Abstract
Cancer is a general name for more than 100 malignant diseases. It is postulated that all cancers start from a single abnormal cell that grows out of control. Untreated cancers can cause serious consequences and deaths. Great progress has been made in cancer research that has significantly improved our knowledge and understanding of the nature and mechanisms of the disease, but the origins of cancer are far from being well understood due to the limitations of suitable model systems and to the complexities of the disease. In view of the fact that cancers are found in various species of vertebrates and other metazoa, here, we suggest that cancer also occurs in parasitic protozoans such as Trypanosoma brucei, a blood parasite, and Toxoplasma gondii, an obligate intracellular pathogen. Without treatment, these protozoan cancers may cause severe disease and death in mammals, including humans. The simpler genomes of these single-cell organisms, in combination with their complex life cycles and fascinating life cycle differentiation processes, may help us to better understand the origins of cancers and, in particular, leukemias.
Keywords: malignancy, single-cell organisms, mammals, evolution, transmissible
Cancer is a collection of important human and animal diseases where cells, the building blocks of our bodies, fail to correctly work genetic instructions that are present in every cell of the human body and control the behavior and destiny of these cells. These instructions control when and how cells grow, reproduce, and die. If the instructions become confused, a single cell might change its behavior and reproduce in an uncontrolled way. This situation is what we call cancer, and each type of cancer may be different depending on which type of cell becomes faulty. All cancers are clonal proliferations that arise owing to mutations that confer selective growth advantage on dividing cells (1). A large body of work has been carried out on cancers, leading to extraordinary advances in our knowledge about the nature and mechanisms of these diseases (2–4). However, the origin of cancers is, with some exceptions, far from being well understood, due to the limited number of suitable models (5, 6). Gaining an understanding of the occurrence and establishment of cancer in humans or other metazoa is not straightforward (7). The cancer processes may take a long time to develop, usually over many years or decades. Our understanding might be greatly accelerated if such processes could be investigated in protozoa (single-cell organisms) as a model system. A key premise for this investigation is to determine whether cancer exists in protozoan species.
General Characteristics of Various Kinds of Cancers Are Uncontrolled Proliferation and Accumulation of Genetic Mutations
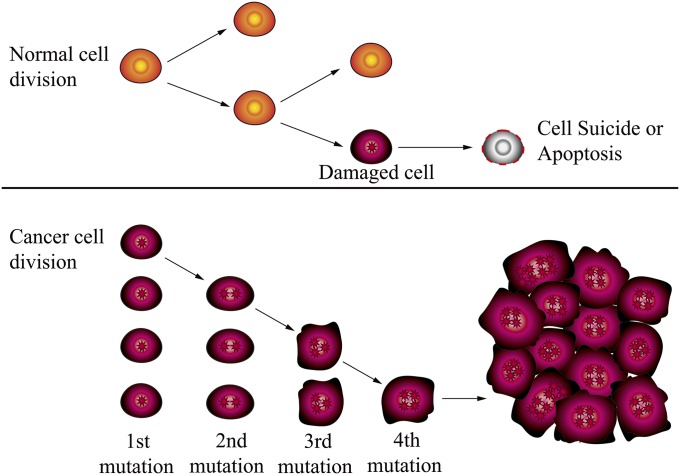
The multiplication of cells is precisely regulated and responsive to specific needs of the human (or animal) body. The number of divisions of all cell types in the human body is genetically controlled and limited (Fig. 1). Different cell types, such as epithelial cells, skeletal muscle cells, macrophages, neurons, granulocytes, etc. carry out specific functions in the body, but all are ultimately differentiated from pluripotent stem cells. These stem cells are embryonic stem cells ultimately derived from the single fertilized zygote. The timing and outcome of each cell multiplication and differentiation event are carefully controlled by complex signaling pathway networks (1, 4). Without precise control of these differentiation processes, cell types will change behavior and may proliferate in disorder. Disruption of these control mechanisms can be caused by mutations occurring within the mitochondrial (mtDNA) or nuclear (nDNA) genomes or by epigenetic changes that modify the expression of control genes (7, 8). The lack of differentiation from stem cells to somatic cells or the lack of the normal limited division of somatic cells in the body will result in the loss of the physiological function of the cells and the destruction of the tissues or organs surrounding any errant cell. The generation of malignant cancers is a complex process involving internal cellular processes, such as the division rate of mutated or cancerous cells, and modulation by other external signals such as cell–cell contact (communication) and circulating signals. Solid tissue-based cancers may differ in behavior from “free-living” blood cells, which can result in leukemias, with a faster division rate. The more rapidly the cancer (malignant) cells multiply and the more rapid is the destruction of organs and tissues, the faster will be the death of the host. Recent studies in leukemia demonstrate the importance of the genetic control of this process (9).
Fig. 1.
Differences between normal and cancer cells. Normal cells arise from stem cells that differentiate to specific cell types that carry out specific functions. Cancer cells lose the division limitation owing to various reasons (e.g., nDNA or mtDNA mutations or damage) and lose the ability to differentiate into specific cell types, as well as being unable to carry out specific physiological functions. During the evolution of a cancer, several mutations in genomes can occur and accumulate. Each factor increases the possibility of occurrence of mutations, insertions, deletions, or epigenetic effects that are linked to a cancer. Adapted from the National Cancer Institute (www.cancer.gov).
A large number of mutations in genes (for example, DNMT3A, ASXL1, TET2, and PPDM1D in blood cancer; ERG gene fusion, NKX3.1 on 8p21, and PTEN on 10q23, SPOP, FOXA1 in prostate cancer; and P53, PIK3CA, and GATA3 in breast tumors) are considered to be related to the occurrence of cancers (9–12). In fact, it is well known that a naturally occurring cancer is most often a consequence of multiple factors that interact over a long period (13). Each factor increases the possibility of occurrence of mutations, insertions, deletions, or epigenetic effects that are linked to a cancer (Fig. 1).
The origin or occurrence of acute promyelocytic leukemia (APL) is well investigated, but acute myeloid leukemia (AML) is a more complex process and has been suggested to be mainly associated with the accumulation of mutations within mtDNA or nDNA that have occurred before the initiation of cancer cell proliferation (14) (Table S1). In other words, many preexisting predisposing mutations may have allowed the initiating event to cause a cell to differentiate into a cancer cell. The loss of differentiation is considered one of the most important events in the pathogenesis of many types of cancers, including leukemia (15). Interestingly, it is becoming increasingly apparent that mutations of mtDNA are strongly associated with human cancers (7, 14, 16). A wide range of mechanisms required for precise communication between the mitochondria and the nucleus are key points required for normal cells to perform their specific functions within the body. Although functioning mitochondrial genes are crucial in cancer cells, some mutations in mitochondrial genes may not inactivate energy metabolism but instead change the mitochondrial bioenergetic and biosynthetic state (7). Accumulating evidence indicates that most mitochondrial mutations are likely to be “passengers” that do not contribute to oncogenesis (1, 7). By analyzing mutations in 24 AML whole-genome sequences, Welch et al. concluded that only a tiny fraction of all of the mutations in each AML genome are likely to be relevant to pathogenesis, disease classification, and targeted therapy (14). In the future, presumably as more clinical samples are analyzed by whole-genome sequences, more evidence will be obtained for refining the specific origin of cancers and hopefully will lead to more effective treatments.
