Significance
The ubiquitous enzymes called cytochromes P450 catalyze a broad range of chemical reactions using molecular oxygen. For example, in humans, these enzymes are involved in breakdown of foreign compounds, including drugs. The bacterial cytochrome P450cam is thought to open up to allow substrate to enter the active site, and then to close during catalysis to keep reactive intermediates inside. Surprisingly, recent crystal structures suggested that the enzyme is open during the reaction. We have studied the enzyme in solution using paramagnetic NMR spectroscopy, demonstrating that, in fact, the enzyme is closed. This finding indicates that the subtle balance between open and closed is affected by crystallization, which can lead to the wrong conclusions about the protein dynamics.
Keywords: paramagnetic NMR, enzyme, dynamics, crystallography, cytochrome
Abstract
The energy landscapes of proteins are highly complex and can be influenced by changes in physical and chemical conditions under which the protein is studied. The redox enzyme cytochrome P450cam undergoes a multistep catalytic cycle wherein two electrons are transferred to the heme group and the enzyme visits several conformational states. Using paramagnetic NMR spectroscopy with a lanthanoid tag, we show that the enzyme bound to its redox partner, putidaredoxin, is in a closed state at ambient temperature in solution. This result contrasts with recent crystal structures of the complex, which suggest that the enzyme opens up when bound to its partner. The closed state supports a model of catalysis in which the substrate is locked in the active site pocket and the enzyme acts as an insulator for the reactive intermediates of the reaction.
During catalysis, an enzyme will traverse a conformational energy landscape that can be highly complex and easily changed by external factors, such as temperature, ionic strength, and crystallization. Cytochromes P450 (CYPs) are b-type heme-containing monooxygenases found throughout the three domains of life. These enzymes catalyze the regiospecific and stereospecific hydroxylation of various aliphatic and aromatic compounds and are involved in a considerable number of metabolic processes, such as steroid biosynthesis and metabolism of xenobiotics in mammals. The CYP superfamily has been extensively studied over the past five decades, and an understanding of its mechanism is crucial for the development of pharmaceutical compounds that do not inhibit these enzymes. Under other circumstances, it is desirable to inhibit a CYP for therapeutic reasons. For example, compounds that inhibit CYP1B1 and CYP1A2 in humans have been proposed as promising anticancer agents (1). The archetypal member of this superfamily is CYP101A1 (more commonly known as P450cam) from Pseudomonas putida, which was the first CYP for which the 3D structure was determined by X-ray crystallography (2). P450cam catalyzes the hydroxylation of d-camphor to 5-exohydroxycamphor using two electrons, two protons, and molecular oxygen. Putidaredoxin (Pdx) reductase oxidizes NADH and transfers the electrons to Pdx, which, in turn, shuttles electrons to P450cam in two consecutive steps (3–5). This complex reaction is controlled by the enzyme to ensure an efficient coupling between dioxygen reduction and substrate hydroxylation and to avoid side reactions. P450cam must go through a cycle of several conformational and electronic states to perform its catalytic task (6).
The first X-ray crystal structures of P450cam showed that the enzyme occupied a closed conformation, both in the presence and absence of substrate; therefore, it was unclear how the substrate was able to bind into the active site (2). Moreover, structures of the intermediates of the catalytic cycle were also in a closed conformation (7), leading to the idea that during the catalytic cycle, P450cam opens to bind camphor, closes to perform hydroxylation, and then reopens to release the product. The first structures of a more open state of P450cam were obtained by using ligands that forced a channel open (8, 9), and, subsequently, an open structure of P450cam in the absence of substrate was published (10). These structures showed that the F- and G-helices and the F/G loop of P450cam moved to reveal an opening through which the active site could be accessed by the substrate.
The conformational landscape of P450cam is, however, more complex, owing to the fact that the catalytic cycle also comprises two electron transfer steps involving binding of its redox partner Pdx, several spin state changes of the heme iron, and a conformational change upon the binding of substrate. A resonance Raman study published by Sjodin et al. (11) demonstrated that additional conformational substates of the oxy-heme were present when Pdx is bound to P450cam, and they argued that these states may be linked to the electron donation properties of the cysteinate ligated to the heme. NMR spectroscopy studies have revealed changes in the chemical shifts of P450cam when Pdx binds (12, 13).
Recently, a structure of the complex of oxidized P450cam tethered to oxidized Pdx obtained using X-ray crystallography has been reported, and it shows P450cam in the open state in this complex [Protein Data Bank (PDB) ID code 4JWS (14)]. It was proposed that oxidized P450cam favors the open conformation when Pdx binds. A crystal structure of the native complex, without cross-links, was simultaneously but independently obtained using X-ray crystallography, and it suggests that P450cam occupies an intermediate state when Pdx binds [PDB ID code 3W9C (15)]. It is somewhat counterintuitive that P450cam would be in the open state when Pdx binds. Changing to the open state during the catalytic cycle could cause the release of the substrate and reactive oxygen species from the enzyme. However, a recent EPR study supported the finding that oxidized Pdx induces oxidized P450cam to adopt an open conformation, and the authors argue that this observation eliminates the possibility that crystal contacts are the reason why P450cam adopts the open state in the complex with Pdx (16). However, all of the EPR measurements were performed at 50 K or less. Kinetic experiments at ambient temperatures reported in the same article (16) show that the rate constant for camphor dissociation is low and hardly affected by Pdx binding, at least at the concentrations of potassium ions present in the EPR solution and the crystallization buffers. Because it is generally assumed that the dissociation rate of camphor is much lower in the closed state than in the open state of the enzyme, these results suggest that the enzyme remains closed at ambient temperatures upon binding to Pdx, implying the presence of a subtle balance between open and closed states that can be affected by crystallography and low temperature.
