Significance
Mycoviruses generally contain dsRNA genomes but ssRNA and ssDNA examples are known. Mycovirus diversity is increasing, and here we describe a unique example that contains four dsRNA elements nominated Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1). We show for the first time (to our knowledge) that both purified AfuTmV-1 and its dsRNA are infectious for protoplasts and that the virus genome is not conventionally encapsidated and has a unique organization. Separation of the genes encoding the RNA-dependent RNA polymerase enzyme responsible for copying the viral genome and an S-adenosyl methionine-dependent methyltransferase capping enzyme on different dsRNAs is also previously unreported for a mycovirus. AfuTmV-1 appears to be intermediate between dsRNA and positive-strand ssRNA viruses, as well as between encapsidated and capsidless RNA viruses.
Keywords: mycovirus, infectious dsRNA, protoplast transfection, capping enzyme, virus evolution
Abstract
We report the discovery and characterization of a double-stranded RNA (dsRNA) mycovirus isolated from the human pathogenic fungus Aspergillus fumigatus, Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1), which reveals several unique features not found previously in positive-strand RNA viruses, including the fact that it represents the first dsRNA (to our knowledge) that is not only infectious as a purified entity but also as a naked dsRNA. The AfuTmV-1 genome consists of four capped dsRNAs, the largest of which encodes an RNA-dependent RNA polymerase (RdRP) containing a unique GDNQ motif normally characteristic of negative-strand RNA viruses. The third largest dsRNA encodes an S-adenosyl methionine–dependent methyltransferase capping enzyme and the smallest dsRNA a P-A-S–rich protein that apparently coats but does not encapsidate the viral genome as visualized by atomic force microscopy. A combination of a capping enzyme with a picorna-like RdRP in the AfuTmV-1 genome is a striking case of chimerism and the first example (to our knowledge) of such a phenomenon. AfuTmV-1 appears to be intermediate between dsRNA and positive-strand ssRNA viruses, as well as between encapsidated and capsidless RNA viruses.
Mycoviruses with increasingly diverse genomes have been described in a wide range of fungi covering all four phyla of the true fungi: Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota. The more abundant mycoviruses with double-stranded RNA (dsRNA) genomes are generally classified into five major families: Totiviridae (nonsegmented, 4.6–7 kbp), Partitiviridae (2 segments, 1.4–2.3 kbp), Chrysoviridae (4 segments, 2.4–3.6 kbp), Reoviridae (10–12 segments, 0.7–5 kbp), all of which are conventionally encapsidated, Megabirnaviridae (2 segments, 7–9 kbp), and the proposed families “Quadriviridae” (4 segments, 3.7–4.9 kbp) and “Alternaviridae” (4 segments, 1.4–3.6 kbp) (1–8). The remaining six families (Alphaflexiviridae, Barnaviridae, Endornaviridae, Gammaflexiviridae, Hypoviridae, and Narnaviridae) accommodate single-stranded RNA (ssRNA) genomes, of which only three families (Alphaflexiviridae, Barnaviridae, and Gammaflexiviridae) form virus particles, whereas members of the remaining three virus families are unencapsidated and do not form typical virions. Two families (Metaviridae and Pseudoviridae) accommodate RNA reverse-transcribing genomes. A negative-strand RNA mycovirus (9) and a geminivirus-related DNA mycovirus (10) have also been identified, characterized, and sequenced.
During routine screening for RNA viruses in the human fungal pathogen Aspergillus fumigatus, we discovered three dsRNA profiles in 366 clinical and environmental isolates ranging in size from ∼1.1 to 3.6 kbp (11). Two of the profiles were found to be representatives of respectively a chrysovirus nominated Aspergillus fumigatus chrysovirus (AfuCV) (12) and a partitivirus nominated Aspergillus fumigatus partitivirus 1 (AfuPV-1) (13). Here, we report the sequence and characterization of the remaining profile of four uncharacterized dsRNA segments, which constitute the genome of a completely novel mycovirus nominated Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1). In common with a number of recently described dsRNAs, AfuTmV-1 is an unconventional virus, in that it is not encapsidated but is apparently coated with a virus-encoded protein and may also be associated with or enveloped in colloidal cellular components (e.g., refs. 14 and 15). For the first time for a mycovirus, to our knowledge, we demonstrate that both AfuTmV-1 and its isolated dsRNA are infectious for fungal protoplasts. The unique sequences of the four AfuTmV-1 dsRNAs and the proposed roles of the proteins predicted from them in virus replication, including the combination of a capping enzyme and the RNA-dependent RNA polymerase (RdRP) on separate dsRNAs, strengthen the suggestion for the assignation of a new virus family where AfuTmV-1 would represent the prototype member.
Results and Discussion
RNA Elements Associated with the Af293 Prototype Isolate of Aspergillus fumigatus.
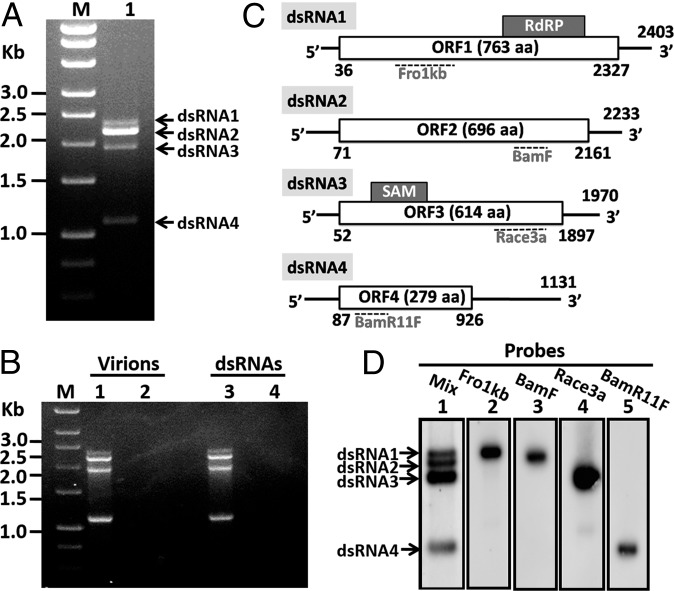
Following phenol/chloroform extraction, AfuTmV-1 was found to contain four dsRNA species, referred to as dsRNA 1 (∼2.4 kbp), dsRNA 2 (∼2.2 kbp), dsRNA 3 (∼1.9 kbp), and dsRNA 4 (∼1.1 kbp) in order of increasing mobility in 1.0% agarose gels (Fig. 1A, lane 1). A BLASTN search of the A. fumigatus Af293 genome sequence with all four dsRNA sequences revealed no significant similarity. We therefore conclude that the A. fumigatus genome does not have a DNA copy of any of the mycovirus dsRNAs described. The double-stranded nature of the RNAs was confirmed by insensitivity to S1 nuclease and sensitivity to RNase III (Fig. 1B, lanes 3 and 4). Purified AfuTmV-1 was also sensitive to RNase III (Fig. 1B, lanes 1 and 2), suggesting the absence of a conventional capsid protein. Interestingly, the lack of a conventional particle may be linked to the unusual asymmetry of the dsRNA quantities, because the necessity for maintaining the stoichiometry between the dsRNAs in order for them to replicate in a confined space or fit into a capsid would not exist.
