Abstract
Domestic animals can be cloned using techniques such as embryo splitting and nuclear transfer to produce genetically identical individuals. Although embryo splitting is limited to the production of only a few identical individuals, nuclear transfer of donor nuclei into recipient oocytes, whose own nuclear DNA has been removed, can result in large numbers of identical individuals. Moreover, clones can be produced using donor cells from sterile animals, such as steers and geldings, and, unlike their genetic source, these clones are fertile. In reality, due to low efficiencies and the high costs of cloning domestic species, only a limited number of identical individuals are generally produced, and these clones are primarily used as breed stock. In addition to providing a means of rescuing and propagating valuable genetics, somatic cell nuclear transfer (SCNT) research has contributed knowledge that has led to the direct reprogramming of cells (e.g., to induce pluripotent stem cells) and a better understanding of epigenetic regulation during embryonic development. In this review, I provide a broad overview of the historical development of cloning in domestic animals, of its application to the propagation of livestock and transgenic animal production, and of its scientific promise for advancing basic research.
Keywords: SCNT, cloning, nuclear transfer, embryo, livestock
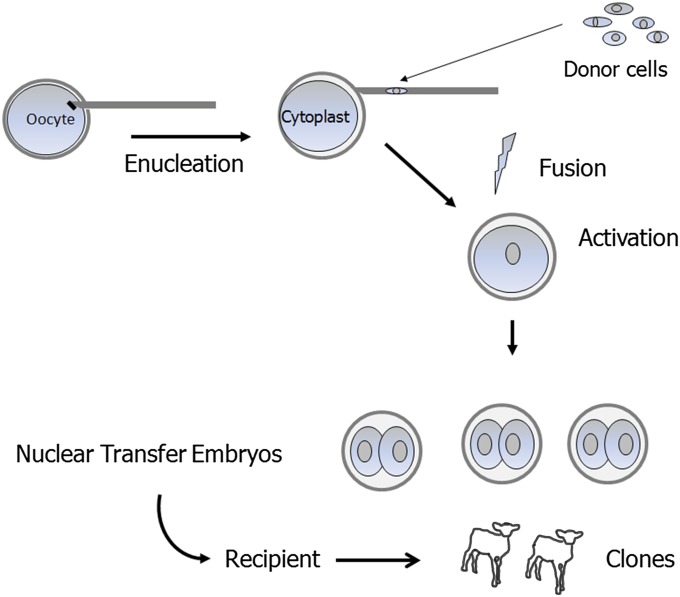
The word clone can mean different things to different people. In molecular biology, it refers to the process of making identical copies of DNA. In cell biology, it is the propagation of a progenitor cell to obtain a population of genetically identical cells whereas, in animal biology, cloning refers to the production of genetic copies of individual animals using nuclear transfer. Advanced reproductive methods involving microsurgery, embryo culture, and transfer into recipients (surrogate mothers) are required to produce animal clones (Fig. 1). More specifically, a nucleus from a cell of the donor individual is inserted into an oocyte whose own nuclear DNA has been removed (enucleation). This reconstructed oocyte is activated to continue embryonic development. Embryos resulting from this procedure can result in the production of a live, genetically identical individual after transfer into a recipient, although at a relative low efficiency (Table 1). The fact that such a complex procedure works at all is amazing and is the result of decades of pioneering research. In this review, the historical work in domestic species leading up to the development of somatic cell nuclear transfer (SCNT), along with the practical applications of this technology, will be discussed. As illustrated below, basic research regarding the biological mechanisms of SCNT has led to scientific advances in the areas of reprogramming, cell fate determination, and epigenetic regulation during development. Moreover, as discussed in the following sections, SCNT in domestic animals will continue to provide promising scientific and practical insights through its application to transgenic and biomedical models.
Fig. 1.
Somatic cell nuclear transfer process.
Table 1.
Cloning efficiencies in domestic livestock
| Species | Efficiency per reconstructed oocyte, % | Efficiency per transferred SCNT embryo, % | Refs. |
| Cattle | 1.7 | 11.5 | 1 |
| Goat | 6 | 6 | 2 |
| Horse | 0.8 | 19 | 3 |
| Pig | 0.3 | 5–13 | 4, 5 |
| Sheep | 0.3 | 3.4–5.9 | 6–8 |
Estimates based on selected publications that provide sufficient datasets to determine cloning efficiencies based both on a per reconstructed oocyte and a transferred embryo basis. Transferred embryos are often highly selected and can represent a very small subset of SCNT embryos produced.