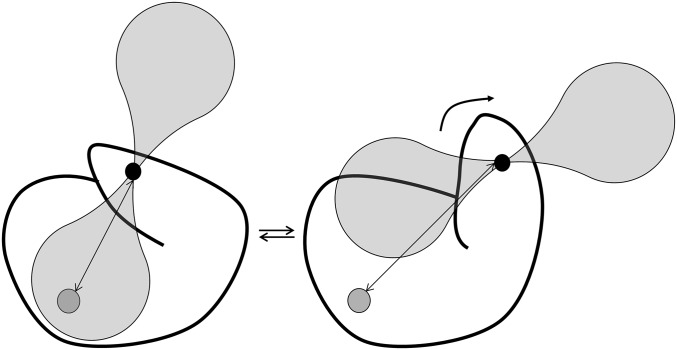
In this work, we studied the state of P450cam in complex with Pdx in solution at ambient temperature (25 °C) using paramagnetic NMR spectroscopy. A pseudocontact shift (PCS) is the difference in chemical shift observed for a nucleus in paramagnetically and diamagnetically tagged protein (17, 18). PCSs can provide long-range distance information and are highly sensitive to structural changes within a protein. The PCS depends on the distance of the nucleus to the paramagnetic center, as well as on the position of the nucleus in the frame defined by the anisotropic component of the magnetic susceptibility of the paramagnetic center, described by the Δχ tensor. The PCS can be used to aid in protein structure determination (19–21), for example, as recently reported for the structure of the seven-transmembrane phototactic receptor sensory rhodopsin II (22). This approach complements the use of paramagnetic relaxation enhancements in structure determination using both liquid and solid-state NMR spectroscopy, as has been demonstrated by various research groups (23–26). Furthermore, PCSs are very useful to obtain long-range distance information for protein docking (17, 27). Recently, the use of PCSs in the refinement of X-ray crystal structures was described, as shown for the catalytic domain of matrix metalloproteinase 1, the third IgG-binding domain of protein G, ubiquitin, free calmodulin, and calmodulin–peptide complexes (28). Similar to residual dipolar couplings (RDCs) (29–32), which are also caused by anisotropic interactions, PCSs can also be used to study conformation changes in proteins, for example, as demonstrated for calmodulin–peptide complexes (33). When a paramagnetic probe is attached to part of a protein that moves between two conformations, the vector that connects the nucleus under observation and the probe will change (Fig. 1). As a consequence, a nucleus experiences distinct distances to the paramagnetic center and positions in the Δχ tensor frame in the two states, and the ensuing differences in PCSs provide a means of differentiating between the two states. We used this approach by mutating residues E195 and A199 to Cys for attachment of the two-armed caged lanthanoid NMR probe-7 (CLaNP-7) (15, 34). This mutation site is at the top of the G-helix, which undergoes a significant conformational change when the protein transitions from the closed state to the open state. Our data demonstrate that Pdx binding does not induce opening of P450cam in solution, in contrast to what is observed in the crystalline state. A closed state of the enzyme is better in line with a model of catalysis in which the substrate and the reactive intermediates remain enclosed within the enzyme during the reaction.
Fig. 1.
Use of PCSs to distinguish conformational states. The paramagnetic center (small black circle) is attached to a part of a protein that exists in two conformations. The nucleus (large gray circle) experiences a distinct PCS in each state because the distance to the center (double-pointed arrows) and the orientation relative to the direction of the anisotropic component of the magnetic susceptibility tensor (transparent shapes) differ.
Results and Discussion
Free P450cam in the Presence of Substrate.
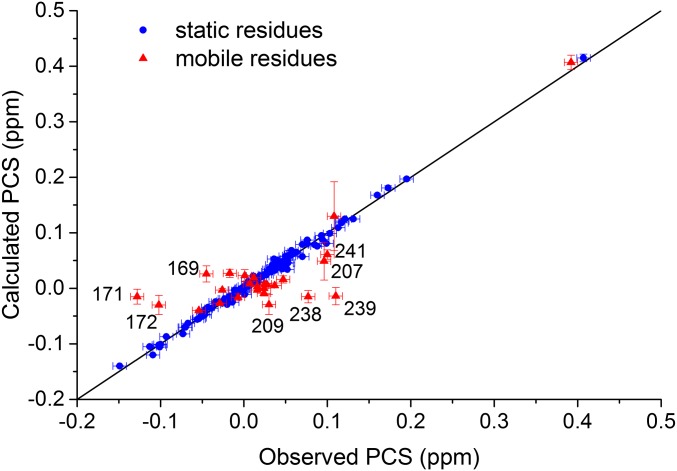
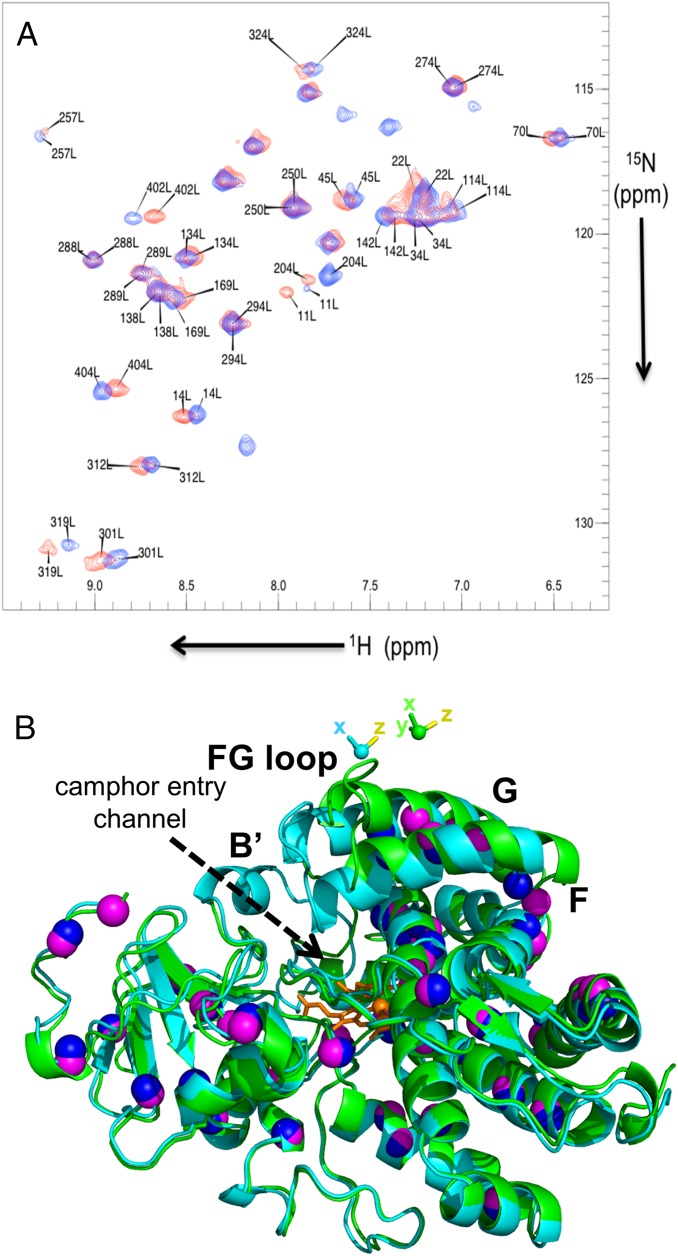
The structure of P450cam in the presence of camphor and KCl has been solved by several groups (2, 7, 10, 35), and the closed state was observed in all structures. We first determined the state of P450cam under these conditions in solution by attaching CLaNP-7 to the double-Cys mutant of oxidized P450cam. The natural substrate, camphor, still binds to this tagged form of the protein (Fig. S1) in the same way as to the WT protein. To obtain PCSs, Lu–CLaNP-7 and Yb–CLaNP-7 were attached to 15N, 2H-labeled samples of the E195C/A199C double-Cys mutant of P450cam in a solution containing 1 mM camphor (Table S1, samples 2 and 3), and backbone nuclei were assigned using previously published data [Biological Magnetic Resonance Data Bank (BMRB) accession code 19038 (15)]. With these data, 139 PCSs could be observed reliably in the transverse relaxation optimized spectroscopy (TROSY) spectrum. The experimental PCSs were used to determine the position of the paramagnetic center and the size and orientation of the Δχ tensor. To determine whether the enzyme with camphor bound is in the open or closed state, the data were fitted using the structures of either the closed [PDB ID code 3L63 (10)] or open [PDB ID code 3L62 (10)] state of P450cam, and only the PCSs from 117 amides that do not move more than 1 Å between the open and closed states. The correlation plot of observed vs. calculated PCSs is shown in Fig. S2. The magnitudes of the axial and rhombic parts of the tensor were determined to be 6.5 ± 0.1 × 10−32 m3 and 5.5 ± 0.1 × 10−32 m3, respectively, using structures of both the open and closed states of P450cam. These values are in line with previously published values for CLaNP-7 (34). As expected, the position of the paramagnetic centers and the orientation of the Δχ tensor are also identical within error for both states. In the structure of the closed state, the lanthanoid is positioned ∼8 Å from the Cα atoms of the two Cys residues (Fig. S3A), consistent with the results of other studies using CLaNP tags (34, 36–38). In contrast, in the fit of the PCS data to the structure of the open state, the paramagnetic center is placed far from the expected position, due to the moved G-helix (Fig. S3B). The position of the lanthanoid is skewed toward Cys195 (5.7 Å to the Cα atom) compared with Cys199 (10.4 Å to Cα atom). It is sterically impossible to place the CLaNP-7 tag to match these distances. These results indicate that the closed state is the better model to describe the structure of camphor-bound P450cam in solution, which is in line with the previously published crystallographic data. Interestingly, the back-predicted PCSs of some of the residues that were left out of the fitting because they are involved in the structural change between open and closed states (169, 171, 172, 207, 209, 238, 239, and 241) fit relatively poorly with the observed PCSs for both open and closed structures (Fig. S2). These residues are located at the base of the F- and G-helices, around the H-helix. This part of the protein is involved in crystal contacts and harbors several loops with high B-factors. Our results suggest that the (average) structure in this region in solution differs slightly from the crystal structures. More NMR data would be required, however, to determine the conformation of this part of the protein with accuracy.
Fig. S1.
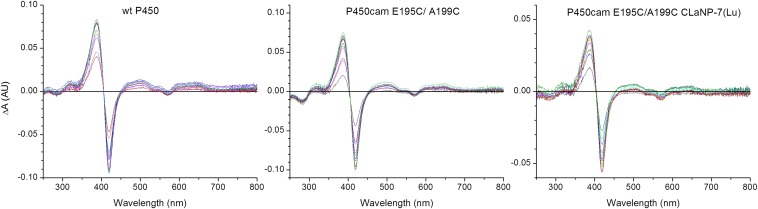
Camphor titrations. Camphor was titrated in WT P450cam (1.6 μM), P450cam E195C/A199C (1.4 μM), and P450cam E195C/A199C CLaNP-7 (1.0 μM), and the changes in the electronic absorbance spectrum were measured under conditions of 20 mM KPi and 100 mM KCl (pH 7.4) at room temperature. The Kd value derived from fitting the absorbance change at 392 nm for all samples is 0.2 ± 0.1 μM. AU, absorption units.
Table S1.
Details of P450cam samples used for NMR experiments
| No. | Labeling scheme | Tag | Percentage assigned, % | [P450cam], μM | Camphor* | Buffer conditions |
| 1 | 15N 13C 2H | — | 74 | 530 | + | 20 mM Hepes (pH 7.5), 100 mM NaCN |
| 2 | 15N 2H | Lu–CLaNP-7 | 42 | 100 | + | 50 mM Tris⋅HCl (pH 7.4), 100 mM KCl |
| 3 | 15N 2H | Yb–CLaNP-7 | 42 | 100 | + | 50 mM Tris⋅HCl (pH 7.4), 100mM KCl |
| 4 | 15N-Leu | Lu–CLaNP-7 | 64 | 175 | + | 20 mM Hepes (pH 7.5), 200 mM KCl |
| 5 | 15N-Leu | Yb–CLaNP-7 | 45 | 300 | + | 20 mM Hepes (pH 7.5), 200 mM KCl |
| 6 | 15N 2H | — | 55 | 340 | − | 20 mM Hepes (pH 7.5), 100 mM KCN |
| 7 | 15N-Leu | Lu–CLaNP-7 | 55 | 175 | − | 20 mM Hepes (pH 7.5) |
| 8 | 15N-Leu | Yb–CLaNP-7 | 40 | 175 | − | 20 mM Hepes (pH 7.5) |
Presence/absence of camphor [1 mM, 1% (vol/vol) methanol].
Fig. S2.
Plot of calculated vs. observed PCSs. The blue circles represent the PCSs of the nuclei that move less than 1 Å between the open and closed states of P450cam, which were used to fit the position of the paramagnetic center and the size and orientation of the Δχ tensor. The data for the closed structure [PDB ID code 3L63 (10)] are shown. The red triangles represent the PCSs of nuclei that move more than 1 Å between the open and closed states, which were not used in the fitting. The labeled points all locate around the H-helix. The results suggest that the structure of this part of the protein in solution differs slightly from the crystal structures of both the open and closed states. Errors on calculated PCSs are based on the precision of the Δχ parameters (many are within the symbols). The errors in the observed PCSs were estimated to be ±0.008 ppm. The solid line represents a perfect correlation. The Q value for the blue circles is 0.041.
Fig. S3.
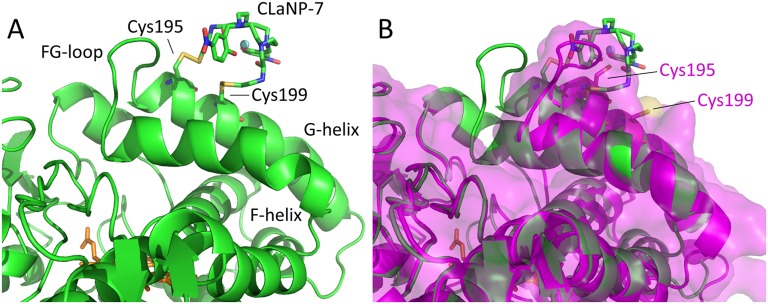
(A) Model of P450cam [camphor-bound, closed state; PDB ID code 3L63 (10)] with mutations 195/199C (cartoon), linked to CLaNP-7 (sticks). The Yb3+ ion is shown as a sphere. The heme is shown in orange sticks. CLaNP-7 was modeled to have the Yb3+ as close as possible to the origin of the experimentally determined tensor, with the z axis along the twofold symmetry axis of the CLaNP molecule. (B) Structure of A overlaid with the open state of camphor-free P450cam [PDB ID code 3L62 (10)] shown in magenta cartoons and a semitransparent surface, with mutations 195/199C modeled (sticks). CLaNP-7 shows severe steric clashes with the open state and is situated far from Cys199. It is concluded that the experimentally determined position of the probe shows that camphor-bound P450cam is in the closed state.