Fig. 1.
Genomic characteristics and organization of Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1). (A) AfuTMV-1 was purified from Af293 mycelia, and dsRNA isolated using phenol/chloroform was fractionated on a 1% agarose gel, and the positions of dsRNAs 1–4 in lane 1 are shown. Lane M contains HyperLadder I kb DNA marker. (B) RNase III sensitivity of purified AfuTmV-1 and AfuTmV-1 dsRNA was investigated, respectively, and untreated samples of each are shown following agarose gel electrophoresis in lanes 1 and 3, with RNase III digestions of each shown in lanes 2 and 4. (C) Genome organization of AfuTMV-1 dsRNAs 1–4 showing putative ORFs and UTRs. The positions of motifs characteristic for RdRP and methyltransferase (SAM) are shown as gray boxes on dsRNA 1 and dsRNA 3, respectively. (D) Assignation of cDNA clones specific to dsRNAs 1–4 by northern hybridization. Purified viral dsRNAs were fractionated by electrophoresis in 1% agarose gel in 1× TAE, denatured, blotted onto nylon membrane, and probed with clones specific for each dsRNA as shown in C. Lane 1 shows the AfuTmV-1 dsRNA profile, and lanes 2–5, individual transfers hybridized with probes specific for dsRNAs 1–4, respectively.
Biological Comparison of Virus-Cured and Virus-Infected Fungal Strains.
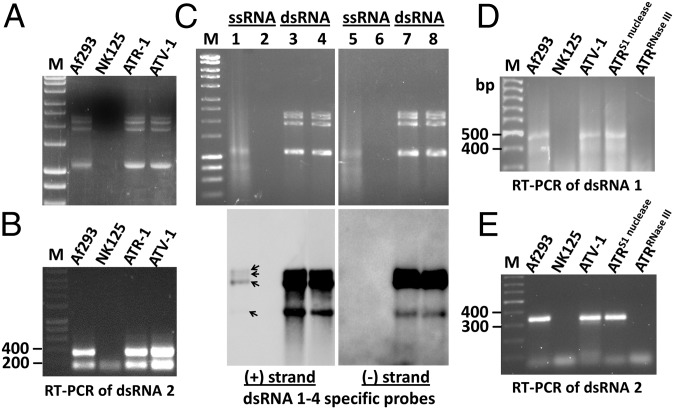
Following cycloheximide treatment of A. fumigatus A293, one isogenic, virus-cured isolate, nominated AfuNK125, was confirmed as being virus-free by electrophoretic analysis of dsRNA, northern blotting, and RT-PCR amplification assay of a 337-bp fragment of AfuTmV-1 dsRNA 2 (Fig. S1A). The phenotypes of the virus-infected Af293 and virus-free AfuNK125 A. fumigatus isolates (Fig. S1B), together with their growth rate and biomass were, respectively, observed and measured over a 5-d incubation period in both solid and liquid Aspergillus minimal medium (MM) and Aspergillus complete medium (ACM) media (Fig. S1C). Apart from a minor reduction in the intensity of blue green pigmentation in AfuNK125, there were no significant differences in phenotype, growth rates, or biomass between A. fumigatus Af293 and AfuNK125 in either medium. Additionally, the greater wax moth Galleria mellonella infection model was used to compare the virulence of A. fumigatus Af293 and AfuNK125 as described previously (16). Statistically significant differences in the survival rates of G. mellonella larvae infected with Af293 and AfuNK125 (Fig. S1D), but not in the fungal burden as assessed by measuring the expression levels of fungal β-tubulin (Fig. S1E), were noted.
Infectivity of Purified AfuTmV-1 and Its dsRNA and Effects on the Host.
Protoplasts from virus-free A. fumigatus isolate AfuNK125 were transfected with both AfuTmV-1 and AfuTmV-1 dsRNA using a standard procedure. Mycelial agar discs were then taken randomly from regenerated A. fumigatus-transfected protoplasts and transferred into ACM broth and also plated onto fresh ACM agar plates and incubated at 37 °C for 4 d. Pooled regenerated colonies, transfected with purified AfuTmV-1 and AfuTmV-1 dsRNA, were nominated ATV-1 and ATR-1, respectively. Colony morphology and hyphal growth of both ATV-1 and ATR-1 were identical to those of isolate Af293 but differed from AfuNK125 in terms of enhanced pigmentation in the former. The presence of AfuTmV-1 dsRNAs from both ATV-1 and ATR-1 was confirmed by electrophoretic analysis using LiCl-purified samples (Fig. 2A), and compared with virus-infected and virus-free isolates. These samples were subsequently subjected to RT-PCR amplification using primers specific to dsRNA 2, yielding the 337-bp PCR product (Fig. 2B). Northern analysis of RNA enriched in dsRNA or ssRNA of both ATV-1 and ATR-1 was performed with positive- and negative-strand specific DIG-labeled probes derived from all four AfuTmV-1 dsRNAs (Fig. 2C). This analysis coupled with pretreatment of the RNA extracts with DNase I alone or in combination with S1 nuclease revealed that, under the conditions of the electrophoresis used here, the single-stranded and double-stranded forms of all four AfuTmV-1 RNAs had similar electrophoretic mobilities (Fig. 2C). The ssRNA and dsRNA were distinguished by their capability to hybridize with only the positive-strand specific probes or both positive-strand and negative-strand specific probes (Fig. 2C). The results indicated that nucleic acid extracts from AfuTmV-1 contain, for all four RNAs, both dsRNA and positive-stranded ssRNA. Because the amounts of ssRNA and dsRNA analyzed were derived from the same amount of total nucleic acids, the relatively strong signals of dsRNA and weak signals of ssRNA from blotting indicated that each of the RNAs exists predominantly in the dsRNA form. No significant amounts of negative-stranded ssRNAs were found for all four RNAs. Additionally, AfuTmV-1 dsRNA treated with proteinase K and the ssRNA-specific S1 nuclease, but not the dsRNA-specific RNase III, was also found to be infectious for A. fumigatus by RT-PCR amplification using primers specific to dsRNA 1, yielding a 467-bp PCR product (Fig. 2D), and dsRNA 2, as described above (Fig. 2E), thus refuting the possibility that contaminating virus or viral ssRNA is the actual infectious agent. To examine whether similarly purified dsRNAs from a negative control dsRNA fungal virus were uninfectious and that we were dealing with a previously unreported phenomenon with AfuTmV-1, we successfully transfected Af-237y protoplasts with AfuPV-1 virions but not a mixture of AfuPV-1 dsRNAs 1 and 2. Successful transfection of protoplasts with AfuPV-1 has been demonstrated previously (17), and this result was repeated here following positive RT-PCR amplification of the entire RdRP ORF on AfuPV-1 dsRNA 1 with no amplicons being produced in regenerated colonies transfected with a mixture of AfuPV-1 dsRNAs 1 and 2 (Fig. S2A). These experiments were performed on three separate occasions with identical results and in total confirm that the genome of AfuTmV-1 is indeed dsRNA and that both purified AfuTmV-1 and AfuTmV-1 dsRNA are infectious for A. fumigatus. The low quantity of positive-stranded ssRNA detected probably serves as mRNA for the production of the viral proteins, whereas no negative-sense ssRNA intermediate is required for the virus replication cycle. This is the norm for dsRNA viruses, whereas in most positive-stranded ssRNA viruses, such as mitoviruses, the quantity of the ssRNA appears to be much greater than that of dsRNA (18), although exceptions have been noted (19).