Setting the Stage—Historical and Evolutionary Perspective
How much have we diverged from nature’s method of cloning? Even omitting most other forms of plant and animal life and focusing on vertebrates—animals with backbones—examples of clones abound in nature. Identical twins are the obvious examples, but perhaps more intriguing are armadillos, in which the offspring in a litter are all clones derived from one zygote (9). The simplest form of artificial cloning is embryo splitting—separating the blastomeres of an early embryo and forming two or more smaller embryos. Initial studies were performed to ask key questions regarding control of lineage development: When is a cell’s fate set and how plastic is that fate? Studies in amphibians, rabbits, and mice suggested that the very early cleavage stages (two-cell to four-cell) were flexible and that each blastomere could yield a viable blastocyst. At later stages, the blastomeres could no longer independently form a viable blastocyst due to the loss of mass as each blastomere underwent cleavage division. That did not mean that the blastomere nucleus was incapable of directing full development, but rather that it was unable to stop the developmental clock and replace the lost mass before continuing. Prior to the blastocyst stage, cells in the early embryo, called blastomeres, divide without increasing in mass between each division: thus the term cleavage divisions—each cell cleaves in half. This constraint led to the obvious question: If you provide additional mass, can later staged blastomeres—or more appropriately—can the nucleus of a later staged blastomere direct complete development to the blastocyst stage and be capable of continued development resulting in a normal, live offspring? Pioneering studies in the 1950s and 1960s in frogs demonstrated that nuclei from embryos up to the tadpole stage were capable of directing normal development, resulting in adult individuals, but that nuclei from adult tissues were able to direct development only to the tadpole stage (10, 11). Despite the failure to obtain adult individuals after nuclear transfer of adult cells, the studies did demonstrate the developmental plasticity of differentiated, somatic cell nuclei. The unknown (at that time) regulatory mechanisms controlling cell-specific gene expression could be reset back to the embryo stage.
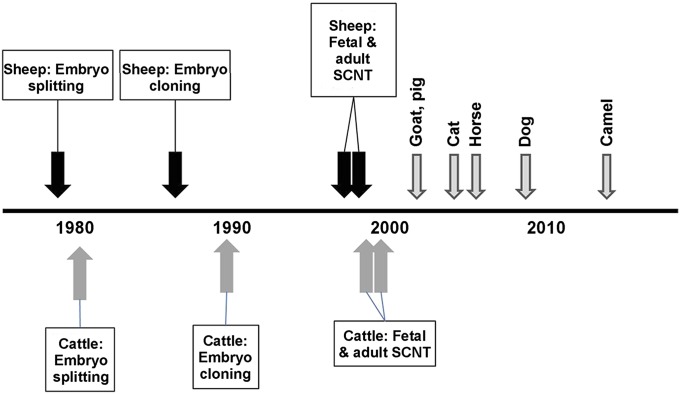
Systematic examinations of mammal embryonic plasticity could not be conducted until appropriate in vitro culture conditions had been established in the 1960s and 1970s (12–14). Subsequently, controversial studies in the 1970s suggested that nuclei from cells that had undergone the first lineage differentiation (that is, cells that had formed the inner cell mass) could direct normal development if substituted for the zygotic nucleus (15). However, failure of other research groups to replicate these studies led some scientist to state that mammalian nuclei after embryonic gene activation were unable to direct development due to irreversible programming changes (16). At this time, advances in reproductive technologies involving farm animals, primarily sheep and cattle, allowed animal scientists to adapt such techniques as embryo splitting and blastomere cloning with a focus on improving production efficiencies and genetic advancement, in addition to asking questions about developmental plasticity. Because developmental biologists, focused on more traditional models (e.g., mouse and frogs), tend not to read the more agricultural-related science journals, the advances made by animal scientists were largely unrecognized until the production of Dolly merited publication in Nature (6). Although the fact that an adult nucleus could indeed direct normal development (resulting in a live offspring) was revolutionary for developmental biology, it followed a series of discoveries that suggested such a possibility (Fig. 2).
Fig. 2.
Timeline of key points during development of SCNT in domestic livestock (6, 17–33).
