Significance
Tuberculosis (TB) infection induces up-regulation of T cell-inhibitory molecules on CD8+ T cells, which may induce impairment of CD8+ T-cell immunity. However, how T cell-inhibitory molecules regulate CD8+ T-cell immune responses during TB infection remains unclear. Here, we demonstrate that CD244, a T cell-inhibitory molecule, mediates inhibition of IFN-γ and TNF-α expression through inducing expression of a long noncoding RNA (lncRNA)-CD244. lncRNA-CD244 physically interacts with a chromatin-modification enzyme, enhancer of zeste homolog 2 (EZH2), and mediates modification of a more repressive chromatin state in infg and tnfa loci. Knock down of lncRNA-CD244 significantly enhances IFN-γ and TNF-α expression and improves protective immunity of CD8+ T cells. This study therefore uncovers a previously unknown mechanism for T-cell immune responses regulated by lncRNA during TB infection.
Keywords: tuberculosis, lncRNA, CD8+ T cells
Abstract
Molecular mechanisms for T-cell immune responses modulated by T cell-inhibitory molecules during tuberculosis (TB) infection remain unclear. Here, we show that active human TB infection up-regulates CD244 and CD244 signaling-associated molecules in CD8+ T cells and that blockade of CD244 signaling enhances production of IFN-γ and TNF-α. CD244 expression/signaling in TB correlates with high levels of a long noncoding RNA (lncRNA)-BC050410 [named as lncRNA-AS-GSTT1(1-72) or lncRNA-CD244] in the CD244+CD8+ T-cell subpopulation. CD244 signaling drives lncRNA-CD244 expression via sustaining a permissive chromatin state in the lncRNA-CD244 locus. By recruiting polycomb protein enhancer of zeste homolog 2 (EZH2) to infg/tnfa promoters, lncRNA-CD244 mediates H3K27 trimethylation at infg/tnfa loci toward repressive chromatin states and inhibits IFN-γ/TNF-α expression in CD8+ T cells. Such inhibition can be reversed by knock down of lncRNA-CD244. Interestingly, adoptive transfer of lncRNA-CD244–depressed CD8+ T cells to Mycobacterium tuberculosis (MTB)-infected mice reduced MTB infection and TB pathology compared with lncRNA-CD244–expressed controls. Thus, this work uncovers previously unidentified mechanisms in which T cell-inhibitory signaling and lncRNAs regulate T-cell responses and host defense against TB infection.
Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) infection remains a leading public health threat with high morbidity and mortality around the world (1, 2). CD4+ T cells, CD8+ T cells, and γδ T cells played critical roles in mounting adaptive immune response against MTB infection (3–8). Deciphering the molecular mechanisms for host responses linked to TB pathogenesis and prognosis is of great importance for developing new vaccines and therapeutics and for diagnosis.
Activation and effector functions of T cells are regulated by CD3/T-cell receptor (TCR) signal upon antigenic engagement and by a group of signals from costimulatory molecules, including CD28, cytotoxic T-lymphocyte–associated protein 4 (CTLA4), inducible T-cell costimulator (ICOS), programmed death-1 (PD-1), T cell immunoglobulin mucin-3 (Tim-3), and CD244 (2B4) (9–14). Accumulating evidence suggests that a variety of pathogens, including HIV, simian immunodeficiency virus, hepatitis C virus (HCV), lymphocytic choriomeningitis virus, and Plasmodium, induce immune evasion by up-regulating costimulatory molecules such as PD-1, CTLA4, and Tim-3 as a result of repeated antigenic stimulation of T cells. However, T-cell immune responses regulated by these “inhibitory receptors” during TB infection appear to be more complex than what have been observed in chronic viral infections. In fact, we have recently reported that up-regulation of the T cell-inhibitory molecule Tim-3 would lead to an enhanced but not inhibitory anti-TB effector function during active human TB (15), and others have also found that Tim-3 signaling may benefit innate immunity against intracellular MTB (16). Thus, it is necessary to elucidate the mechanisms by which T cell-inhibitory molecules regulate T-cell effector functions producing cytokines during active microbial infection.
Although heritable changes in gene regulation that occur via modification of the DNA without changes to the DNA sequence are often referred to as epigenetic programming, noncoding RNA (ncRNA)-mediated transcriptional or posttranscriptional regulation is one of the major regulation mechanisms for epigenetic programming (17–30). Recent studies have identified thousands of long ncRNAs (lncRNAs) (17, 23) in mammalian genomes that regulate gene expression in a variety of immunological processes (31–35), such as differentiation of T cells (33, 36) and dendritic cells (DCs) (35). Thus, diverse functions among T-cell subpopulations may manifest through highly dynamic changes in lncRNA-regulated epigenetic programming, and lncRNA-regulated epigenetic reprogramming is emerging as a novel mechanism to explain functional plasticity and diversity of T cells. Despite these advances, it remains unclear whether and how lncRNAs act as regulators of T-cell immune response during TB infection.
CD244 (2B4) is a costimulatory receptor regulating immune functions of natural killer (NK) cells (37), and it may also provide a negative signal that counters the activation signal provided by TCR engagement in CD8+ T cells (38). It has recently been shown that CD244 is expressed on virus-specific CD8+ T cells (9, 39–41) and that CD244 signaling correlates with viral persistence of hepatitis B virus (HBV) and HCV in humans (9, 39, 40). However, little is known about the molecular mechanism and consequence for CD244 regulation of T-cell effector functions during active TB infection. In this study, we examine whether and how CD244 signaling regulates T-cell effector function and impacts homeostasis and host defense against MTB infection. We hypothesize that sustained CD244 signaling directly or indirectly induces epigenetic changes to regulate the expression of proinflammatory cytokines by TB-specific T cells. Our data provide previously unidentified insights into the mechanism by which T cell-inhibitory signaling-derived lncRNA acts as an epigenetic regulator of IFN-γ and TNF-α production in CD8+ T cells and impacts CD8+ T-cell immunity against active MTB infection.
Results
Active TB Infection Induces CD244 Signaling Cascades in CD8+ T Cells.
To determine whether CD244 signaling is involved anti-TB immune responses, we first examined CD244 expression levels in CD4+ T cells, CD8+ T cells, and NK cells. Flow cytometric analysis showed that, compared with healthy controls, active TB infection induced significant increases in CD244+CD8+ T cells but not CD244+CD3−CD56Bright NK cells (Fig. 1 A and B). In addition, percentages of CD244+CD8+ T cells were much higher than those of CD244+CD4+ T cells in peripheral blood mononuclear cells (PBMCs) from either healthy controls or patients with active TB, regardless of ex vivo stimulation with MTB lysates (Fig. 1 A and B). Furthermore, ex vivo stimulation with MTB lysate induced further increases in percentages of CD244+CD8+ T cells during active TB infection, suggesting that up-regulation of CD244 in CD8+ T cells is at least partially TB-driven (Fig. 1 A and B). These data collectively suggested the importance of CD244 signaling in regulating CD8+ T-cell immune responses during active TB infection. The role for CD8+ T cells in anti-TB immunity (5) led us to examine the expression of molecules associated with the CD244 signaling pathway in CD8+ T cells during active TB infection. Because signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) and EWS-Fli1-activated transcript 2 (EAT-2) are potential downstream molecules associated with CD244 signaling (42), we sought to determine whether SAP and EAT-2 could be affected by CD244 signaling in CD8+ T cells during active TB infection. To address this, PBMCs from patients with active TB were transfected with siRNA targeting CD244 (siRNA-CD244), siRNA control (siRNA-Ctrl), or transfection medium. Although we detected significant percentages of SAP+CD8+ T-cell subsets and EAT-2+CD8+ T-cell subsets in CD8+ T cells from patients with active TB (Fig. S1), siRNA-CD244 transfection significantly decreased percentages of CD244+CD8+ T, CD244+SAP+CD8+ T, and CD244+EAT-2+CD8+ T-cell subsets in total CD8+ T cells (Fig. S1). The data support the idea that SAP and EAT-2 (42–44) may be associated with CD244 signaling in CD8+ T cells during active TB infection.
Fig. 1.
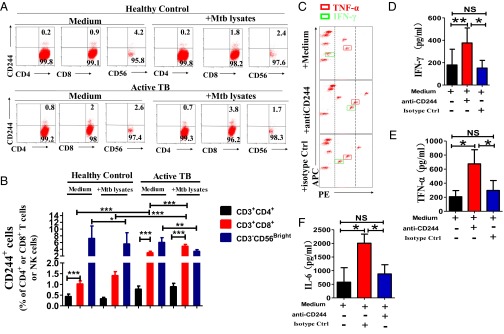
CD244 is preferentially up-regulated on CD8+ T cells during active MTB infection, and blockade of CD244 signaling enhances production of IFN-γ and TNF-α by CD8+ T cells. (A) Representative flow cytometric dot plots show the ex vivo expression of CD244 on CD4+ and CD8+ T cells and CD3-CD56Bright NK cells from one healthy control and one patient with active TB. Data were gated on CD3+CD4+, CD3+CD8+, and CD3-CD56Bright. Percentages of CD244+ T (or NK) cells are shown in the upright quadruple in each dot plot. PBMCs were treated either with or without ex vivo restimulation with MTB lysates. (B) Pooled data show the percentages of CD244+CD4+ T cells, CD244+CD4+ T cells, or CD244+NK cells among total CD3+CD4+ T cells, CD3+CD8+ T cells, and NK cells (n = 15). Error bars represent SEM. (C) Representative CBA assays of a patient with active TB showing that treatment of anti-CD244 mAb induced significant increase of concentration of IFN-γ and TNF-α in culture supernatants of CD8+ T cells purified from PBMCs of patients with active TB. The red and green squares mark the TNF-α and IFN-γ, respectively. The dashed lines mark relative fluorescent intensity of TNF-α and IFN-γ. Treatment of anti-CD244 mAb increased the concentrations of TNF-α and IFN-γ (i.e., the fluorescent intensity of phycoerythrin (PE) increased, and squares shift toward right). (D–F) Pooled data show the concentrations of IFN-γ, TNF-α, and IL-6 in the presence of indicated antibody treatment (n = 7). *P < 0.05; **P < 0.01; NS, no statistical significance. Error bars represent SEM from three independent experiments.
Fig. S1.
