Abstract
Research over the past two decades shows that both recombination and clonality are likely to contribute to the reproduction of all fungi. This view of fungi is different from the historical and still commonly held view that a large fraction of fungi are exclusively clonal and that some fungi have been exclusively clonal for hundreds of millions of years. Here, we first will consider how these two historical views have changed. Then we will examine the impact on fungal research of the concept of restrained recombination [Tibayrenc M, Ayala FJ (2012) Proc Natl Acad Sci USA 109 (48):E3305–E3313]. Using animal and human pathogenic fungi, we examine extrinsic restraints on recombination associated with bottlenecks in genetic variation caused by geographic dispersal and extrinsic restraints caused by shifts in reproductive mode associated with either disease transmission or hybridization. Using species of the model yeast Saccharomyces and the model filamentous fungus Neurospora, we examine intrinsic restraints on recombination associated with mating systems that range from strictly clonal at one extreme to fully outbreeding at the other and those that lie between, including selfing and inbreeding. We also consider the effect of nomenclature on perception of reproductive mode and a means of comparing the relative impact of clonality and recombination on fungal populations. Last, we consider a recent hypothesis suggesting that fungi thought to have the most severe intrinsic constraints on recombination actually may have the fewest.
Keywords: fungi, reproduction, recombination, clonality, population genomics
Easily the most famous claim of strict clonality in fungi is the “ancient, asexual scandal” of the Glomales (1), home to the fungi that form arbuscular mycorrhizae with the great majority of land plants (2). They are the oldest fungi with a solid fossil record, in terms of both fossils of their clonal spores at 460 Ma (3) and fossils of their association with the underground organs of plants at 400 Ma (4). They are massively multinucleate (Fig. 1), and no one has ever reported a stage in the life cycle in which there is only one nucleus per cell, let alone a single cell with a single nucleus. Nor has anyone reported sexual reproduction in these fungi. To explain their apparent avoidance of the genome decay that should accompany exclusive clonality (5), nuclei within an individual have been hypothesized, based on cytological investigation, to be genetically different (Fig. 1) and capable of internuclear genetic exchange (6). This interpretation was challenged by comparison of the nuclei of parents and progeny and by analysis of individual nuclei (7), but the matter remained unresolved because of the difficulty of experimenting with these obligate plant symbionts. Recently, however, the genome sequence of the most famous of these fungi, Rhizophagus intraradices (syn. Glomus intraradices) (8–10), has shown that the genomes are 10-fold larger than previously estimated (11), that there is no evidence for multiple, diverged genomes in an individual, that genes known to be involved in sexual reproduction in other fungi are intact in R. intraradices, and that, in some cases, the number of these genes is expanded. Based on these results, one must conclude that arbuscular mycorrhizal fungi are not ancient asexual scandals. However, it still is true that no one has reported sex in these fungi or observed a stage in the life cycle in which these fungi exist with only a single nucleus. Clearly, mycologists have more work to do with these extremely important fungi.
Fig. 1.
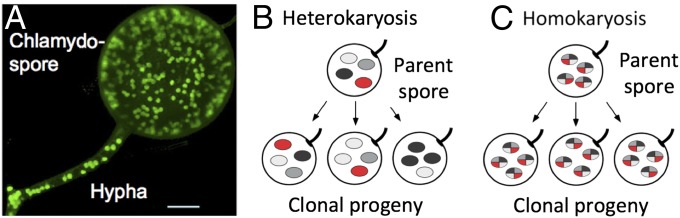
Arbuscular mycorrhizal fungi in the Glomales. (A) All known phases of the life cycle are multinucleate, as shown here in a hypha and chlamydospore. (Scale bar: 10 µm.) Reproduced from ref. 8. (B and C) Alternative interpretations of how genetic variation is distributed among multiple nuclei in arbuscular mycorrhizal Glomales. (B) Heterokaryosis with genetic variation spread among different nuclei, so that nuclei in a single individual are quite different. (C) Homokaryosis with genetic variation present in every nucleus, so that nuclei are essentially identical. B and C reproduced with permission from ref. 7.
Population Genetic Evidence for Recombination
Evidence that clonality was not limited to Glomales but instead was widespread in fungi came from the oft-cited statistic that 20% of fungi are asexual (12). This fraction represented the number of fungi for which sexual reproduction had not been observed or was rarely observed. At a time when observation of the sexual morphology of a fungus was required for its classification, these fungi were classified in the Deuteromycota, apart from sexual fungi. Two decades later, with DNA providing the variable characters required for phylogenetic classification, all fungi now can be classified in one Kingdom, Fungi, and the Deuteromycota classification has been officially abandoned (13). In those two decades, DNA variation also was applied to population studies, some of which addressed the reproductive mode of fungi with no observed sexual morphology. Here we present two examples of these studies, one with Coccidioides posadasii and the other with Aspergillus fumigatus.
C. posadasii is known to mycologists and physicians because it causes disease in otherwise healthy humans (14). It is limited to hot, dry areas of the Southwestern United States and Mexico and to similar locations in Central and South America, where it is adapted to life with desert mammals. The same environments are occupied by its sister species, Coccidioides immitis, which is found in California and northern Mexico. These species make abundant clonal spores, but no one has reported sex in either species. The first evidence for recombination in C. immitis, and the first for any morphologically asexual fungus, came from a population study using randomly amplified markers that showed electrophoretic variation that could be confirmed by sequencing. With only 25 individuals and 14 markers, the null hypothesis of recombination could not be rejected, unlike the hypothesis for clonality, which could be rejected (15). The discovery of a recombining population structure was followed by the discovery by two independent groups of intact mating-type genes in the two Coccidioides species (16) and the observation that the mating-type regions for the two mating types (which are so diverged in fungi that they are termed “idiomorphs” instead of “alleles”) have been in equal proportion over time, as would be expected of a sexual population (17). Additional evidence for genetic exchange and recombination has come from population genomic research. Population genetics had shown that the two Coccidioides species (18) harbor genetically differentiated populations (19), and this finding stimulated the sequencing individuals from five populations in the two species (20). Genome scans for exceptional levels of divergence between the two populations (Fig. 2) showed that as much as 7% of the genomes of one species originated from the other species, most plausibly by interspecific hybridization and subsequent introgression. Thus, although no one has yet reported sexual reproduction in this biological safety level 3 pathogen, studies of reproductive mode, genetic control of sex, and population genomics all support recombination, most parsimoniously explained by sexual reproduction.
Fig. 2.
Scan of divergence, measured as the Wright's fixation index (Fst), between genomes of two Coccidioides species. The Fst reports on genetic isolation from none (0) to strong (1). Most of chromosome 1 is well diverged, but one region (asterisk) shows no divergence caused by introgression of regions from one Coccidioides species into the other. Figure based on data from ref. 20. Image courtesy of Dan Neafsey (Broad Institute, Cambridge, MA).