Fig. 2.
Viral dsRNAs extracted from transfected cultures, RT-PCR amplification and northern detection of ssRNA and dsRNA from A. fumigatus isolate NK125 following transfection with AfuTmV-1. (A) AfuTmV-1 dsRNAs extracted from transfected cultures (ATV-1 and ATR-1), wild-type (Af293) and virus-free (NK125) isolates. (B) RT-PCR amplification of a 337-bp segment from dsRNA 2. Lane M contains HyperLadder I kb DNA marker. (C) Amounts of viral ssRNA and dsRNA, equivalent to 10 mg of total nucleic acids before LiCl fractionation, were electrophoresed in 1.0% nondenaturing agarose gels and blotted onto nylon membranes. The samples were treated with DNase I (lanes 1, 3, 5, and 7) or DNase I and S1 nuclease (lanes 2, 4, 6, and 8). Hybridization was carried out using positive-strand specific and negative-strand specific riboprobes for all four dsRNAs. The positions of the single-stranded forms of each RNA are indicated on the blots by arrows. RT-PCR amplification of a 467-bp segment from dsRNA 1 (D) and a 337-bp segment from dsRNA 2 (E) from equal amounts of RNA extracted from transfected cultures (ATV-1, ATRS1 nuclease, and ATRRNase III), wild-type (Af293) and virus-free (NK125) isolates. Lane M contains GeneRuler 100-bp DNA ladder.
Cloning and Sequencing of the Full-Length cDNA Clones of AfuTmV-1 dsRNAs 1–4, Their Phylogeny and Putative Roles in Replication.
The complete nucleotide sequences of the four AfuTmV-1 dsRNAs were determined by assembling and combining a series of cDNA clones, including those generated by RNA linker-mediated (RLM)–RACE, which spanned the entire length of each dsRNA and whose genetic organization is shown in Fig. 1C. Contiguous sequence assemblies were assigned to dsRNA segments by northern blotting using representative cDNA fragments as probes for individual RNA species where each probe facilitated the detection of a single dsRNA band (Fig. 1D). The full-length genomic cDNA sequences of AfuTmV-1 dsRNAs 1–4 were deposited in the GenBank database with accession numbers HG975302–HG975305, respectively (Table S1). The GC content of all four AfuTmV-1 dsRNAs (average value of 63.40%) and their coding regions (average value of 63.53%) are considerably higher than those of most characterized mycoviruses including dsRNA partitiviruses and totiviruses, and also most ssRNA plant viruses.
The 5′-untranslated regions (UTRs) of AfuTmV-1 dsRNAs 1–4 were similar in length and sequence and ranged in size from 35 to 86 nt, where the extreme 5′-terminal nucleotides showed sequence heterogeneity (U/C), whereas the next nine 5′ nucleotides of all four dsRNAs were identical (Fig. S3A). Sequence heterogeneity at both the 5′ and 3′ termini of mycovirus dsRNAs has been reported previously (5). The 3′-terminal sequences of AfuTmV-1 dsRNAs 1–3 were also highly conserved and comprised 18/22 identical nucleotides, including 4 G residues at the extreme 3′ terminus of each dsRNA (Fig. S3B). The lengths of the AfuTmV-1 dsRNAs 1–3 3′-UTRs were similar and ranged in size from 74 to 76 nt, but the 205-nt AfuTmV-1 dsRNA 4 3′-UTR was considerably longer with a poorly conserved sequence compared with the other three dsRNAs apart from the very 3′-terminal 4 G residues (Fig. S3C). The 5′- and 3′-UTRs of all four AfuTmV-1 dsRNAs were predicted to be highly structured following analysis with the mfold program (20); they mostly fold independently of the rest of the sequence and consist of long stem-loops resembling panhandle structures, especially at the 3′ terminus (Fig. S3D). It has been suggested that the potential for RNA to be folded into a secondary panhandle structure at the 5′ and 3′ ends may be a diagnostic feature for ssRNA mitoviruses (21) and that these structures may act as promoters for RNA replication. It is possible that they may also function in a similar fashion for AfuTmV-1.