Table S1.
Gene mutations found in some human cancers
| Mutated genes | Function | Refs. |
| Acute myeloid leukemia | ||
| SMC3 | Structural maintenance of chromosomes 3 | (14) |
| SMC1A | Structural maintenance of chromosomes 1A | (14) |
| STAG2 | Stromal antigen 2 | (14) |
| RAD21 | RAD21 homolog | (14) |
| FLT3 | Fms-related tyrosine kinase 3 | (68) |
| NPM1 | Nucleophosmin (nucleolar phosphoprotein B23 | (68) |
| DNMT3A | DNA (cytosine-5-)-methyltransferase 3 alpha | (68) |
| IDH1 | Isocitrate dehydrogenase 1 | (68) |
| IDH2 | Isocitrate dehydrogenase 2 | (68) |
| TET2 | Tet methylcytosine dioxygenase 2 | (68) |
| RUNX1 | Runt-related transcription factor 1 | (68) |
| TP53 | Tumor protein p53 | (68) |
| NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | (68) |
| CEBPA | CCAAT/enhancer binding protein | (68) |
| WT1 | Wilms tumor 1 | (68) |
| PTPN11 | Protein tyrosine phosphatase | (68) |
| KIT | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | (68) |
| U2AF1 | U2 small nuclear RNA auxiliary factor 1 | (68) |
| KRAS | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | (68) |
| SMC1A | Structural maintenance of chromosomes 1A | (68) |
| PHF6 | PHD finger protein 6 | (68) |
| STAG2 | Stromal antigen 2 | (68) |
| FAM5C | Family with sequence similarity 5 | (68) |
| EZH2 | Enhancer of zeste homolog 2 | (68) |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K | (68) |
| Breast cancer | ||
| TP53 | Tumor protein p53 | (69) |
| PIK3CA | Phosphatidylinositol-4 | (69) |
| RB1 | Retinoblastoma 1 | (69) |
| PTEN | Phosphatase and tensin homolog | (69) |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | (70) |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | (70) |
| TBX3 | T-box 3 | (70) |
| RUNX1 | Runt-related transcription factor 1 | (70) |
| LDLRAP1 | Low density lipoprotein receptor adaptor protein 1 | (70) |
| STMN2 | Stathmin-like 2 | (70) |
| MYH9 | Myosin | (70) |
| AGTR2 | Angiotensin II receptor | (70) |
| STMN2 | Stathmin-like 2 | (70) |
| SF3B1 | Splicing factor 3b | (70) |
| CBFB | Core-binding factor | (70) |
| AKT2 | V-akt murine thymoma viral oncogene homolog 2 | (71) |
| ARID1B | AT rich interactive domain 1B (SWI1-like) | (71) |
| CASP8 | Caspase 8 | (71) |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | (71) |
| MAP3K13 | Mitogen-activated protein kinase kinase kinase 13 | (71) |
| NCOR1 | Nuclear receptor corepressor 1 | (71) |
| SMARCD1 | SWI/SNF related | (71) |
| GATA3 | GATA binding protein 3 | (12) |
| MAP2K4 | Mitogen-activated protein kinase kinase 4 | (12) |
| Prostate cancer | ||
| SPOP | Speckle-type POZ protein | (12) |
| FOXA1 | Forkhead box A1 | (12) |
| MED12 | Mediator complex subunit 12 | (12) |
| CHD1 | Chromodomain helicase DNA binding protein 1 | (73) |
Some Cancers Can Naturally Be Infectious
Interestingly, there are two known examples of (nonvirally causative) mammalian cancers that have the innate ability to be communicable and pass horizontally from host to host. The endangered Tasmanian devil can acquire an infectious cancer, devil facial tumor disease (DFTD), from individuals biting one another (17). This disease was first reported in 1996 and is 100% fatal to the devils. Genome sequencing of clones of the infectious cancer shows that it probably arose from a single female Tasmanian devil but subsequently evolved to form two dominant subclones that have swept through the devil populations (18). Genetic data are consistent with observations of the disease appearance in 1996, suggesting that this infectious cancer might have arisen recently (since the 1990s). Canine transmissible venereal tumor (CTVT) is another known example. This disease has been ascertained to have originated as a single cancer cell clone from a single animal ∼11,000 y ago (19, 20) and seems to be relatively benign, in that it does not cause death of the dog. Both of the diseases DFTD and CTVT, although different in their respective pathogenicity, are caused by cancer cells that behave as allografts (21), but have developed the ability to escape from surveillance and destruction by the host immunity (22). This avoidance may occur by epigenetic down-regulation of cell surface MHC receptors (23). The difference in pathogenicity in the two diseases may reflect the differences in evolutionary time during which the hosts may have developed countermeasures to mediate the effects. In essence, both of these cancers are actually behaving as autonomous organisms; their relationships with their hosts may shed light on the evolution of host–pathogen interactions (24), and they may even be taken as asexually duplicated unicellular pathogens (25). By analysis of experimental evolution, this hypothesis is supported by current results from Chen et al. (26).
In addition to these two cases of mammalian horizontally transmitted infectious cancers, there have been rare cases of vertical transmission (mother to offspring) of cancers reported over the past 100 y. Until recently, it was unclear whether these cancers were really infectious from the mother. A recent genetic study has demonstrated the ability of a leukemic cancer cell clone to pass from mother to fetus where it was able to spread and form tumors (27). This puzzling phenomenon indicates that the cancer clone was not detected and destroyed by the fetal immune system. A deletion of HLA alleles in the clone, which was not present in the fetal cells, may have enabled the clone not to be recognized as foreign matter, suggesting a possible mechanism of immune evasion by these cells (27). Very recently, another infectious cancer has been reported in invertebrates. Metzger et al. used forensic DNA markers to demonstrate that leukemia cells have a clonal origin and seem to be transmitted among the soft-shell clam (Mya arenaria) through sea water along the Atlantic coast of North America (28). When we carefully think about the evolution (or occurrence) of transmissible cancer in humans, then we understand that transmission can certainly happen, but such transmission from one person to another person is rare before the host dies. We suggest that most cancers can be transmitted mechanically if there are enough opportunities to be passed on to a suitable host. Strong evidence from recent studies on the reverse evolution from multicellularity to unicellularity supports this hypothesis (26). The parallels between these infectious cancers (transmission via immune evasion) will be discussed below in the light of parasitic infections.