A. fumigatus is known to mycologists and physicians because it causes disease in humans whose immune systems are suppressed as a prerequisite for or consequence of medical interventions such as bone marrow or solid organ transplant or cancer therapy. It is cosmopolitan and ubiquitous in soil and composting plant material. As with Coccidioides species, sex had not been observed when two research groups, each using a different, global collection of strains and each using different variable genetic markers, independently demonstrated a recombining population structure in A. fumigatus (21, 22). Both mating-type idiomorphs were described, and a balance of mating type idiomorphs was found throughout the range of this fungus (21). Therefore, all population genetics studies support recombination in nature. The story was completed when strains of each mating type from an Irish population were crossed and, after 6 mo in the dark on a diet of oatmeal, produced recombined progeny (Fig. 3) (23).
Fig. 3.
Evidence for sexual reproduction in the fungus A. fumigatus, formerly thought to be asexual. Experimental mating between individuals of opposite mating type from a population of A. fumigatus recognized by multilocus sequence typing. (A) Rows of sexual structures where colonies of the two individuals met. (B) Meiotically produced ascospores. (Scale bars: A, 1 cm; B, 2 µm.) Reproduced with permission from ref. 23.
These two examples are typical of a number of other studies of fungi that lacked the morphology of sexual reproduction, among them species of the famous “asexual” genus Penicillium (24, 25). The progression of the discovery of evidence for recombination in these fungi also is typical, beginning with evidence from population genetics, progressing to demonstrations of intact mating-type genes, and in some cases culminating in laboratory demonstration of successful sexual reproduction. We would argue that the population genetics evidence is the evidence needed to show that recombination has an impact in nature, but we recognize that a demonstration of sex in the laboratory is the evidence that convinces most biologists. Although only a few of the many fungi that lack the morphology of sex have been studied in the detail described for Coccidioides and Aspergillus, these studies shift the default assumption of fungal reproduction in nature away from clonality. It is now clear that fungi reproduce both sexually and clonally and that restrictions to recombination can be involved. These restrictions will be the subject of the next section. We would be remiss if we did not mention a by-product of the population genetics studies that demonstrated recombination in natural populations of fungi, that is, to provide the data to show that fungi have biogeography complete with endemic populations and species (26, 27). Before the use of nucleic acid variation, this situation was not obvious, as it was for plants, animals, and other macrobes, because fungi and other microbes offer few morphological characters to distinguish among species.
Restricted Recombination
Tibayrenc and Ayala (28) introduced the very useful concept of restricted recombination. They view the matter from the angle of pervasive clonal reproduction, stating that “… clonality does not mean total absence of recombination, but that it is too rare to break the prevalent pattern of clonal population structure” (28). For fungi, it may be useful to view the matter from the angle of recombination, i.e. that recombination does not mean the total absence of clonality but rather that recombination can be constrained by extrinsic and intrinsic means to diminish the pattern of a recombining population structure. No matter the viewpoint, it is clear that recombination has been restricted in fungi. Arguably, the fungi most commonly subjected to restraints on recombination are plant pathogens. The reproduction of these fungi has been reviewed thoroughly (29–33), leading us to focus on animal pathogenic fungi and model fungi. Also notable is a recent review on the impact of genomics on the study of all aspects of fungal reproductive biology (34).
Extrinsic Restrictions to Recombination.
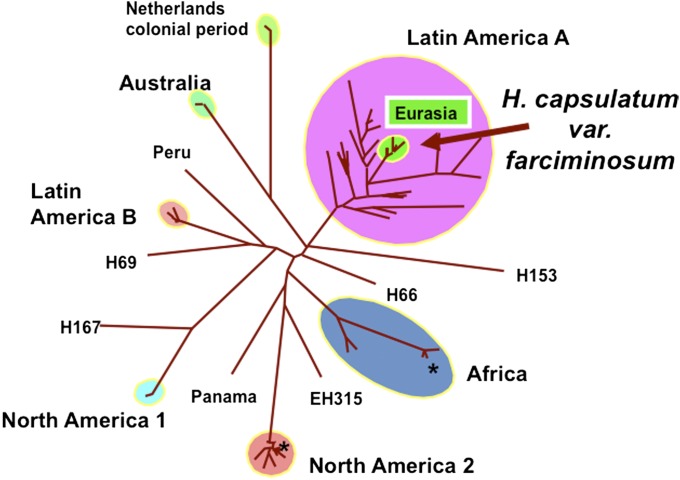
There are several examples of severe restraints to recombination in fungi. Histoplasma farciminosum has long been considered to be a close relative of Histoplasma capsulatum. H. capsulatum is capable of causing systemic infections in otherwise healthy humans, whereas H. farciminosum was known to cause superficial infections in Equidae—horses, mules, and donkeys. H. capsulatum is broadly distributed, in the New World, Africa, and Australia, is known to reproduce both by sexual and clonal spores, and is acquired by inhaling spores produced in nature. H. farciminosum is restricted to Eurasia and is passed from host to host by spores produced in the skin lesions. Like Coccidioides species, Histoplasma species were recognized by genealogical concordance (35, 36) based on phylogenetic analysis of several sequenced genes or regions, in what the bacterial world calls “multilocus sequence typing” (MLST) (37). Using fungal MLST with four regions, 137 Histoplasma isolates could be resolved into seven species-level clades (Fig. 4) with the most diverse clade composed almost entirely of South America isolates (38). Nested within this South American clade was one clade embracing 10 of the 14 H. farciminosum strains, all of which had been isolated in Eurasia and all of which had the same multilocus genotype. In this large South American clade, tests of reproductive mode indicated recombination, but in H. farciminosum, the structure was clonal. The likely explanation is that H. farciminosum represents South American strains that were transported to Europe beginning no earlier than the Spanish conquest of the New World in the late 1400s and early 1500s. The H. farciminosum strains, which now are known as “H. capsulatum var. farciminosum,” have been subjected to severe restrictions to recombination by several extrinsic means. First, their genetic diversity was severely restricted (i.e., bottle necked) when one or a few individuals were taken to Europe. Second, the balance of mating types is likely to be skewed, reducing the opportunities for mating. Third, the mode of transmission lends itself to clonal reproduction, that is, by clonal spores being passed from host to host rather than being acquired from the environment from recombined individuals. Fourth, the spread from South America to Europe would not have been successful without an available niche, which was provided by the abundance of domesticated equids in Eurasian civilization. The absence of a resident European Histoplasma population may have had a role in preserving the H. capsulatum var. farciminosum lineage in Europe.
Fig. 4.
Populations and varieties of H. capsulatum showing that the clonal fungus H. capsulatum var. farciminosum emerges from the most diverse and sexually recombining clade, H. capsulatum, Latin America A. The restrictions on recombination borne by H. capsulatum var. farciminosum stem from its dispersal from its population of origin to Eurasia and its transmission to naive hosts from infected hosts rather than being acquired from the environment. Isolates in the Netherlands colonial period clade are likely from Indonesia, not from Europe. Reproduced with permission from ref. 38.
