Significance
Relatedness-dependent behavior modification is common among social organisms and has been a major feature of social evolution theory for decades. However, the evolutionary causes of kin discrimination are often unclear. Here, we document many spontaneous origins of kin discrimination in a social microbe that appear to arise as indirect byproducts of adaptation at other traits and we show that kin discrimination evolves by diverse genetic mechanisms.
Keywords: social evolution, nonself recognition, kin recognition, cooperation, territoriality
Abstract
Diverse forms of kin discrimination, broadly defined as alteration of social behavior as a function of genetic relatedness among interactants, are common among social organisms from microbes to humans. However, the evolutionary origins and causes of kin-discriminatory behavior remain largely obscure. One form of kin discrimination observed in microbes is the failure of genetically distinct colonies to merge freely upon encounter. Here, we first use natural isolates of the highly social bacterium Myxococcus xanthus to show that colony-merger incompatibilities can be strong barriers to social interaction, particularly by reducing chimerism in multicellular fruiting bodies that develop near colony-territory borders. We then use experimental laboratory populations to test hypotheses regarding the evolutionary origins of kin discrimination. We show that the generic process of adaptation, irrespective of selective environment, is sufficient to repeatedly generate kin-discriminatory behaviors between evolved populations and their common ancestor. Further, we find that kin discrimination pervasively evolves indirectly between allopatric replicate populations that adapt to the same ecological habitat and that this occurs generically in many distinct habitats. Patterns of interpopulation discrimination imply that kin discrimination phenotypes evolved via many diverse genetic mechanisms and mutation-accumulation patterns support this inference. Strong incompatibility phenotypes emerged abruptly in some populations but strengthened gradually in others. The indirect evolution of kin discrimination in an asexual microbe is analogous to the indirect evolution of reproductive incompatibility in sexual eukaryotes and linguistic incompatibility among human cultures, the commonality being indirect, noncoordinated divergence of complex systems evolving in isolation.
The behavior of many vertebrates (1, 2), invertebrates (3, 4) and microbes (5–7), as well as some plants (8), is altered by social encounters with conspecifics in a relatedness-dependent manner. Such kin discrimination is commonly interpreted in the context of inclusive fitness theory to have resulted from positive selection for kin discrimination per se (7, 9–11) specifically because such discrimination promotes preferential cooperation among kin that share cooperation alleles (12–14). However, despite any theoretical plausibility of kin selectionist explanations, the actual evolutionary origins of kin discrimination in natural populations are often difficult to infer due to their temporal remove (11, 15). Kin discrimination, broadly defined (SI Appendix, Methods), might also arise indirectly as a byproduct of alternative evolutionary forces (15–20). Despite decades of rigorous empirical and theoretical investigation of kin discrimination and its causes, a biological system that allows both the documentation of kin discrimination origins and causally proximate analysis of the evolutionary forces responsible for those origins has been lacking.
Among motile microbes, one form of biological kin discrimination is reduced merger of genetically distinct swarming colonies upon encounter relative to “self–self” controls. This phenomenon was first documented in 1946 with the observation of “Dienes lines” that formed between distinct nonmerging colonies of the bacterium Proteus mirabilis (21) and has since been found in several other species (20, 22). For example, a large number of such colony-merger incompatibilities evolved among closely related genotypes of the cooperative bacterium Myxococcus xanthus in a natural centimeter-scale population (20). Irrespective of their original evolutionary cause(s), the emergence of colony-merger incompatibilities in nature is likely to have profound implications for the distribution of social interactions among genotypes during cooperative processes. For example, barriers to colony merger may promote the maintenance of genotypes that are inferior social competitors in chimeric groups and promote cooperation by hindering the territorial spread of socially defective cheaters (20), as might other forms of kin discrimination (23).