Possibilities for Using Single-Cell Protozoans as Model Systems for Investigating the Origins of Cancers
Although it might not seem immediately evident that single-cell organisms could play a role in unveiling the origins of human (or metazoan) cancers, a number of common aspects suggest that this try may not be a wild idea. In metazoan cells, there is significant intercellular communication that directs the differentiation status of a cell, and, of course, such interactive control would not be expected to occur between independent organisms. However, as will be seen later, “quorum sensing”—the communication between single-cell organisms—has recently been described in detail in the single-cell blood parasite Trypanosoma brucei (29). In any case, protozoan parasites resemble normal functioning cells in terms of cell multiplication and differentiation from one life cycle stage to another, which are necessitated for successful growth and survival in the metazoan hosts and vectors (30). For example, the protozoan parasite must have genes that function to control the cell cycle to ensure correct coordination of multiplication. In the case of T. brucei, the molecular components of cell cycle control are well characterized (31). At least 45 genes, mostly encoding protein kinases, have been identified that are essential for parasite growth and replication. These genes include a family of cdc2-related protein kinase genes recognizable as homologs of the cdc2-related genes of higher eukaryotic cells. Furthermore, cellular differentiation in this parasite also occurs to prepare itself for survival on transition from the mammalian host to the insect vector, and the control of this process is governed by quorum sensing within the population of parasite cells (29). Both processes of multiplication and differentiation require tight control for accurate development, and both are, as with cancer cells, under the close scrutiny of the immune system. Any failure to complete stage transitions in the correct manner will lead to elimination of the protozoan by the host. However, it must be recognized that a coevolutionary journey between host and parasite should ensure no death of the host, upon which it depends, before finding their way to a new host.
Despite the dependence of a protozoan parasite on the host, the innate cellular mechanisms, such as control of the cell cycle and differentiation, are analogous in protozoan, metazoan, or human cells (31). Failure of these processes (uncontrolled parasite growth and incorrect differentiation) could lead to further adverse pathogenesis to the host caused by an increased parasite load, causing cell and tissue destruction—a similar outcome to that generated by invasive cancer cells. Incorrect differentiation could also result in a lower success rate for the parasite if, for example, transmission or parasite evasion from the host immunity was impaired.
In the case of human or other host cells, control of cellular processes like replication and differentiation is complex. Because they are dependent on complex signals such as hormones, cellular signals, and cell–cell contact, there are many different interactions that need to be considered when tackling the mechanisms of cancer generation. In protozoan parasites, these interactions are much simpler. Furthermore, a comprehensive range of tools are available. These include the complete genome sequences for many protozoans and gene analysis tools such as transfection with foreign genes, gene knockdown using RNAi, RNASeq, microarray analysis, and techniques for epigenetic analysis (32). This reduced complexity and availability of tools make protozoan parasites a good model system for investigating at least some aspects of cancer.
Although the genomes of many protozoans are now available, sequence analysis alone is not sufficient to enable meaningful functional comparisons to be made between human and parasite genomes. The T. brucei genome is well-characterized, and ∼10,000 genes are predicted, with 1,700 of those unique to trypanosomes (33). There are many similar families of genes compared with humans: for example, metabolic enzymes, protein kinase genes (albeit a reduced 30% of the protein kinase genes found in humans), cell division cycle genes, DNA repair and synthesis genes, and many others. However, there are also notable differences, such as the absence of tyrosine protein kinases. Functional analysis is still ongoing, and the overall functional similarities and differences remain to be determined.
Examples of Parallels Between Cancer Cells and Protozoan Parasites
T. brucei.
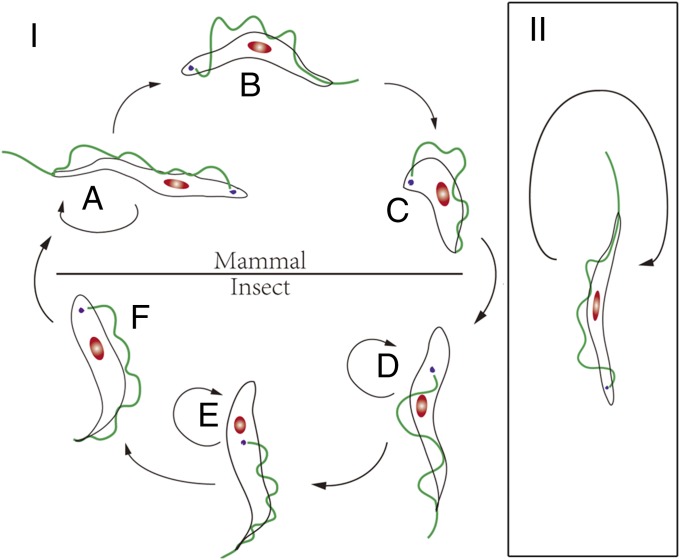
Taking the two typical characteristics of cancer (that is, uncontrolled growth and dedifferentiation) will enable us to define cancer in reference to protozoans. Many protists have only one cell type without regulation of cell growth. Therefore, we will focus on the protozoans that have various cell types requiring regulation: e.g., differentiation. A relevant feature of many important parasitic protozoans is the requirement at least of one or more hosts to complete their life cycles by transmission from one host to another (e.g., insect to human). During the life cycle, the morphological and physiological status of these parasites changes drastically so that they can carry out specific functions required to adapt to the new environment in the new host. This differentiation process is similar to the differentiation of one cell type into another in the human body—for example, stem cells differentiating into specialized cells. T. brucei, a hemoflagellate (a protozoan blood parasite), is the pathogen of human sleeping sickness and animal Nagana disease in Africa. It is an extracellular parasite that circulates within the blood in the same manner as resident white blood cells. The life cycle of this parasite (Fig. 2I) requires a mammalian host (such as humans) and a blood-sucking insect (tsetse fly) for completion (34, 35). In these distinct hosts, massive alterations in morphological, physiological, and molecular features are observed (34, 36–38) (Table 1). In the mammalian host, T. brucei shows three distinct cell types: the slender, intermediate, and stumpy forms (39) (Fig. 2I). The slender form is the only cell type capable of division in the blood circulation of the mammalian host. However, it can be differentiated into the stumpy form via the intermediate form, using a number of mechanisms that are driven by both the host and the parasite itself. Among them, the density of the parasite population seems to be detected (“quorum sensing”) by the parasite itself (29). Other factors include the pH of the blood, immune responses including hydrolysis products of cAMP analogs (40), a specific induction signal [stumpy induction factor (SIF)] (41), and changes in the cytokine(s) and chemokine(s) activity of the host (42). Earlier research also shows that mammalian growth factors like EGF and other factors may influence trypanosome growth and differentiation in the host (43–47) although the lack of an identifiable EGF receptor gene sequence in the T. brucei genome suggests a form of molecular mimicry (48). Accumulative studies indicate, although it has not been fully established yet, that the growth and differentiation of the single-cell T. brucei is, at least partly, under external control and that communication between parasite–parasite and parasite–host may be occurring. Further work is required to establish the complete process of differentiation from the slender to the stumpy form.
Fig. 2.
Life cycles of T. brucei and T. evansi. (I) Life cycle of T. brucei (with fully functioning kDNA). Blood slender (normal) forms (stage A) differentiate, via an intermediate stage (stage B), into the stumpy form (stage C), which is the only stage with the necessary specific functions for infecting the insect vector. Once in the insect host (stages D–F), only the metacyclic form (stage F) in the vector can infect the mammalian host and differentiate into the slender form. The infected host can survive longer due to the regulated parasite growth and differentiation. (II) Life cycle of T. evansi (considered a mutant strain of T. brucei). Only the blood slender form (stage A) is found. It replicates directly (stages A and B), but it cannot differentiate into the stumpy form (similar to the dedifferentiation in a cancer cell). It loses the ability to infect the insect vector (cannot survive and differentiate within the insect vector). Some natural hosts can be killed by the infection of this parasite in a short time, depending on the mammalian species.