A likely human equivalent of H. capsulatum var. farciminosum is the athlete’s foot fungus, Trichophyton rubrum, in that it is passed from infected host to infected host via clonal spores (39). Although no population genetics studies have been reported, molecular phylogenetic studies indicate that close relatives of T. rubrum have sexual morphologies (40, 41), making it possible that the athlete’s foot fungus is clonal because of the severe restrictions to recombination caused by its method of transmission, host to host rather than acquisition from nature. This method of transmission, like that for H. farciminosum, probably has allowed one or a few genotypes with restricted variation and mating type to migrate away from the area of origin. Also like H. farciminosum, an expanding niche was available to facilitate the spread, in this case provided by human feet enclosed in shoes.
Hybridization also can restrict recombination in fungi. A well-studied example of hybridization suppressing recombination is found in the plant pathogenic fungus, Epichloë (42). These fungi are well studied because they make toxins that adversely affect both small insect herbivores and large mammalian ones. This behavior increases plant fitness but injures and kills livestock. Epichloë species are capable of both sexual and clonal modes of reproduction when they infect grasses. In the sexual mode, Epichloë produces meiospores that spread the infection to other grass individuals; in the asexual mode, Epichloë can spread vertically as hyphae that grow into the embryos of grass seeds. In both modes, Epichloë species make clonal spores that also cause horizontal spread. There is a remarkable, reciprocal symmetry in Epichloë infections; when the fungus reproduces sexually, it sterilizes the plant host, preventing it from flowering or setting seed. When the fungus reproduces asexually, the plant host is sexual, a necessity for the vertical transmission of Epichloë into the next generation.
Severe restrictions to recombination become apparent when Epichloë species hybridize. The hybrids live as endophytes in the grasses, have no sexual reproduction, and are transmitted vertically through seeds. Parentage of hybrids has been determined through analysis of DNA variation, but researchers have not been able to create the hybrids in the laboratory, leading to speculation that the hybrids form when two, or in some cases three, different species that exist as endophytes in the same host plant exchange nuclei by hyphal fusions in a form of parasexuality (42).
Another notable example of hybrid fungi is found in the human pathogenic Basidiomycota, Cryptococcus neoformans. In this case, individuals of C. neoformans var. neoformans have formed hybrids in nature with C. neoformans var. grubi (43–45). These hybrids clearly can reproduce clonally by budding, but it is not clear if they can reproduce sexually or if the hybrid would be preserved if they did reproduce sexually (46). The two examples of Epichloë and Cryptococcus raise a larger question: Does hybridization lead to as widespread a barrier to recombination in fungi as it can in animals (47) and plants (48)? Judging from current knowledge, the answer would be no. Indeed, it also is possible that the extent of interspecific hybridization has not yet been realized. As mentioned above, population genomics studies of well-diverged Coccidioides species found evidence for interspecific gene flow that likely involved interspecific hybridization (20), and another population genomics study of Neurospora crassa found evidence for hybridization with well-diverged but as yet unidentified populations of this fungus (49). A more extensive study of 10 lineages of Neurospora tetrasperma also found evidence for interspecific hybridizations leading to movement of mating regions (50), of all things, between species, as did previous studies of other Neurospora species (51) and Stemphylium (52). Recently, heterogeneity seen in rDNA repeat regions has been linked to hybridization in mushrooms (53), where hybrids can be recognized more easily because parental nuclei coexist as dikaryons for long periods in these Basidiomycota. The same coexistence of parental genomes is seen in diploid Ascomycota yeasts, such as Saccharomyces and Candida, but not in filamentous, haploid Ascomycota. In terms of clonality, the important question is whether recombination is broadly restricted in the reproduction of hybrid fungi, but there are too few studies to provide a satisfying answer.
Cryptococcus and Candida.
In formulating their concepts of restrictions to recombination, Tibayrenc and Ayala (28) feature several fungi, among them Cryptococcus gattii and Candida albicans. C. gattii causes human disease. It is a Basidiomycota with two mating types, a and alpha. As a haploid, it reproduces clonally by budding as a yeast. After mating, it can produce hyphae, basidia, and recombined basidiospores. The human disease is acquired from the environment, by inhaling sexually produced spores or airborne yeast cells. Mating typically requires partners of different mating type, but sex has been reported between partners of the same mating type (54). C. gattii is not as large a threat to public health as its sister species C. neoformans, but several recent outbreaks of cryptococcosis caused by C. gattii in otherwise healthy humans in British Columbia, Canada and the Pacific Northwest of the United States have resulted in the infection of more than 200 people with mortality ranging from 8% to 33% (55).
The outbreak began in 1999 (56), and by 2005 the first population genetics research was published (57). The strains cultivated from patients were all of one mating type (alpha) and could be sorted into two types based on variation in the mating region: the major genotype, restricted to Vancouver and the minor genotype found in both Australia and Vancouver. Based on the relationship of these individuals to other C. gattii genotypes, it was hypothesized either that the major and minor genotypes were siblings from one alpha–alpha mating or that one was a parent of the other, again in a single-sex mating (57).
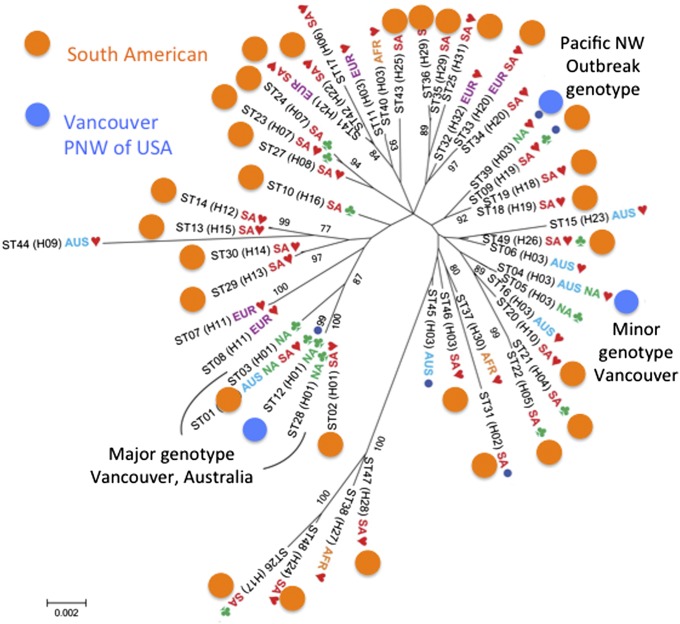
Eight years later, with many more strains available for study, it was discovered that the major and minor genotypes and a third Pacific Northwest genotype could be interpreted as independent migrations from an outbreeding population of C. gattii found in South America (Fig. 5) (58, 59). Thus, the major, minor, and Pacific Northwest genotypes had been subjected to restricted recombination resulting from transport away from their home population, events that limited the genetic diversity of the dispersed strains in general and of the mating type in particular. Locally, C. gattii can appear to be exclusively clonal, but globally it also is recombining.