In this study, we first use natural isolates of M. xanthus to test whether kin discrimination affects patterns of cooperation during fruiting body development when migrating social groups encounter one another. To do so, we quantify the frequency of chimerism among fruiting bodies that form near the border of colony territories, both for pairings of distinct colony-merger allotypes and self–self controls. Subsequently, we use experimentally evolved laboratory populations to test whether (i) generic adaptation by migrating populations (irrespective of variable selective conditions) tends to generate colony-merger incompatibilities toward an ancestral genotype and (ii) whether mere independence of the adaptive process causes allopatric populations to evolve trait differences that generate kin discrimination phenotypes upon secondary contact. We also characterize temporal patterns of kin discrimination evolution and test whether kin discrimination evolved by one or rather multiple molecular mechanisms.
Results and Discussion
Naturally Evolved Kin Discrimination Reduces Chimerism at Colony-Territory Borders.
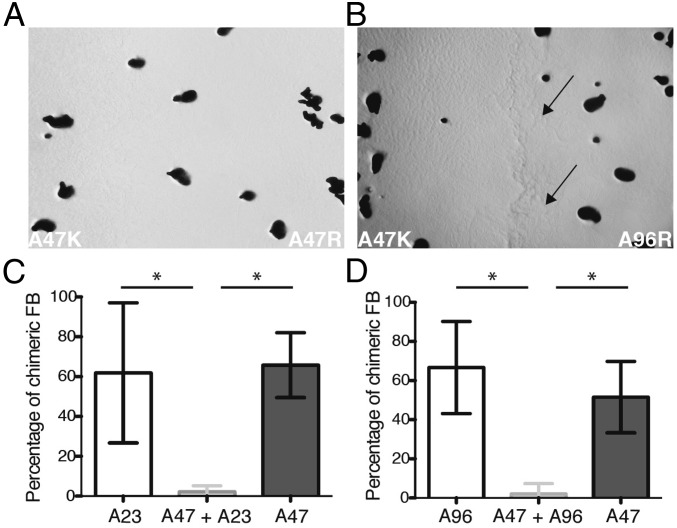
In response to nutrient deprivation, M. xanthus cells cooperatively construct multicellular, spore-bearing fruiting bodies (24). We tested whether colony-merger incompatibilities between natural isolates of M. xanthus affect the spatio-genetic structure of social interactions by reducing chimerism within fruiting bodies that form near the interface of oncoming colonies. We examined three distinct isolates (A23, A47, and A96) sampled from a centimeter-scale natural soil population in which many colony-merger incompatibilities were previously documented (20). Colonies were initiated from separate locations and allowed to swarm toward one another on a low-nutrient agar surface on which colonies grow and swarm but eventually deplete growth substrates and initiate fruiting body development (SI Appendix, Fig. S1). Colonies of the same genotype marked with distinct antibiotic-resistance markers were used as self–self controls in which no lines of demarcation between colonies were visible (e.g., Fig. 1A and SI Appendix, Fig. S1B). In contrast, nonself encounters between distinct natural isolates resulted in a visually evident line of demarcation between colonies (Fig. 1B).
Fig. 1.
Kin discrimination between natural isolates reduces chimerism among fruiting bodies near territory borders. (A) Self–self encounter control of oncoming colonies of the same natural isolate (A47) that differ only in their antibiotic-resistance marker (K and R indicate kanamycin- and rifampicin-resistant strain variants, respectively). No interface demarcation line was visible for any self–self encounters. Dark spots are individual fruiting bodies. (B) Kin discrimination phenotype between distinct natural isolates. A visible line of demarcation formed between colonies of A47 and A96. Similar barriers formed during encounters between A47 and A23. Arrows indicate the colony-interface line. (C and D) Percentage of fruiting bodies (FB) chimeric for A23 and A47 (C) or A47 and A96 (D) sampled from near the interfaces of genetically distinct colonies (center, light gray) and from self–self encounter controls [white, A23 (C) or A96 (D); dark gray, A47]. Average chimerism across independent experimental replicates is shown. Error bars represent 95% confidence intervals. *P < 0.05; two-tailed t test.