Inspection of the coding potential of the AfuTmV-1 dsRNAs revealed a large ORF on the positive strands of all four dsRNAs (Fig. 1C and Table S1). Additionally, in vitro rabbit reticulocyte lysate translation assays confirmed that each of the four denatured AfuTmV-1 dsRNAs is monocistronic, as each major ORF was translated into a single major product of the size predicted from its deduced amino acid sequence (Fig. S2B), viz. AfuTmV-1 dsRNAs 1–4; 84, 76, 67, and 29 kDa, respectively (Table S1). Examination of the deduced amino acid sequence of the AfuTmV-1 dsRNA 1 ORF (nucleotides 36–2,327: 763 aa; molecular mass, 84 kDa) revealed the presence of three conserved motifs (Fig. S4A) characteristic of RdRPs of dsRNA viruses of simple eukaryotes (22), classified into supergroup 1 of the viral RdRP families (RdRP_4, pfam02123) (23). However, the sequence of motif VI (23) contains a GDN triplet rather than G/ADD, which is normally invariant for positive-strand RNA viruses. The GDN triplet in RdRP genes followed immediately by a Q residue is a characteristic sequence motif normally found in the L genes of rhabdoviruses and paramyxoviruses within the order of Mononegavirales (24), which also includes the recently described negative-stranded mycovirus of Sclerotinia sclerotiorum (9), and this motif GDNQ is also present in the AfuTmV-1 dsRNA 1 ORF. There is experimental evidence that the GDN motif of negative-strand, nonsegmented RNA viruses is a variant of the GDD motif of positive-strand RNA viruses, compatible with the notion that the second D is not essential, but its discovery in the sequence of a dsRNA virus is unique. For positive-strand RNA viruses, a different D can be used for Mg2+ ion chelation so the RdRP is assumed to be active. The AfuTmV-1 RdRP is a derivative of the picorna-like superfamily with, technically, the highest similarity to comoviruses and potyviruses and less similarity to cryptoviruses (Fig. S4B). However, when AfuTmV-1 RdRP was used as a query sequence in database searches, it returned as a subject three RdRP conserved core domains of a walrus calicivirus with an E value of 8e−06 (29% identity, 49% similarity) as the most related sequence. The RdRP sequences of representative members of the virus families described above with less significant E values compared with AfuTmV-1 RdRP were then aligned with the PSI-Coffee, version 10.0, program (25) (Table S2 and Fig. S4B). Mycovirus RdRPs are known to be related to the RdRPs of viruses in the picorna-like superfamily of eukaryotic, positive-strand RNA viruses (26), and numerous examples can be found in the literature including Ustilaginoidea virens nonsegmented virus 1 (27), a number of viruses in the Partitiviridae family (28), and Aspergillus foetidus virus S2, which is closely related to several members of the Caliciviridae family and Comovirinae subfamily (29). Our phylogenetic analysis results described here (Fig. S4B) support the contention that AfuTmV-1 should also be classified as a picorna-like virus.
The 5′ sequence of AfuTmV-1 dsRNA 2 contains two potential AUG codons, and the upstream one is in a more favorable context (30) to initiate translation of a putative protein of 696 aa (molecular mass, 76 kDa), predicted from the ORF between nucleotides 71 and 2,161. Following a search of the public databases, the deduced amino acid sequence of the AfuTmV-1 dsRNA 2 ORF was found to be significantly similar to proteins of unknown function putatively encoded by, respectively, a 2,233-bp dsRNA isolated from Alternaria sp. FA0703 (AltR1; accession no. ACL80752: 9e−156; 39% identical; 57% similar) and a ∼2.3-kbp mycovirus dsRNA, nominated B2, isolated from the flax rust, Melampsora lini (Fig. S4C, RZB2; 6e−109; 35% identical; 53% similar). The only interesting feature of these three putative proteins is the presence of a zinc finger-like motif as shown in the alignment in Fig. S4C. No function can be ascribed to the putative protein encoded by AfuTmV-1 dsRNA 2.
For AfuTmV-1 dsRNA 3, the second in-frame AUG in the predicted ORF was placed in a favorable context to potentially initiate translation. The deduced amino acid sequence of the predicted ORF (nucleotides 52–1,897: 614 aa; molecular mass, 66 kDa) contained a high proportion of positively charged amino acids in an APC basic domain that between amino acids 110 and 250 had clear similarities with domains associated with methyltransferase activity (methyltransf_25, Pfam 13649; nucleotides 485–766; 1.05e−03). Further analysis of the amino acid sequences of the ORF predicted from AfuTmV-1 dsRNA 3 showed it to be significantly similar to a protein predicted from the sequence of another M. lini mycovirus dsRNA element, 1,932 bp long, nominated B3 (31) (accession no. X64371; 2e−120: 40% identical; 54% similar), which contained motifs characteristic of S-adenosyl methionine–dependent methyltransferases. The RNA-capping methyltransferase domain is conserved in the alphavirus-like superfamily and present in some but not all picorna-like mycoviruses, e.g., Botrytis virus F (32) and some endornaviruses (33). Methyltransferases add a guanylyl residue to the 5′ end of RNAs to form a G (5′) ppp (5′) N cap structure. Mycoviruses with dsRNA genomes are not naturally capped but do furnish ssRNA transcripts with a cap structure by a cap-snatching mechanism from the host in at least one case in the totivirus Saccharomyces cerevisiae virus L-A (34). During capping, the guanylyltransferase (GTP:mRNA guanylyltransferase) is reversibly and covalently guanylylated. In this enzyme-GMP intermediate, GMP is linked to the epsilon-amino group of a lysine residue via a phosphoamide bond. This principal lysine residue has been identified in the AfuTmV-1 dsRNA 3 ORF sequence as part of a characteristic catalytic motif for viral GTPs, which is also present in M. lini mycovirus dsRNA B3, several DNA viruses, including Vaccinia virus, African swine fever virus (Fig. S5A), and some bacteria but with no clear relationship with the capping enzymes of any other RNA viruses, making its origin potentially unexpected. Oligo-cap analysis of AfuTmV-1 genomic dsRNAs, as performed previously (35), clearly indicates the presence and absence of cap structures on the positive and negative strands of all four dsRNAs, respectively (Fig. S5 B and C). In confirmation of the absence of 5′-cap structures on the negative strands of AfuTmV-1 dsRNAs and confirmation that they are phosphorylated, it was possible to label the 5′ terminus of all four genomic dsRNAs with no prior treatment with [γ-32P]ATP using polynucleotide kinase. A combination of a capping enzyme with a picorna-like RdRP in the AfuTmV-1 genome is a striking case of chimerism and the first example to our knowledge of such a phenomenon.
Inspection of the amino acid sequence predicted from the only reasonably sized ORF on AfuTmV-1 dsRNA 4 (nucleotides 87–926: 279 aa; molecular mass, 29 kDa) revealed it contained an overrepresentative number of proline (P), alanine (A), and serine (S) residues in a PAS-rich protein (rp). These PASrps have been documented as being potentially encoded by a number of dsRNA-containing, unclassified insect, plant, and fungal viruses (Fig. S6 and Table S3). It was also possible to predict the presence of a hexosaminidase domain from amino acids 35–110 in the putative protein encoded by AfuTmV-1 dsRNA 4. However, BLAST searches failed to return subjects with significant E values to any known proteins in global databases, and any similarities to hexosaminidase may be spurious. The presence of PASrps was first noted in the mycovirus Phlebiopsis gigantea large virus 1 (14), but their role in dsRNA replication in any of the examples described thus far (Fig. S6 and Table S3) is obscure. For instance, it is known that P-rich domains can mediate protein–protein interactions, interact with membrane components, and might thus serve as scaffold proteins anchoring replication complexes. However, these domain motifs are present in some but not all cases (Table S3) and, as indicated by Cai et al. (36), have a high possibility of random occurrence.