The initial attempts to artificially clone domestic animals involved embryo splitting. Steen Willadsen demonstrated that twins could be produced in sheep (17) and cattle (18) after splitting of cleavage-staged embryos and transfer of the demi-embryos into recipients. These studies demonstrated that triplets and even quadruplets could be obtained, albeit at lower frequencies due to the loss of cellular mass. To overcome this limitation, Willadsen used a modified version of the nuclear transfer technique that had been used in the earlier amphibian cloning studies described above (Fig. 1). In brief, the oocyte’s DNA was removed by aspirating the portion of ooplasm containing the chromosomes, thus forming a cytoplast; the donor cell was placed abutting the ooplasmic membrane; and the cytoplast and donor cell were fused together. Using this procedure, Willadsen obtained live lambs (19) and calves (20) after transfer of the reconstructed embryos into surrogates. Subsequently, several other research groups with ties to the agricultural industry began exploring the possibilities of embryo and embryonic cell nuclear transfer, achieving success with progressively later stages of embryos (Table 2). In 1996, researchers at the Roslin Research Institute reported successful production of live lambs using long-term cultured embryonic (21) and even transgenic (22) cells. These achievements were soon followed by the report of the production of a lamb (Dolly) using cultured somatic cells that had been obtained from an adult (6). Although much has been made of the low efficiency of somatic cell nuclear transfer (SCNT)—Dolly was the single live offspring that resulted from 29 transferred reconstructed embryos for which 247 oocytes had been manipulated—the fact that a live lamb was produced is still astounding.
Table 2.
Use of progressively more advances staged nuclei for SCNT in cattle
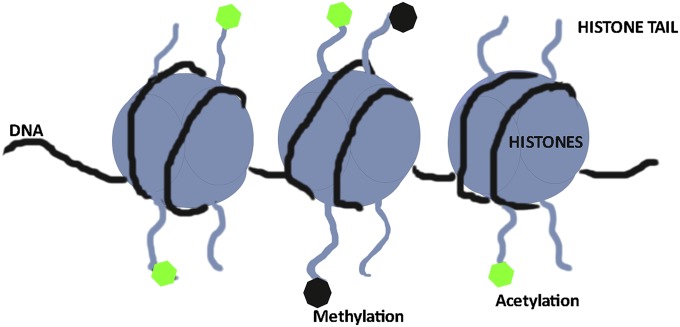
To fully comprehend the barriers to artificial cloning, one must first understand the processes of gametogenesis and fertilization. During mammalian development, the primordial germ cells in the developing fetus migrate to the gonadal ridges and, depending on the sex of the fetus, form either oogonia or spermatogonia. DNA methylation patterns are established such that sperm are hypermethylated and oocytes are hypomethylated compared with somatic cells (34). After fertilization, the sperm DNA undergoes active demethylation, and the maternal DNA undergoes passive demethylation. Some genes maintain a paternal or maternal imprint such that active transcription occurs only from one parental chromosome; these imprinted genes often play critical roles in placentation: e.g., the imprinted growth factor, IGF2, is expressed by the paternal and not the maternal allele whereas the receptor, IGF2R, is maternally expressed (35). Although these imprints are generally described by their DNA methylation patterns, the complex mechanisms and levels of imprinting, including histone modifications, are still being deciphered. Why is the imprint important during artificial cloning? After all, the donor cells have both maternally and paternally derived chromosome sets. During development, as cells undergo lineage differentiation, epigenetic changes are continually being established that alter the genetic program for cell-specific gene expression. These changes include multiple levels of epigenetic alterations, including DNA methylation and posttranslational modifications of histone proteins (Fig. 3). During reprogramming by the ooplasm, these patterns must be reset back to the zygotic patterns. This requirement places a strain on the capabilities of the reprogramming factors within the ooplasm as they have evolved to reset the paternal and maternal gametic epigenetic patterns, not that of a somatic cell. Therefore, it is not surprising that oocytes are not able to fully and correctly reprogram the somatic epigenome. In fact, it is surprising that they are capable of achieving a sufficiently appropriate epigenome that allows full development. As remarked by other authors, nature allows a certain amount of flexibility in the epigenome and gene expression during growth and development (28). As might be anticipated, clones that fail during gestation and/or have physiological abnormalities have been found to have abnormal epigenetic patterns whereas those that thrive have a comparatively normal pattern (28, 36). It is also interesting to note that the gestational losses and abnormalities observed in SCNT were also noted during the development of in vitro embryo production and culture techniques in domestic species in the 1990s. In sheep and cattle, affected offspring were typically larger than normal; thus, the term “large offspring syndrome” was coined (37). It was determined that exposure to serum and coculture altered embryonic epigenetic methylation patterns (38, 39). With improvements in culture media, the incidence of large offspring after in vitro embryo production no longer seems to be the issue it once was although the more subtle epigenetic changes that may have long-term consequences on offspring health are of great interest. A number of research groups are exploring these subtle epigenetic changes that can occur during gamete, embryo, and early pregnancy with potentially long-term consequences on offspring (40, 41).