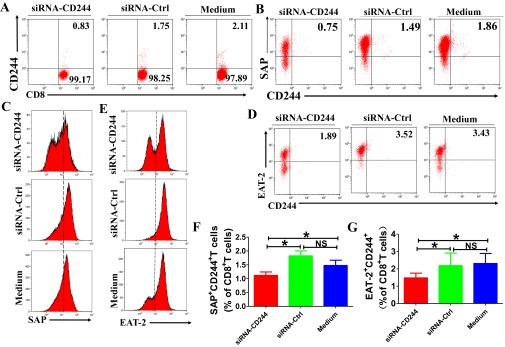
SAP and EAT-2 are downstream signaling molecules of CD244 in CD8+ T cells during active TB infection. PBMCs from patients with active TB were transfected with siRNA targeting CD244 (siRNA-CD244) or siRNA-Ctrl (si-Ctrl) or transfection medium for 48 h and cultured for another 3 d. Cells were then harvested and analyzed for the expression of CD244, SAP, and EAT-2 in CD8+ T cells using ICS/flow cytometry. (A and B) Representative flow cytometric dot plots shows that siRNA-CD244 but not siRNA-Ctrl or transfection medium decreased expression of CD244 (A), SAP (B), and EAT-2 (D) in CD8+ T cells. (C and E) Representative flow cytometric histogram analysis shows that siRNA-CD244 but not siRNA-Ctrl or transfection medium decreased expression of SAP (C) and EAT-2 (E) in CD8+ T cells. Pooled data show the frequency of SAP+CD8+ T cells (F) and EAT-2+CD8+ T cells (G) among total CD8+ T cells after transfection with indicated siRNA. Error bars, SEM (n = 8). Error bars represent SEM from two independent experiments.
Anti-CD244 mAb Modulation of CD244 Signaling in CD8+ T Cells from TB Patients Leads to Increased Production of IFN-γ and TNF-α.
We then examined the role of CD244 signaling in mediating the effector function of CD8+ T cells. We found that anti-CD244 mAb but not control IgG significantly increased concentration of IFN-γ, TNF-α, and IL-6 in supernatants of cultured CD8+ T cells from patients with active TB (Fig. 1 C–F). These data suggested that antibody modulation of CD244 on CD8+ T cells of patients with active TB could signal an increase in effector function for cytokine production.
CD244 Signaling Epigenetically Inhibits EZH2 Expression, but the Differential EZH2 Expression Itself Is Not Efficient Enough to Inhibit IFN-γ and TNF-α Production in CD8+ T Cells.
We then sought to explore whether CD244 expression or signaling in CD8+ T cells correlated with altered expression of regulatory molecules in active TB infection. We purified CD244+CD8+ T cells and CD244−CD8+ T cells from PBMCs of patients with active TB for differential expression of genes, and the hierarchical clustering analysis revealed differential gene expression profiles between CD244+CD8+ T and CD244−CD8+ T cells (Fig. 2A). Particularly, two genes encoding histone-modification enzymes, including polycomb protein enhancer of zeste homolog 2 (EZH2) and histone deacetylases 11 (HDAC11), were differentially expressed between CD244+CD8+ T and CD244−CD8+ T cells (Fig. 2B). Confocal microscopic images of immunofluorescence staining of EZH2 verified that CD8+ T cells expressed significant amounts of EZH2 in the nucleus (Fig. 2C).
Fig. 2.
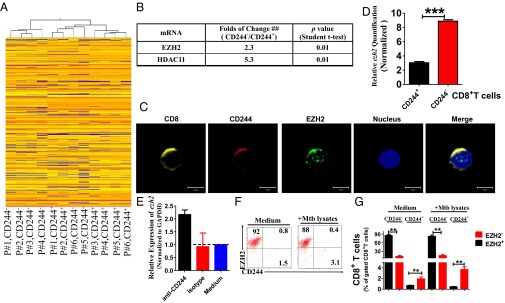
EZH2 correlates negatively with CD244 signaling. (A) Unsupervised clustering analysis of differentially expressed genes between CD244+CD8+ T cells and CD244−CD8+ T cells that were purified from PBMCs of patients with active TB. Individual squares represent the relative gene expression intensity of the given genes (rows) in each of six patients (columns), with red indicating an increase in expression and blue a decrease. (B) A table shows the fold of changes of expression of EZH2 and HDAC11 in CD244+CD8+ T cells overexpression in CD244−CD8+ T cells. (C) Typical confocal microscopic images show the expression of EZH2 in CD244high CD8+ T cells in PBMCs derived from patients with active TB. (Scale Bar: 5 μm.) (D) qPCR validation of differential expression of EZH2 gene between CD244+CD8+ T cells and CD244−CD8+ T cells. (E) qPCR analysis of the EZH2 gene in PBMCs from patients with active TB treated with anti-CD244 mAb or control antibody for 5 d. Data are presented as relative expression levels of ezh2 in PBMCs treated with anti-CD244 mAb or control antibody over expression levels of ezh2 in PBMCs treated with medium (n = 7). Data were normalized to GAPDH. (F) Representative flow cytometric dot plot data showing expression of EZH2 and CD244 in CD8+ T cells of PBMCs with or without ex vivo restimulation with MTB lysates. Data were gated on CD8+ T cells. (G) Pooled data show the frequency of EZH2+CD244−, EZH2-CD244+, and EZH2+CD244+ subpopulations of CD8+ T cells over total CD8+ T cells (n = 6). **P < 0.01; NS, no statistical significance. Except for A, error bars represent SEM from two independent experiments.
It has been suggested that EZH2 forms two closely related PRC2 complexes that can trimethylate H3K27 (45, 46); this process marks a repressive chromatin state coinciding with gene silencing (47). We thus examined whether differential EZH2 expression between CD244+ and CD244− subpopulations might impact effector functions of CD8+ T cells during active TB. Indeed, quantitative (q)PCR validation indicated that CD244−CD8+ T cells expressed much higher levels of EZH2 than their CD244+ counterparts (Fig. 2D). Consistently, ex vivo anti-CD244 mAb modulation of CD244 signaling in PBMCs of patients with active TB significantly enhanced ezh2 gene expression (Fig. 2E). In addition, immune costaining of CD244 and EZH2 in CD8+ T cells and flow cytometric analysis showed that percentages of EZH2+CD244−CD8+ T-cell subsets or EZH2−CD244+CD8+ T-cell subsets are much higher than those of EZH2+CD244+CD8+ T-cell subsets regardless of MTB lysate ex vivo restimulation, suggesting that EZH2 and CD244 tend to be expressed in distinct CD8+ T-cell subpopulations during active TB infection (Fig. 2 F and G). Thus, these results suggest that CD244 signaling negatively regulates EZH2 expression in CD8+ T cells during active TB infection.
We then determined whether differential expression of EZH2 contributes to inhibition of IFN-γ and TNF-α production by CD8+ T cells during active TB infection. PBMCs from patients with active TB were transfected with siRNA targeting EZH2 (siRNA-EZH2), control siRNA (siRNA-Ctrl), or transfection medium only. Compared with PBMCs transfected with siRNA-Ctrl or medium, transfection of siRNA-EZH2 did not significantly increase production of IFN-γ and TNF-α in PBMC culture supernatants or percentages of IFN-γ+CD8+ T cells and TNF-α+CD8+ T cells among total CD8+ T cells (Fig. S2 A–G). Consistently, ChIP-qPCR also showed that, compared with siRNA-Ctrl, siRNA-EZH2 was not able to induce a significant decrease in the amounts of H3K27Me3 at promoter regions of infg and tnfa loci (Fig. S2H). Thus, the differential expression of EZH2 itself in CD244+ T cells was not efficient enough to depress production of IFN-γ and TNF-α by CD8+ T cells in active TB.
Fig. S2.
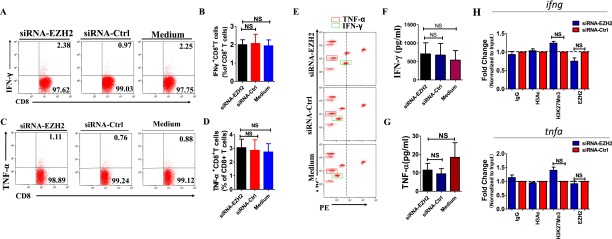
EZH2 differential expression is not efficient enough to enhance CD8+ T-cell effector function. (A and C) Representative flow cytometric dot plots showed that, compared with transfection with siRNA-Ctrl or medium, siRNA-EZH2 did not induce significant changes of percentages of IFN-γ– or TNF-α–producing CD8+ T cells. Data were gated on CD8+ T cells. PBMCs from patients with active TB were transfected with siRNA targeting EZH2 (siRNA-EZH2) or siRNA-Ctrl or transfection medium for 48 h and cultured for another 3 d. Cells were then harvested and analyzed for the expression of EZH2 in CD8+ T cells and intracellular IFN-γ and TNF-α responses of CD8+ T cells using ICS/flow cytometry. (B and D) Pooled data show the percentages of IFN-γ– or TNF-α–producing CD8+ T cells in response to knock down of indicated siRNA or medium (n = 10). (E) Representative CBA assay of PBMC of a patient with active TB showing that, compared with transfection with siRNA-Ctrl or medium, siRNA-EZH2 did not induce significant changes of concentration of TNF-α and IFN-γ. The red and green squares mark the TNF-α and IFN-γ, respectively. (F and G) Pooled data showing the concentrations of IFN-γ and TNF-α upon transfection of indicated siRNA or medium (n = 10). (H) ChIP-qPCR analysis of H3K27Me3, EZH2, and H3Ac at the promoters of IFN-γ and TNF-α in CD8+ T cells transfected with siRNA-EZH2 and siRNA-Ctrl. NS, no statistical significance. Error bars represent SEM from two independent experiments.
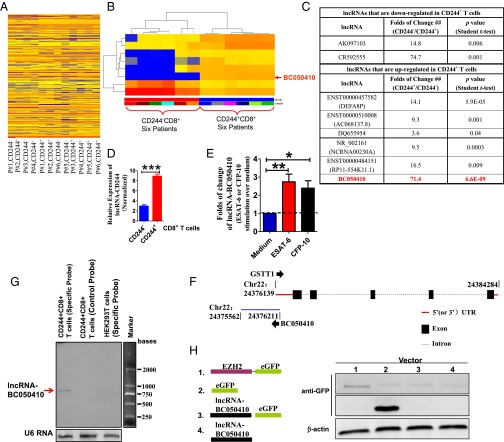
CD244 Signaling Positively Correlates with High-Level Expression of TB-Specific lncRNA-BC050410 [lncRNA-AS-GSTT1(1-72) or lncRNA-CD244] in CD8+ T Cells.