Structural Aspects of AfuTmV-1.
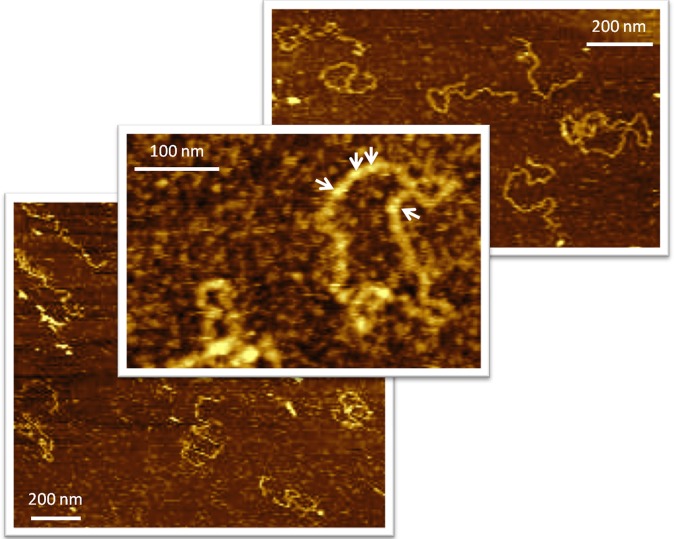
Several attempts to isolate AfuTmV-1 virions, using procedures routinely used for mycoviruses, failed to recover discrete, virus-like particles as evidenced by electron microscopy, although some amorphous electron-dense material was visible. This observation suggests that AfuTmV-1 is not conventionally encapsidated in virions and is consistent with the fact that none of the AfuTmV-1 dsRNAs encodes a protein identified in BLASTX searches as a capsid protein. Following atomic force microscopy (AFM) of purified AfuTmV-1, chain-like molecules were visualized in virtually every scanned area and example images are shown here (Fig. 3). Numerous linear strands of different lengths were observed with structures very similar to AFM images of dsRNA observed previously by Magae (37) of an unusual, apparently unencapsidated mycovirus of Lentinula edodes, which appears to be associated with a protein that has some similarities to the AfuTmV-1 PAS-rich protein (Table S3 and Fig. S6). The lengths of a number of RNA molecules were measured and processed using the ImageJ program, and, allowing for the occurrence of damaged RNA strands, representative dsRNAs of all four AfuTmV-1 dsRNAs based on a value of 1 μm = ∼3-kb dsRNA were found.
Fig. 3.
AFM images of purified AfuTmV-1. AfuTmV-1 is visualized as numerous chain-like, linear nucleic acids of different lengths corresponding to those predicted from the genomic size of the four genomic dsRNAs. In the middle panel, arrows indicate the AfuTmV-1 PAS-rich ORF 4-encoded protein putatively associated with dsRNA 1.
Because dsRNA was recovered from pellets obtained after ultracentrifugation, it is assumed that, as described previously for a number of unusual, apparently unencapsidated infectious dsRNAs that are pathogenic for insects, plants, and fungi (15), AfuTmV-1 dsRNAs are associated with or enveloped in colloidal proteinaceous components. In confirmation of this, peptide mass fingerprinting of purified AfuTmV-1 yielded nine peptide fragments (25% of the peptides identified) that matched the deduced polypeptide sequence encoded by AfuTmV-1 dsRNA 4 with ion scores higher than 46 (95% confidence), accounting for 53% of the entire coverage (279 aa), whereas no other viral proteins were found to be associated with AfuTmV-1 in detectable quantities. Interestingly, proteins with a similar PAS-rich profile are apparently encoded by most if not all of the infectious dsRNA agents described thus far that are apparently unencapsidated in conventional virus particles (Tables S2 and S3, and Fig. 3).
In this investigation, we demonstrate by peptide mass fingerprinting for the first time (to our knowledge) that the unusually small AfuTmV-1 PASrp, compared with the other examples (Table S3), is associated with and possibly coats the dsRNA genome of the virus in an unconventional manner. We consider that it is not insignificant that, thus far, all dsRNA viruses that encode PASrps appear to be unencapsidated, including a M. lini mycovirus (31, 38), which has a similar dsRNA profile to AfuTmV-1 and two sequenced components that are significantly related to AfuTmV-1 dsRNAs 2 and 3, respectively (see above). These observations, the finding of a homolog for AfuTmV-1 dsRNA 2 in AltR1 and our recent discovery of four dsRNAs in several isolates of Beauveria bassiana and in Cladosporium cladosporioides (accession nos. YP_009052470–009052474) that are similar in profile and size but not sequence to their AfuTmV-1 counterparts, suggest that tetraviruses are prevalent in Ascomycota but are also present in the Basidiomycota such as M. lini.
In conclusion and in emphasis of the increasing diversity of mycovirus-related dsRNA viruses, our discovery of a segmented dsRNA virus with a genome derived in part from unsegmented viral ancestors with reference to a capping enzyme and RdRP activity is unique as is the fact that the dsRNA is infectious. How AfuTmV-1 replicates is unknown, but it is conceivable that the AfuTmV-1 dsRNA 4 gene product may be intimately involved in perhaps assisting a host RNA helicase to partially unwind the dsRNA, and this allow some translation. In vitro rabbit reticulocyte lysate translation assays showed that each of the four AfuTmV-1 major ORFs was translated into the anticipated product; hence, as described above, in vivo protein translation may be feasible. AfuTmV-1 appears to be intermediate between dsRNA and positive-strand ssRNA viruses, as well as between encapsidated and capsidless RNA viruses.
Materials and Methods
Fungal Strains and Culture Media.
A. fumigatus (Nakazawa) strain Af293, Fungal Genetics Stock Center A1100, is a clinical isolate that contains four uncharacterized viral dsRNA elements (11). Strain NK125 is a virus-free strain obtained by cycloheximide treatment of strain Af293 as described by Bhatti et al. (17), as verified by RT-PCR and northern hybridization analysis. Strain AF-237y is a virus-free, hygromycin-resistant yellow strain (17). A. fumigatus Δpaba is the mutated yellow strain (39) used as an avirulent control in the G. mellonella infectivity assays. All strains, maintained in 20% (vol/vol) glycerol, were grown on ACM agar (40) or MM agar (41) for 5 d. Spores were harvested by decanting 20 mL of autoclaved dH2O onto the lawn of fungal spores and resuspending the spores, giving a typical yield of 5 × 109 spores. The spore suspension was filtered through Miracloth (Calbiochem). Following spore enumeration with a hemocytometer, the filtrate was used directly to inoculate 500 mL of ACM broth cultures, which were then grown in 1-L flasks and incubated with shaking at 130 rpm at 37 °C. After 7 d of growth, the mycelium was harvested by gravity filtration on Miracloth, washed with water, dried briefly on filter paper, snap frozen with liquid nitrogen, and stored at −80 °C until further processing.