Fig. 3.
Epigenetic factors regulate DNA availability to transcriptional machinery (transcription factors, polymerases, etc.) and are involved in the control of cell tissue-specific gene expression.
Propagation of Genetics
Once lambs and calves had been produced using embryonic and fetal cells, publications dealing with SCNT research escalated from a few studies a year to hundreds. At this time, more than twenty different species have been cloned by SCNT techniques although not all offspring have survived long-term. Of particular interest were the advantages that embryo cloning and SCNT offered for the propagation of valued genomes, whether for animal production purposes or rescue of rare genotypes. Animal genetics companies that sell semen and embryos for genetic improvement of dairy and beef herds could take advantage of SCNT to expand their products. Clones of valued dams and sires can be produced by SCNT, thus extending their reproductive output potential. SCNT can also be used to propagate hybrids, to increase the speed of genetic gain through selection of animals with superior phenotype and genotype, and even to replicate animals with advantageous genotypes whether or not that animal is fertile (e.g., steers and geldings). Examples include the cloning of a prized Texas Longhorn steer (42) and racing mules (43). Moreover, animals that have died can be cloned as long as viable cell samples were collected and stored. A resurrected prized bull, Starbuck II, produced daughters that had normal chromosome stability (telomere lengths) and hematological, physiological, and reproductive parameters (44) although his semen was never sold commercially due to Canadian governmental regulations. In Texas, beef cattle have been resurrected based on their carcass traits (45). Unlike the steer from which the carcass was obtained, these cloned calves can look forward to siring offspring that are expected to have superior meat production traits.
In addition to propagation and/or replication of domestic species, SCNT has been used to propagate genetics of endangered species with mixed success because often this process involves interspecies SCNT (46). In fact, most efforts involving interspecies transfers, in which an oocyte from one species is used as the recipient for a nucleus from another species, have not been successful. Only a few cases of interspecies SCNT between closely related species have resulted in the actual production of offspring. Frequently, these animals do not thrive and die relatively soon after birth. As a case in point, the first gaur calf that was produced by transplantation of a gaur nucleus into a bovine oocyte died shortly after birth (47) although more recent attempts seem to have been more successful (48). Success has also been achieved between endangered cats using oocytes from domestic cat as recipient (46). Studies suggest that the frequent failures of interspecies SCNT are due to a number of factors including incomplete activation of the embryonic genome and nuclear–mitochondrial incompatibilities (49).
SCNT and Transgenic Animal Production
SCNT may provide the most advantages for the production of transgenic animals. Although transgenic animal production is an efficient process in mice in which multiple methods can be used, including pronuclear microinjection of DNA constructs, chimera production using transfected embryonic stem cells (ESCs), or SCNT using transgenic donor cells, only the last method has any practical application in domestic species. Pronuclear injection has resulted in transgenic pigs, sheep, goats, and cattle, but at much lower efficiencies than mice and at much greater costs (50, 51). SCNT allows production of transgenic offspring after selection and characterization of donor cells. This process ensures that the offspring are transgenic and have the appropriate number of copies of the transgene, and virtually ensures that the animal contains and expresses the transgene. Transgenic cattle, goats, pigs, and sheep have been produced that express industrial proteins (e.g., spider silk), biopharmaceuticals (e.g., antithrombin), and human polyclonal antibodies (51). Moreover, animals have been produced with modified production traits, including increased casein in the milk (52), altered fatty acid composition (53), and resistance to mastitis (54). Prion protein, the causative agent in mad cow disease, has been knocked out in cattle using SCNT (55). Despite advantages conferred by these transgenic modifications to production traits or disease resistance, none of these animals will be used for food production due to regulatory roadblocks (56, 57). Thus far, only one product from a transgenic animal has been approved in the United States and Europe for human use, and that is the biotherapeutic protein antithrombin (ATryn). Whether transgenic animals ever fulfill the animal production-related promise researchers envisaged will depend on societal acceptance and revised regulatory guidelines. More likely is progress in establishing medical models of human and animal disease for biomedical research. SCNT in domestic animals has been used to study the potential of regenerative medicine. For example, cloned pigs have served as both controls and recipients for neural stem cells, demonstrating the potential for spinal cord repair (58). SCNT has also been used to produce medical models for cystic fibrosis, diabetes, retinitis pigmentosa, cancer, amyotrophic lateral sclerosis, and other diseases (58–60). These newly developed models hold great promise for providing insight into diseases and should lead to new therapeutic treatments.