Table 1.
Mutations that may contribute to the loss of differentiation into stumpy and insect forms in T. evansi and T. equiperdum by comparison with T. brucei
| Gene groups (T. brucei) | Difference (T. evansi/T. equiperdum) | Refs. |
| Mitochondrion | ||
| Respiratory chain complexes (15 genes) | Absence of all genes/absence of 11 genes and one truncated in some isolates | (36) |
| Mitochondrial ribosomal genes (3 genes) | Absence of all genes/one absent and one truncated in some isolates | (36) |
| RNA editing guide RNA genes (>100 genes) | Absence of most genes/absence of most genes | (36) |
| Nucleus | ||
| Respiratory chain complexes (>50 genes) | Mutations found in at least 3 genes/mutations found in at least 1 gene | (36, 37) |
| Procyclin-associated genes (7 genes) | Absence of PAG3 and mutations in others/absence of PAG3 and mutations in other genes in some isolates | (38) |
| Proteins associated with differentiation (PADs, 8 genes) | Mutations and frame shifts in 7 genes/mutations and frame shifts in 7 genes in some isolates | (38) |
The stumpy form is the terminal stage of this parasite in the mammalian host and the only stage that can infect the insect vector—thus, it is analogous to the terminally differentiated mammalian cell. Before infecting the insect host, the stumpy parasites arrest cell proliferation but will die if they are trapped in the mammalian host for a significant period (34). The slender and stumpy forms of T. brucei have been well studied for their distinct mitochondrial functions. In nutrition-rich mammalian blood, some of the tricarboxylic acid (TCA) cycle genes in the slender form are silenced. These metabolic functions need to be reactivated when differentiation from the slender to stumpy form occurs. This reactivation prepares the trypanosome for further development in the insect gut, where limited nutrition is provided (34). Therefore, the trypanosome cannot complete the entire life cycle without a fully functional mitochondrion (11, 49–52). An interesting parallel can also be drawn between this differentiation process and cancer. It has long been known that prolonged passage of the slender form in experimental animals has generated a monomorphic strain of T. brucei that is unable to differentiate into the stumpy form (53, 54). These cells have uncontrolled growth in the host, in the same manner as cancer cells, and they lack control at the G0 restriction point in the cell cycle (53). Recent studies, using transgenic trypanosomes, are beginning to identify some of the genetic components that can induce these monomorphic trypanosomes to reactivate a degree of pleomorphism (ability to transform into the stumpy form) (54).
The origin of these monomorphic forms is currently unclear but is presumably caused by mutations of genes that interfere with the differentiation process, perhaps in a way analogous to the generation of cancerous cells. Recent research has shown that T. brucei has mechanisms of DNA proofreading and correction analogous to those in human or metazoan cells. A striking example is the presence of a BRCA2 gene ortholog in T. brucei (55). In humans, mutations in the BRCA2 gene have been shown to be associated with an increased risk of breast cancer, which is thought to be due to impaired DNA repair (56). In T. brucei, the ortholog is also involved in DNA repair (55), and studies have shown that it interacts with the DNA replication protein CDC45 (57). Furthermore, silencing or mutation studies have revealed the inhibition of DNA repair (58) by the trypanosome BRCA2 gene, leading to aberrant cell division (59). Although no link has yet been established between the role of this gene and the establishment of these monomorphic T. brucei mutants, there could be a striking parallel with the perceived role of the BRCA2 gene in increasing the risk of establishment of breast cancer cells in humans. These developmental mutants of T. brucei offer an interesting prospect for investigating cancer-like behavior in trypanosomes and alternative approaches to cancer studies.
Trypanosoma evansi (Fig. 2II) is considered a subspecies of T. brucei (38, 39), or even a strain of T. brucei, because they are indistinguishable on the basis of morphological and molecular evidence (36). It is generally accepted that T. evansi relatively recently evolved from T. brucei (36). However, T. evansi has completely lost the ability to differentiate from slender to stumpy form, and it cannot be further differentiated or developed into the stages in the insect vector. Thus, T. evansi fails to infect the blood-sucking insect and can be mechanically transmitted only from host to host between mammals (51, 52). Thus, due to lack of differentiation, T. evansi has only the uncontrolled proliferating stage in the mammalian hosts, suggesting a parallel with the status of AML in humans. This lack of differentiation and unchecked growth also invokes parallels with DFTD cancer cells. Therefore, we could consider T. evansi as a typical cancer of T. brucei. In fact, mice are killed in a few days by T. evansi whereas they can survive much longer when infected with WT T. brucei. The recent evolutionary origin of T. evansi and its relatively narrow host range (for example, lack of infection in humans) suggest that evolution has enabled the parasite to colonize only suitably susceptible hosts. The lack of coevolutionary history between hosts and parasite may explain the higher degrees of pathogenicity observed in T. evansi. Furthermore, the lack of constraint in having to differentiate to facilitate transmission (as seen in T. brucei) may have taken the constraints off proliferation. Nothing is known about quorum sensing in T. evansi; perhaps this growth regulatory mechanism is also absent. The recent evolution of the DFTD cancer cells, which are highly pathogenic to devils, may also explain this parallel.
Mutations in T. evansi, similar to those in human cancers, have also been identified. For example, mutations of kinetoplast DNA (kDNA) in the mitochondrion (52, 60) and in the genome (nDNA) have long been observed (36, 38). The kDNA in the Kinetoplastidae species is a type of mitochondrial DNA (mtDNA), with functions similar to those found in metazoans. It consists of two kinds of circular DNAs: minicircle and maxicircle molecules. Hundreds of types of the heterogeneous minicircles are used as guide RNAs (gRNAs) for the transcription of RNA whereas the maxicircles (20–50 copies in a single kinetoplast) carry out functions similar to those found in the mitochondria of metazoan cells (49, 61). The majority of kDNA maxicircle mRNAs undergo RNA editing to insert or delete uridine residues, as specified by template gRNAs, in a process catalyzed by multiprotein complexes called editosomes (62–64). Interestingly, T. evansi, unlike its ancestor T. brucei, completely lacks maxicircle DNA molecules and has only homologs of minicircles in the kDNA. Loss of minicircle complexity in T. evansi has been considered the key reason for its inability to develop through the insect developmental stages, which requires the activation of TCA enzymes. Evidence indicates that one kDNA-encoded transcript that requires editing is the F1FO-ATPase subunit 6 (A6), which is essential in the slender form of T. brucei (60, 65–67). However, recently it has been shown that mutations found in the nuclear-encoded ATPase subunit γ of the T. evansi strain can compensate for the loss of kDNA, accounting for their viability without A6 (37). Nevertheless, the key question as to why there is no differentiation into the stumpy form in T. evansi is still an enigma. Recently, genome sequence data have identified the mutations and genes lost in T. evansi STIB 805, indicating a complicated mechanism behind the lack of differentiation from T. brucei to T. evansi (38). Similar situations have also been found in human leukemias and other cancers (Table S1) (12, 14, 68–73). Interestingly, infectious cancers are characterized by the loss of significant portions of the genome with significant aneuploidy or rearrangements, such as the loss of 646 genes in CTVT (20), and diverse chromosome rearrangements, indels, and gene loss in DFTD (18). Additionally, in the case of the maternal–fetal cancer, significant deletion of HLA alleles has been observed (27). This gene loss and subsequent uncontrolled growth, seen in T. evansi, are not the only examples of a cancerous form of T. brucei.