State of the Free P450cam in Absence of Substrate.
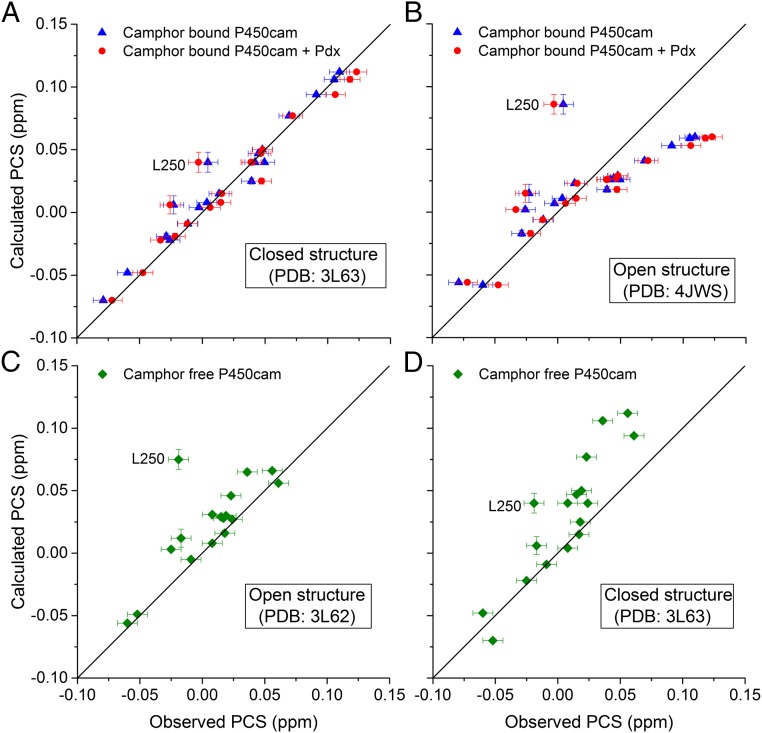
In the absence of substrate, P450cam is only stable for a few hours, particularly when a paramagnetic tag, such as CLaNP-7, has been attached. As a result, it turned out to be impossible to obtain sufficient uniformly 15N, 2H-labeled, CLaNP-7–tagged sample of this form. It was possible to obtain a nondeuterated sample, so we resorted to selectively 15N-Leu–labeled samples to reduce spectral overlap, allowing unambiguous identification of nuclei under various experimental conditions. First, 15N-Leu–labeled P450cam samples were tagged with Lu–CLaNP-7 and Yb–CLaNP-7, and PCSs were measured in the presence of camphor and KCl (Table S1, samples 4 and 5 and Fig. 2). Then, PCSs for substrate-free oxidized P450cam were obtained from spectra of the same samples in the absence of KCl and camphor (Table S1, samples 7 and 8). Because of the instability of substrate-free P450cam, these spectra were assigned on the basis of the camphor-bound P450cam NMR assignments in combination with a camphor titration (Table S1, sample 6). The 15N-Leu PCSs measured in the presence and absence of camphor were compared with the predicted values for both the closed and open states of P450cam. For the prediction of the closed-state PCSs, the tensor obtained for that state (discussed above) was used. PCSs for the open state were predicted using a model of the probe in the open state, obtained by translation of the probe with the G-helix to the open-state position, keeping the tensor orientation and metal position fixed relative to the two Cys residues on the G-helix (Fig. 2). The PCSs observed for P450cam in the presence of camphor and KCl are predicted very well by the closed structure and poorly by the open-state structure (Fig. 3 A and B, respectively; blue triangles). On the other hand, the measured PCSs for substrate-free P450cam fit better to the PCSs predicted for the open state than to the PCSs for the closed state (Fig. 3 C and D). In Fig. 3C, one outlier is observed, Leu250. According to the data of Miao et al. (39), Leu250 is mobile in the camphor-free form of P450cam, so its position may not be well defined. Therefore, we attribute the deviation of the PCSs from the expected value to dynamic behavior. It is concluded that P450cam is in the closed state in solution in the presence of substrate and KCl and in the open state in their absence. This result indicates that probe attachment to the G-helix does not prevent opening of the protein. We note that the possibility to distinguish between the states is a consequence of the high precision with which PCSs can be measured. Most other NMR observables are less precise. According to our calculations, RDC resulting from alignment of the protein by the CLaNP that would be significantly different for the open and closed states would only be observed for one or two Leu amides.
Fig. 2.
(A) Overlay of [15N-1H]-HSQC spectra recorded of 15N-Leu–labeled Lu– (blue) and Yb– (red) CLaNP-7–tagged P450cam with camphor bound. Labels indicate assignment at 298 K, 20 mM Hepes (pH 7.5), 1 mM camphor, 200 mM KCl (Table S1, samples 4 and 5). (B) Overlay of open and closed states of P450cam. In a cartoon representation, the open state (PDB ID code 3L62) is shown in green and the closed state (PDB ID code 3L63) is shown in cyan. The spheres indicate the N atoms of Leu residues (purple and blue for the open and closed states, respectively). The heme is shown in orange. The magnetic susceptibility tensors of CLaNP-7 attached to 195/199Cys are shown as a sphere for the metal and as sticks for the axes for the two states in the same colors as in the cartoons. The yellow sticks mark the z direction. The tensor position for the closed structure is based on a fit to the experimentally observed PCS for the camphor-bound state of P450cam. The position for the open state was modeled by fixing it relative to the G-helix, following the movement of this helix going from the closed state to the open state. The bold letters label the relevant helices and F/G loop.
Fig. 3.
PCSs for 15N-1H backbone amides in Leu residues in P450cam. Calculated PCSs based on the closed structure (A; PDB ID code 3L63) or the open structure (B; PDB ID code 4JWS) are plotted against the observed PCSs for camphor-bound P450cam (blue triangles) and Pdx-bound, camphor-bound P450cam (red circles). Calculated PCSs based on the open structure (C; PDB ID code 3L62) or the closed structure (D; PDB ID code 3L63) are plotted against the observed PCSs for camphor-free P450cam (green diamonds). Errors on calculated PCSs are based on the precision of the Δχ parameters. The errors in the observed PCSs were estimated to be ±0.008 ppm. The solid line represents a perfect correlation. The PCSs for Leu250 are labeled.