Because differential EZH2 expression was not efficient enough to inhibit production of IFN-γ and TNF-α by CD8+ T cells, we then investigated the possibility that EZH2 might be recruited to the promoters of IFN-γ and TNF-α in CD8+ T cells to induce repressive chromatin states at infg and tnfa loci in CD244+CD8+ T cells. This consideration was supported by the finding that lncRNA might mediate targeted recruitment of repressive histone-modifying activities to epigenetically silence transcription (48–52). We used human lncRNA microarray and hierarchical clustering analyses to compare lncRNA expression in CD244+CD8+ T cells and CD244−CD8+ T cells. The comparative analysis between these two subsets allowed us to display a distinct lncRNA expression profile in CD244+CD8+ T cells (Fig. 3A). The supervised hierarchal clustering segregation analysis then identified dominant groups of lncRNAs selectively expressed in CD244+CD8+ T cells (Fig. 3B). Interestingly, lncRNA-BC050410 [named as lncRNA-AS-GSTT1(1-72) based on its genomic context (53) and termed as lncRNA-CD244 here for simplicity] with genomic overlapping to 5′ UTR of GST θ1 (GSTT1) was one of the eight lncRNAs that could distinguish the CD244+CD8+ T-cell subpopulation from its CD244− counterpart, with a largest-fold difference and a most-significant P value (Fig. 3 C and E and Fig. S3 A and B). Note that lncRNA-CR592555, a lncRNA mostly down-regulated in CD244+CD8+ T cells, was located between 79,946,861 bp and ∼79,947,776 bp in chromosome 5 (Fig. S4). Such differential expression of lncRNA-CD244 in CD244+CD8+ T cells in active TB was also validated by qPCR (Fig. 3D). Furthermore, stimulation with peptide pools of 6-kDa early secretory antigenic target (ESAT-6) or 10-kDa culture filtrate protein (CFP-10) of MTB induced higher expression levels of lncRNA-CD244 in purified CD244+CD8+ T cells, suggesting that expression of lncRNA-CD244 is at least partially TB-specific (Fig. 3E). Also, expression of full-length lncRNA-CD244 was confirmed by Northern blot analysis (Fig. 3F). We then assessed whether lncRNA-CD244 has protein-coding capability. As with lncRNA-DC (35), analysis based on coding potential calculator (CPC) (Fig. S3 C–E) suggests that the overall protein-coding potential of lncRNA-CD244 is weak [e.g., the hit score of lncRNA-CD244 (∼23.56) is as small as that of lncRNA-DC (∼24.35), which suggests that lncRNA-CD244, like lncRNA-DC (35), is unlikely protein-coding; lncRNA-CD244 has five potential ORFs (Fig. S3F), but ORF coverage of lncRNA-CD244 (∼33.54%) is much smaller than that of lncRNA-DC (∼50.72%), which suggests that the ORF quality of lncRNA-CD244 is worse than that of lncRNA-DC]. In addition, analysis of the ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site (Ka/Ks or dN/dS) between two different species of Hominoidea (e.g., dN/dS ratio between human and chimpanzee is 0.48; P = 0.068 > 0.05) (Fig. S3G) suggests that the ORFs of lncRNA-CD244 lack negative selection in species of Hominoidea. Thus, no bioinformatics evidence implicates lncRNA-CD244 as having protein-coding capability. Furthermore, a vector construct comprising full-length lncRNA-CD244 and EGFP tag was developed and assessed for expression using immunoblotting (IB) and fluorescent imaging. The lncRNA-CD244 did not have any detectable protein-coding ability because neither lncRNA-CD244-EGFP vector nor lncRNA-CD244 vector expressed EGFP (Fig. 3H and Fig. S5). Thus, lncRNA-CD244 preferentially expressed in CD244+CD8+ T cells during active human TB infection.
Fig. 3.
lncRNA-CD244 is highly expressed in CD244+CD8+ T cells during active TB. (A) Unsupervised clustering analysis of differentially expressed lncRNAs between CD244+CD8+ T cells and CD244−CD8+ T cells that were purified from PBMCs of patients with active TB. Individual squares represent the relative lncRNA expression intensity of the given lncRNAs (rows) in each of the patients (columns), with red indicating an increase in expression and blue a decrease. (B) Supervised clustering analysis using differentially expressed lncRNAs that can distinguish CD244+CD8+ T cells from CD244−CD8+ T cells. (C) A table shows the folds of change and P values (Student t test) of eight lncRNAs that could distinguish CD244+CD8+ T-cell subpopulation from CD244−CD8+ T-cell subpopulation of six patients with active TB. (D) qPCR validation of differential expression of lncRNA-CD244 between CD244+CD8+ T cells and CD244−CD8+ T cells. (E) qPCR analysis of lncRNA-CD244 expression in CD244+CD8+ T cells purified from PBMCs of patients with active TB upon stimulation with MTB ESAT-6 (10 μg/mL) or CFP-10 (10 μg/mL) peptide pools for 5 d in vitro. (F) Schematic diagram of the lncRNA-CD244 genomic locus in human chromosome 22. The bars represent exons, dashed lines represent intron, red line represents 5′ (or 3′) UTR, and arrows indicate the direction of transcription. The length of lncRNA-CD244 is 796 bases, which overlapped 5′ UTR of GST θ1 (GSTT1) by 72 bases. No homologs of lncRNA-CD244 were found in mouse (Fig. S3 A and B). (G) Northern blot analysis show the expression of full-length lncRNA-CD244 in CD244+CD8+ T cells from patients with active TB using specific probe but not antisense control probe. No lncRNA-CD244 was detected in HEK293T cells. U6 RNA served as a control. (H) Plasmids as schematically shown at Left were transfected to HEK293T cells (Right). Immunoblotting using antibody specific to EGFP and fluorescent imaging (Fig. S5) showed that lncRNA-CD244-EGFP plasmid and lncRNA-CD244 plasmid did not express GFP. Error bars represent SEM. Data shown in D, E, G, and H are representative of at least two independent experiments.
Fig. S3.
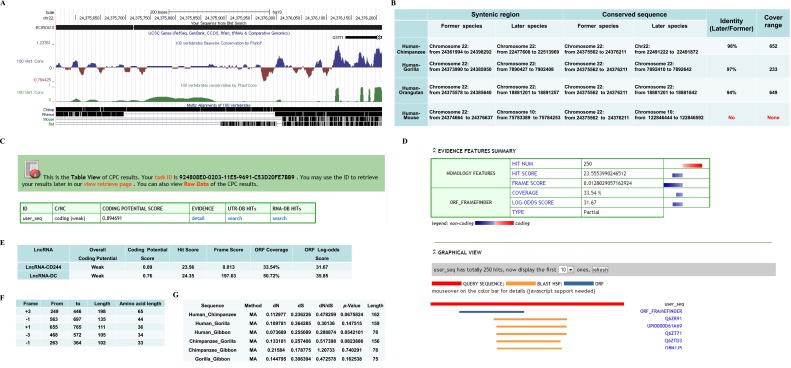
Bioinformatics analyses of evolutional conservation and protein-coding potential of lncRNA-BC050410. (A) The conservation track of lncRNA-BC050410 analyzed by the UCSC Genome Browser (genome.ucsc.edu). No mouse or rat homolog was found. (B) Conservation analysis in the syntenic region. Identity was calculated by alignment between later (e.g., chimpanzee) and former (e.g., human) species in the cover range (e.g., 652) indicated in the table. Note that no identity between human and mouse was found. (C and D) The analysis of protein-coding potential of lncRNA-BC050410 using tools provided by the Peking University Center for Bioinformatics (cpc.cbi.pku.edu.cn/programs/run_cpc.jsp) shows lack of protein-coding capability. (E) Comparison between lncRNA-BC050410 and the variant-1 transcript of lncRNA-DC (35) for parameters to determine protein-coding potential using the same tools provided by the Peking University Center for Bioinformatics (cpc.cbi.pku.edu.cn/programs/run_cpc.jsp). The larger hit score and frame score indicate that a transcript is more likely protein-coding, and larger ORF coverage or log-odds score indicates the better ORF quality. (F) The lengths of five potential ORFs in lncRNA-BC050410. (G) dN/dS analyses between two different species of Hominoidea indicated in the table. Data were calculated by the KaKs_Calculator software (https://code.google.com/p/kaks-calculator/wiki/KaKs_Calculator) using pair-wised DNA sequences (lengths are indicated in the table) from homologous amino acid sequences aligned between two different species of Hominoidea. “MA” indicates model averaging, which assigns each candidate model a weight value and engages more than one model to estimate average parameters across models (64). A value of P > 0.05 was considered as no negative or positive selection.
Fig. S4.

Genome location analysis of human lncRNA-CR592555 using UCSC Genome Browsers showed that lncRNA-CR592555 is located between 79,946,861 bp and ∼79,947,776 bp in chromosome 5.
Fig. S5.
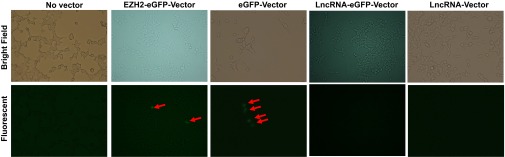
Representative fluorescent images of HEK293T cells transfected with vectors as indicated in Fig. 3 showed that only cells transfected with EZH2-EGFP vector and EGFP vector expressed GFP. Data shown are representative of at least two independent experiments.
CD244 Signaling Drives lncRNA-CD244 Expression via Sustaining a More Permissive Chromatin State in lncRNA-CD244 Locus.
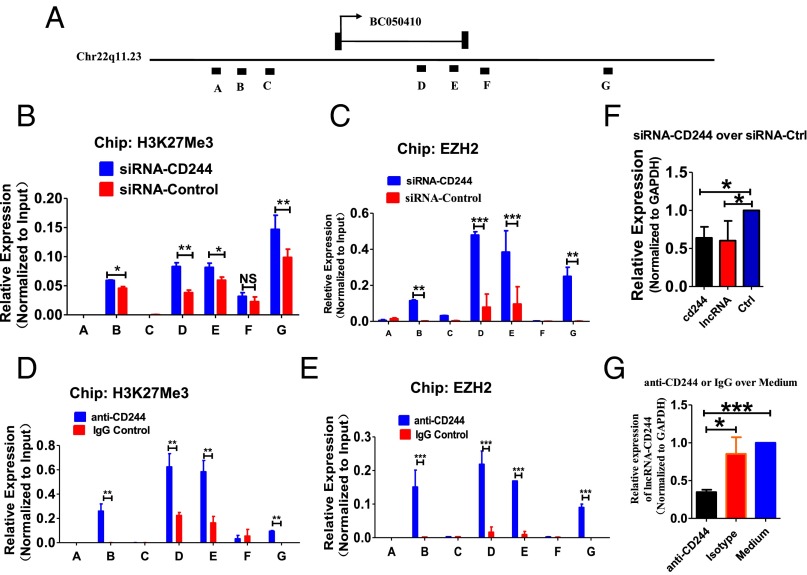
To determine the mechanisms underlying the preferential expression of lncRNA-CD244 mediated by CD244 signaling, PBMCs of patients with active TB were transfected with siRNA targeting CD244 (siRNA-CD244) and control siRNA (siRNA-Ctrl) or treated with anti-CD244 and control IgG. ChIP-qPCR analysis showed that EZH2 and trimethylation at H3K27, a histone modification that negatively regulates transcription, markedly increased in lncRNA-CD244 loci after treatment with siRNA-CD244 but not siRNA-Ctrl (Fig. 4 A–C). Consistently, siRNA-CD244, not siRNA-Ctrl, decreased expression of the cd244 gene and lncRNA-CD244 (Fig. 4F). Furthermore, anti-CD244 but not IgG control induced significant increases of H3K27Me3 and EZH2 in lncRNA-CD244 loci (Fig. 4 A, D, and E), and anti-CD244 but not IgG significantly decreased the expression of lncRNA-CD244 (Fig. 4G). Taken together, knock down of CD244 or blockade of CD244 signaling induces a more repressive chromatin state in lncRNA-CD244 locus and reduces expression of lncRNA-CD244. Thus, these results implicate CD244 signaling as driving expression of lncRNA-CD244 in CD8+ T cells most likely through sustaining a more permissive chromatin state in the lncRNA-CD244 locus during active TB infection.