The parasite Trypanosoma equiperdum is a pathogen that causes the widely distributed disease “Dourine” in horses and other members of the Equidae group (39). Like T. evansi, T. equiperdum is deemed to be genetically similar to T. brucei and considered as a subspecies or strain (51, 52). Analogously, transmission bypasses the developmental cycles in the tsetse fly (39), and it has similar gene deletions in mitochondrial DNA, which seem to underlie this process (36). T. equiperdum has adapted to a different mode of transmission—directly from horse to horse by venereal transfer during coitus—which has enabled it to escape the geographical constraints encountered by T. brucei (51). Without the constraints of cellular differentiation through the tsetse fly, this parasite behaves as a further example of a cancer of T. brucei. Interestingly, its transmission route mirrors that of CTVT in dogs, raising the interesting concept of evolutionary convergence of cancer cells derived from both mammalian and protozoan ancestors.
Toxoplasma gondii.
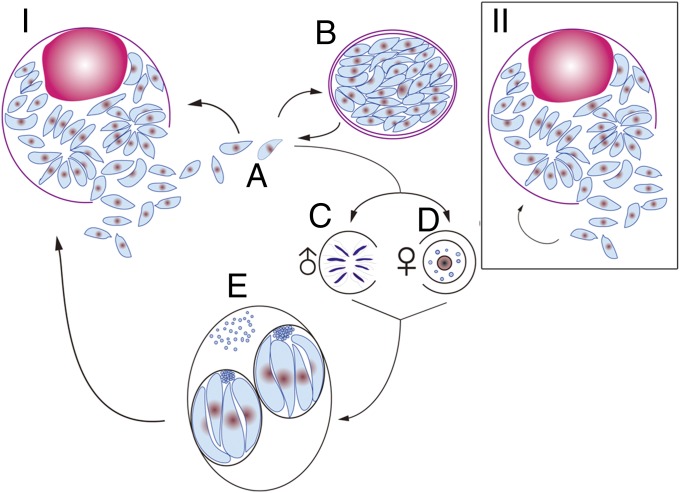
In addition to T. brucei, Toxoplasma gondii is also a potential cell model for studying the origin/occurrence of cancer. In contrast to T. brucei, T. gondii is an obligate intracellular parasitic protozoan. Like T. brucei, T. gondii has more than one host to complete its life cycle. Feline species (cats) are the definitive host where fertilization occurs, and large numbers of oocysts are generated and released to the environment with the feces (Fig. 3I). Mammals (including humans) and birds are the intermediate hosts of T. gondii and are infected by eating undercooked meat containing the cysts of the parasite, or through ingesting water or food contaminated with the oocysts shed by the felid (74). The complete life cycle of the parasite in the host requires normal differentiation from stage to stage, with specific functions being switched on and off appropriately. In nature, the parasite first multiplies as the tachyzoite (rapidly dividing stage) before the immune response occurs. Later, the cells differentiate into the bradyzoite (slowly dividing stage), forming a cyst with the cyst wall being generated by the immune response of the host (including humans). These two distinct stages are very important factors enabling the parasite to complete its life cycle and cell multiplication, and to retain a permanent presence of the parasite in the intermediate host. However, in some laboratory strains, for example the RH strain of T. gondii, mutations have occurred that prevent this strain of the parasite from differentiation of the tachyzoite into the bradyzoite stage (Fig. 3II). The lack of differentiation results in the parasite’s losing the ability to form cysts in the intermediate hosts or to generate oocysts in cats (75, 76). In some cases, both the host and the parasite die when an animal host is infected with the RH strain. For example, mice will die very quickly when they are infected with the RH strain without any treatment whereas rats can survive without any observed clinical signs because the RH strain is unable to grow in these animals due to innate immunity (77–79).
Fig. 3.
(I) Life cycle of WT T. gondii. Tachyzoites (stage A) can differentiate into bradyzoites (stage B) in definitive or intermediate hosts and can differentiate into oocysts (stage E) in the definitive host (feline animals, cats) after sexual reproduction (stages C and D). Both parasite and host can survive in most cases depending on the life span of the host. (II) RH strain, a mutant strain of WT T. gondii. The tachyzoite (stage A) cannot differentiate into the bradyzoite in either the intermediate or the definitive host and cannot differentiate into oocysts in the definitive host. It reproduces by reinfection of host cells (stage B). The hosts will normally be killed by the parasites or vice versa.
Although little has been done on the mutations of mtDNA, mutations in the RH strain of T. gondii have been detected in the genome by transcriptome analysis. Radke et al. have reported that they found no difference in the specificity and expression levels of mRNAs of RH strain populations collected from different phases of cultivation, suggesting that the development from tachyzoite to the bradyzoite stage of RH may be arrested (80). In contrast, strains of the type II-Me49B7 and type III-VEGmsj (both of which are capable of differentiating into bradyzoite stages) contain SAGE tags corresponding to bradyzoite genes. This suggests that the priming of developmental expression is likely to play a greater role in the capacity of these strains to complete bradyzoite development (80). Through comparative genome and gene expression analysis, Yang et al. (81) have identified a list of candidate genes that could be responsible for the phenotypic differences between different type I strains of T. gondii. They have shown that polymorphisms in GRA2 and GRA15 determined type I strain differences in the survival in IFN-γ–stimulated cells and in the activation of NF-κB, respectively. Thus, despite there being a reported 1,394 SNPs or indels that differentiate the type I strain from others, only a small number of genes were directly involved in phenotypic differences (Table 2) (81). These situations are similar to those found in cancers where cell transformation can usually be tracked down to a small number of genes. Again, as with the case in many cancers, although a great deal of work has been done on identifying the gene mutations linked to the lack of differentiation of the RH strain from the tachyzoite to bradyzoite stages, the process is still far from completely understood. Because this conversion is a key stage from acute (tachyzoite) to chronic (bradyzoite) infection in the host—a process that normally limits infection in the host—to all intents and purposes, the RH strain of T. gondii behaves as the equivalent of a T. gondii cancer.
Table 2.
Mutations that may contribute to loss of the ability to form cysts in the T. gondii RH strain, in comparison with another type I strain, GT1
| Gene groups | Difference (RH strain) | Refs. |
| Nucleus | ||
| 3′−5′ cyclic nucleotide phosphodiesterases | Mutations and/or indels in 3 genes | (81) |
| Protein kinases | Mutations and/or indels in 6 genes | (81) |
| Proteins with ATP binding and metal ion binding activities | Mutations and/or indels in 3 genes | (81) |
| Mitochondrial | ||
| No related data |
Indels, inserts or deletions.