State of P450cam Bound to Pdx.
A Pdx titration was performed with 15N-Leu–labeled P450cam in the presence of camphor and KCl, enabling us to measure PCSs for the oxidized P450cam–Pdx complex in the presence of substrate. The Kd for the Pdx–P450cam complex is 2.2 μM (40), and 2.5 molar equivalents of Pdx were titrated into the NMR samples, which corresponds to over 99% of P450cam in the samples being bound to Pdx. Significant chemical shift perturbations were observed due to the binding of Pdx, as has been reported before (13). By comparing the difference between the chemical shifts for the Pdx–P450cam complex tagged with CLaNP-7 (Yb) and (Lu), the Leu amide PCSs were obtained. The PCSs are very similar to those PCSs observed for free P450cam under the same conditions, suggesting that no major change in the position of the paramagnetic center occurs upon binding of Pdx. In accord with that finding, PCSs measured for the Pdx-bound P450cam agree well with the predicted PCSs for the closed state of P450cam and fit poorly with PCSs calculated for the open state of P450cam in complex with Pdx [PDB ID code 4JWS (14)], as demonstrated by the Q values of 0.094 and 0.322 for the fits to the closed and open states, respectively (Fig. 3 A and B, red circles). L250 was not included in the Q-value calculations. All Q values are listed in Table S2. Thus, we conclude that upon binding of Pdx, oxidized P450cam remains in the closed state in solution at 25 °C.
Table S2.
Q values for fits of predicted Leu amide PCSs against observed PCSs under different conditions
| Structure used for calculation | Camphor-bound P450cam | Camphor-bound P450cam + Pdx | Camphor-free P450cam |
| Open (PDB ID code 3L62) | 0.281 | 0.322 | 0.228 |
| Closed (PDB ID code 3L63) | 0.085 | 0.094 | 0.355 |
All studies on the structure of P450cam in the presence of Pdx used samples containing between 50 and 200 mM KCl, and potassium ions have been implicated in stabilizing the closed form of P450cam when camphor is bound (41). Therefore, in this study, the Leu-labeled samples used to determine the state of P450cam in the presence of Pdx (Table S1, samples 4 and 5) also contained 200 mM KCl.
The open state observed for P450cam in complex with Pdx by X-ray crystallography and EPR spectroscopy may be an artifact of the methodologies, such that the protein was trapped in the open state during crystallization for the X-ray experiments or freezing to 30–50 K for EPR spectroscopy. At first sight, it may appear unlikely that an open state detected by these techniques does not exist in solution at ambient temperatures, but it should be realized that a delicate equilibrium must be present between open and closed states. Camphor binding can shift that equilibrium from mostly open to mostly closed, and the binding free energy difference for this substrate is only about 8 kcal⋅mol−1 (42). Thus, large temperature changes or crystallization could cause a similar shift. It is noted that the resonance Raman spectroscopy experiments that have been performed on this system were carried out at 213 K; therefore, these results could also have been influenced by the low temperature.
Poulos and coworkers (14) also solved the structure of the P450cam–Pdx complex after reduction with dithionite and found four complexes per asymmetrical unit, of which one contained the substrate, camphor, and the other three contained the product, 5-exohydroxycamphor. The structure of each complex was solved and found to be in the open form, which could result from crystal packing effects, because the structure was open in the crystal before reduction. Interestingly, by EPR spectroscopy, the open state was only detected for the oxidized complex. For the reduced complex, the closed state was reported (16). This result emphasizes the subtleties in the energy difference between the different states. Reducing conditions did not allow for the measurement of PCSs because of the loss of the CLaNP upon reduction of the disulfide linkages. We speculate, however, that P450cam in this state also remains closed upon Pdx binding. It has been established that Pdx is specifically required as a donor for the second electron transfer step, somehow signaling the start of the catalytic reaction in the active site (43). This signal is called the effector activity of Pdx. In this light, the molecular mechanism of the effector activity of Pdx remains unclear. Significant chemical shift perturbations were observed for P450cam upon titration with Pdx. These perturbations could be due to a change in the spin state of the heme or to local conformational changes akin to those changes reported by Pochapsky and coworkers (44). It is concluded that opening of P450cam is not part of the effector activity, so further work is required to identify the structural or electronic changes that represent the effector activity of Pdx and are responsible for starting the catalytic reaction inside P450cam.
Conclusions
In this work, we have studied the behavior of P450cam under different conditions in the solution state at ambient temperatures. The data presented show that substrate-free P450cam occupies an open state, because the PCSs measured for substrate-free P450cam fit best to the X-ray crystal structures of substrate-free P450cam, which showed the open state of the enzyme (10). Moreover, the PCSs measured for substrate-bound P450cam fit the X-ray crystal structures of substrate-bound P450cam, with the enzyme in the closed state (2, 10). Our findings for P450cam bound to both camphor and Pdx give evidence for the protein being in the closed state, in disagreement with previously published EPR and X-ray crystallographic data, which show the open form (14–16). In summary, we believe that the equilibrium between the open and closed forms of P450cam is subtly balanced and can easily be perturbed by changes in its environment.
Materials and Methods
Cloning and Mutagenesis.
The pET28a plasmid harboring the cDNA of P450cam C334A was used as a template for site-directed mutagenesis. As described in previous studies, the surface-exposed Cys residue at position 334 on P450cam was substituted to Ala to prevent dimerization (45), as well as interference with CLaNP tagging (34). Site-directed mutagenesis was carried out using the Quik-Change protocol (Stratagene). The primers used for the substitution of Cys residues at the positions 195 and 199 were 5′-cttcgcatgcgccaaggagtgtctctacgactatctgatac-3′ and its complement.
Sample Preparation.