Fig. 4.
Knock down of CD244 or blockade of CD244 signaling induces a more repressive chromatin state in lncRNA-CD244 locus and inhibits expression of lncRNA-CD244. Seven regions (capital letters A to G) across lncRNA-CD244 locus, as shown in A, were analyzed in ChIP-qPCR analyses for H3K27Me3 (B and D) histone modification and EZH2 (C and E) in PBMCs from patients with active TB. PBMCs were transfected with siRNA-CD244 or siRNA-Ctrl or treated with anti-CD244 mAb or IgG control as indicated in each of subfigure. Values derived from three independent experiments were normalized by background signals and input chromatin. (F and G) qPCR analysis of lncRNA-CD244 and/or the cd244 gene in PBMCs from patients with active TB transfected (or treated) with indicated siRNAs (F) or antibodies (G). Data are presented as relative expression levels of lncRNA-CD244 (or cd244) (normalized to GAPDH) in siRNA-CD244–transfected (or anti-CD244–treated) PBMCs over expression levels of lncRNA-CD244 (or cd244) in siRNA-Ctrl–transfected (or IgG-treated) PBMCs (n = 7). *P < 0.05; **P < 0.01; ***P < 0.001; NS, no statistical significance. Error bars represent SEM from three independent experiments.
lncRNA-CD244 Epigenetically Regulates Expression of IFN-γ and TNF-α by CD8+ T Cells.
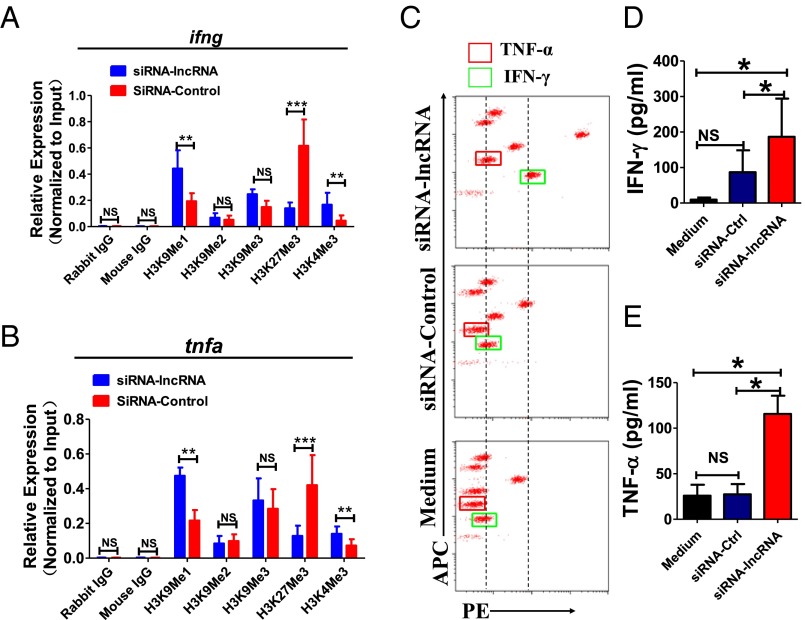
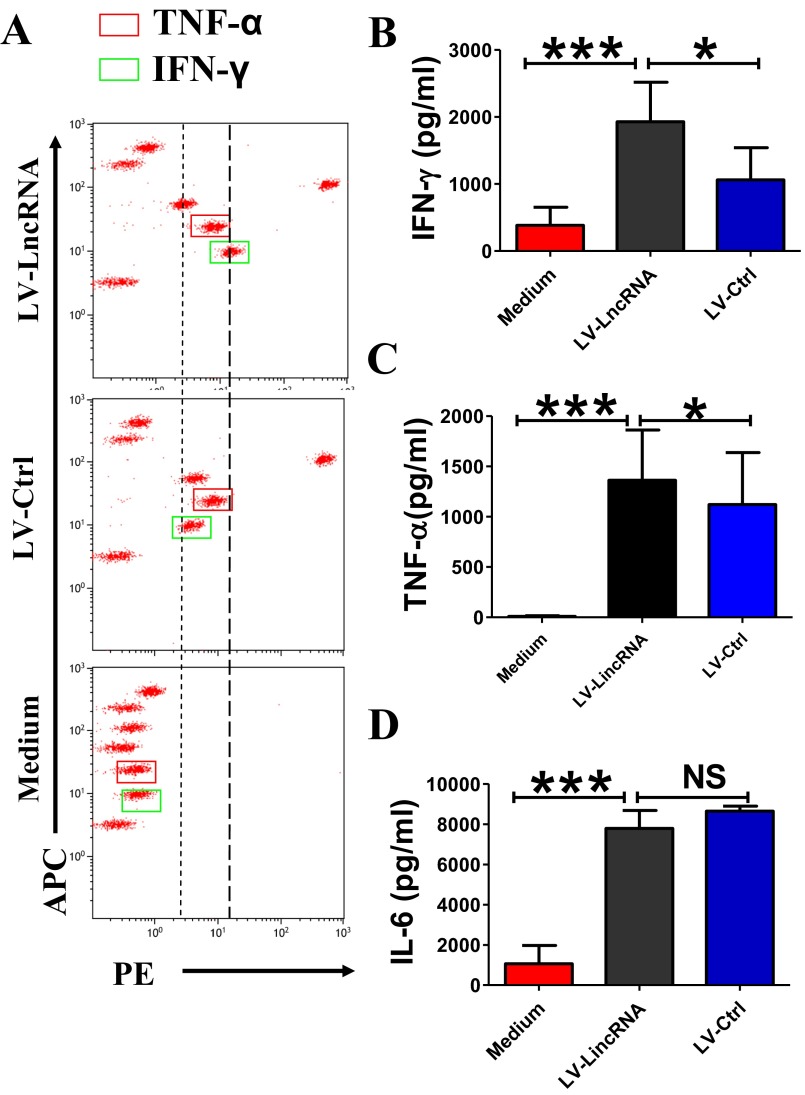
We then determined whether lncRNA-CD244 could mediate epigenetic regulation of IFN-γ and TNF-α expression in CD8+ T cells during active human TB infection. Accumulative evidence suggests that H3 trimethylation on lysine4 (H3K4me3) and histone H3 methylation on lysine9 (H3K9Me1) are indicative of permissive chromatin states (54). In contrast, histone H3 trimethylation on lysine 27 (H3K27Me3) and histone H3 methylation and dimethylation on lysine9 (H3K9Me1, H3K9Me2) are indicative of repressive chromatin states (54). We hypothesized that lncRNA-CD244 might affect IFN-γ and TNF-α production at the transcriptional level by epigenetic regulation of histone modification states at infg and tnfa loci. To address this hypothesis, CD8+ T cells isolated from PMBCs of patients with active TB were transfected with siRNA-lncRNA-CD244 to knock down the expression of lncRNA, with transfection of siRNA-Ctrl as a control. The amounts of H3K9Me1, H3K9Me2, H3K9Me3, H3K27Me3, and H3K4Me3 at the promoters of infg and tnfa were determined by ChIP-qPCR. Interestingly, transfection with siRNA-lncRNA, but not siRNA-Ctrl, led to a significant increase in H3K9Me1 and H3K4Me3 and decrease in H3K27me3 at infg and tnfa loci (Fig. 5A and B). Consistently, much greater concentrations of IFN-γ and TNF-α were detected via cytometric bead array (CBA) analysis in culture supernatants of PBMCs transfected with siRNA-lncRNA compared with siRNA-Ctrl or medium controls (Fig. 5 C–E). Also, CD8+ T cells purified from PBMCs of patients with active TB were transduced with lentiviral (LV) vector encoding GFP and shRNA targeting lncRNA-CD244 (LV-lncRNA) or LV vector encoding GFP only (LV-Ctrl). We found that transduction with LV-lncRNA, but not LV-Ctrl or medium, in CD8+ T cells induced significant increases in IFN-γ and TNF-α in culture supernatants (Fig. S6), suggesting that lncRNA-CD244 indeed regulated expression of IFN-γ and TNF-α by CD8+ T cells. Moreover, such enhanced production of IFN-γ and TNF-α upon knock down of lncRNA-CD244 was unlikely caused by the impact of lncRNA-CD244 on GSTT1 expression, because we found no significant changes in GSTT1 expression upon knock down of lncRNA-CD244 (Fig. S7A) and no significant enhancement of IFN-γ and TNF-α production upon GSTT1 knock down by shRNA (Fig. S7 B and C). Furthermore, such enhanced production of IFN-γ and TNF-α was not attributable to changes in viability of CD8+ T cells because CD8+ T cells did not exhibit enhanced apoptosis after lncRNA-CD244 knockdown (Fig. S7 D and E). Thus, these results collectively suggest that whereas TB-driven up-regulation of lncRNA-CD244 might influence repressive chromatin states at infg and tnfa loci, silence or down-regulation of lncRNA-CD244 could confer permissive chromatin states at infg and tnfa loci and enhance expression of IFN-γ and TNF-α.
Fig. 5.
lncRNA-CD244 regulates IFN-γ and TNF-α expression in active TB infection. (A and B) ChIP-qPCR analysis of H3K9Me1, H3K9Me2, H3K9Me3, H3K4me3, H3K27Me3, and control antibodies at the promoters of IFN-γ (A) and TNF-α (B) in CD8+ T cells transfected with indicated siRNAs. (C) Representative CBA assays of PBMCs from a patient with active TB showing that, compared with siRNA-Ctrl or transfection medium, siRNA-lncRNA-CD244 induced significant increases in concentrations of IFN-γ/TNF-α. (D and E) Pooled data showing the concentrations of IFN-γ/TNF-α upon transfection of indicated siRNAs or medium (n = 7). *P < 0.05; **P < 0.01; ***P < 0.001; NS, no statistical significance. Error bars represent SEM from three independent experiments.
Fig. S6.
CD8+ T cells with inhibited expression of lncRNA-CD244 show greater production of INF-γ and TNF-α. (A) Representative CBA assays of CD8+ T cells purified from PBMCs of a patient with active TB showing that, compared with transduction of LV-Ctrl or treatment with transduction medium, transduction of LV vector encoding GFP and shRNA targeting lncRNA-CD244 (LV-lncRNA-CD244) in CD8+ T cells induced a significant increase of concentration of IFN-γ and TNF-α. (B–D) Pooled data showing the concentrations of IFN-γ, TNF-α, and IL-6 upon transfection of indicated LV vectors (n = 8). Error bars represent SEM. Shown is a representative of at least three independent experiments.