Metastasis in Cancer and Spread of T. gondii and T. evansi
Metastasis of cancerous cells can be compared with the transmission of pathogenic microorganisms from an original site to one or more sites elsewhere in the body, usually by way of the blood vessels or lymphatics. In the case of the infectious cancers CTVT and DFDT, this metastasis is transmitted horizontally to different individual animals before spreading throughout the body. Although many factors can affect metastasis in cancer or the spread/distribution of T. evansi and T. gondii in the body, the natural immunity of the host may be one of the key factors. Tumors arise with high frequency, especially in older animals and humans, but most pose little risk to their host because they are localized. We call such tumors benign: e.g., warts. Tumors become life-threatening if they spread throughout the body. Such tumors are called malignant and are the cause of cancer (13). The process of metastasis in cancer is a complex one (4). In free ranging cells (e.g., blood cells), this process may be largely dependent on the growth rates of the errant cells. However, in solid tissue based cancers, hormonal, cellular, and contact-based signals may influence the degree and effects of metastasis on local tissue damage (4). T. evansi and the T. gondii RH strain will be killed by macrophages and other immune cells if they are restricted to the localized site where they were inoculated. Thus, spreading to other sites of the body can protect the parasite but may cause pathological damage. T. evansi, being an extracellular pathogen, freely ranges through the host and eventually resides extravascularly—usually in neurological tissue where it causes damage (82). T. evansi might be considered analogous to a free-living cancer such as leukemia. T. gondii is an intracellular pathogen and evades the host immune system by cellular invasion. In this way, the parasite can induce tissue damage depending on the location or cell type that has been invaded. For example, recent studies have demonstrated impaired anion secretion as a possible mechanism for pathology in infected airway epithelia leading to possible airway blockage and pneumonia (83). In this respect, T. gondii might be analogous to a solid tissue cancer where the outcome is influenced by the parasite’s final destination(s). In fact, the spreading and invasion of tachyzoites from pseudocyst or cyst to other places mirror the metastasis of a solid malignant tumor.
Transmission
Transmission is normally associated with parasites but not cancer cells (with the exception of the infectious cancers DFDT and CTVT). However, even noninfectious cancers can be transmitted in the laboratory by mechanical transmission using syringes. As mentioned above, T. brucei is transmitted by a blood-sucking insect vector, the tsetse fly, when T. brucei undergoes its life cycle (Fig. 2I). However, T. evansi, called a cancer of T. brucei, loses this complex life cycle and is only mechanically transmitted from individual to individual by a blood-sucking insect or vampire bat in the field or by a syringe in the laboratory (34, 38) (Fig. 2II). Interestingly, the loss of the maxicircle kDNA by T. evansi has two evolutionary consequences on transmission. Firstly, this cancerous form of T. brucei has a restricted host range (primarily in camels, horses, cattle, and, very rarely, humans) in nature due to its inability to complete the life cycle through the tsetse fly. However, mechanical transmission, bypassing the tsetse, has enabled it to increase its geographical range. Now, it is found in Asia and South America, continents outside the range of tsetse flies. Thus, T. evansi’s geographical range has far transcended that of its ancestor, T. brucei, which remains confined to Africa. A similar situation has evolved around T. equiperdum. Rather like the relatively benign nature of the infectious cancer CTVT, with its host the dog, both T. evansi and T. equiperdum seem to have evolved alongside their natural hosts (particularly the camel and the horse) to induce disease while maintaining conditions that ensure future transmission.
Similarly, mutational changes and loss of gene function are also found in the T. gondii RH strain, causing it to lose its ability to form cysts in the mammalian host, and only the tachyzoite stage is found in its life cycle (74, 84). Therefore, the T. gondii RH strain can only be mechanically transmitted in the laboratory from individual to individual (Fig. 3II). In many hosts, such as the mouse, the lack of cellular control and differentiation results in a high degree of virulence and host death. This situation is analogous to the infectious cancer DFDT, which is transmitted mechanically between Tasmanian devils and is highly virulent. An evolutionary consequence of this degree of virulence is that highly virulent strains of T. gondii are rarely seen in nature. Although DFDT is commonly seen in nature at present, the high virulence (100% mortality) found in infected hosts will ultimately ensure the extinction of the endangered Tasmanian devil. Being a recent evolutionary event, unless cancer-resistant variant devils evolve rapidly or humans intervene, DFDT is likely to eliminate its host and its transmission cycle, thereby consigning itself to rarity in nature.
These examples of horizontal transmission of parasites and cancers can also be complemented by examples of vertical transmission. T. gondii undergoes vertical transmission (mother to fetus) as one of its means of transmission (85). To enable a vertical transmission, it must have evolved mechanisms to facilitate transplacental transmission in the mother and immune evasion in the fetus. An analogous situation must occur in the case of the rare transplacentally vertically transmitted cancers (27).
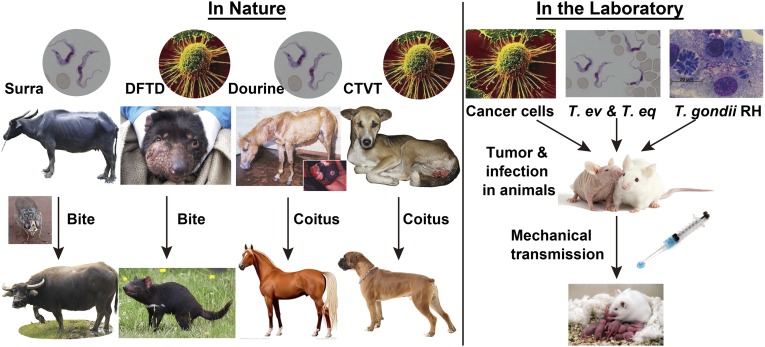
In these cases of cancerous parasites and infectious cancers, mechanical transmission from individual to individual seems to be the key transmission route. This route is also the most widely used method for the researchers who are working on cancer to inoculate human and mouse cancer cells into a mouse model (SCID or nude mouse) in the laboratory. The parallels in the transmission of T. evansi, T. gondii, and cancer cells (Fig. 4), taken alongside the other similarities discussed in this paper, lead us to conclude that consideration of some protozoan parasites as cancers is a genuine comparison. The wide diversity of protozoan parasites in nature suggests that many other examples of cancers of parasites may exist and be eventually discovered.
Fig. 4.
Transmission routes of protozoan parasites and infectious or noninfectious cancers in nature and the laboratory. Naturally, T. evansi, the cancer of T. brucei we considered, and the devil facial tumor disease (DFTD) can be mechanically transmitted by the biting of insects or the devil itself. T. equiperdum, the other cancer of T. brucei and canine transmissible venereal tumor (CTVT) can be naturally transmitted by intercourse between hosts. All strains of T. evansi, T. equiperdum, T. gondii RH, and most cancers can be mechanically transmitted from individual to individual by syringe in laboratory.