Basic Research Questions
SCNT is also being used to answer basic questions in developmental and reproductive biology. The majority of publications on SCNT describe efforts to overcome the inefficiencies of the process itself. These reports include attempts to identify the best source and treatment of donor cells, improved oocyte activation protocols, and methods of chemically assisted reprogramming. In the latter case, either donor cells before SCNT or the reconstructed embryo afterward are exposed to chemicals thought to either stimulate or inhibit various enzymes involved in remodeling chromatin. Many of these publications report conflicting findings, and rarely are a sufficient number of embryo transfers performed to establish credible evidence of any real improvements in efficiencies. Nevertheless, chemically assisted reprogramming during SCNT does hold promise (61).
Due to the cost and extended generation times for domestic species, most of these studies, so far, have focused on early embryonic and fetal development. As with epigenetic patterns described above, many of these studies report either the normal expression of key genes after a reconstructed embryo passes a developmental checkpoint (e.g., blastocyst formation) or abnormal gene expression after failure to pass such checkpoints (28, 36). These experiments focused mainly on how SCNT worked or did not work, and few use SCNT to explore questions regarding developmental regulation of genes. One exception includes the use of SCNT to explore the differential regulation of POU5f1 (OCT4), a key transcription factor involved in trophectoderm formation, in bovine and mouse blastocysts (62). This study provided insight into species-specific gene regulation during early development that would not have been achievable without SCNT.
In a limited number of cases, SCNT has been used to test hypotheses that could not be easily answered through other methods. For example, reproductive immunologists had long questioned the role of foreign paternal antigens in the establishment and maintenance of pregnancy. Cesare Galli et al. used SCNT to investigate this question. Cultured somatic cells from a mare were used as nuclear donors in SCNT. Two of the resulting cloned embryos were then transferred into the same mare, resulting in the establishment of a full-term pregnancy and the birth of a live foal (63). This birth demonstrated that a mare could successfully carry a pregnancy initiated by her own identical clone, which implied that foreign paternal antigens are not necessary for establishing a viable, full-term pregnancy (63). Additional studies using SCNT could help further decipher the role of paternal antigens during pregnancy (64).
Owing to time and cost commitments, only a few studies have looked at the long-term consequences of cloning on such physiological parameters as reproductive performance or health. A large-scale project involving 96 cow clones and 40 corresponding genetic donors, as comparative controls, was carried out over a 6-y period. In this longitudinal study, Polejaeva et al. (65) determined that the ability of clones to produce transferrable-quality embryos after artificial insemination or in vitro embryo production was not different from that of their genetic comparatives that had been produced through normal breeding practices. Other work has focused on the health of clones: these studies suggest that clones that survive the critical neonatal period are generally normal physiologically. Cattle clones surviving greater than 200 d were found to be essentially equivalent in terms of animal health and milk and meat production performance as conventionally bred cattle (66). These studies support the feasibility of using clones in various comparative studies. For example, multiple copies of genetically identical embryos produced by SCNT can be frozen and subsequently transferred at predetermined intervals, resulting in genetically identical individuals of different ages. This approach was used in a recent publication in which the method of ovarian stimulation, but not maternal age, was found to be associated with lower mitochondrial copy number in oocytes obtained from genetically identical cow clones of different ages (67). Future studies taking advantage of such unique research opportunities provided by SCNT may help answer questions and solve technical issues in reproductive medicine and regenerative medicine.
Conclusion
In general, SCNT efficiencies have improved only marginally over the past decade, with the generally accepted rate of 5–15% of transferred embryos resulting in live offspring (28). Direct comparisons of efficiencies reported by various research groups are often difficult because only subsets of embryos may have been transferred or reported. Strict selection of embryos for transfer can result in improved pregnancy rates, per cloned embryo transferred, that do not reflect the true viability of the full set of reconstructed embryos. Nevertheless, SCNT research has contributed knowledge that has led to the direct reprogramming of cells (e.g., inducing pluripotent stem cells) and to better understanding of epigenetic regulation during embryonic development and has provided means of propagating and rescuing valuable genetics and establishing large-animal biomedical models.
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IX: Clonal Reproduction: Alternatives to Sex,” held January 9–10, 2015, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_IX_Clonal_Reproduction.
This article is a PNAS Direct Submission.