Fig. S7.
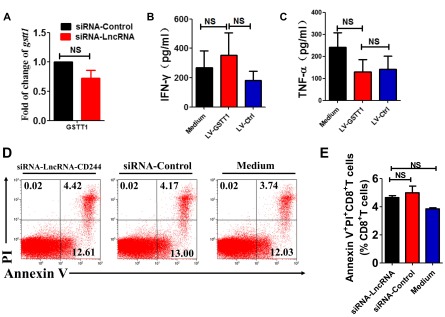
Effect of lncRNA-CD244 knockdown on apoptosis of CD8+ T cells and GSTT1 expression and analysis of the association between GSTT1 expression and IFN-γ/TNF-α expression by CD8+ T cells. (A) qPCR analysis of GSTT1 mRNA expression in CD8+ T cells of patients with active TB that were transfected with siRNA-lncRNA and siRNA-Ctrl. (B and C) Pooled data showing the concentrations of IFN-γ and TNF-α determined by CBA analysis upon transduction of indicated LV containing shRNAs targeting GSTT1 (LV-GSTT1), control LV (LV-Ctrl), or medium. (D and E) Representative flow cytometric analysis (D) and pooled data of five TB patients (E) of CD8+ T cells transfected with siRNA-lncRNA-CD244 or siRNA-Ctrl at day 5 and then analyzed for apoptosis based on the intracellular expression of Annexin V and propidium iodide (n = 5). NS, no statistically significant difference. Error bars represent SEM from three independent experiments.
lncRNA-CD244 Associates Physically with EZH2 and Mediates Recruitment of EZH2 to ifng and tnfa Loci for Repressive Chromatin States.
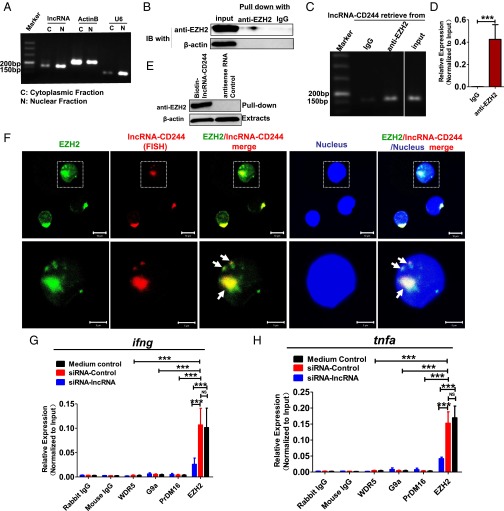
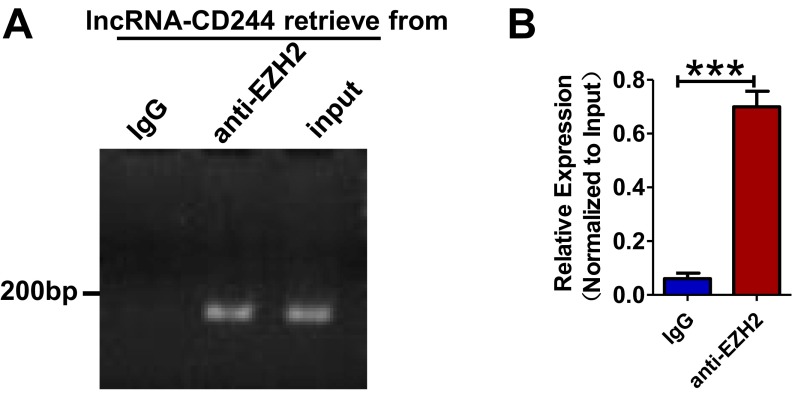
Next, we sought to examine the interrelation of lncRNA-CD244, repressive chromatin states at infg and tnfa loci, and altered expression of EZH2 (Fig. 2). We presumed that lncRNA-CD244 could mediate the recruitment of the histone-modifying enzyme EZH2, which catalyzed the trimethylation modification of H3K27 at promoters of infg and tnfa. Of note, while performing gel electrophoresis of RNA extracts of nuclear and cytoplasmic fractions of CD8+ T cells from patients with active TB, we found that most of the lncRNA-CD244 localized in the nuclear fraction of CD8+ T cells (Fig. 6A). In addition, when we performed immunoprecipitation (IP) of EZH2 in extracts of CD8+ T cells from TB patients or in HEK293T cells with exogenous expression of EZH2 and full-length lncRNA-CD244, we found that EZH2-specific mAb, but not control IgG, could actually coprecipitate lncRNA-CD244 molecules, as detected by qRT-PCR(Fig. 6 B–D and Fig. S8). Furthermore, biotinylated lncRNA-CD244 and an antisense control RNA were incubated with nuclear extracts of CD8+ T cells from patients with active TB, and Western blotting showed that lncRNA-CD244 but not antisense control RNA specifically bound to EZH2 (Fig. 6E). Consistently, when we performed confocal microscope-based FISH analysis of lncRNA-CD244 and immunofluorescent analysis of EZH2, we found that significant amounts of EZH2 colocalized with lncRNA-CD244 in the nucleus of CD8+ T cells from patients with active TB (Fig. 6F). These results suggest that lncRNA-CD244 is physically associated with EZH2 during active TB. Furthermore, transfection of CD8+ T cells with siRNA-lncRNA-CD244, but not siRNA-Ctrl, led to significant decreases in EZH2, but not WDR5, Prdm16, and G9a, at either the infg or tnfa promoter (Fig. 6 G and H). ChIP-qPCR analysis also demonstrated that EZH2 accumulated at promoters of infg or tnfa at much greater levels than WDR5, PRDM16, and G9a in CD8+ T cells from patients with active TB (Fig. 6 G and H). These results suggest that EZH2 and lncRNA-CD244 complex formation may lead to trimethylation of H3K27, which contributes to inducing repressive chromatin states at infg or tnfa loci in CD8+ T cells during active TB infection. The data also suggest a hypothetical model in which expression of lncRNA-CD244 may physically recruit EZH2 to control H3K27Me3 at the ifng and tnfa loci and therefore allow chromatin to program repressive states and inhibit transcription of infg and tnfa genes in CD244+CD8+ T cells.
Fig. 6.
lncRNA-CD244 interacts directly with EZH2 and recruits EZH2 to ifng and tnfa promoters. (A) Gel electrophoresis of lncRNA-CD244 extracted from nucleus and cytoplasm of CD8+ T cells purified from PBMCs of patients with active TB. As controls, more actin B expressed in cytoplasm and more U6 expressed in nucleus, respectively. (B) IB analysis of EZH2 in the IP by IgG or anti-EZH2–specific antibody from CD8+ T-cell lysates of patients with active TB. (C and D) Gel electrophoresis (C) and qPCR analysis (D) of lncRNA-CD244 retrieve in IP by IgG or anti-EZH2–specific antibody from CD8+ T-cell lysates of patients with active TB. The levels of qRT-PCR products were expressed as a percentage of input RNA. (E) Biotinylated lncRNA-CD244 or antisense RNA control was incubated with nuclear extracts of CD8+ T cells from patients with active TB and targeted with streptavidin-conjugated magnetic beads (MB), and associated proteins were assessed with Western blot using anti-EZH2–specific antibody. (F) Confocal microscopic images of RNA FISH assay of lncRNA-CD244 and immunofluorescence analysis of EZH2 show that EZH2 colocalizes with lncRNA-CD244 in nucleus of CD8+ T cells from patients with active TB. Lower images were cropped from the squares in the upper images. (Scale bars: 10 μm in Upper and 5 μm in Lower.) More than 30 cells were examined and had similar results. White arrowheads mark the EZH2/lncRNA-CD244 colocalization. (G and H) ChIP-qPCR analysis of WDR5, G9a, Prdm16, EZH2, and control antibodies at the promoters of IFN-γ and TNF-α in CD8+ T cells transfected with siRNA-lncRNA and siRNA-Ctrl (n = 7). ***P < 0.001. Error bars represent SEM from three independent experiments.
Fig. S8.
RIP analysis shows that lncRNA-CD244 interacts with EZH2 upon exogenous expression in HEK293T cells. lncRNA-CD244 vector and EZH2-EGFP vector were cotransfected into HEK293T cells. Extracts of HEK293T cells were prepared, and EZH2 protein was immunoprecipitated by either IgG control antibody or anti-EZH2–specific antibody and tested for association with lncRNA-CD244 by qRT-PCR. (A and B) Gel electrophoresis (A) and qPCR analysis (B) of lncRNA-CD244 retrieve in IP by IgG or anti-EZH2–specific antibody from lysates of HEK293T cell. The levels of qRT-PCR products were expressed as a percentage of input RNA. ***P < 0.001. Error bars represent SEM from three independent experiments.
lncRNA-CD244 Is a Major Regulator of CD8+ T-Cell Immune Response During in Vivo MTB Infection.
Finally, we sought to determine whether the lncRNA-CD244 could regulate immune response of CD8+ T cells in vivo. Because no mouse homolog of lncRNA-CD244 was found (Fig. 3F and Fig. S3 A and B), we developed an adoptive transfer strategy to determine the in vivo role of lncRNA-CD244 in regulating CD8+ T-cell immune response during MTB infection (Fig. 7A). CD8+ T cells and CD14+ monocytes were first purified from PBMCs of patients with active TB. CD8+ T cells were then transduced with LV-lncRNA to knock down the expression of lncRNA-CD244 and enhance production of IFN-γ and TNF-α (Fig. S6). LV-lncRNA–transduced (lncRNA-CD244–depressed) CD8+ T cells were then adoptively transferred to SCID mice on the third day after MTB infection (Fig. 7A). LV-Ctrl–transduced (lncRNA-CD244–expressed) CD8+ T cells and medium-treated (lncRNA-expressed) CD8+ T cells served as controls, respectively. At the same time of CD8+ T-cell transfer, each mouse also received autologous monocytes from TB patients to facilitate MTB infection (Fig. 7A). Interestingly, mice infused with LV-lncRNA–transduced (lncRNA-CD244–depressed) CD8+ T cells showed significantly lower MTB bacterial burdens in lungs and blood compared with control mice infused with LV-Ctrl–transduced (lncRNA-CD244–expressed) CD8+ T cells or medium-treated (lncRNA-CD244–expressed) CD8+ T cells (Fig. 7B). The control mice infused with LV-Ctrl–transduced CD8+ T cells or medium-treated CD8+ T cells exhibited severer lung necrosis (Fig. 7C) and histopathology characterized by significant hemorrhages or infiltration of RBCs in alveoli and apparent damage of pulmonary structures (Fig. 7D). In contrast, only mild changes in alveoli were observed in lung sections of mice infused with LV-lncRNA-CD244–transduced CD8+ T cells (Fig. 7D). Thus, lncRNA-CD244–depressed CD8+ T cells can more potently control in vivo MTB infection than lncRNA-CD244–expressed CD8+ T cells in SCID mice, suggesting that lncRNA-CD244 may play a role in regulating in vivo immune response of CD8+ T cells during MTB infection.