By reducing developmental coaggregation of distinct genotypes along intercolony borders, colony-merger incompatibilities should benefit genotypes that compete poorly within chimeric social groups in which mean relatedness among interactants is low relative to their performance within high-relatedness groups (e.g., pure cultures) (20). One such genotype is A23, which sporulates at a high level (similar to that of A47) in pure groups, but which makes very few spores when forcibly mixed with A47 at a 1:1 ratio before development (SI Appendix, Table S1) (20). In contrast, A47 sporulates at high levels in forced 1:1 mixes with A23 as well as in pure culture, giving A47 an extreme fitness advantage in those mixed groups.
To test the above hypothesis, we staged motility-driven encounters between colonies of A47 and A23 and assessed the frequency of chimerism among fruiting bodies that formed on both sides of the interface of oncoming colonies growing on low-nutrient medium. If a colony of A47 were capable of freely merging with an A23 colony and coaggregating into fruiting bodies that form near the colony-interface border on the A23 side of the initial encounter zone at nearly a 1:1 ratio, A23 should suffer a great fitness cost in this region due to its poor competitive performance within mixed groups containing A47 (SI Appendix, Table S1). We first quantified chimerism in self–self encounter controls between differently-marked variants of A23 and A47 and found that more than half of all sampled fruiting bodies across all experimental replicates were chimeric for both controls [14 of 25 (56%) and 31 of 59 (53%) for A23 and A47, respectively]. In contrast, in nonself encounter assays between A23 and A47, chimerism was rare among fruiting bodies that formed near colony-interface borders with only 2 of 58 fruiting bodies sampled from both sides of the demarcation border being chimeric across all replicates (Fig. 1C). On the A23 side of the demarcation line, the strong mixed-group competitor A47 was present in far fewer fruiting bodies [2 of 23 fruiting bodies sampled (9%)] than was predicted from control experiments (P < 0.0001 for deviation from expectation of 13 chimeric fruiting bodies based on A47 controls, two-tailed binomial test).
Similar results were obtained in nonself colony encounters between isolates A47 and A96. Like A23, A96 loses to A47 during developmental competitions on low nutrient medium when populations are forcibly mixed at a 1:1 ratio before development, although to a lesser degree (SI Appendix, Table S1). In the colony-encounter assays, only one fruiting body among 24 (4%) isolated near intercolony borders from the A96 side (across four replicate experiments) was found to contain A47 as well as A96 (Fig. 1D, P < 0.0001 for deviation from expectation of 12 chimeric fruiting bodies based on A47 controls, two-tailed binomial test). Like for A23, the colony-merger incompatibility between A96 and A47 benefited A96 by nearly completely preventing A47 from being present in fruiting bodies on the A96 side of the colony interface that A47 would have entered in the absence of kin discrimination. Additionally, because A96 did produce substantial numbers of spores in forced mixes (despite making fewer than A47, SI Appendix, Table S1), chimerism patterns in self–self controls would predict that, in the absence of kin discrimination, A96 should penetrate into some fruiting bodies on the A47 side of A47–A96 colony borders. However, no such chimerism was found among 21 fruiting bodies sampled.
These results demonstrate that naturally evolved social incompatibilities in M. xanthus can reduce interactions between nonkin during a cooperative process, irrespective of their evolutionary origins. The natural isolates examined here and others from the same locale diverged relatively recently (20), indicating that kin discrimination evolves over short evolutionary periods in nature, but the relative contributions of various evolutionary forces to maintaining kin discrimination traits at high frequencies in natural populations are unknown. We thus turned to a more defined evolutionary system and tested whether similar forms of kin discrimination might evolve rapidly among laboratory populations of M. xanthus with known evolutionary histories and defined selective regimes. Specifically, we asked whether replicate experimental populations that evolved in several distinct environments independently evolved colony-merger incompatibilities toward their common ancestor and/or toward one another.
Independent Adaptation Pervasively Generated Kin Discrimination.