15N, 13C, 2H-labeled CLaNP-7–tagged P450cam and 15N, 2H-labeled CLaNP-7–tagged P450cam (Table S1, samples 2, 3, and 6) were produced according to a previously published protocol (15). 15N, 13C, 2H-labeled P450cam (Table S1, sample 1) was produced according to the same protocol with the following modifications. d-glucose-13C6 (2 g/L, 99 atom % 13C; Sigma) was used as a carbon source in M9 medium containing 15NH4Cl and D2O. The expression of the protein was carried on for 72 h at 22 °C after induction. Approximately, 5 mg of P450cam was obtained from 1 L of culture. 15N-Leu–labeled P450cam (E195C/A199C/C334A) (Table S1, samples 4, 5, 7, and 8) was also produced following the protocol of Hiruma et al. (15), with the following modifications to protein production from the work of Tong et al. (46). During expression, 0.1 g/L of all unlabeled amino acids except Leu was added to the M9 medium, as well as 0.125 g/L Guo, Ado, Cyt, and Ura; 0.05 g/L Thy; 2 g/L succinic acid; and 50 mg/L nicotinic acid according to Senn et al. (47). The bacteria were incubated at 37 °C until an OD600 of 0.6 was reached. Subsequently, 1 g/L all amino acids except Leu was added to the medium, along with 0.1 g/L 15N-Leu (98%; Cambridge Isotope Laboratories). Induction was performed with isopropyl-β-d-thiogalactopyranoside (0.5 mM), and δ-aminolevulinic acid (0.4 mM), camphor (1.0 mM), and FeCl3 (100 μM) were added. The cells were harvested after 4 h of further incubation at the same temperature. CLaNP-7 tagging was performed as previously described (15). To obtain camphor-free P450cam (Table S1, samples 7 and 8), camphor was removed by repetitive (three times) buffer exchange using a PD-10 column (GE Healthcare) equilibrated with 20 mM Hepes (pH 7.4). All samples contained 6%/94% (vol/vol) D2O/H2O. Pdx was obtained as described (15).
NMR Spectroscopy and Spectral Assignment.
The resonances in the spectra of samples 2 and 4 (Table S1) were assigned using previously published assignments [BMRB accession code 19038 (15)]. The resonances in the spectra of samples 3 and 5 (Table S1) were assigned by iterative Δχ tensor fitting and assignment (discussed below). To investigate complex formation between Pdx and P450cam, microliter aliquots of a 4 mM Pdx stock solution were titrated into samples 4 and 5 (Table S1) to ∼2.5 molar equivalents, and the effects on the diamagnetically and paramagnetically tagged proteins were observed. After each addition of protein, 15N-1H heteronuclear single quantum coherence (HSQC) spectra were recorded on a Bruker AVIIIHD 850 spectrometer with a TCI-Z-GRAD cryoprobe at 298 K.
1H, 13C, and 15N sequential resonance assignments were obtained on a sample containing camphor as well as cyanide (Table S1, sample 1). Cyanide binding changes the Fe(III) to a low-spin state, which reduces the paramagnetic relaxation broadening in the vicinity of the heme. Cyanide and camphor can bind simultaneously in the active site. This form of P450cam is much more stable than the oxidized camphor-free form, which, in our hands, denatures over the course of several hours at room temperature. The 3D TROSY-HNCACB, 3D TROSY-HNcaCO, and 3D TROSY-HNCO experiments were performed on a Bruker Avance III spectrometer equipped with a TCI-Z-GRAD cryoprobe operating at a Larmor frequency of 950 MHz, along with a 3D TROSY-HNcoCACB experiment performed on a Bruker Avance I spectrometer equipped with a TCI-Z-GRAD cryoprobe operating at a Larmor frequency of 800 MHz at a temperature of 298 K. [15N,1H]-TROSY-HSQC spectra were recorded between each 3D experiment to ensure samples were stable, and an example of one of these spectra is shown in Fig. S4. The numbers of complex points and spectral widths used to record the experiments for backbone assignment are shown in Table S3. To obtain the assignments of camphor-free P450cam, camphor was removed from cyanide-bound P450cam and a 15N-TROSY-HSQC spectrum was acquired on a Bruker AVIII 600 with a TCI-Z-GRAD cryoprobe at a temperature of 298 K (Table S1, sample 6). Then, camphor was titrated back to a final concentration of 680 μM, and spectra were measured at each titration point. In this way, the assignments for camphor-free, cyanide-bound P450cam were obtained. Finally, a 15N-1H HSQC spectrum taken of P450cam tagged with Lu–CLaNP-7 (Table S1, sample 7) in the absence of both camphor and cyanide (Bruker AVIIIHD 850 with a TCI-Z-GRAD cryoprobe, at 298 K) was assigned by comparison with the assignments obtained for sample 6 (Table S1). The assignments for the equivalent paramagnetic sample (Table S1, sample 8; tagged with Yb–CLaNP-7) were obtained using predicted PCSs for the open state of P450cam (discussed below). All data were processed using Topspin 2.1 (Bruker Biospin) or NMRPipe (48), and spectral assignment and analysis were carried out using CCPN Analysis 2.1.5 (49). The chemical shift assignments of the camphor/CN-bound and camphor-free/CN-bound samples were deposited in the BMRB with accession codes 19740 and 19763, respectively.
Fig. S4.
[15N,1H] TROSY-HSQC spectrum of P450cam with NaCN bound at 298 K, 20 mM Hepes (pH 7.5), 1 mM camphor (Table S1, sample 1).
Table S3.
Numbers of complex points and spectral width in the indirect dimensions used to acquire the 3D spectra used for the assignments of sample 1 (Table S1)
| 15N | 13C | ||||
| Experiment | Complex points | Spectral width, kHz | Complex points | Spectral width, kHz | Acquisition time, h |
| TROSY-HSQC | 256 | 4.05 | 1 | ||
| TROSY-HNcoCACB | 128 | 3.00 | 76 | 12.82 | 129 |
| TROSY-HNCACB | 160 | 3.57 | 128 | 15.38 | 200 |
| TROSY-HNcaCO | 160 | 3.57 | 64 | 3.33 | 151 |
| TROSY-HNCO | 160 | 3.57 | 64 | 3.33 | 50 |
All experiments were acquired with 1,024 complex points in the direct dimension.
Determination of the Δχ Tensor and PCS Prediction.
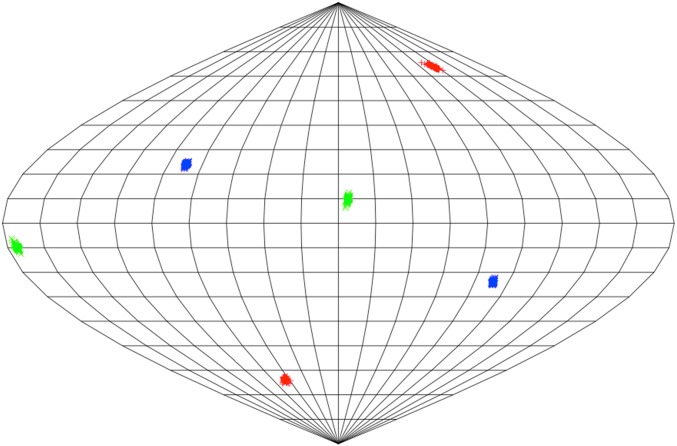
The Δχ tensor for the E195C/A199C/C334A mutant of P450cam (Table S1, samples 2–5) was calculated using an X-ray crystal structure of the closed form of P450cam (PDB ID code 3L63) (10) and Numbat software (50). The position and orientation of the lanthanoid ion were predicted according to the protocol of Skinner et al. (51), and estimates of the magnitudes of the Δχ tensor for CLaNP-7 were taken from Hiruma et al. (15). PCSs were predicted on the basis of these estimates in Numbat, and resonances in the spectra of samples 3 and 5 (Table S1) were assigned based on these predictions. Using the PCSs derived from these assignments, the magnitudes, position, and orientation of the Δχ tensor were recalculated, and further resonance assignments were obtained based on predictions from this new tensor. PCSs for any residue that moved more than 1 Å between the open and closed structures were excluded from the fitting process. This process of tensor calculation and resonance assignment yielded the final dataset used for tensor calculation containing 117 PCSs from samples 2 and 3 (Table S1), corresponding to residues that did not change position significantly between the open and closed structures of P450cam. The validity of the tensors was verified using jack-knife error analysis in Numbat, wherein 10% of the data were removed and the tensor was recalculated. The results of this analysis are shown in Fig. S5. To validate the predicted PCSs for the closed state of P450cam, the obtained magnetic susceptibility tensor was used to calculate PCSs using another X-ray crystal structure of the closed form of P450cam [PDB ID code 2CPP (2)], and the PCSs obtained were in complete agreement with the PCSs determined using the closed form of P450cam (PDB ID code 3L63).
