to clean up this confusing terminology, because there is no reason, other than tradition, to give different names to similar evolutionary entities.
Conclusion
The PCE model makes it possible to reconsider the issues of N. meningitidis population structure and of species definition in the N. meningitidis complex of species. MLST has made a considerable contribution to this field of research. However, it has limitations, as does any molecular tool. WGS analysis has shown that N. meningitidis exhibits deep phylogeny (12), challenging the MLST results (14, 35, 50). In the future, as new technologies such as microarrays, WGS, and SNP analysis become increasingly routine, their contributions probably will revise the population genetics of N. meningitidis and other pathogens.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IX: Clonal Reproduction: Alternatives to Sex,” held January 9–10, 2015, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_IX_Clonal_Reproduction.
This article is a PNAS Direct Submission.
References
- 1.Selander RK, Levin BR. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 2.Ørskov F, Ørskov I. From the national institutes of health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983;148(2):346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- 3.Tibayrenc M, Cariou ML, Solignac M, Carlier Y. Arguments génétiques contre l’existence d’une sexualité actuelle chez Trypanosoma cruzi; implications taxinomiques. C R Acad Sci Paris III. 1981;293:207–209. [Google Scholar]
- 4.Tibayrenc M, Ward P, Moya A, Ayala FJ. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci USA. 1986;83(1):115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tait A. Evidence for diploidy and mating in trypanosomes. Nature. 1980;287(5782):536–538. doi: 10.1038/287536a0. [DOI] [PubMed] [Google Scholar]
- 7.Caugant DA. Genetics and evolution of Neisseria meningitidis: Importance for the epidemiology of meningococcal disease. Infect Genet Evol. 2008;8(5):558–565. doi: 10.1016/j.meegid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Caugant DA, et al. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169(6):2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiden MCJ. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 10.Turner KME, Feil EJ. The secret life of the multilocus sequence type. Int J Antimicrob Agents. 2007;29(2):129–135. doi: 10.1016/j.ijantimicag.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Tibayrenc M, Ayala FJ. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci USA. 2012;109(48):E3305–E3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budroni S, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA. 2011;108(11):4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiden MCJ. Population genomics: Diversity and virulence in the Neisseria. Curr Opin Microbiol. 2008;11(5):467–471. doi: 10.1016/j.mib.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel U, Schoen C, Elias J. 2010. in Bacterial Population Genetics in Infectious Disease, eds Robinson DA, Falush D, Feil EJ (Wiley-Blackwell, Hoboken, NJ), pp. 247–267.
- 15.Tibayrenc M, Ayala FJ. How clonal are Trypanosoma and Leishmania? Trends Parasitol. 2013;29(6):264–269. doi: 10.1016/j.pt.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Avise JC. 2004. Molecular Markers, Natural History and Evolution (Chapman and Hall, New York), 2nd Ed.