One hundred and four populations of M. xanthus were established from independent clones of two laboratory ancestors [GJV1 (25), and GJV2, a rifampicin-resistant derivative of GJV1] and each population was allowed to evolve in one of twelve distinct agar environments that varied in surface type (hard or soft agar), nutrient level (high or low Casitone), and/or nutrient type (Casitone and/or prey bacteria) (SI Appendix, Table S2). In each treatment, either eight or twelve independent replicate populations of swarming colonies were allowed to expand outward for two week intervals, after which a sample from the leading colony edge was transferred to the center of a new plate, as described in SI Appendix, Methods and Fig. S2. This transfer protocol was shared by all treatments and selected for numerical dominance at the leading swarm edge.
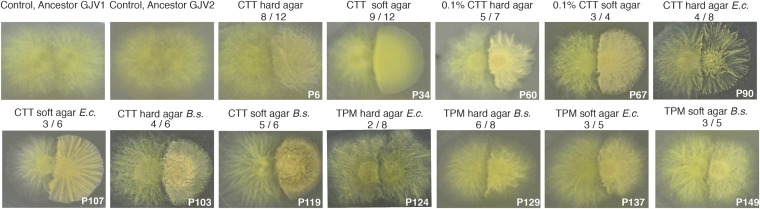
We screened for kin discrimination (hereafter also “KD”) phenotypes among 88 of these evolved populations in the form of merger incompatibilities between swarming colonies growing on soft nutrient-rich agar (Fig. 2). Evolved populations were first scored for kin discrimination phenotypes in encounters with their own experimental ancestor (hereafter “KD-A”). A majority of colonies grown from samples of evolved populations [55 of 88 examined (63%), SI Appendix, Table S2] did not merge with ancestral colonies in the same manner as paired colonies of the ancestor in self–self encounters that freely merge into a continuous swarm lacking territorial demarcation (Fig. 2). KD-A phenotypes evolved independently in at least half of all replicate populations examined within every evolutionary treatment except one (TPM hard agar, predation on Escherichia coli), in which KD-A evolved in only two populations (Fig. 2 and SI Appendix, Table S2). No significant effects of evolutionary surface type, nutrient level, nutrient type, or prey type on the frequency of KD-A evolution were evident (P > 0.05 in all cases).
Fig. 2.
Kin discrimination phenotypes evolved de novo in all evolutionary treatments. Examples of kin discrimination phenotypes between colonies of evolved populations from each evolutionary treatment and their ancestor (KD-A) on high-nutrient agar are shown. White inset text depicts respective labels of evolved populations. Evolutionary treatments are stated above each picture. The two first panels show colony-merger phenotypes of self–self encounter controls for the two ancestral variants. Ratios show the proportion of examined populations within each evolutionary treatment that exhibited consistent KD-A phenotypes (SI Appendix, Table S2). B.s., B. subtilis; CTT, media containing Casitone; E.c., E. coli; TPM, starvation buffer.
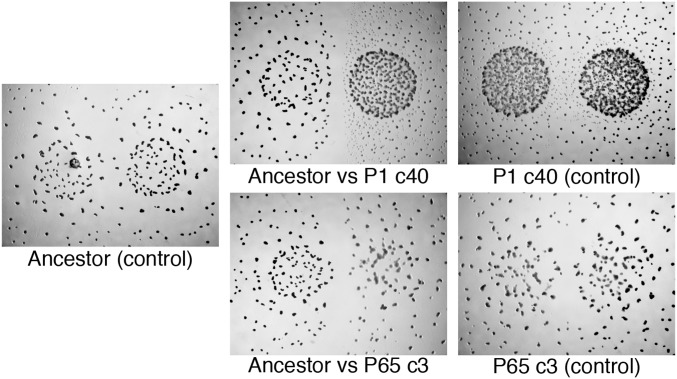
We tested whether, like natural isolates, experimentally evolved populations also exhibit kin discrimination phenotypes at colony borders during multicellular fruiting body development under low-nutrient conditions. Out of 28 populations examined, 15 (54%) were found to exhibit clear KD-A phenotypes under these conditions (e.g., Fig. 3). For several evolved populations examined, fruiting bodies were absent from an area along the territorial interface of evolved vs. ancestor colonies, whereas fruiting bodies did form in the same respective regions of self–self encounter controls with both ancestral and evolved populations (Fig. 3).