Fig. S5.
Precision of the Δχ tensor. One hundred Δχ tensors, generated from jackknife analysis within Numbat for the fit of 117 PCSs of static nuclei in P450cam, were placed at the center of a sphere. The locations where the x, y, and z components pierce the sphere are recorded as blue, green, and red marks, respectively. The surface of the sphere is shown as a Sanson–Flamsteed equivalent projection (sometimes called a Mercator equal area projection) with gridlines at every 20° (vertical) and 10° (horizontal).
To obtain PCS predictions for the open state, residues 195 and 199 in the structures of two open-state structures [PDB ID codes 4JWS (14) and 3L62 (10)] were superposed onto the same residues in the structure of the closed state, such that the position and orientation relative to the attachment site were the same in all three structures. These residues were selected because they form the attachment site for the CLaNP-7 probe. In this way, the paramagnetic center remains fixed relative to the G-helix, moving with it when going from the closed structure to the open structure (Fig. 2B). The two open-state structures with the paramagnetic center modeled in this way were imported into Numbat; using the same values of magnitude and orientation derived for the Δχ tensor in the closed form, PCSs of the Leu amides for the two open states were predicted.
The Q scores of the fits of the PCSs were calculated as previously described (52), using the formula:
Acknowledgments
We thank Betül Ölmez for help with mutagenesis. We thank Prof. Harald Schwalbe and Dr. Frank Löhr for help with NMR spectroscopy (Center for Biomolecular Magnetic Resonance). Measurement time was supported by the European Union-funded project Bio-NMR (Grant 261863). This work was supported financially by The Netherlands Organization for Scientific Research, VICI Grant 700580441.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.d.V. is a guest editor invited by the Editorial Board.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession codes BMRB 19740 and BMRB 19763).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502351112/-/DCSupplemental.
References
- 1.Liu J, Sridhar J, Foroozesh M. Cytochrome P450 family 1 inhibitors and structure-activity relationships. Molecules. 2013;18(12):14470–14495. doi: 10.3390/molecules181214470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260(30):16122–16130. [PubMed] [Google Scholar]
- 3.Sevrioukova IF, Poulos TL. Structural biology of redox partner interactions in P450cam monooxygenase: A fresh look at an old system. Arch Biochem Biophys. 2011;507(1):66–74. doi: 10.1016/j.abb.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewer CB, Peterson JA. Single turnover kinetics of the reaction between oxycytochrome P-450cam and reduced putidaredoxin. J Biol Chem. 1988;263(2):791–798. [PubMed] [Google Scholar]
- 5.Unno M, Shimada H, Toba Y, Makino R, Ishimura Y. Role of Arg112 of cytochrome p450cam in the electron transfer from reduced putidaredoxin. Analyses with site-directed mutants. J Biol Chem. 1996;271(30):17869–17874. doi: 10.1074/jbc.271.30.17869. [DOI] [PubMed] [Google Scholar]
- 6.Ortiz de Montellano PR, De Voss JJ. Substrate oxidation by cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism and Biochemistry. 3rd Ed. Springer; Boston: 2005. pp. 183–245. [Google Scholar]
- 7.Schlichting I, et al. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287(5458):1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 8.Hays A-MA, et al. Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires. J Mol Biol. 2004;344(2):455–469. doi: 10.1016/j.jmb.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 9.Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci USA. 2001;98(22):12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49(16):3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjodin T, et al. Resonance Raman and EPR investigations of the D251N oxycytochrome P450cam/putidaredoxin complex. Biochemistry. 2001;40(23):6852–6859. doi: 10.1021/bi002510b. [DOI] [PubMed] [Google Scholar]
- 12.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450cam. Structure. 2008;16(6):916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Pochapsky SS, Pochapsky TC, Jain NU. Solution NMR structure of putidaredoxin-cytochrome P450cam complex via a combined residual dipolar coupling-spin labeling approach suggests a role for Trp106 of putidaredoxin in complex formation. J Mol Biol. 2008;384(2):349–363. doi: 10.1016/j.jmb.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi S, Li H, Poulos TL. Structural basis for effector control and redox partner recognition in cytochrome P450. Science. 2013;340(6137):1227–1230. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- 15.Hiruma Y, et al. The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography. J Mol Biol. 2013;425(22):4353–4365. doi: 10.1016/j.jmb.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Myers WK, Lee Y-T, Britt RD, Goodin DB. The conformation of P450cam in complex with putidaredoxin is dependent on oxidation state. J Am Chem Soc. 2013;135(32):11732–11735. doi: 10.1021/ja405751z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hass MAS, Ubbink M. Structure determination of protein-protein complexes with long-range anisotropic paramagnetic NMR restraints. Curr Opin Struct Biol. 2014;24:45–53. doi: 10.1016/j.sbi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Koehler J, Meiler J. Expanding the utility of NMR restraints with paramagnetic compounds: Background and practical aspects. Prog Nucl Magn Reson Spectrosc. 2011;59(4):360–389. doi: 10.1016/j.pnmrs.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertini I, Luchinat C, Parigi G. Paramagnetic constraints: An aid for quick solution structure determination of paramagnetic metalloproteins. Concepts Magn Reson. 2002;14(4):259–286. [Google Scholar]