References
- 1.Wells DN, et al. Coordination between donor cell type and cell cycle stage improves nuclear cloning efficiency in cattle. Theriogenology. 2003;59(1):45–59. doi: 10.1016/s0093-691x(02)01273-6. [DOI] [PubMed] [Google Scholar]
- 2.Keefer CL, et al. Production of cloned goats after nuclear transfer using adult somatic cells. Biol Reprod. 2002;66(1):199–203. doi: 10.1095/biolreprod66.1.199. [DOI] [PubMed] [Google Scholar]
- 3.Hinrichs K, Choi YH, Varner DD, Hartman DL. Production of cloned horse foals using roscovitine-treated donor cells and activation with sperm extract and/or ionomycin. Reproduction. 2007;134(2):319–325. doi: 10.1530/REP-07-0069. [DOI] [PubMed] [Google Scholar]
- 4.Mao J, et al. Oxamflatin treatment enhances cloned porcine embryo development and nuclear reprogramming. Cell Reprogram. 2014;17(1):28–40. doi: 10.1089/cell.2014.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callesen H, Liu Y, Pedersen HS, Li R, Schmidt M. Increasing efficiency in production of cloned piglets. Cell Reprogram. 2014;16(6):407–410. doi: 10.1089/cell.2014.0053. [DOI] [PubMed] [Google Scholar]
- 6.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher CJ, Roberts CT, Hartwich KM, Walker SK, McMillen IC. Somatic cell nuclear transfer in the sheep induces placental defects that likely precede fetal demise. Reproduction. 2007;133(1):243–255. doi: 10.1530/rep.1.01203. [DOI] [PubMed] [Google Scholar]
- 8.Rathbone AJ, Fisher PA, Lee JH, Craigon J, Campbell KH. Reprogramming of ovine somatic cells with Xenopus laevis oocyte extract prior to SCNT improves live birth rate. Cell Reprogram. 2010;12(5):609–616. doi: 10.1089/cell.2010.0015. [DOI] [PubMed] [Google Scholar]
- 9.Prodöhl PA, Loughry WJ, McDonough CM, Nelson WS, Avise JC. Molecular documentation of polyembryony and the micro-spatial dispersion of clonal sibships in the nine-banded armadillo, Dasypus novemcinctus. Proc Biol Sci. 1996;263(1377):1643–1649. doi: 10.1098/rspb.1996.0240. [DOI] [PubMed] [Google Scholar]
- 10.Briggs R, King TJ. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc Natl Acad Sci USA. 1952;38(5):455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurdon JB. From nuclear transfer to nuclear reprogramming: The reversal of cell differentiation. Annu Rev Cell Dev Biol. 2006;22:1–22. doi: 10.1146/annurev.cellbio.22.090805.140144. [DOI] [PubMed] [Google Scholar]
- 12.McLaren A, Biggers JD. Successful development and birth of mice cultivated in vitro as early as early embryos. Nature. 1958;182:877–878. doi: 10.1038/182877a0. [DOI] [PubMed] [Google Scholar]
- 13.Whitten WK. Culture of tubal mouse ova. Nature. 1956;177(4498):96. doi: 10.1038/177096a0. [DOI] [PubMed] [Google Scholar]
- 14.Whitten WK, Biggers JD. Complete development in vitro of the pre-implantation stages of the mouse in a simple chemically defined medium. J Reprod Fertil. 1968;17(2):399–401. doi: 10.1530/jrf.0.0170399. [DOI] [PubMed] [Google Scholar]
- 15.Illmensee K, Hoppe PC. Nuclear transplantation in Mus musculus: Developmental potential of nuclei from preimplantation embryos. Cell. 1981;23(1):9–18. doi: 10.1016/0092-8674(81)90265-8. [DOI] [PubMed] [Google Scholar]
- 16.McGrath J, Solter D. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro. Science. 1984;226(4680):1317–1319. doi: 10.1126/science.6542249. [DOI] [PubMed] [Google Scholar]
- 17.Willadsen SM. A method for culture of micromanipulated sheep embryos and its use to produce monozygotic twins. Nature. 1979;277(5694):298–300. doi: 10.1038/277298a0. [DOI] [PubMed] [Google Scholar]
- 18.Willadsen SM, Polge C. Attempts to produce monozygotic quadruplets in cattle by blastomere separation. Vet Rec. 1981;108(10):211–213. doi: 10.1136/vr.108.10.211. [DOI] [PubMed] [Google Scholar]
- 19.Willadsen SM. Nuclear transplantation in sheep embryos. Nature. 1986;320(6057):63–65. doi: 10.1038/320063a0. [DOI] [PubMed] [Google Scholar]
- 20.Willadsen SM. Cloning of sheep and cow embryos. Genome. 1989;31(2):956–962. doi: 10.1139/g89-167. [DOI] [PubMed] [Google Scholar]
- 21.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380(6569):64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 22.Schnieke AE, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278(5346):2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 23.Stice SL, Keefer CL. Multiple generational bovine embryo cloning. Biol Reprod. 1993;48(4):715–719. doi: 10.1095/biolreprod48.4.715. [DOI] [PubMed] [Google Scholar]