- 17.Agrawal AF. Evolution of sex: Why do organisms shuffle their genotypes? Curr Biol. 2006;16(17):R696–R704. doi: 10.1016/j.cub.2006.07.063. [DOI] [PubMed] [Google Scholar]
- 18.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18(9):405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 19.Avise J. Clonality. The Genetics, Ecology, and Evolution of Sexual Abstinence in Vertebrate Animals. Oxford Univ Press; Oxford, UK: 2008. [Google Scholar]
- 20.Feretzaki M, Heitman J. Unisexual reproduction drives evolution of eukaryotic microbial pathogens. PLoS Pathog. 2013;9(10):e1003674. doi: 10.1371/journal.ppat.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibayrenc M, et al. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88(12):5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S, Day KP. A strain theory of malaria transmission. Parasitol Today. 1994;10(12):476–481. doi: 10.1016/0169-4758(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 24.Tibayrenc M. Antigenic diversity and the transmission dynamics of Plasmodium falciparum: The clonality/sexuality debate revisited. Parasitol Today. 1994;10(12):456–457. doi: 10.1016/0169-4758(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez M, Alvar J, Tibayrenc M. Leishmania infantum is clonal in AIDS patients too: Epidemiological implications. AIDS. 1997;11(5):569–573. doi: 10.1097/00002030-199705000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira GEM, et al. The genetic structure of Leishmania infantum populations in Brazil and its possible association with the transmission cycle of visceral leishmaniasis. PLoS ONE. 2012;7(5):e36242. doi: 10.1371/journal.pone.0036242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibayrenc M, Kjellberg F, Ayala FJ. A clonal theory of parasitic protozoa: The population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87(7):2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckee CO, et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc Natl Acad Sci USA. 2008;105(39):15082–15087. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph B, et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS ONE. 2011;6(4):e18441. doi: 10.1371/journal.pone.0018441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang JF, et al. Clonal and antigenic analysis of serogroup A Neisseria meningitidis with particular reference to epidemiological features of epidemic meningitis in the People’s Republic of China. Infect Immun. 1992;60(12):5267–5282. doi: 10.1128/iai.60.12.5267-5282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bart A, et al. The population structure of Neisseria meningitidis serogroup A fits the predictions for clonality. Infect Genet Evol. 2001;1(2):117–122. doi: 10.1016/s1567-1348(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 32.Dunning Hotopp JC, et al. Comparative genomics of Neisseria meningitidis: Core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152(Pt 12):3733–3749. doi: 10.1099/mic.0.29261-0. [DOI] [PubMed] [Google Scholar]
- 33.Guttman DS, Stavrinides J. 2010. in Bacterial Population Genetics in Infectious Disease, eds Robinson DA, Falush D, Feil EJ (Wiley-Blackwell, Hoboken, NJ), pp. 121–151.
- 34.Falush D. Toward the use of genomics to study microevolutionary change in bacteria. PLoS Genet. 2009;5(10):e1000627. doi: 10.1371/journal.pgen.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didelot X, Urwin R, Maiden MCJ, Falush D. Genealogical typing of Neisseria meningitidis. Microbiology. 2009;155(Pt 10):3176–3186. doi: 10.1099/mic.0.031534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanage WP, Spratt BG, Turner KME, Fraser C. Modelling bacterial speciation. Philos Trans R Soc Lond B Biol Sci. 2006;361(1475):2039–2044. doi: 10.1098/rstb.2006.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maynard Smith J, Smith NH. Detecting recombination from gene trees. Mol Biol Evol. 1998;15(5):590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 38.Feil EJ, Smith JM, Enright MC, Spratt BG. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics. 2000;154(4):1439–1450. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci USA. 2005;102(6):1968–1973. doi: 10.1073/pnas.0406993102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schurko AM, Logsdon JM, Jr, Eads BD. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol. 2009;9:78. doi: 10.1186/1471-2148-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infect Genet Evol. 2008;8(3):267–285. doi: 10.1016/j.meegid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 43.De Meeûs T, Prugnolle F. In: Genetics and Evolution of Infectious Diseases. Tibayrenc M, editor. Elsevier Insights; Amsterdam: 2011. pp. 133–146. [Google Scholar]
- 44.Caugant DA, Maiden MCJ. Meningococcal carriage and disease—population biology and evolution. Vaccine. 2009;27(Suppl 2):B64–B70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cracraft J. 1983. in Johnson RF Current Ornithology, ed Johnson RF (Plenum, New York), pp 159–187.
- 46.Tibayrenc M, Ayala FJ. 2014. Cryptosporidium, Giardia, Cryptococcus, Pneumocystis genetic variability: Cryptic biological species or clonal near-clades? PLoS Pathogens 10(4):e1003908.
- 47.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6(6):431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 48.Bennett JS, et al. Species status of Neisseria gonorrhoeae: Evolutionary and epidemiological inferences from multilocus sequence typing. BMC Biol. 2007;5:35. doi: 10.1186/1741-7007-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feil EJ. 2010. in Bacterial Population Genetics in Infectious Disease, eds Robinson DA, Falush D, Feil EJ (Wiley-Blackwell, Hoboken, NJ), pp. 19–35.