Growth and Virulence Assays.
Mycelial growth and colony morphology were evaluated according to the procedures described by Bhatti et al. (17). To assess growth rate, equal numbers of spores (n = 500) of isogenic lines of virus-infected A. fumigatus Af293 and virus-free NK125 were centrally inoculated onto MM and ACM agar on Petri plates and incubated at 37 °C. The colony diameters of the isolates were measured every 24 h over a period of 5 d and growth rate per time interval used to calculate the average growth rate per day. Time points were selected such that fungal growth had initiated before the first measurement, and the last measurement was made before the mycelium reached the Petri dish edge. This ensured that the measurements can be considered as estimates of axial growth rates in the exponential growth phase. All experiments were performed in triplicate. To assess biomass production, equal numbers of spores (n = 3 × 108) of the two isolates were inoculated into 100-mL flasks containing 60 mL of ACM or MM broth and incubated at 37 °C on a rotary shaker (130 rpm) over a period of 5 d. The mycelium from individual cultures was harvested daily by filtration through Miracloth and the pellets dried at 37 °C until their weights were constant and biomass produced per time interval used to calculate the average growth rate per day. All experiments were performed in triplicate. Student’s t test was used to analyze all of the data for significant differences in growth rate and biomass production. G. mellonella infectivity assays, fungal burden assays, and statistical analyses were performed as described by Özkan and Coutts (16). All experiments were performed in triplicate.
Nucleic Acid Extraction and Northern Hybridization Analyses.
To obtain mycelial samples of strains Af293 and NK125, mycelia were grown in ACM broth as above and total RNA samples were prepared either using a TRIzol kit (Invitrogen) or a modification of the procedure described by Covey and Hull (42). LiCl fractionation of dsRNA and ssRNA fractions was carried out according to Diaz-Ruiz and Kaper (43). Isolation of RNA from purified AfuTmV-1 was performed as described previously using phenol/chloroform (12) with or without prior treatment with proteinase K at 65 °C (Sigma Chemicals) to achieve 100% activity as described by the manufacturer. Gel separation, denaturation, neutralization, and electrophoretic blotting of dsRNA and ssRNA fractions, pretreated with RNase-free DNase1 with or without S1 nuclease, were as described before (18). RNase III (New England Biolabs) treatment of purified AfuTmV-1 and AfuTmV-1 dsRNA was performed according to the manufacturer’s instructions. Blots were hybridized with strand-specific riboprobes for each viral dsRNA element prepared by in vitro transcription of template DNA in the presence of digoxigenin-UTP, using T7 RNA polymerase (DIG Northern Starter Kit; Roche), followed by immunological detection using alkaline phosphatase-conjugated, anti-digoxigenin antibody (Roche).
Molecular Cloning and Sequencing.
After electrophoretic separation on agarose gels, dsRNAs 1–4 were used, either collectively or individually, for reverse transcription, PCR amplification, cloning, and sequencing as described previously (44). The cDNA clones of dsRNAs 1–4 were initially obtained by random priming of methyl mercuric hydroxide-denatured dsRNA using the Froussard procedure (45) with further DNA manipulations being performed according to standard protocols (46). For the synthesis of additional cDNAs covering their complete sequence, purified dsRNAs 1–4 were denatured with methyl mercuric hydroxide and subjected to a single-primer, genome-walking RT-PCR protocol as described previously (44). An RLM-RACE PCR procedure was used to determine the 5′- and 3′-terminal sequences of the dsRNAs (47). All clones from at least four separate experiments were sequenced in triplicate.
Sequence and Phylogenetic Analyses.
Sequence similarity searches of the GenBank databases were conducted using the BLAST program (48). Searches for amino acid signatures and protein motifs were conducted using the programs included in the ExPASy proteomics tools (www.expasy.org/tools/). Sequence alignments were performed using the PSI-Coffee, version 10.0, program (21), and phylogenetic analysis was performed using the fast Fourier transform MAFFT program L9INS-1 (49). A bootstrap test was conducted with 1,000 resamplings to construct neighbor-joining trees.
Mycovirus Purification and Characterization of Associated Protein.
Approximately 50 g of frozen fungal mycelia were homogenized in 2 vol (wt/vol) of TE (0.5 mM Tris⋅HCl and 1.0 mM EDTA, pH 7.5) buffer for 3 min in a blender. The homogenate was filtered through Miracloth, transferred into precooled, sterile tubes, and subjected to low speed centrifugation (10,000 × g for 20 min at 4 °C). The supernatant was collected, and colloidal material precipitated with 10% (wt/vol) PEG-6000 and 0.6 M NaCl after overnight stirring at 4 °C. Following low-speed centrifugation, the supernatant was discarded and the colloidal material were resuspended in 60 mL of TE buffer and clarified by further low-speed centrifugation. Colloidal material was pelleted by ultracentrifugation at 105,000 × g for 90 min at 4 °C, resuspended in 1 mL of TE buffer, and clarified by low-speed centrifugation at 10,000 rpm for 20 min (AccuSpin Micro; Fisher Scientific). Further fractionation was achieved by gradient centrifugation in a SW 50.1 rotor at 55,000 × g for 90 min at 4 °C of 0.5 mL of colloidal material through a cushion of 4.5-mL CsCl, density of 1.45 g/cm3. A layer of colloidal material below the interface was then collected with a finely drawn Pasteur pipette in a volume of ∼1.0 mL and repelleted by ultracentrifugation at 105,000 × g for 90 min at 4 °C before resuspension in the smallest volume of TE buffer. Purified AfuTmV-1 was negatively stained with 1% uranyl acetate on carbon-coated 400-mesh copper grids and examined in a transmission electron microscope (LEO 906E; Zeiss), or examined by AFM (see below), before storage at −80 °C. Purified AfuTmV-1 was also subjected to SDS/PAGE, and a single Coomassie blue-stained protein, 29 kDa in size, was analyzed by peptide mass fingerprinting broadly as described previously (5). Viral dsRNA was isolated from the final sample by SDS/phenol extraction (18).
AFM.