- 20.Yagi H, et al. Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites. Structure. 2013;21(6):883–890. doi: 10.1016/j.str.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz C, Vernon R, Otting G, Baker D, Huber T. Protein structure determination from pseudocontact shifts using ROSETTA. J Mol Biol. 2012;416(5):668–677. doi: 10.1016/j.jmb.2011.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crick DJ, et al. Integral membrane protein structure determination using pseudocontact shifts. J Biomol NMR. 2015;61(3-4):197–207. doi: 10.1007/s10858-015-9899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottstein D, Reckel S, Dötsch V, Güntert P. Requirements on paramagnetic relaxation enhancement data for membrane protein structure determination by NMR. Structure. 2012;20(6):1019–1027. doi: 10.1016/j.str.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta I, Nadaud PS, Jaroniec CP. Protein structure determination with paramagnetic solid-state NMR spectroscopy. Acc Chem Res. 2013;46(9):2117–2126. doi: 10.1021/ar300360q. [DOI] [PubMed] [Google Scholar]
- 25.Sengupta I, Nadaud PS, Helmus JJ, Schwieters CD, Jaroniec CP. Protein fold determined by paramagnetic magic-angle spinning solid-state NMR spectroscopy. Nat Chem. 2012;4(5):410–417. doi: 10.1038/nchem.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131(4):756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ubbink M, Ejdebäck M, Karlsson BG, Bendall DS. The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure. 1998;6(3):323–335. doi: 10.1016/s0969-2126(98)00035-5. [DOI] [PubMed] [Google Scholar]
- 28.Rinaldelli M, et al. Simultaneous use of solution NMR and X-ray data in REFMAC5 for joint refinement/detection of structural differences. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 4):958–967. doi: 10.1107/S1399004713034160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J Am Chem Soc. 2003;125(30):9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 30.Tian F, Valafar H, Prestegard JH. A dipolar coupling based strategy for simultaneous resonance assignment and structure determination of protein backbones. J Am Chem Soc. 2001;123(47):11791–11796. doi: 10.1021/ja011806h. [DOI] [PubMed] [Google Scholar]
- 31.Chou JJ, Li S, Klee CB, Bax A. Solution structure of Ca(2+)-calmodulin reveals flexible hand-like properties of its domains. Nat Struct Biol. 2001;8(11):990–997. doi: 10.1038/nsb1101-990. [DOI] [PubMed] [Google Scholar]
- 32.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nat Struct Biol. 1997;4(4):292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 33.Bertini I, et al. Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: Calmodulin-peptide complexes as examples. J Am Chem Soc. 2009;131(14):5134–5144. doi: 10.1021/ja8080764. [DOI] [PubMed] [Google Scholar]
- 34.Liu WM, et al. A pH-sensitive, colorful, lanthanide-chelating paramagnetic NMR probe. J Am Chem Soc. 2012;134(41):17306–17313. doi: 10.1021/ja307824e. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai K, Shimada H, Hayashi T, Tsukihara T. Substrate binding induces structural changes in cytochrome P450cam. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 2):80–83. doi: 10.1107/S1744309108044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keizers PHJ, Saragliadis A, Hiruma Y, Overhand M, Ubbink M. Design, synthesis, and evaluation of a lanthanide chelating protein probe: CLaNP-5 yields predictable paramagnetic effects independent of environment. J Am Chem Soc. 2008;130(44):14802–14812. doi: 10.1021/ja8054832. [DOI] [PubMed] [Google Scholar]
- 37.Keizers PHJ, et al. A solution model of the complex formed by adrenodoxin and adrenodoxin reductase determined by paramagnetic NMR spectroscopy. Biochemistry. 2010;49(32):6846–6855. doi: 10.1021/bi100598f. [DOI] [PubMed] [Google Scholar]
- 38.Liu WM, et al. A two-armed lanthanoid-chelating paramagnetic NMR probe linked to proteins via thioether linkages. Chemistry. 2014;20(21):6256–6258. doi: 10.1002/chem.201400257. [DOI] [PubMed] [Google Scholar]
- 39.Miao Y, et al. Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation. Biophys J. 2012;103(10):2167–2176. doi: 10.1016/j.bpj.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiruma Y, et al. Hot-spot residues in the cytochrome P450cam-putidaredoxin binding interface. ChemBioChem. 2014;15(1):80–86. doi: 10.1002/cbic.201300582. [DOI] [PubMed] [Google Scholar]
- 41.OuYang B, Pochapsky SS, Pagani GM, Pochapsky TC. Specific effects of potassium ion binding on wild-type and L358P cytochrome P450cam. Biochemistry. 2006;45(48):14379–14388. doi: 10.1021/bi0617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atkins WM, Sligar SG. The roles of active site hydrogen bonding in cytochrome P-450cam as revealed by site-directed mutagenesis. J Biol Chem. 1988;263(35):18842–18849. [PubMed] [Google Scholar]
- 43.Kuznetsov VY, Poulos TL, Sevrioukova IF. Putidaredoxin-to-cytochrome P450cam electron transfer: Differences between the two reductive steps required for catalysis. Biochemistry. 2006;45(39):11934–11944. doi: 10.1021/bi0611154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asciutto EK, Madura JD, Pochapsky SS, OuYang B, Pochapsky TC. Structural and dynamic implications of an effector-induced backbone amide cis-trans isomerization in cytochrome P450cam. J Mol Biol. 2009;388(4):801–814. doi: 10.1016/j.jmb.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450cam: Practical consequences and engineering of a monomeric enzyme. Protein Eng. 1997;10(12):1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 46.Tong KI, Yamamoto M, Tanaka T. A simple method for amino acid selective isotope labeling of recombinant proteins in E. coli. J Biomol NMR. 2008;42(1):59–67. doi: 10.1007/s10858-008-9264-0. [DOI] [PubMed] [Google Scholar]
- 47.Senn H, Eugster A, Otting G, Suter F, Wüthrich K. 15N-labeled P22 c2 repressor for nuclear magnetic resonance studies of protein-DNA interactions. Eur Biophys J. 1987;14(5):301–306. doi: 10.1007/BF00254895. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 50.Schmitz C, Stanton-Cook MJ, Su XC, Otting G, Huber T. Numbat: An interactive software tool for fitting Deltachi-tensors to molecular coordinates using pseudocontact shifts. J Biomol NMR. 2008;41(3):179–189. doi: 10.1007/s10858-008-9249-z. [DOI] [PubMed] [Google Scholar]
- 51.Skinner SP, Moshev M, Hass MAS, Keizers PH, Ubbink M. PARAssign—Paramagnetic NMR assignments of protein nuclei on the basis of pseudocontact shifts. J Biomol NMR. 2013;55(4):379–389. doi: 10.1007/s10858-013-9722-1. [DOI] [PubMed] [Google Scholar]
- 52.Bashir Q, Volkov AN, Ullmann GM, Ubbink M. Visualization of the encounter ensemble of the transient electron transfer complex of cytochrome c and cytochrome c peroxidase. J Am Chem Soc. 2010;132(1):241–247. doi: 10.1021/ja9064574. [DOI] [PubMed] [Google Scholar]