Fig. 7.
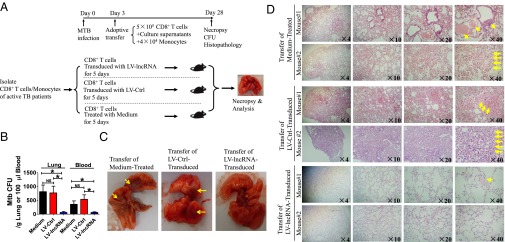
Adoptive transfer of lncRNA-CD244–depressed CD8+ T cells to MTB-infected SCID mice reduced MTB infection and TB pathology compared with lncRNA-CD244–expressed controls. (A) Schematic diagram shows the experimental strategy of adoptive transfer of CD8+ T cells with inhibited lncRNA-CD244 expression, culture supernatants of indicated CD8+ T cells, and monocytes in MTB-infected mice. (B) Bacterial load (CFU, H37Rv) in the lungs and peripheral blood of mice indicated in A (n = 8 mice per group). (C) Representative digital camera images of lungs collected from mice receiving transfer of indicated cells with or without knock down of lncRNA-CD244. Yellow arrows mark the necrosis. (D) H&E-stained lung sections derived from two representative mice in each group of mice indicated in A. Yellow arrows mark the infiltration of red blood cells or damage of pulmonary structure. The magnification is shown at the lower right of each image. *P < 0.05. Error bars represent SEM from two independent experiments.
Discussion
Molecular mechanisms for CD244 modulation of T-cell responses in infections remain unknown, although CD244 appears to regulate NK cell function that correlates with HBV or HCV persistence in humans (40, 41). In this study, we find that human TB can up-regulate CD244 and CD244 signaling-related molecules in CD8+ T cells. Whereas the CD244+CD8+ T-cell subset from TB patients expresses much higher levels of lncRNA-CD244 than its CD244−CD8+ T-cell subset counterpart, siRNA or shRNA knock down of lncRNA-CD244 leads to increased production of IFN-γ and TNF-α. The blockade of CD244 signaling by anti-CD244 mAb can similarly lead to in vitro enhancement of IFN-γ and TNF-α production. Thus, we demonstrate the interrelation between CD244 signaling, lncRNA-CD244 expression, and regulation of effector function in CD8+ T cells at epigenetic levels in TB patients.
The current study provides a previously unidentified mechanism by which CD244 signaling regulates IFN-γ and TNF-α production in human CD8+ T cells in MTB infection. CD244 signaling during active MTB infection can exploit lncRNA and histone-modifying enzymes to regulate the effector functions of CD244+CD8+ T cells. Often, CD244 expression/signaling in CD244+CD8+ T cells from TB patients leads to remarkable increases in expression of lncRNA-CD244. Interestingly, lncRNA-CD244 appears to physically interact with a polycomb protein, EZH2. This interaction mediates recruitment of EZH2 to infg and tnfa loci and trimethylates H3K27 at promoters of infg and tnfa toward repressive chromatin states and suppression of infg and tnfa expression. Our findings support the hypothesis that ncRNA can interact with chromatin and mediate targeted recruitment of repressive histone-modifying activities to epigenetically silence transcription (49, 51). In fact, it has been recently shown that a number of intronic RNA sequences are capable of binding to the core component EZH2 and regulating the transcriptional output of its genomic counterpart (55). In addition, the ncRNAs HOTAIR (56), Xist (57), and RepA (58) might recruit the polycomb complex to the HoxD locus or the X chromosome, respectively, where they mediate trimethylation of H3K27 and induce heterochromatin formation and repression of gene expression. Furthermore, EZH2 is also shown to bind strongly to genes encoding the transcription factors T-bet, Eomes, and Gata3; controls differentiation into Th1 and Th2 effector cells; and regulates plasticity of these subsets after differentiation (59). However, we cannot rule out the possibility that lncRNA-CD244 in our model may regulate the repression of infg and tnfa via other undefined epigenetic mechanisms (48, 50).
The data from the current study also suggest that down-regulation of lncRNA-CD244 in CD8+ T cells from TB patients can yield a favorable in vivo consequence during MTB infection at SCID model. Adoptive transfer of lncRNA-CD244–depressed CD8+ T cells with the enhanced IFN-γ/TNF-α production could attenuate MTB infection and TB pathology in SCID mice compared with lncRNA-CD244–expressed controls. Enhanced CTL activity and IFN-γ/TNF-α production of lncRNA-CD244–depressed CD8+ T cells may contribute to inhibition of intracellular MTB replication in infused autologous monocytes/macrophages. This notion is supported by the previous studies demonstrating CD8+ T-cell-mediated anti-TB immunity (5). Thus, the identification of the CD244 and lncRNA-CD244 axis in the modulation of IFN-γ and TNF-α expression provides an lncRNA-driven epigenetic program of T-cell immunity against MTB infection.
lncRNAs have recently been in the spotlight for their critical roles in human biology and diseases (21, 22, 34, 50, 60, 61). They have been explored as biomarkers for cancers and potential targets for disease or dysregulated expression of genes or phenotypes (49). Here, we demonstrate that lncRNA-CD244 serves as one of the mechanisms by which CD244 signaling regulates the ability of human CD8+ T cells to produce IFN-γ and TNF-α at epigenetic levels in MTB infection. To our knowledge, this is the first evidence that lncRNA is one of the major epigenetic factors modulating Th1 immune response in human TB.
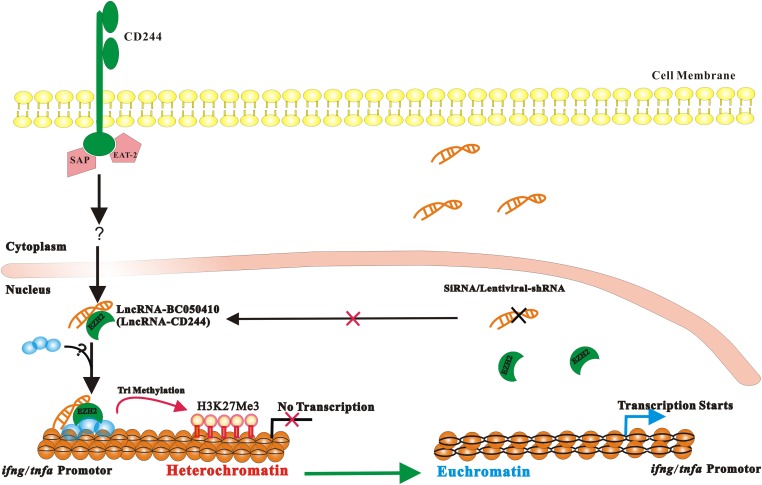
Thus, this study allows us to demonstrate that CD244 signaling in active human TB regulates repression of IFN-γ and TNF-α through a mechanism in which lncRNA-CD244 modulates recruitment of EHZ2 to promoters of IFN-γ and TNF-α for potential trimethylation of H3K27 and repression of infg and tnfa expression (Fig. S9). The CD244 signaling and lncRNA-CD244 modulation of IFN-γ and TNF-α expression presents an lncRNA-driven epigenetic program of T-cell immunity against microbial infection. Our findings also suggest that lncRNA-CD244 may be a potential target for therapeutic intervention of TB.
Fig. S9.
A proposed model of ifng and tnfa expression regulated by lncRNA-CD244 in CD8+ T cells during active TB infection. TB infection induces up-regulation of CD244 on CD8+ T cells, which drives expression of lncRNA-CD244. lncRNA-CD244 that localizes in the nucleus mediates recruitment of polycomb protein EZH2 to trimethylate H3K27 at promoter of IFN-γ and TNF-α, which therefore induces a repressive chromatin (heterochromatin) in ifng and tnfa locus, and therefore transcription of ifng and tnfa is inhibited. Knock down of lncRNA-CD244 using siRNA or LV vector encoding shRNA targeting lncRNA-CD244 results in failure of recruitment of EZH2 to promoter of IFN-γ and TNF-α, and therefore heterochromatin is changed to euchromatin and transcription of ifng and tnfa starts. It remains unclear whether lncRNA-CD244 mediates recruitment of EZH2 alone or with other histone modification enzymes. SAP and EAT-2 are associated with CD244 signaling in active TB, but it remains unknown which molecules are downstream of CD244-SAP/EAT-2 signaling cascades.
Materials and Methods
Study Subjects.
Active TB infections were confirmed based on clinical symptoms, chest radiography, sputum staining for acid-fast bacilli (AFB), and laboratory culture and PCR for MTB that were carried out in the Institute for Chronic Diseases Prevention of Huadu District in Guangzhou, China. All patients were not receiving anti-TB therapy at the time of analysis. Patients with active TB were recruited in our study and gave written informed consent according to the protocols approved by the institutional review and the ethics boards of the Zhongshan School of Medicine of Sun Yat-sen University (SYSU).
Animal study protocols were also reviewed and approved by the SYSU Institutional Animal Care and Use Committee.
Statistics.
Statistical analysis was performed using GraphPad Prism. Statistical significance was determined with Student t test. A value of P < 0.05 was considered significant. Asterisks in the figures represent the following: *P < 0.05, **P < 0.01, and ***P < 0.001. “NS” in the figures indicates no statistical significance.
Full materials and methods and any associated references for following experiments are described in detail in SI Materials and Methods: isolation of PBMCs; antibodies for flow cytometry; CBA analysis of the cell culture supernatants; blocking experiments using anti-CD244 mAb; intracellular cytokine staining (ICS) and flow cytometric assay; siRNA transfection; purification of monocytes, CD8+CD244− T cells, and CD8+CD244+ T cells; mRNA and lncRNA microarray analysis, data analysis, and statistics; lentivirus-mediated knock down of lncRNA-CD244 or GSTT1; qPCR; ChIP-qPCR; RNA IP (RIP)-qPCR; nuclear and cytoplasmic extraction of lncRNA; RNA FISH and immunofluorescence microscopy; confocal microscopic analysis; bioinformatics analyses of evolutionary conservation and coding potential of lncRNA-CD244 and plasmid constructions; Northern blot assay of lncRNA; lncRNA pull-down assay; MTB infection of mice; adoptive transfer; and histopathological, bacterial, and immune analyses of MTB-infected mice.
SI Materials and Methods
Isolation of PBMCs.
PBMCs were isolated from freshly collected lithium heparin blood by Ficoll-Paque Plus, density gradient centrifugation before analysis. Briefly, blood was loaded with Ficoll-Paque Plus and centrifuged at 1,500 rpm (Beckman Coulter, Allegra X-12, SX4750 rotor) for 20 min at 20 °C. Isolated mononuclear cells were washed with pH 7.4 PBS (Gibco) and finally suspended into 10% (vol/vol) FBS/RPMI-1640 (Gibco) media and cultured for further study.
Antibodies for Flow Cytometry.