Conclusion
Although there are many hypotheses regarding the origin of cancer, it is extremely difficult to study the exact origin of a cancer cell in metazoans and particularly in humans. It is difficult to establish which cell type will lose the ability to correctly control cell division although significant progress has been made to demonstrate the clonal evolution of cancer (14). However, it might be easier to carry out such experiments with single-cell parasites, both in vitro and in vivo. T. brucei and T. gondii provide many advantages for the investigation of the relationship between mutations of mtDNA/nDNA and the process of differentiation. These advantages include (i) well-known and well-established models, developed over many decades, for cellular, molecular, and immunological studies; (ii) good model systems to study the function of mtDNA/nDNA; (iii) available complete genome sequences and, in general, simpler genomes; (iv) ease of handling in vivo and in vitro; (v) well-established tools for gene manipulation; (vi) consistent differentiation profiles occurring during the life cycle; and (vii) organisms of medical and veterinary importance. With these advantages, T. brucei and T. gondii are ideal potential model systems to develop future studies in this area. There are clearly very strong parallels between our example parasites: the two cases of infectious cancers and free-ranging cancer cells such as the leukemias. There may be a very strong case for exploring protozoan cancers as a possible model system in these cases. Clearly, with solid tissue cancers, there are greater challenges—the effects of soluble and cell-based signaling systems add a further complexity. The trypanocidal drug α-difluoromethylornithine (DFMO) is widely used to cure late-stage sleeping sickness in people in West Africa, despite being a failed candidate anticancer drug (86). Perhaps taking a broader evolutionary perspective will promote a greater understanding of the origins of both protozoan and metazoan cancers and lead to the development of new cures for both cancers and parasitic infections. The war against cancer is far from completion although progress toward understanding the nature and logical basis of cancer has been impressive (87). In fact, few cancers can be cured without early detection and surgical excision; consequently, it has been proposed that current anticancer strategies need to be reconsidered and that totally new approaches need to be discussed (87). Perhaps protozoan parasites offer one such novel avenue of research.
Acknowledgments
We thank Mr. Xia-Bing Dai for his excellent skill in drawing the figures. This work was supported by National Base Research Program (973 Project) Grant 2010CB530000; National Natural Science Foundation of China Grants 31272305, 31472058, 31301876, and 31402029; and Academic Degrees Committee of Guangdong Province Grant SYBZZXM201011.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution IX: Clonal Reproduction: Alternatives to Sex,” held January 9–10, 2015, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/ILE_IX_Clonal_Reproduction.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502599112/-/DCSupplemental.
References
- 1.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawyers C. Targeted cancer therapy. Nature. 2004;432(7015):294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 3.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371(18):1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Ostrow SL, Barshir R, DeGregori J, Yeger-Lotem E, Hershberg R. Cancer evolution is associated with pervasive positive selection on globally expressed genes. PLoS Genet. 2014;10(3):e1004239. doi: 10.1371/journal.pgen.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. A decade of exploring the cancer epigenome: Biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carreira S, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014;6(254):ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnaufer A, Domingo GJ, Stuart K. Natural and induced dyskinetoplastic trypanosomatids: How to live without mitochondrial DNA. Int J Parasitol. 2002;32(9):1071–1084. doi: 10.1016/s0020-7519(02)00020-6. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodish H, et al. In: Molecular Cell Biology. 3rd Ed Lodish H, et al., editors. Freeman; New York: 1995. [Google Scholar]
- 14.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3(2):89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25(34):4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 17.Pearse AM, et al. Evolution in a transmissible cancer: A study of the chromosomal changes in devil facial tumor (DFT) as it spreads through the wild Tasmanian devil population. Cancer Genet. 2012;205(3):101–112. doi: 10.1016/j.cancergen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Murchison EP, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148(4):780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126(3):477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murchison EP, et al. Transmissible [corrected] dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343(6169):437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearse AM, Swift K. Allograft theory: Transmission of devil facial-tumour disease. Nature. 2006;439(7076):549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 22.Siddle HV, Kaufman J. Immunology of naturally transmissible tumours. Immunology. 2015;144(1):11–20. doi: 10.1111/imm.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddle HV, et al. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc Natl Acad Sci USA. 2013;110(13):5103–5108. doi: 10.1073/pnas.1219920110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murchison EP. Clonally transmissible cancers in dogs and Tasmanian devils. Oncogene. 2008;27(Suppl 2):S19–S30. doi: 10.1038/onc.2009.350. [DOI] [PubMed] [Google Scholar]
- 25.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Origins and evolution of a transmissible cancer. Evolution. 2009;63(9):2340–2349. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Lin F, Xing K, He X. The reverse evolution from multicellularity to unicellularity during carcinogenesis. Nat Commun. 2015;6:6367. doi: 10.1038/ncomms7367. [DOI] [PubMed] [Google Scholar]
- 27.Isoda T, et al. Immunologically silent cancer clone transmission from mother to offspring. Proc Natl Acad Sci USA. 2009;106(42):17882–17885. doi: 10.1073/pnas.0904658106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzger MJ, Reinisch C, Sherry J, Goff SP. Horizontal transmission of clonal cancer cells causes leukemia in soft-shell clams. Cell. 2015;161(2):255–263. doi: 10.1016/j.cell.2015.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mony BM, et al. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature. 2014;505(7485):681–685. doi: 10.1038/nature12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. 2011;331(6021):1149–1153. doi: 10.1126/science.1198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones NG, et al. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS Pathog. 2014;10(1):e1003886. doi: 10.1371/journal.ppat.1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews KR. 25 years of African trypanosome research: From description to molecular dissection and new drug discovery. Mol Biochem Parasitol. 2015 doi: 10.1016/j.molbiopara.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 34.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull. 1985;41(2):105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 35.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 36.Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukes J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci USA. 2008;105(6):1999–2004. doi: 10.1073/pnas.0711799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110(36):14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carnes J, et al. Genome and phylogenetic analyses of trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis. 2015;9(1):e3404. doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoare CA. The Trypanosomes of Mammals: A Zoological Monograph. Blackwell Scientific Publications; Oxford: 1972. [Google Scholar]