Because AFM is a nondestructive procedure, we chose to use it to examine purified AfuTmV-1 (50 ng/μL), which was diluted 50-fold in ultrapure water and 100 μL incubated on poly-l-lysine–coated mica for 1 min, rinsed with water (3 × 150 μL), and blown dry with argon. The deposited sample was then placed in the liquid cell of the AFM and filled up with 300 μL of butanol. Butanol enables minimization of the loading force so that the molecules can be resolved in contact mode. Observations were performed on an East Coast Scientific microscope operated in contact mode (also known as DC mode and constant-force mode) using a silicon nitride tip on a V-shaped cantilever (100-μm-long Budget Sensor SiNi; k = 0.4 N/m).
Determination of the 5′-Capped Status of AfuTmV-1 dsRNAs.
The oligo-cap method (RLM-RACE) was used with purified AfuTmV-1 dsRNAs, which was treated with calf intestinal phosphatase (CIP) and tobacco acid pyrophosphatase (TAP), to remove the cap structure from the positive strand and denatured in DMSO before ligation of an adaptor-oligonucleotide primer (5′-GCUGAUGGCGAUGAAUGAACAC-UGCGUUUGCUGGCUUUGAUGAAA-3′) to the 5′ terminus and RT-PCR using nested primers and known internal sequence primers on the AfuTmV-1 dsRNAs. The procedure was modified from the protocol provided with the Ambion First Choice RLM-RACE kit as described previously (35). Three different preparations of AfuTmV-1 dsRNAs (those treated with CIP and TAP, those treated with CIP only, and those untreated) were subjected to adaptor ligation and RT-PCR.
In Vitro Translation of AfuTmV-1 dsRNAs.
AfuTmV-1 dsRNAs were isolated from purified virus and treated with DNase and S1 nuclease as described above. Total dsRNAs, denatured at 95 °C for 5 min and immediately cooled on ice, were translated in vitro using the TNT Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]methionine as described by the manufacturer. In vitro translation products were analyzed by SDS/PAGE and autoradiography.
Protoplast Preparation and Transfection.
Protoplasts of A. fumigatus strain NK125, which is an isogenic line of isolate A. fumigatus Af293 cured of virus infection following cycloheximide treatment (17) and Af-237y, were generated from hyphae using a similar procedure to that originally described by Tang et al. (50) as modified by Szewczyk et al. (51) and Bhatti et al. (17). Purified AfVTmV-1 or AfVTmV-1 dsRNAs 1–4, AfuPV-1 and AfuPV-1 dsRNAs 1 and 2 were filtered through a Millipore filter and used to transfect protoplasts using PEG 6000 (10 μL at 0.05 μg/mL/2 × 107 protoplasts) in a procedure similar to one described previously for a mycoreovirus (52). Spore-producing colonies were then rescued and regenerated on an agar-based medium containing 5 mM ammonium tartrate, 1% glucose, 1 M sucrose, and ACM salt solution (17). Following incubation at 37 °C for 36–48 h, pooled colonies were inoculated into ACM broth and then onto agar plates and incubated at 37 °C for 4 d. To verify that cultures were transfected with AfuTmV-1 or AfuPV-1, the colony morphology of newly transfected isolates were compared with that of strains NK125, Af293, and Af-237y. Following transfection, dsRNA and ssRNA fractions were isolated by LiCl fractionation and analyzed as described above. Further confirmation of successful transfection with AfuTmV-1 was made following northern hybridization and probing of total RNA extracts prepared from the liquid cultures using the RNeasy Plant Mini Kit (Qiagen) and RT-PCR amplification of viral amplicons 337 and 467 bp in size using gene-specific primer pairs that were designed based on the sequences of AfuTmV-1 dsRNAs 1 and 2, respectively (AfuTmV-1 dsRNA 1 RdRP FOR, 5′-CTTACGGAGAC-AACCAGCTCTTC-3′, and AfuTmV-1 RdRP REV, 5′-CGCCCTGTAGACGGCGAGC-AG-3′; AfuTmV-1 dsRNA 2 gap FOR2, 5′-ATGTGCGGGAACCAGGACGTCGT-3′, and gap REV2, 5′-CGAACAGTGTATTGAGGGTGTC-3′). To confirm successful transfection with AfuPV-1, total RNA extracts were made as above and RT-PCR amplification of the entire RdRP ORF present in dsRNA 1 was attempted (17).
Supplementary Material
Acknowledgments
We thank Ren Zhang (Commonwealth Scientific and Industrial Research Organisation) and Matt Dickinson (University of Nottingham) for sequence data of flax rust Melampsora lini dsRNAs, and Profs. Eugene Koonin and Valerian Dolja for their advice on phylogenetic analyses. L.K. and S.O. thank the Thai and the Turkish Government education programs, respectively, for supporting their PhD programs. R.H.A.C. thanks The Leverhulme Trust for an Emeritus Fellowship award, and I.K.-L. acknowledges the support of the Erasmus European Union Programme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.S. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HG975302–HG975305).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419225112/-/DCSupplemental.