Fig. 5.
C. gattii genotypes from the outbreak in Vancouver and the Pacific Northwest of the United States (blue) in the context of genotypes from South America (orange). Until South American individuals were collected and analyzed, it was impossible to understand the relationship of the several independent, clonally reproducing outbreak strains to the recombining South American strains. A complete legend can be found in ref. 58. The heart symbol designates a clinical origin; the club symbol designates an environmental origin; a circle designates a veterinary origin. Reproduced from ref. 58.
C. albicans is a normal part of the skin and gut mycota of humans but can cause disease of mucous membranes in otherwise healthy people and all too frequently causes a fatal sepsis in immunocompromised hosts (60). Natural isolates are typically diploid and reproduce clonally by budding, and these cells function as spores that can be spread from host to host. In the laboratory, C. albicans can be mated to produce 4N progeny (61–65), and the 2N condition can be restored to these cells by aneuploidy. There would seem to be many restrictions to recombination in C. albicans, including host-to-host transmission, and no one has reported meiosis in this fungus (although its meiotic genes are intact). A very thorough population genetic study with 1,391 strains and an MLST using seven genes found that C. albicans has clear population structure in the form of 17 distinct clades and that, perhaps surprisingly, recombination in the largest of these clades is sufficient to “remove what would otherwise be expected to be a generally clonal evolutionary pattern in an asexually reproducing species” (66). This result supports a very early report that also found recombination in this species (67) but does not reduce the importance of clonal reproduction to the spread of C. albicans, e.g., in clinics and hospitals. The recombination detected by Odds and colleagues (66) could be caused by parasexuality (68), in which nuclear fusion results in recombination without the benefit of meiosis, as suggested by the absence in the C. albicans genomes of the major regulator of meiosis in Saccharomyces cerevisiae, IME1 (60). It also could be caused by meiosis, in which case mycologists simply have not found the right conditions to facilitate C. albicans sex in their laboratories.
Let us return to the concept of restricted recombination. The studies of fungi described above show how fungal populations have a basis in recombination but may be restricted from recombining by a variety of means. Dispersal is likely the most common means of restricting recombination, greatly facilitated by the transmission of fungi from host to host by clonal spores and by the emergence of a previously untapped but abundant niche, such as feet enclosed in shoes or immunocompromised humans. The history of research in this area shows how difficult it can be to recognize that recombination has been restricted when the full distribution of a fungus is unknown. With H. farciminosum, for example, the effect of dispersal on recombination could not be appreciated until it was recognized that H. farciminosum was found in Europe but originated in South America. The situation is almost the same with C. gattii, in which restrictions to recombination imposed by the migration of a few, similar genotypes, each with the same mating type, could not be appreciated until the diversity in South America was characterized. Similarly, current research on another fungal pathogen, the scourge of amphibians, worldwide, Batrachochytrium dendrobatidis, is likely hampered by our not having found the population from which the global, pathogenic strain originated (69, 70). Finding populations of origin can be difficult because in the area of origin the fungus and its hosts will have coevolved, so that outbreaks are unlikely to occur. Given the need to devote all available resources to combating the outbreak, there will be no reason to sample in geographic regions where there is no outbreak. The examples given above are focused on pathogens because the resources for population genomics have been obtained most easily for fungi that cause problems relevant to society, such as human health and amphibian decline. However, the point that sampling is the key to successful population genetics or genomics applies to any fungus.
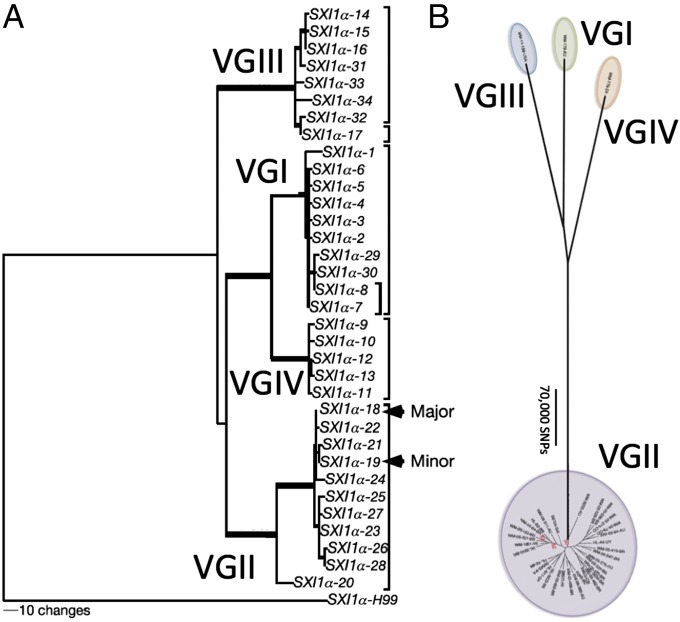
One other point illuminated by Tibayrenc and Ayala (71) should be raised: the multitude of different names used to identify evolutionary units in all organisms, fungi included. The confusion caused by not having a common framework for naming these evolutionary units can impede scientific progress. For example, C. gattii is divided into different clades, each of which is approaching genetic isolation from the other (59). If each of the four clades were accorded species status, then the scant variation seen in the clade with the outbreak isolates in 2005 (57) might have warned researchers that more samples were needed to put the outbreak isolates in a global context (Fig. 6).
Fig. 6.
How the presentation of population data can affect their interpretation. (A) Inclusion of a distant outgroup minimizes the differences between C. gattii clades (VGI–VGIV) and creates the illusion of broad sampling in the species. Reproduced with permission from ref. 57. (B) The absence of an outgroup emphasizes the differences among C. gattii clades (VGI–VGIV) and shows that the clades are as distinct as are species of other fungi. This distinction calls attention to the sparse sampling of three of the clades, a level of sampling similar to that for clade VGII (A) before the inclusion of South American strains. Reproduced from ref. 59.
Intrinsic Restrictions to Recombination.
The examples of restricted recombination noted above have focused on extrinsic restrictions to recombination, but there also are intrinsic restrictions. To explore these restrictions, we need to consider the full range of fungal reproduction, which can be explored with two genera of model fungi, Saccharomyces and Neurospora.