- 40.MacGregor P, Matthews KR. Identification of the regulatory elements controlling the transmission stage-specific gene expression of PAD1 in Trypanosoma brucei. Nucleic Acids Res. 2012;40(16):7705–7717. doi: 10.1093/nar/gks533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domenicali Pfister D, et al. A Mitogen-activated protein kinase controls differentiation of bloodstream forms of Trypanosoma brucei. Eukaryot Cell. 2006;5(7):1126–1135. doi: 10.1128/EC.00094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc Natl Acad Sci USA. 2006;103(50):19194–19199. doi: 10.1073/pnas.0608971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hide G, Gray A, Harrison CM, Tait A. Identification of an epidermal growth factor receptor homologue in trypanosomes. Mol Biochem Parasitol. 1989;36(1):51–59. doi: 10.1016/0166-6851(89)90199-0. [DOI] [PubMed] [Google Scholar]
- 44.Sternberg JM, McGuigan F. Trypanosoma brucei: Mammalian epidermal growth factor promotes the growth of the African trypanosome bloodstream form. Exp Parasitol. 1994;78(4):422–424. doi: 10.1006/expr.1994.1047. [DOI] [PubMed] [Google Scholar]
- 45.Bakhiet M, et al. Human and rodent interferon-gamma as a growth factor for Trypanosoma brucei. Eur J Immunol. 1996;26(6):1359–1364. doi: 10.1002/eji.1830260627. [DOI] [PubMed] [Google Scholar]
- 46.Hide G. Mammalian epidermal growth factor stimulates G-protein activity in Trypanosoma brucei. Parasitol Res. 1998;84(2):143–146. doi: 10.1007/s004360050372. [DOI] [PubMed] [Google Scholar]
- 47.Ghansah TJ, Ager EC, Freeman-Junior P, Villalta F, Lima MF. Epidermal growth factor binds to a receptor on Trypanosoma cruzi amastigotes inducing signal transduction events and cell proliferation. J Eukaryot Microbiol. 2002;49(5):383–390. doi: 10.1111/j.1550-7408.2002.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 48.Ludin P, Nilsson D, Mäser P. Genome-wide identification of molecular mimicry candidates in parasites. PLoS ONE. 2011;6(3):e17546. doi: 10.1371/journal.pone.0017546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuart K. RNA editing in trypanosomatid mitochondria. Annu Rev Microbiol. 1991;45(2):327–344. doi: 10.1146/annurev.mi.45.100191.001551. [DOI] [PubMed] [Google Scholar]
- 50.Lun ZR, Desser SS. Is the broad range of hosts and geographical distribution of Trypanosoma evansi attributable to the loss of maxicircle kinetoplast DNA? Parasitol Today. 1995;11(4):131–133. doi: 10.1016/0169-4758(95)80129-4. [DOI] [PubMed] [Google Scholar]
- 51.Brun R, Hecker H, Lun ZR. Trypanosoma evansi and T. equiperdum: Distribution, biology, treatment and phylogenetic relationship (a review) Vet Parasitol. 1998;79(2):95–107. doi: 10.1016/s0304-4017(98)00146-0. [DOI] [PubMed] [Google Scholar]
- 52.Lun ZR, Lai DH, Li FJ, Lukes J, Ayala FJ. Trypanosoma brucei: Two steps to spread out from Africa. Trends Parasitol. 2010;26(9):424–427. doi: 10.1016/j.pt.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 53.Diffley P, Mama K. Fixed and temporary fluctuations in the cell cycle of monomorphic lines of Trypanosoma brucei gambiense. Mol Biochem Parasitol. 1989;32(1):1–5. doi: 10.1016/0166-6851(89)90123-0. [DOI] [PubMed] [Google Scholar]
- 54.MacGregor P, et al. High-throughput chemical screening for antivirulence developmental phenotypes in Trypanosoma brucei. Eukaryot Cell. 2014;13(3):412–426. doi: 10.1128/EC.00335-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68(5):1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkitaraman AR. The breast cancer susceptibility gene, BRCA2: At the crossroads between DNA replication and recombination? Philos Trans R Soc Lond B Biol Sci. 2000;355(1394):191–198. doi: 10.1098/rstb.2000.0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oyola SO, Bringaud F, Melville SE. A kinetoplastid BRCA2 interacts with DNA replication protein CDC45. Int J Parasitol. 2009;39(1):59–69. doi: 10.1016/j.ijpara.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Hall M, 3rd, Misra S, Chaudhuri M, Chaudhuri G. Peptide aptamer mimicking RAD51-binding domain of BRCA2 inhibits DNA damage repair and survival in Trypanosoma brucei. Microb Pathog. 2011;50(5):252–262. doi: 10.1016/j.micpath.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trenaman A, et al. Trypanosoma brucei BRCA2 acts in a life cycle-specific genome stability process and dictates BRC repeat number-dependent RAD51 subnuclear dynamics. Nucleic Acids Res. 2013;41(2):943–960. doi: 10.1093/nar/gks1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24(23):4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: The replication of kinetoplast DNA. Trends Parasitol. 2005;21(8):363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 62.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30(2):97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 63.Hajduk S, Ochsenreiter T. RNA editing in kinetoplastids. RNA Biol. 2010;7(2):229–236. doi: 10.4161/rna.7.2.11393. [DOI] [PubMed] [Google Scholar]
- 64.Aphasizhev R, Aphasizheva I. Mitochondrial RNA processing in trypanosomes. Res Microbiol. 2011;162(7):655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61(5):885–894. doi: 10.1016/0092-8674(90)90199-o. [DOI] [PubMed] [Google Scholar]
- 66.Hashimi H, et al. The assembly of F(1)F(O)-ATP synthase is disrupted upon interference of RNA editing in Trypanosoma brucei. Int J Parasitol. 2010;40(1):45–54. doi: 10.1016/j.ijpara.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Zíková A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5(5):e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ley TJ, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephens PJ, et al. Oslo Breast Cancer Consortium (OSBREAC) The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev. 1998;11(2):267–299. doi: 10.1128/cmr.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frenkel JK, Dubey JP, Hoff RL. Loss of stages after continuous passage of Toxoplasma gondii and Besnoitia jellisoni. J Protozool. 1976;23(3):421–424. doi: 10.1111/j.1550-7408.1976.tb03799.x. [DOI] [PubMed] [Google Scholar]
- 76.Villard O, Candolfi E, Ferguson DJ, Marcellin L, Kien T. Loss of oral infectivity of tissue cysts of Toxoplasma gondii RH strain to outbred Swiss Webster mice. Int J Parasitol. 1997;27(12):1555–1559. doi: 10.1016/s0020-7519(97)00144-6. [DOI] [PubMed] [Google Scholar]
- 77.Chinchilla M, Guerrero OM, Solano E. Lack of multiplication of Toxoplasma in macrophages of rats in vitro. J Parasitol. 1982;68(5):952–955. [PubMed] [Google Scholar]
- 78.Cavaillès P, et al. The rat Toxo1 locus directs toxoplasmosis outcome and controls parasite proliferation and spreading by macrophage-dependent mechanisms. Proc Natl Acad Sci USA. 2006;103(3):744–749. doi: 10.1073/pnas.0506643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z, et al. Differences in iNOS and arginase expression and activity in the macrophages of rats are responsible for the resistance against T. gondii infection. PLoS ONE. 2012;7(4):e35834. doi: 10.1371/journal.pone.0035834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Radke JR, et al. The transcriptome of Toxoplasma gondii. BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang N, et al. Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics. 2013;14:467. doi: 10.1186/1471-2164-14-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Desquesnes M, et al. Trypanosoma evansi and surra: A review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int. 2013;2013:194176. doi: 10.1155/2013/194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo HM, et al. Infection by Toxoplasma gondii, a severe parasite in neonates and AIDS patients, causes impaired anion secretion in airway epithelia. Proc Natl Acad Sci USA. 2015;112(14):4435–4440. doi: 10.1073/pnas.1503474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992;359(6390):82–85. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- 85.Hide G, et al. Evidence for high levels of vertical transmission in Toxoplasma gondii. Parasitology. 2009;136(14):1877–1885. doi: 10.1017/S0031182009990941. [DOI] [PubMed] [Google Scholar]
- 86.Eperon G, et al. Treatment options for second-stage gambiense human African trypanosomiasis. Expert Rev Anti Infect Ther. 2014;12(11):1407–1417. doi: 10.1586/14787210.2014.959496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanahan D. Rethinking the war on cancer. Lancet. 2014;383(9916):558–563. doi: 10.1016/S0140-6736(13)62226-6. [DOI] [PubMed] [Google Scholar]