- 24.Peura TT, Lane MW, Lewis IM, Trounson AO. Development of bovine embryo-derived clones after increasing rounds of nuclear recycling. Mol Reprod Dev. 2001;58(4):384–389. doi: 10.1002/1098-2795(20010401)58:4<384::AID-MRD5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Prather RS, et al. Nuclear transplantation in the bovine embryo: Assessment of donor nuclei and recipient oocyte. Biol Reprod. 1987;37(4):859–866. doi: 10.1095/biolreprod37.4.859. [DOI] [PubMed] [Google Scholar]
- 26.Collas P, Barnes FL. Nuclear transplantation by microinjection of inner cell mass and granulosa cell nuclei. Mol Reprod Dev. 1994;38(3):264–267. doi: 10.1002/mrd.1080380306. [DOI] [PubMed] [Google Scholar]
- 27.Keefer CL, Stice SL, Matthews DL. Bovine inner cell mass cells as donor nuclei in the production of nuclear transfer embryos and calves. Biol Reprod. 1994;50(4):935–939. doi: 10.1095/biolreprod50.4.935. [DOI] [PubMed] [Google Scholar]
- 28.Long CR, Westhusin ME, Golding MC. Reshaping the transcriptional frontier: Epigenetics and somatic cell nuclear transfer. Mol Reprod Dev. 2014;81(2):183–193. doi: 10.1002/mrd.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemann H, Tian XC, King WA, Lee RS. Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning. Reproduction. 2008;135(2):151–163. doi: 10.1530/REP-07-0397. [DOI] [PubMed] [Google Scholar]
- 30.Cibelli JB, et al. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280(5367):1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 31.Forsberg EJ, et al. Production of cloned cattle from in vitro systems. Biol Reprod. 2002;67(1):327–333. doi: 10.1095/biolreprod67.1.327. [DOI] [PubMed] [Google Scholar]
- 32.Kato Y, et al. Eight calves cloned from somatic cells of a single adult. Science. 1998;282(5396):2095–2098. doi: 10.1126/science.282.5396.2095. [DOI] [PubMed] [Google Scholar]
- 33.Wells DN, Misica PM, Tervit HR, Vivanco WH. Adult somatic cell nuclear transfer is used to preserve the last surviving cow of the Enderby Island cattle breed. Reprod Fertil Dev. 1998;10(4):369–378. doi: 10.1071/r98109. [DOI] [PubMed] [Google Scholar]
- 34.Seisenberger S, et al. Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh C, et al. The non-viability of uniparental mouse conceptuses correlates with the loss of the products of imprinted genes. Mech Dev. 1994;46(1):55–62. doi: 10.1016/0925-4773(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 36.Li G, et al. Dysregulation of genome-wide gene expression and DNA methylation in abnormal cloned piglets. BMC Genomics. 2014;15:811. doi: 10.1186/1471-2164-15-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3(3):155–163. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- 38.Hill JR. Incidence of abnormal offspring from cloning and other assisted reproductive technologies. Annu Rev Anim Biosci. 2014;2:307–321. doi: 10.1146/annurev-animal-022513-114109. [DOI] [PubMed] [Google Scholar]
- 39.Young LE, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27(2):153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 40.Gardner DK, Lane M. Ex vivo early embryo development and effects on gene expression and imprinting. Reprod Fertil Dev. 2005;17(3):361–370. doi: 10.1071/rd04103. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair KD. Assisted reproductive technologies and pregnancy outcomes: Mechanistic insights from animal studies. Semin Reprod Med. 2008;26(2):153–161. doi: 10.1055/s-2008-1042954. [DOI] [PubMed] [Google Scholar]
- 42.Leford A. A breed apart. Nature. 2006;444(7116):137. doi: 10.1038/444137a. [DOI] [PubMed] [Google Scholar]
- 43.Woods GL, et al. A mule cloned from fetal cells by nuclear transfer. Science. 2003;301(5636):1063. doi: 10.1126/science.1086743. [DOI] [PubMed] [Google Scholar]
- 44.Ortegon H, et al. Genomic stability and physiological assessments of live offspring sired by a bull clone, Starbuck II. Theriogenology. 2007;67(1):116–126. doi: 10.1016/j.theriogenology.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 45.Hawkins D, Lawrence T. Proceedings of the 23rd Range Beef Cow Symposium. Angus Association; St. Joseph, MO: 2013. From imagination to reality: Using DNA from an exceptional carcass to produce a sire or donor cow; pp. 109–111. [Google Scholar]