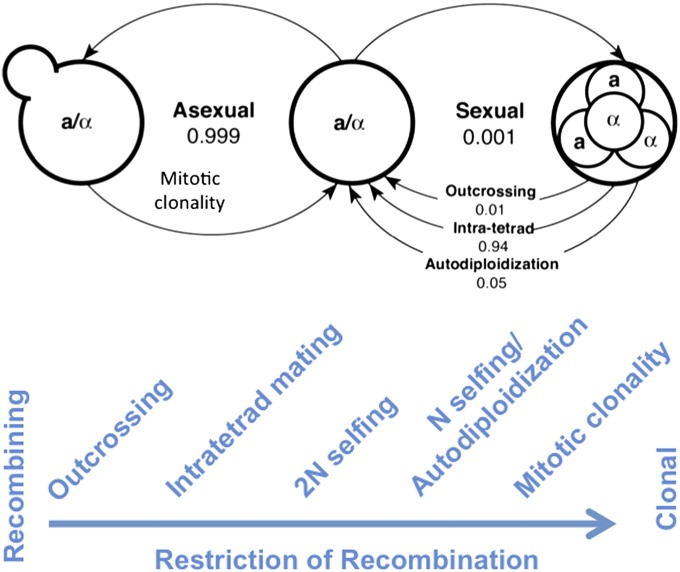
S. cerevisiae and its sister species S. paradoxus are two of the most morphologically simple of all fungi but also share one of the most complete of all fungal life cycles. These species can reproduce asexually and clonally by budding to make daughter cells both as haploids and as diploids. They can reproduce sexually to make meiotic progeny in the form of ascospores, which can be clonal, freely recombined, or restrictedly recombined. Clonal ascospores are the result of haploid selfing resulting from mating-type switching. Freely recombined ascospores arise from outcrossing. Ascospores that fall between the clonal and freely recombined types are the result of two types of inbreeding, one between progeny of different meioses (diploid selfing, which reduces heterozygosity of the zygote by half) and the other between progeny of the same meiosis (intratetrad mating or one type of automixis, which reduces heterozygosity by one-third) (Fig. 7).
Fig. 7.
The range of reproductive modes in fungi as exemplified by Saccharomyces species. Asexual reproduction is simple and exemplified by mitotic clonality. Sexual reproduction is far more complex than simply outbreeding because of various restrictions to recombination caused by N selfing (mating between mitotic siblings), 2N selfing (mating between any meiotic progeny of the same two parents), and intratetrad mating (mating between meiotic progeny of a single zygote) as described in the text. The numbers in large type represent the fraction of nuclear divisions that are mitotic or are meiotic with only 1 in 1,000 estimated to be meiotic. Numbers in small type represent the fraction of diploidizations caused by outcrossing, intratetrad mating, and autodiploidization with only 1 in 100 estimated to be outcrossing. Reproduced with permission from refs. 72 and 87.
Tsai and colleagues (72) used the DNA sequence from one chromosome of 19 strains from two natural S. paradoxus populations to estimate the relative importance of the various asexual and sexual modes of reproduction of this yeast. For every 100,000 reproductive events, 99,900 were asexually mitotic, and 100 were sexually meiotic. Of the 100 sexual reproductive events, 94 were from diploids originating through inbreeding by diploid selfing or intratetrad mating, both of which reduce the heterozygosity in the progeny compared with the parent. Five were from diploids originating by autodiploidization (i.e., haploid selfing) events as a result of mating-type switching, which ensures homozygosity in the progeny at all loci except the mating locus. Only 1 in 100,000 reproductive events was estimated to be meiosis from a diploid that had originated by outbreeding. Despite the relative paucity of outbreeding, S. paradoxus is considered a sexual fungus. On one hand, population genetics studies of S. paradoxus and S. cerevisiae detect recombination as the underlying mode of reproduction, as is consistent with the routine observation of sexual reproduction in the laboratory. On the other hand, these yeast species demonstrate two major intrinsic restrictions to recombination in the form of intratetrad mating and haploid selfing.
The recombination restrictions seen by Saccharomyces can be compared with those of N. crassa. As with Saccharomyces species, N. crassa is considered a recombining fungus, and it is routinely mated in the laboratory. In nature Neurospora exists as a haploid, either as haploid ascospores resting in soil between wildfires or as haploid mycelium-producing reproductive structures in colonies that emerge from heat-killed vegetation following fires (73). Fertilization results in rapid karyogamy, meiosis, and ascospore production. The ascospores then are shot into the soil, where they await the next fire. Just how the ascospores, which require heat to germinate, end up producing colonies on cooked vegetation after a fire is not understood, but endophytism is a possibility (74).
Asexual reproduction by the clonal spores would seem to be a certainty, given the ease with which N. crassa can contaminate laboratories or bakeries, but population genetics and genomics studies have failed to find evidence of clones in nature (75, 76). Haploid selfing does not seem possible, because there is no mating-type switching in N. crassa, nor is intratetrad mating possible, because the asci eject the spores before they germinate. It may be that the only restriction to recombination comes from diploid selfing. However, even this restriction to reproduction may be unlikely if the individuals must wait years until the next fire allows them the opportunity to mate.
Tibayrenc and Ayala (28) note that it would be useful to have a metric for the relative contribution of recombination or clonality compared with microbial reproduction to assess restrictions to recombination. In the case of Saccharomyces and N. crassa, such a metric is available from population genomics studies in each species. The metric is linkage disequilibrium (LD) decay, that is, the genetic distance over which LD decays to half its maximum value. Fungi in which recombination is restricted will have larger LD decay distances than fungi in which recombination is unrestricted. LD decay is 9 kb in S. paradoxus and is 3 kb in S. cerevisiae. In contrast, it is 0.85 kb in a N. crassa population from the Caribbean and is 0.70 kb in a N. crassa population from Louisiana. The difference between S. paradoxus and N. crassa is at least an order of magnitude, 10.5- to 12.8-fold, and between S. cerevisiae and N. crassa the difference is 3.5- to 4.2-fold (49).
In comparing the reproductive mode in N. crassa (outcrossing) with those of S. cerevisiae or S. paradoxus (clonal, haploid selfing, intratetrad mating, and diploid selfing in addition to outcrossing), one gets the impression that the reproductive repertoire in Neurospora is very limited. However, the variation in reproductive mode in Neurospora is seen among species, not within them. For example, intratetrad mating is dominant in N. tetrasperma (77, 78), and haploid selfing is the dominant mode of reproduction in a number of Neurospora species (79–81).