The following antibodies were used for surface and ICS for flow cytometry: CD3-FITC (OKT3; eBioscience); CD4-PECY7, CD4-AF647, CD4-AF488, or CD4-V450 (RPA-T4; BD); CD8-PE, CD8-AF647, CD8-FITC, or CD8-PECY7 (RPA-T8; BD); IFN-γ–APC, IFN-γ–PECY7, IFN-γ–AF488 (4S.B3; eBioscience); TNF-α–FITC, TNF-α–PECY7, TNF-α–AF488, or TNF-α–APC (MAb11; eBioscience); CD244-PE (Clone 2-69; BD); SAP (Clone EPR3168; Abcam); and EAT-2 (polyclonal rabbit IgG; Thermo Scientific).
ICS and Flow Cytometric Assay.
PBMCs isolated from the research subjects were stained directly without ex vivo antigenic restimulation or were stimulated ex vivo using MTB supersonic lysates (0.75 µg/mL), as we previously described (15, 62, 63). Briefly, PBMCs were incubated with or without restimulation with MTB supersonic lysates for 6 h in the presence of brefeldin A (5 µg/mL; BD). Cells in 96-well plates were transferred into polystyrene round bottom tubes (BD) for surface staining. After staining for cell-surface markers for 20 min, cells were then permeabilized for 30 min (Cytofix/Cytoperm; BD) and stained 45 min for intracellular molecules such as IFN-γ and TNF-α before fixation with 2% formalin/PBS. Cells were then analyzed using polychromatic flow cytometry. To ensure the specific immune staining of ICS, isotype IgG served as negative controls for staining surface markers or intracellular cytokines. Data were acquired with a Beckman Coulter Gallios (Beckman) and analyzed with Kaluza 1.2 software (Beckman).
CBA of the Cell Culture Supernatants.
Culture supernatants of cells were analyzed for the production of following cytokines: IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ, and IL-17A using the CBA Human Th1/Th2/Th17 Cytokine Kit (BD) according to the manufacturer’s instructions. Data were acquired on the Beckman Coulter Gallios (Beckman). The concentration of that cytokine was revealed by the fluorescence intensity of each supernatant sample. Cytokine concentrations were calculated relative to the standard dilution curve.
Blocking Experiments Using Anti-CD244 mAb.
Cells were cultured in the presence or absence of anti-CD244 Ab (clone: 2B4; 10 µg/mL; eBioscience) or IgG control for 5 d. Each group of culture has the same number of cells and the same volume of culture medium; 100-µL cultural supernatant was then collected for further CBA analysis.
Confocal Microscopic Imaging.
PBMCs derived from patients with active TB were stained with anti-CD244, anti-EZH2, CD8, and DAPI for nucleus and then imaged with a confocal microscope (LSM710; Zeiss). The details for the sample preparation and confocal microscopic image analysis were described previously by us (63).
siRNA Transfection.
siRNA transfection were performed as we previously described (15). Cells were transfected with 50 nM siRNA-targeting CD244, EZH2, or lncRNA, as indicated in Results or 50 nM siRNA-Ctrl using Lipofectamine 2000 (Invitrogen/Life). siRNAs were synthesized by RiboBio. Cells were then analyzed by qRT-PCR and/or flow cytometry to analyze the expression of CD244, EZH2, GSTT1, or lncRNA 48 h posttransfection to determine the silence efficiency.
siRNA Targets of lncRNA-BC050410.
The targets of siRNA are underlined in the following sequence of lncRNA-BC050410: GTAAAATGCTAGTGCCTTTGAAAGCAAAATTCAGACATACTTGCATAGCTTAAGGTAAATTGATGTATGAGAGTTATTTGTAATTAGATATTTGTGTATATATATATTTTTGAGACAGTTTCGCTCTTCCAGCCTGGGCAACAGAGCAAGACACTATCTCAAAAAAAAAAAAAAAAAAGTTTGTTTTGAAATCCTGGACTCAAGCAATCCCCCCAACCTGGACCTCCCAAAGTACTAGGATTATAAGTATGAGCTACCACACTCAGCCACTTTTTTTTTTGAGATGGAGTCTCGCTCGGTTGCCCAGGCTGGAGTGCAGTGGCGCGATCTCGGCTCATTGCAAGGTCCGCCTCCTGGGTTCATGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTACAGGCACCCA.
CCACCACGCCCGGCTAATTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCTCCTGACCTCGTGATCCCCCTGCCCCGGCCTCCCAAAGTGCTGGGATTACAAGCGTGAGCCACCGCACCCGGCCTCAGCCACATTTATTAATTTCACTCTTGGCAAACATCAGGGGGAAGCTGACCCACACGGCCTGGGAAGGGGGTTGTCTTTTGCATAGAGACCATGACCAGGTCTGGGACAGAGGAAAGTCAAATAAATCACACATTAGAGTTAGAAGCAGAGGCTCAGGCTGAGCCCAGGTTTATTATCCAAAATCAAAATGAAATGCAGTGATTAAAGGACAAAAAAAAAAAAAAAAAAAAAA.
Purification of Monocytes, CD8+CD244− T Cells, and CD8+CD244+ T Cells.
Cells isolation was carried out as previously described (15, 63). Briefly, PBMCs isolated from TB patients were cultured in plastic plates. After 2 h of adherence, monocytes adhered to the bottom of the plate and detached using cold 2% FBS/PBS, whereas suspension cells containing T cells were collected and CD8+ T cells were purified by positive selection with magnetic beads. Cells were stained with anti-CD8-FITC (eBioscience), followed by anti-FITC magnetic bead (Miltenyi Biotec). The stained cells were then loaded to the purification column following the instructions of the manufacturer. The passing fraction was collected as CD8− (negative) T cells that did not bear CD8, whereas the fraction that was captured by magnetic beads was CD8+ (positive) T cells and was released by releasing buffer (Miltenyi Biotec). The isolated CD8+ T cells were stained again with anti–CD244-PE (2B4; BD), followed by anti-PE magnetic microbeads for secondary purification.
mRNA and lncRNA Microarray Analysis, Data Analysis, and Statistics.
CD8+CD244+ T cells and CD8+CD244− T cells from PBMCs that were derived from six individuals with active TB disease were purified by positive selection with magnetic beads as described above. Total RNA of CD8+CD244+ T cells and CD8+CD244− T cells were isolated with TRIzol reagent (Invitrogen Life Technologies). After RNA cleanup and quality analysis, total RNA was used for human mRNA and lncRNA microarray analysis. Briefly, the RNAs were first labeled with fluorescent probes using the Quick Amp Labeling Kit (Agilent) and purity and quality of the labeled RNA was examined by RNeasy Mini Kit (Qiagen) and NanoDrop ND-1000. RNA was then hybridized using the Agilent Gene Expression Hybridization Kit (Agilent). After careful washing using Gene Expression Wash Buffer (Agilent), the hybridized ChIPs were then scanned to examine fluorescent intensity and to determine expression profiles of mRNA and lncRNA in CD8+CD244+ T cells and CD8+CD244− T cells using Agilent Microarray Scanner (Agilent) and Agilent Feature Extraction (FE) software. The median of endogenous controls was used as the computed control and the change in cycling threshold (ΔCT) was calculated. Unpaired Student t test was used for analysis of mRNA and lncRNA expression between CD8+CD244+ T cells and CD8+CD244− T cells. If higher expression levels of some mRNAs and lncRNA were observed in CD8+CD244+ T cells, the ratio of ΔCT of CD8+CD244+ T cells to ΔCT of CD8+CD244− T cells was calculated to determine the folds of differentiated expression of these mRNAs and lncRNAs. Only those mRNAs and lncRNA with folds of differentiated expression larger than two (or smaller than 0.5) and P < 0.05 were chosen for further bioinformatics analysis.
Lentivirus-Mediated Knockdown of lncRNA-CD244 or GSTT1.
CD8+ T cells derived from individuals with active TB disease were transduced with either LV vector encoding GFP and shRNA targeting lncRNA-CD244 (LV-lncRNA-CD244) or GSTT1 (LV-GSTT1) or control LV vector encoding GFP only (LV-Ctrl) at multiplicities of infection of 5 (2 × 107 transducing units per mL) in the presence of Polybrene (Santa Cruz Biotechnology) (5 µg/mL). CD8+ T cells were then observed by inverted fluorescence microscope (Leica) 5 d posttransduction. Transduction efficiency was also assessed by qPCR and flow cytometry through determination of the frequency of cells positive for GFP. After 10 d of culture, the concentration of Th1/Th2/Th17 cytokines in PBMC culture supernatants were analyzed by CBA.
qPCR.
Total RNA was isolated, converted to cDNA, and analyzed by real-time qPCR amplification on a Bio-Rad iCycler by using Bio-Rad IQ5 (Bio-Rad). Primers for lncRNCA-CD244, gstt1, infg, and tnfa were purchased from Invitrogen Life Technologies.
ChIP-qPCR.
ChIP analysis was performed on 2 × 105 cells using the ChIP Assay Kit (Millipore) following the manufacturer’s instructions. The antibodies used were HDAC2 (51-5100; Invitrogen), G9a (ab-40542; Abcam), EZH2 (AB3748; Abcam), H3Ac (06-599; Millipore), H3K4me3 (17-614; Millipore), H3K9me1 (AB9045; Abcam), H3K9me2 (ab-1220; Abcam), H3K9me3 (17-625; Millipore), or H3K27me3 (ab-6002; Abcam). Real-time qPCR amplification on a Bio-Rad iCycler by using Bio-Rad IQ5 was performed with 2 µL from a total of 50 µL of the immunoprecipitated DNA. As a control, input DNA purified from chromatin before IP was used.
RIP-qPCR.
RIP was performed using the Magna RIP-Binding Protein Immunoprecipitation Kit (Millipore). Briefly, cells were crosslinked with 1% formaldehyde, incubated with anti-EZH2 (AB3748; Abcam) or control antibody overnight, recovered with protein G Dynabeads (Life Technologies), and washed with radioimmunoprecipitation assay buffer. lncRNA-CD244 and control RNAs were then analyzed by qPCR.
lncRNA Pull-Down Assay.
Biotin-labeled lncRNA-CD244 were transcribed with Biotin RNA Labeling Mix (47) and T7 RNA polymerase (Promega), treated with RNase-free DNase I (Promega), and purified using RNeasy Mini Kit (Qiagen). Biotin-labeled lncRNA-CD244 was mixed and incubated with nuclear extracts of CD8+ T cells purified from PBMCs of patients with active TB. Streptavidin-conjugated magnetic beads (Invitrogen) were added to each binding reaction and further incubated. Beads were washed thoroughly. Then the retrieved proteins were detected by Western blot.
Nuclear and Cytoplasmic Extraction of lncRNA.
CD8+ T cells isolated from PBMCs of patients with active TB were subject to nuclear and cytoplasmic extraction of RNA using the Cytoplasmic & Nuclear RNA purification kit, following the instructions of the manufacturer (Norgen Biotek). The nuclear and cytoplasmic fractions of RNA of CD8+ T cells were then subjected to qPCR analysis of lncRNA and electrophoresis analysis of PCR products.