- 46.Gómez MC, et al. Cloning endangered felids using heterospecific donor oocytes and interspecies embryo transfer. Reprod Fertil Dev. 2009;21(1):76–82. doi: 10.1071/rd08222. [DOI] [PubMed] [Google Scholar]
- 47.Vogel G. Endangered species: Cloned gaur a short-lived success. Science. 2001;291(5503):409. doi: 10.1126/science.291.5503.409a. [DOI] [PubMed] [Google Scholar]
- 48.Srirattana K, et al. Full-term development of gaur-bovine interspecies somatic cell nuclear transfer embryos: Effect of trichostatin A treatment. Cell Reprogram. 2012;14(3):248–257. doi: 10.1089/cell.2011.0099. [DOI] [PubMed] [Google Scholar]
- 49.Lagutina I, Fulka H, Lazzari G, Galli C. Interspecies somatic cell nuclear transfer: Advancements and problems. Cell Reprogram. 2013;15(5):374–384. doi: 10.1089/cell.2013.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eyestone WH. Challenges and progress in the production of transgenic cattle. Reprod Fertil Dev. 1994;6(5):647–652. doi: 10.1071/rd9940647. [DOI] [PubMed] [Google Scholar]
- 51.Keefer CL. Production of bioproducts through the use of transgenic animal models. Anim Reprod Sci. 2004;82-83:5–12. doi: 10.1016/j.anireprosci.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Brophy B, et al. Cloned transgenic cattle produce milk with higher levels of beta-casein and kappa-casein. Nat Biotechnol. 2003;21(2):157–162. doi: 10.1038/nbt783. [DOI] [PubMed] [Google Scholar]
- 53.Lai L, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006;24(4):435–436. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wall RJ, et al. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat Biotechnol. 2005;23(4):445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- 55.Richt JA, et al. Production of cattle lacking prion protein. Nat Biotechnol. 2007;25(1):132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells KD. Natural genotypes via genetic engineering. Proc Natl Acad Sci USA. 2013;110(41):16295–16296. doi: 10.1073/pnas.1315623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller HI. FDA on transgenic animals: A dog’s breakfast? Nat Biotechnol. 2008;26(2):159–160. doi: 10.1038/nbt0208-159. [DOI] [PubMed] [Google Scholar]
- 58.Piedrahita JA, Olby N. Perspectives on transgenic livestock in agriculture and biomedicine: An update. Reprod Fertil Dev. 2011;23(1):56–63. doi: 10.1071/RD10246. [DOI] [PubMed] [Google Scholar]
- 59.Gong J, et al. Activating the expression of human K-rasG12D stimulates oncogenic transformation in transgenic goat fetal fibroblast cells. PLoS ONE. 2014;9(3):e90059. doi: 10.1371/journal.pone.0090059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chieppa MN, et al. Modeling amyotrophic lateral sclerosis in hSOD1 transgenic swine. Neurodegener Dis. 2014;13(4):246–254. doi: 10.1159/000353472. [DOI] [PubMed] [Google Scholar]
- 61.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg DK, et al. Trophectoderm lineage determination in cattle. Dev Cell. 2011;20(2):244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Galli C, et al. Pregnancy: A cloned horse born to its dam twin. Nature. 2003;424(6949):635. doi: 10.1038/424635a. [DOI] [PubMed] [Google Scholar]
- 64.Noronha LE, Antczak DF. Maternal immune responses to trophoblast: The contribution of the horse to pregnancy immunology. Am J Reprod Immunol. 2010;64(4):231–244. doi: 10.1111/j.1600-0897.2010.00895.x. [DOI] [PubMed] [Google Scholar]
- 65.Polejaeva IA, et al. Longitudinal study of reproductive performance of female cattle produced by somatic cell nuclear transfer. PLoS ONE. 2013;8(12):e84283. doi: 10.1371/journal.pone.0084283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe S. Effect of calf death loss on cloned cattle herd derived from somatic cell nuclear transfer: Clones with congenital defects would be removed by the death loss. Anim Sci J. 2013;84(9):631–638. doi: 10.1111/asj.12087. [DOI] [PubMed] [Google Scholar]
- 67.Cree LM, et al. Maternal age and ovarian stimulation independently affect oocyte mtDNA copy number and cumulus cell gene expression in bovine clones. Hum Reprod. 2015 doi: 10.1093/humrep/dev066. [DOI] [PubMed] [Google Scholar]