Intratetrad mating in N. tetrasperma is enforced by the inclusion of two nuclei, one of each mating type, in each ascospore, rather than the inclusion of only one, as is the ancestral state in Neurospora species. As a result, the mycelium arising from germinated ascospores contains nuclei of both mating partners from the start, obviating the need to find a partner to complete the life cycle. The positioning of the products of meiosis in the developing ascus requires that the mating types segregate in the first division of meiosis (77). This segregation is enforced by a barrier to recombination that extends beyond the mating locus to include the centromere and much of the mating-type chromosome (50, 82, 83). Generation after generation of intratetrad mating would be expected to result in very diverged genes and a high SNP density in the nonrecombining region of the mating-type chromosomes found in each of the two nuclei in ascospores and hyphae. Conversely, almost no SNPs should be found between the autosomes in the paired nuclei because recombination following intratetrad mating would have eliminated all heterozygosity. A comparison of the diversity in the genome sequences of the two nuclei from one individual confirms these expectations: Of the 192,225 SNPs in the two genomes, more than 99% are in the nonrecombining portion of the mating chromosome. The nonrecombining portion of the genome is ∼7.7 Mb, and the recombining portion is ∼31.3 Mb, so the difference in variation per nucleotide is ∼500-fold (82).
The restriction to recombination in the mating chromosome of N. tetrasperma and other fungi with similarly long regions of suppressed recombination on the mating chromosome [e.g., C. neoformans (84) or Microbotryum violaceum (85)] ought to result in degeneration caused by the accumulation of deleterious mutations, that is, Muller’s ratchet. In fact, the problem also should apply to idiomorphs in Ascomycota and Basidiomycota with the more typical short regions of restricted recombination in their mating regions. In this regard, it is fascinating to find that different lineages of N. tetrasperma have obtained copies of the mating region from other species of Neurospora, including N. crassa (50) and that this exchange of mating loci extends to other species of Neurospora (51), a phenomenon first discovered in Stemphylium (52).
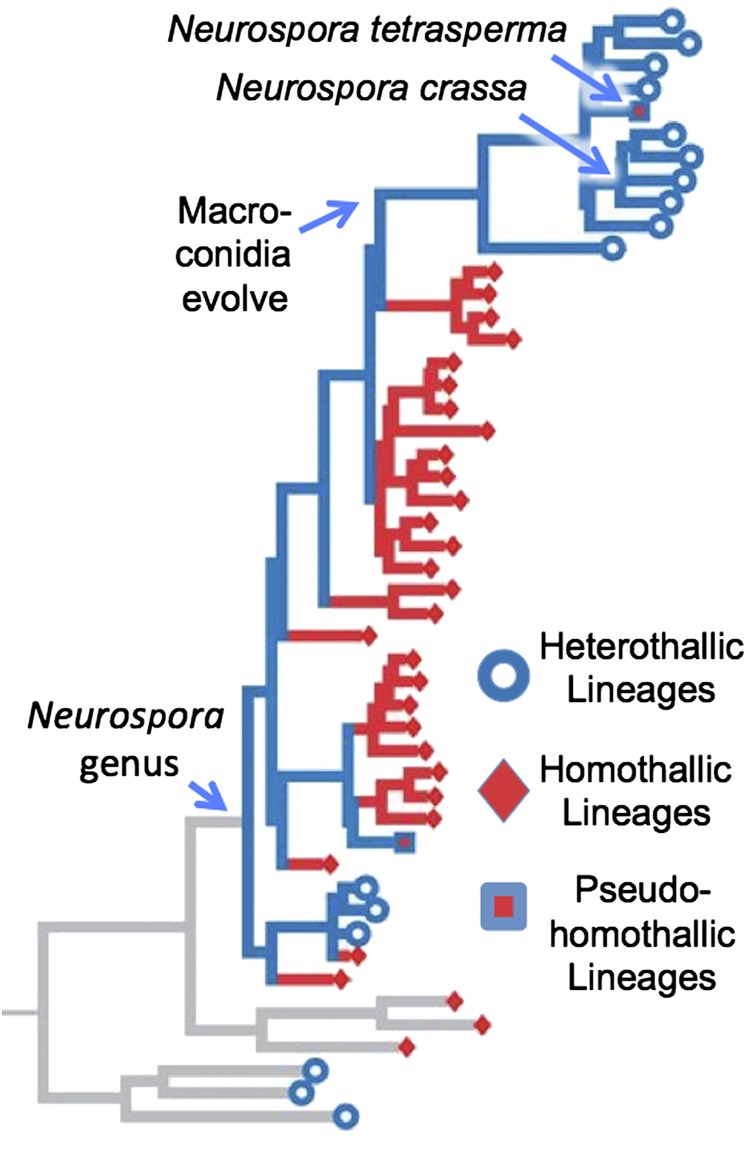
Haploid selfing is the strongest restriction to recombination in fungi that are morphologically sexual, and it is seen in fungi that are termed “homothallic” because they can complete their sexual life cycle without needing any partner. These truly homothallic fungi are a more extreme version of the asci with paired nuclei seen in N. tetrasperma (which is termed “pseudohomothallic”), in that pairs of nuclei are not needed because both mating loci can reside in a single nucleus (79, 81). The genus Neurospora has many of these homothallic species, many known from only a single individual, that branch basal to the derived clade that harbors the well-known, outbreeding (termed “heterothallic”) and pseudohomothallic species (Fig. 8) (14). Given that heterothallism is ancestral in Ascomycota, the derived position of heterothallic species, apparently emerging from a background of homothallic ancestors, poses a puzzle. The explanation of the puzzle involves the presence of characteristic, easily recognized clonal spores (i.e., macroconidia) in the derived clade of heterothallic and pseudohomothallic species and their absence in the basal clades. When mycologists examine fungi isolated from nature, they can recognize new Neurospora individuals in two ways, either from sexual structures (perithecia) made by homothallic or pseudohomothallic species or from clonal spores (macroconidia) made by heterothallic species from the derived clade. If, however, the individual is heterothallic and in the basal region, it will exhibit neither diagnostic conidia nor sexual structures and will be discarded as an unknown, sterile mycelium. Glass et al. (81) discovered that these apparently sterile mycelia are, in fact, aconidial heterothallic Neurospora individuals when they mated them to produce sexual structures and recombined progeny. Recent comparison of the genomes of the basal homothallic species with that of the admittedly distant heterothallic species N. crassa found that basal homothallic species exhibited reductions in purifying selection, transposable element silencing, and codon bias, supporting a hypothesis of reduced adaptive evolution and an evolutionary dead end (86). A very different hypothesis about homothallic fungi was recently advanced by Billiard et al. (87): that homothallic individuals whose genomes contain both mating-type loci might not be genetically isolated from their close heterothallic relatives, as has been assumed. Instead, possessing both mating types, they might be able to mate with all members of their closest heterothallic species, thereby enjoying a higher rate of outcrossing than the heterothallic individuals. By this hypothesis, they would not be an evolutionary dead end. It even is possible that progeny from these hypothesized homothallic–heterothallic matings would displace the heterothallics. Comparison of the genomes of homothallic Neurospora with their close heterothallic relatives could provide a test of these two hypotheses by comparing the architecture of the genomes of hetero- and homothallic individuals.
Fig. 8.
Range of reproductive modes in the genus Neurospora, which include clonal (caused by mitotic conidia), outbreeding (heterothallic), intratetrad mating (pseudohomothallic), and haploid selfing (homothallic and clonal). Modified from ref. 80.