Bioinformatics Analyses of Evolutionary Conservation and Coding Potential of lncRNA-CD244 and Plasmid Constructions.
Evolutionary conservation of lncRNA-CD244 was analyzed by the University of California Santa Cruz (UCSC) Genome Browser (genome.ucsc.edu). The coding potential of lncRNA-CD244 was first analyzed using a program provided by the Peking University Center for Bioinformatics (cpc.cbi.pku.edu.cn/programs/run_cpc.jsp). lncRNA-CD244 or EZH2 expression vector was then constructed by PCR-based amplification of cDNA from human CD8+ T cells of patients with active TB and then subcloned into the ORF of eukaryotic expression vector pReceiver-M29 with EGFP tag. lncRNA (or EZH2) and EGFP share the same promoter, CMV. Vector pReceiver-M02R with only EGFP tag and vector pReceiver-M02R with lncRNA-CD244 but without EGFP tag were served as controls. All constructs were confirmed by DNA sequencing.
Northern Blotting Assay.
A total of 10 μg of the indicated RNA was subjected to formaldehyde gel electrophoresis and transferred to a Biodyne Nylon membrane (Pall). A digoxin-labeled (Roche) lncRNA-CD244 cDNA probe was prepared using PCR. The sequence of the specific probe for Northern blot analysis of lncRNA-BC050410 was as follows: AAGTGAGAACCGTTTGTAGTCCCCCTTCGACTGGGTGTGCCGGACCCTTCCCCCAACAGAAAACGTATCTCTGGTACTGGTCCAGACCCTGTCTCCTTTCAGTTTATTTAGTGTGTAATCTCAATCTTCGTCTCCGAGTCCGAC. After 60 min of prehybridization in ULTRAhybTM-Oligo buffer (Ambion), the membrane was hybridized for overnight at 68 °C in ULTRAhybTM-Oligo buffer containing the denatured probe. Washes were performed as described in the NorthernMax Kit protocol (Ambion), and the membrane was detected using an Odyssey infrared scanner (Li-Cor).
RNA FISH and Immunofluorescence Microscopy.
To detect lncRNA-CD244, CD8+ T cells from patients with active TB were rinsed briefly in PBS and then fixed in 3.6% formaldehyde plus 10% acetic acid in PBS (pH 7.4) for 15 min. CD8+ T cells were permeabilized in PBS containing 0.2% to ∼0.5% Triton X-100 and 5 mM vanadyl ribonucleoside complex (10 mM) (New England BioLabs), washed in PBS, and rinsed once in SSC buffer. Hybridization was carried out using a digoxin-labeled (Roche) lncRNA-CD244 cDNA probes in a moist chamber at 37 °C overnight. After hybridization, cells were incubated with anti–digoxin–rhodamine mAb (Roche) and subjected to confocal microscopic imaging. For colocalization studies, after RNA-FISH, cells were again fixed for 5 min in 2% formaldehyde and subjected to immunofluorescence staining of EZH2 using anti-EZH2 mAb (Abcam). Cells were then observed with a Zeiss (LSM 780) confocal laser scanning microscope, as we previously described (63).
MTB Infection of Mice.
Mice were infected by i.p. injection of 2.5 × 105 cfu MTB (H37Rv) at the Biosafety Level-3 (BSL-3) Laboratory SYSU. Mice were housed in a specific pathogen-free animal laboratory before moving into the BSL-3 Laboratory for MTB infection.
Adoptive Transfer.
CD8+ T cells and CD14+ monocytes were purified from PBMCs of patients with active TB using positive selection using a MACS Pro Separator (Miltenyi Biotech). Cell purity was consistently ≥95%. Three days before transfer, 4-wk-old SCID mice were infected with 2.5 × 105 cfu MTB (H37RV); 5 × 105 CD8+ T cells were transduced with LV-lncRNA-CD244 or LV-Ctrl or treated with transduction medium for 5 d. Culture supernatants for CD8+ T cells and CD8+ T cells transduced with indicated LV vector or medium and 4 × 104 autologous monocytes were transferred into each recipient mouse via i.p. injections.
Histopathological, Bacterial, and Immune Analyses of MTB-Infected Mice.
For bacterial burden analysis, tissues of mice were homogenized and peripheral blood was collected for MTB CFU counting analysis, as we previously described (7). For tissue histopathological analysis, lung tissues of mice were fixed in 10% zinc formalin and embedded in paraffin; 5-μm-thick sections were stained with H&E, and images were obtained using a microscope (BX51WI; Olympus) and a digital camera (DP30BW; Olympus). Matched lobes were used for analysis of bacterial burdens, tissue histopathology, and immune parameters of mice with different adoptive transfers of indicated cells.
Acknowledgments
We thank members of the Sun Yat-sen University Biosafety Level-3 Laboratory for biosafety management; Xiaobo Li for flow cytometric analysis; Dr. Yijun Zhang, Shaoyuan Li, Sufen Zhang, and Dongting Mao for technical assistance; and Jun Liu, Jianguo Wang, and Dr. Xionglei He for help with bioinformatics analysis. This work was supported by National Natural Science Foundation of China–National Institutes of Health (NSFC-NIH) Biomedical Collaborative Research Program Grant 81361120379 (to G.Z.), NSFC Grant 31170847 (to G.Z.), the Guangzhou Municipality Commission for Science and Technology Innovation (Pearl River S&T Nova Program) Grant 201506010034 (to G.Z.), Guangdong Innovative R&D Team Program Grant 2009010058 (to H. Zhang), United States–China Collaborative Research Program Grants NIH AI106590 and NSFC 31129002 (to Z.W.C.), and NIH R01 Grants HL64560 and OD015092 (RR13601) and AI106590 (to Z.W.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501662112/-/DCSupplemental.
References
- 1.Rubin EJ. Troubles with tuberculosis prevention. N Engl J Med. 2014;370(4):375–376. doi: 10.1056/NEJMe1312301. [DOI] [PubMed] [Google Scholar]
- 2.Zumla A, George A, Sharma V, Herbert N. Baroness Masham of Ilton WHO’s 2013 global report on tuberculosis: Successes, threats, and opportunities. Lancet. 2013;382(9907):1765–1767. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 3.Bold TD, Ernst JD. CD4+ T cell-dependent IFN-γ production by CD8+ effector T cells in Mycobacterium tuberculosis infection. J Immunol. 2012;189(5):2530–2536. doi: 10.4049/jimmunol.1200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CY, et al. A critical role for CD8 T cells in a nonhuman primate model of tuberculosis. PLoS Pathog. 2009;5(4):e1000392. doi: 10.1371/journal.ppat.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufmann SH. How can immunology contribute to the control of tuberculosis? Nat Rev Immunol. 2001;1(1):20–30. doi: 10.1038/35095558. [DOI] [PubMed] [Google Scholar]
- 7.Chen CY, et al. Phosphoantigen/IL2 expansion and differentiation of Vγ2Vδ2 T cells increase resistance to tuberculosis in nonhuman primates. PLoS Pathog. 2013;9(8):e1003501. doi: 10.1371/journal.ppat.1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes-Alves C, et al. In search of a new paradigm for protective immunity to TB. Nat Rev Microbiol. 2014;12(4):289–299. doi: 10.1038/nrmicro3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bengsch B, et al. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6(6):e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Ha SJ, et al. Enhancing therapeutic vaccination by blocking PD-1-mediated inhibitory signals during chronic infection. J Exp Med. 2008;205(3):543–555. doi: 10.1084/jem.20071949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Yao S, Chen L. Cell surface signaling molecules in the control of immune responses: A tide model. Immunity. 2011;34(4):466–478. doi: 10.1016/j.immuni.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velu V, et al. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009;458(7235):206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Y, et al. Tim-3-expressing CD4+ and CD8+ T cells in human tuberculosis (TB) exhibit polarized effector memory phenotypes and stronger anti-TB effector functions. PLoS Pathog. 2012;8(11):e1002984. doi: 10.1371/journal.ppat.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaraman P, et al. Tim3 binding to galectin-9 stimulates antimicrobial immunity. J Exp Med. 2010;207(11):2343–2354. doi: 10.1084/jem.20100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: The role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18(9):1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 18.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(4):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(81):145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 24.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monnier P, et al. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc Natl Acad Sci USA. 2013;110(51):20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garzon R, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giovarelli M, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci USA. 2014;111(47):E5023–E5028. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trimarchi T, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15(6):484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 32.Gomez JA, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 36.Pang KC, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182(12):7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 37.Waggoner SN, Taniguchi RT, Mathew PA, Kumar V, Welsh RM. Absence of mouse 2B4 promotes NK cell-mediated killing of activated CD8+ T cells, leading to prolonged viral persistence and altered pathogenesis. J Clin Invest. 2010;120(6):1925–1938. doi: 10.1172/JCI41264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kambayashi T, Assarsson E, Chambers BJ, Ljunggren HG. Cutting edge: Regulation of CD8(+) T cell proliferation by 2B4/CD48 interactions. J Immunol. 2001;167(12):6706–6710. doi: 10.4049/jimmunol.167.12.6706. [DOI] [PubMed] [Google Scholar]
- 39.Raziorrouh B, et al. The immunoregulatory role of CD244 in chronic hepatitis B infection and its inhibitory potential on virus-specific CD8+ T-cell function. Hepatology. 2010;52(6):1934–1947. doi: 10.1002/hep.23936. [DOI] [PubMed] [Google Scholar]
- 40.Sun C, et al. TGF-β1 down-regulation of NKG2D/DAP10 and 2B4/SAP expression on human NK cells contributes to HBV persistence. PLoS Pathog. 2012;8(3):e1002594. doi: 10.1371/journal.ppat.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlaphoff V, et al. Dual function of the NK cell receptor 2B4 (CD244) in the regulation of HCV-specific CD8+ T cells. PLoS Pathog. 2011;7(5):e1002045. doi: 10.1371/journal.ppat.1002045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol. 2007;25:337–379. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 43.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6(1):56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 44.Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27(5):698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Di Meglio T, et al. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339(6116):204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allan RS, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487(7406):249–253. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- 48.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 49.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 50.Martin L, Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest. 2012;122(5):1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guil S, Esteller M. Cis-acting noncoding RNAs: Friends and foes. Nat Struct Mol Biol. 2012;19(11):1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 52.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22(9):1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guil S, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19(7):664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 56.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tumes DJ, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39(5):819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 60.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao S, et al. Differentiation, distribution and gammadelta T cell-driven regulation of IL-22-producing T cells in tuberculosis. PLoS Pathog. 2010;6(2):e1000789. doi: 10.1371/journal.ppat.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng G, et al. Membrane-bound IL-22 after de novo production in tuberculosis and anti-Mycobacterium tuberculosis effector function of IL-22+ CD4+ T cells. J Immunol. 2011;187(1):190–199. doi: 10.4049/jimmunol.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4(4):259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]