References
- 1.Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi. Annu Rev Phytopathol. 2009;47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 2.Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. 2009;10(1):115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mertens P. The dsRNA viruses. Virus Res. 2004;101(1):3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Chiba S, et al. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol. 2009;83(24):12801–12812. doi: 10.1128/JVI.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y-H, et al. A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology. 2012;426(1):42–50. doi: 10.1016/j.virol.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Kozlakidis Z, et al. Molecular characterization of the largest mycoviral-like double-stranded RNAs associated with Amasya cherry disease, a disease of presumed fungal aetiology. J Gen Virol. 2006;87(Pt 10):3113–3117. doi: 10.1099/vir.0.82121-0. [DOI] [PubMed] [Google Scholar]
- 7.Aoki N, et al. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res. 2009;140(1-2):179–187. doi: 10.1016/j.virusres.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Kozlakidis Z, et al. Sequence determination of a quadripartite dsRNA virus isolated from Aspergillus foetidus. Arch Virol. 2013;158(1):267–272. doi: 10.1007/s00705-012-1362-3. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci USA. 2014;111(33):12205–12210. doi: 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA. 2010;107(18):8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatti MF, Jamal A, Bignell EM, Petrou MA, Coutts RHA. Incidence of dsRNA mycoviruses in a collection of Aspergillus fumigatus isolates. Mycopathologia. 2012;174(4):323–326. doi: 10.1007/s11046-012-9556-5. [DOI] [PubMed] [Google Scholar]
- 12.Jamal A, Bignell EM, Coutts RHA. Complete nucleotide sequences of four dsRNAs associated with a new chrysovirus infecting Aspergillus fumigatus. Virus Res. 2010;153(1):64–70. doi: 10.1016/j.virusres.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Bhatti F, Bignell EM, Coutts RH. Complete nucleotide sequences of two dsRNAs associated with a new partitivirus infecting Aspergillus fumigatus. Arch Virol. 2011;156(9):1677–1680. doi: 10.1007/s00705-011-1045-5. [DOI] [PubMed] [Google Scholar]
- 14.Kozlakidis Z, et al. Molecular characterisation of two novel double-stranded RNA elements from Phlebiopsis gigantea. Virus Genes. 2009;39(1):132–136. doi: 10.1007/s11262-009-0364-z. [DOI] [PubMed] [Google Scholar]
- 15.Spear A, Sisterson MS, Yokomi R, Stenger DC. Plant-feeding insects harbor double-stranded RNA viruses encoding a novel proline-alanine rich protein and a polymerase distantly related to that of fungal viruses. Virology. 2010;404(2):304–311. doi: 10.1016/j.virol.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Özkan S, Coutts RHA. Aspergillus fumigatus mycovirus causes mild hypervirulent effect on pathogenicity when tested on Galleria mellonella. Fungal Genet Biol. 2015;76:20–26. doi: 10.1016/j.fgb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Bhatti MF, et al. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet Biol. 2011;48(11):1071–1075. doi: 10.1016/j.fgb.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Hong Y, Dover SL, Cole TE, Brasier CM, Buck KW. Multiple mitochondrial viruses in an isolate of the Dutch Elm disease fungus Ophiostoma novo-ulmi. Virology. 1999;258(1):118–127. doi: 10.1006/viro.1999.9691. [DOI] [PubMed] [Google Scholar]
- 19.Dodds JA. Association of type 1 viral-like dsRNA with club-shaped particles in hypovirulent strains of Endothia parasitica. Virology. 1980;107(1):1–12. doi: 10.1016/0042-6822(80)90267-6. [DOI] [PubMed] [Google Scholar]
- 20.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillman BI, Cai G. The family Narnaviridae: Simplest of RNA viruses. In: Ghabrial SA, editor. Advances in Virus Research 86. Elsevier; New York: 2013. pp. 149–176. [DOI] [PubMed] [Google Scholar]
- 22.Bruenn JA. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 1993;21(24):5667–5669. doi: 10.1093/nar/21.24.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28(5):375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 24.Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: Theoretical assignment of functional domains. J Gen Virol. 1990;71(Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 25.Kemena C, Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25(19):2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol. 2008;6(12):925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 27.Zhang T, Jiang Y, Dong W. A novel monopartite dsRNA virus isolated from the phytopathogenic fungus Ustilaginoidea virens and ancestrally related to a mitochondria-associated dsRNA in the green alga Bryopsis. Virology. 2014;462-463:227–235. doi: 10.1016/j.virol.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Nibert ML, et al. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014;188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Kozlakidis Z, Herrero N, Ozkan S, Bhatti MF, Coutts RHA. A novel dsRNA element isolated from the Aspergillus foetidus mycovirus complex. Arch Virol. 2013;158(12):2625–2628. doi: 10.1007/s00705-013-1779-3. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 31.Dickinson MJ, Zhang R, Pryor A. Nucleotide sequence relationships of double-stranded RNAs in flax rust, Melampsora lini. Curr Genet. 1993;24(5):428–432. doi: 10.1007/BF00351852. [DOI] [PubMed] [Google Scholar]
- 32.Howitt RLJ, Beever RE, Pearson MN, Forster RLS. Genome characterization of Botrytis virus F, a flexuous rod-shaped mycovirus resembling plant “potex-like” viruses. J Gen Virol. 2001;82(Pt 1):67–78. doi: 10.1099/0022-1317-82-1-67. [DOI] [PubMed] [Google Scholar]
- 33.Roossinck MJ, Sabanadzovic S, Okada R, Valverde RA. The remarkable evolutionary history of endornaviruses. J Gen Virol. 2011;92(Pt 11):2674–2678. doi: 10.1099/vir.0.034702-0. [DOI] [PubMed] [Google Scholar]
- 34.Fujimura T, Esteban R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc Natl Acad Sci USA. 2011;108(43):17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki N, Supyani S, Maruyama K, Hillman BI. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J Gen Virol. 2004;85(Pt 11):3437–3448. doi: 10.1099/vir.0.80293-0. [DOI] [PubMed] [Google Scholar]
- 36.Cai G, Krychiw JF, Myers K, Fry WE, Hillman BI. A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb dsRNA genome: The sixth member of a proposed new virus genus. Virology. 2013;435(2):341–349. doi: 10.1016/j.virol.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Magae Y. Molecular characterization of a novel mycovirus in the cultivated mushroom, Lentinula edodes. Virol J. 2012;9:60. doi: 10.1186/1743-422X-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickinson MJ, Pryor AJ. Encapsidated and unencapsidated double-stranded RNAs in flax rust, Melampsora lini. Can J Bot. 1989;67:1137–1142. [Google Scholar]
- 39.Brown JS, et al. Signature-tagged and directed mutagenesis identify PABA synthetase as essential for Aspergillus fumigatus pathogenicity. Mol Microbiol. 2000;36(6):1371–1380. doi: 10.1046/j.1365-2958.2000.01953.x. [DOI] [PubMed] [Google Scholar]
- 40.Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113(1):51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 41.Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 42.Covey SN, Hull R. Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology. 1981;111(2):463–474. doi: 10.1016/0042-6822(81)90349-4. [DOI] [PubMed] [Google Scholar]
- 43.Diaz-Ruiz JR, Kaper JM. Isolation of viral double-stranded RNAs using a LiCl fractionation procedure. Prep Biochem. 1978;8(1):1–17. doi: 10.1080/00327487808068215. [DOI] [PubMed] [Google Scholar]
- 44.Coutts RHA, et al. Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. II. Characterization of a new species in the genus Partitivirus. J Gen Virol. 2004;85(Pt 11):3399–3403. doi: 10.1099/vir.0.80182-0. [DOI] [PubMed] [Google Scholar]
- 45.Froussard P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992;20(11):2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 47.Coutts RHA, Livieratos IC. A rapid method for sequencing the 5′- and 3′-termini of dsRNA viral templates using RLM-RACE. J Phytopathol. 2003;151:525–527. [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang CM, Cohen J, Holden DW. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol Microbiol. 1992;6(12):1663–1671. doi: 10.1111/j.1365-2958.1992.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 51.Szewczyk E, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1(6):3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 52.Hillman BI, Supyani S, Kondo H, Suzuki N. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J Virol. 2004;78(2):892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.