Summary
The “Deuteromycota” and the “ancient asexual scandal of the Glomales,” two influential hypotheses that over the past 20 y have created the impression that fungi are pervasively, and sometime exclusively, clonal, have not stood up to population genetics and genomics testing. Instead, the current paradigm is that fungi reproduce by both recombination and clonality, but there are extrinsic and intrinsic restrictions to recombination that can favor clonal reproduction. Extrinsic restrictions include dispersal of individuals from the native range and hybridization. Dispersal from the native range can be facilitated by a shift in transmission from recombined spores to clonal spores, and recombination can be restricted by population bottlenecks caused by the dispersal of a few individuals or of individuals of only one mating type. Dispersal from the native range can be difficult to detect when sampling is incomplete, as is always the case when dispersal is associated with an emerging fungal disease. Hybridization also can lead to restricted recombination when the parents are so diverged that the hybrid progeny lack normal meiotic recombination and rely on parasexuality. Intrinsic restrictions to recombination include asexual reproduction by mitosis; haploid selfing when single individuals possess both mating loci (with or without mating type switching) and therefore have no need for a mating partner; intratetrad mating when siblings from a single meiosis mate; and diploid selfing when siblings from the same parents mate. The recognition that both recombination and clonality can be important to fungal reproduction calls for a measure of the relative importance of each mode; population genomics may have answered the call with LD decay statistics. Curiously, the observation of sexual morphology, including the production of meiotic progeny, is not a guarantee of recombination, because recombination may be more restricted in homothallic fungi than in any other fungi because of haploid selfing. Or, is it possible that these fungi could enjoy expanded opportunities for recombination because they possess both mating type loci? Phylogenomics and population genomics should provide the answer soon.
Acknowledgments
D. Neafsey is thanked for Fig. 2 image. J.W.T. received support from National Science Foundation Grants BIO 1257528 and DEB 1046115.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IX: Clonal Reproduction: Alternatives to Sex,” held January 9–10, 2015, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_IX_Clonal_Reproduction.
This article is a PNAS Direct Submission.
References
- 1.Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11(2):41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- 2.Smith SE, Read DJ. Mycorrnizal Symbiosis. 3rd Ed Elsevier; Amsterdam: 2008. [Google Scholar]
- 3.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289(5486):1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 4.Remy W, Taylor TN, Hass H, Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91(25):11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106(1):2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414(6865):745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- 7.Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427(6976):733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- 8.Tisserant E, et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci USA. 2013;110(50):20117–20122. doi: 10.1073/pnas.1313452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley R, Corradi N. Searching for clues of sexual reproduction in the genomes of arbuscular mycorrhizal fungi. Fungal Ecol. 2013;6(1):44–49. [Google Scholar]
- 10.Lanfranco L, Young JPW. Genetic and genomic glimpses of the elusive arbuscular mycorrhizal fungi. Curr Opin Plant Biol. 2012;15(4):454–461. doi: 10.1016/j.pbi.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Hijri M, Sanders IR. The arbuscular mycorrhizal fungus Glomus intraradices is haploid and has a small genome size in the lower limit of eukaryotes. Fungal Genet Biol. 2004;41(2):253–261. doi: 10.1016/j.fgb.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Ainsworth and Bisby’s Dictionary of the Fungi. 8th Ed CABI; Wallingford, United Kingdom: 1996. [Google Scholar]
- 13.McNeill J, et al., editors. 2012. International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code), Adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011 (Koeltz Scientific Books, Königstein, Germany), pp 125–126.
- 14.Nguyen C, et al. Recent advances in our understanding of the environmental, epidemiological, immunological, and clinical dimensions of coccidioidomycosis. Clin Microbiol Rev. 2013;26(3):505–525. doi: 10.1128/CMR.00005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burt A, Carter DA, Koenig GL, White TJ, Taylor JW. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93(2):770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser JA, et al. Evolution of the mating type locus: Insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot Cell. 2007;6(4):622–629. doi: 10.1128/EC.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandel MA, Barker BM, Kroken S, Rounsley SD, Orbach MJ. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot Cell. 2007;6(7):1189–1199. doi: 10.1128/EC.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koufopanou V, Burt A, Taylor JW. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94(10):5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher MC, Koenig GL, White TJ, Taylor JW. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia. 2002;94(1):73–84. [PubMed] [Google Scholar]
- 20.Neafsey DE, et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010;20(7):938–946. doi: 10.1101/gr.103911.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paoletti M, et al. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 2005;15(13):1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 22.Pringle A, et al. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 2005;59(9):1886–1899. [PubMed] [Google Scholar]
- 23.O’Gorman CM, Fuller H, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457(7228):471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 24.Ropars J, et al. Induction of sexual reproduction and genetic diversity in the cheese fungus Penicillium roqueforti. Evol Appl. 2014;7(4):433–441. doi: 10.1111/eva.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henk DA, et al. Clonality despite sex: The evolution of host-associated sexual neighborhoods in the pathogenic fungus Penicillium marneffei. PLoS Pathog. 2012;8(10):e1002851. doi: 10.1371/journal.ppat.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: Examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci. 2006;361(1475):1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peay KG, Bidartondo MI, Arnold AE. Not every fungus is everywhere: Scaling to the biogeography of fungal-plant interactions across roots, shoots and ecosystems. New Phytol. 2010;185(4):878–882. doi: 10.1111/j.1469-8137.2009.03158.x. [DOI] [PubMed] [Google Scholar]
- 28.Tibayrenc M, Ayala FJ. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci USA. 2012;109(48):E3305–E3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson JB, Kohn LM. Clonality in soilborne, plant-pathogenic fungi. Annu Rev Phytopathol. 1995;33:369–391. doi: 10.1146/annurev.py.33.090195.002101. [DOI] [PubMed] [Google Scholar]
- 30.Dyer PS, O’Gorman CM. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol Rev. 2012;36(1):165–192. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 31.Kohn LM. Mechanisms of fungal speciation. Annu Rev Phytopathol. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. [DOI] [PubMed] [Google Scholar]
- 32.Rosewich UL, Kistler HC. Role of horizontal gene transfer in the evolution of fungi. Annu Rev Phytopathol. 2000;38:325–363. doi: 10.1146/annurev.phyto.38.1.325. [DOI] [PubMed] [Google Scholar]
- 33.Taylor J, Jacobson D, Fisher M. The evolution of asexual fungi: Reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 34.Gladieux P, et al. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol. 2014;23(4):753–773. doi: 10.1111/mec.12631. [DOI] [PubMed] [Google Scholar]
- 35.Avise JC. Gene trees and organismal histories - a phylogenetic approach to population biology. Evolution. 1989;43(6):1192–1208. doi: 10.1111/j.1558-5646.1989.tb02568.x. [DOI] [PubMed] [Google Scholar]
- 36.Taylor JW, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31(1):21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 37.Taylor JW, Fisher MC. Fungal multilocus sequence typing—it’s not just for bacteria. Curr Opin Microbiol. 2003;6(4):351–356. doi: 10.1016/s1369-5274(03)00088-2. [DOI] [PubMed] [Google Scholar]
- 38.Kasuga T, et al. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol. 2003;12(12):3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 39.Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51(Suppl 4):2–15. doi: 10.1111/j.1439-0507.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 40.Cafarchia C, Iatta R, Latrofa MS, Gräser Y, Otranto D. Molecular epidemiology, phylogeny and evolution of dermatophytes. Infect Genet Evol. 2013;20:336–351. doi: 10.1016/j.meegid.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Martinez DA, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio. 2012;3(5):e00259-e12. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clay K, Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat. 2002;160(Suppl 4):S99–S127. doi: 10.1086/342161. [DOI] [PubMed] [Google Scholar]
- 43.Boekhout T, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147(Pt 4):891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Vilgalys R, Mitchell TG. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9(10):1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 45.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 46.Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun. 2001;69(1):115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avise JC. Clonality: The Genetics, Ecology, and Evolution of Sexual Abstinence in Vertebrate Animals. Oxford Univ Press; New York: 2008. [Google Scholar]
- 48.Hörandl E. The complex causality of geographical parthenogenesis. New Phytol. 2006;171(3):525–538. doi: 10.1111/j.1469-8137.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- 49.Ellison CE, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci USA. 2011;108(7):2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, et al. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous ascomycete Neurospora tetrasperma. PLoS Genet. 2012;8(7):e1002820. doi: 10.1371/journal.pgen.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strandberg R, et al. Conflict between reproductive gene trees and species phylogeny among heterothallic and pseudohomothallic members of the filamentous ascomycete genus Neurospora. Fungal Genet Biol. 2010;47(10):869–878. doi: 10.1016/j.fgb.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. Lateral transfer of mating system in Stemphylium. Proc Natl Acad Sci USA. 2005;102(32):11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes KW, et al. Evolutionary consequences of putative intra-and interspecific hybridization in agaric fungi. Mycologia. 2013;105(6):1577–1594. doi: 10.3852/13-041. [DOI] [PubMed] [Google Scholar]
- 54.Phadke SS, Feretzaki M, Clancey SA, Mueller O, Heitman J. Unisexual reproduction of Cryptococcus gattii. PLoS ONE. 2014;9(10):e111089. doi: 10.1371/journal.pone.0111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen SCA, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin Microbiol Rev. 2014;27(4):980–1024. doi: 10.1128/CMR.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: Implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2(5):1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraser JA, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437(7063):1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 58.Hagen F, et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS ONE. 2013;8(8):e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engelthaler DM, et al. Cryptococcus gattii in North American Pacific Northwest: Whole-population genome analysis provides insights into species evolution and dispersal. MBio. 2014;5(4):e01464-e14. doi: 10.1128/mBio.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49(2):171–177. doi: 10.1007/s12275-011-1064-7. [DOI] [PubMed] [Google Scholar]
- 61.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289(5477):310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 62.Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285(5431):1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 63.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289(5477):307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 64.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110(3):293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 65.Lockhart SR, Daniels KJ, Zhao R, Wessels D, Soll DR. Cell biology of mating in Candida albicans. Eukaryot Cell. 2003;2(1):49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Odds FC, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007;6(6):1041–1052. doi: 10.1128/EC.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gräser Y, et al. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93(22):12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22(10):2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosenblum EB, et al. Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci USA. 2013;110(23):9385–9390. doi: 10.1073/pnas.1300130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olson DH, et al. Bd Mapping Group Mapping the global emergence of Batrachochytrium dendrobatidis, the amphibian chytrid fungus. PLoS ONE. 2013;8(2):e56802. doi: 10.1371/journal.pone.0056802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tibayrenc M, Ayala FJ. Cryptosporidium,Giardia, Cryptococcus, Pneumocystis genetic variability: Cryptic biological species or clonal near-clades? PLoS Pathog. 2014;10(4):e1003908. doi: 10.1371/journal.ppat.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105(12):4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis RH. Neurospora: Contributions of a Model Organism. Oxford Univ Press; Oxford, United Kingdom: 2000. [Google Scholar]
- 74.Kuo HC, et al. Secret lifestyles of Neurospora crassa. Sci Rep. 2014;4:5135. doi: 10.1038/srep05135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powell AJ, Jacobson DJ, Salter L, Natvig DO. Variation among natural isolates of Neurospora on small spatial scales. Mycologia. 2003;95(5):809–819. [PubMed] [Google Scholar]
- 76.Palma-Guerrero J, et al. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 2013;9(8):e1003669. doi: 10.1371/journal.pgen.1003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raju NB, Perkins DD. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora. Dev Genet. 1994;15(1):104–118. doi: 10.1002/dvg.1020150111. [DOI] [PubMed] [Google Scholar]
- 78.Menkis A, Bastiaans E, Jacobson DJ, Johannesson H. Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J Evol Biol. 2009;22(9):1923–1936. doi: 10.1111/j.1420-9101.2009.01801.x. [DOI] [PubMed] [Google Scholar]
- 79.Beatty NP, Smith ML, Glass NL. Molecular characterization of mating-type loci in selected homothallic species of Neurospora, Gelasinospora and Anixiella. Mycol Res. 1994;98(11):1309–1316. [Google Scholar]
- 80.Nygren K, et al. A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Mol Phylogenet Evol. 2011;59(3):649–663. doi: 10.1016/j.ympev.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 81.Glass NL, Metzenberg RL, Raju NB. Homothallic Sordariaceae from nature - the absence of strains containing only the a mating type sequence. Exp Mycol. 1990;14(3):274–289. [Google Scholar]
- 82.Ellison CE, et al. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics. 2011;189(1):55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobson DJ. Blocked recombination along the mating-type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics. 2005;171(2):839–843. doi: 10.1534/genetics.105.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lengeler KB, et al. Mating-type locus of Cryptococcus neoformans: A step in the evolution of sex chromosomes. Eukaryot Cell. 2002;1(5):704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hood ME, Petit E, Giraud T. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics. 2013;193(1):309–315. doi: 10.1534/genetics.112.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gioti A, Stajich JE, Johannesson H. Neurospora and the dead-end hypothesis: Genomic consequences of selfing in the model genus. Evolution. 2013;67(12):3600–3616. doi: 10.1111/evo.12206. [DOI] [PubMed] [Google Scholar]
- 87.Billiard S, López-Villavicencio M, Hood ME, Giraud T. Sex, outcrossing and mating types: Unsolved questions in fungi and beyond. J Evol Biol. 2012;25(6):1020–1038. doi: 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]