Fig. 3.
Kin discrimination phenotypes between colonies of evolved populations and their ancestor (KD-A) on low-nutrient plates. Fruiting bodies failed to form along evolved-ancestor interface zones but do form along the interface zones of self–self controls. “c” (cycle) indicates evolutionary time point.
Temporal Patterns of Kin Discrimination Emergence.
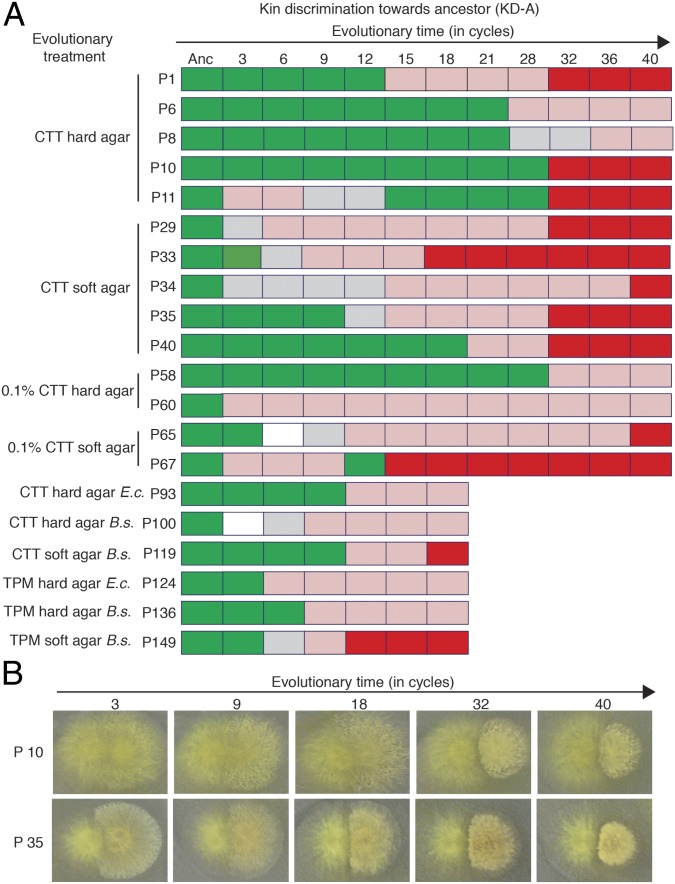
To determine whether temporal patterns of initial KD-A evolution were similar or divergent across populations, we tested for the presence of KD-A in temporally intermediate population samples in 20 populations. The timing of KD-A emergence was found to vary greatly, including across populations from the same evolutionary treatment (Fig. 4). In some cases, KD-A traits were already present within three cycles (6 wk) of selection (e.g., P60 and P67), whereas in other populations KD-A was not evident until after 32 cycles (64 wk, e.g., P10 and P58). Of the 20 populations examined for temporal dynamics of KD-A appearance, one population transitioned directly from merger compatibility to a visually striking form of incompatibility (P10), whereas nine first exhibited incompatibility phenotypes of intermediate visual strength (but that were nonetheless clear and consistent) before evolving more visually conspicuous incompatibilities. Eight populations transitioned just once to incompatibility phenotypes of intermediate strength, and two populations reevolved merger compatibility with their ancestor after having initially evolved intermediate phenotypes but then subsequently transitioned to strong incompatibility phenotypes (P11 and P67) (Fig. 4). Variation in the visual conspicuousness of incompatibility phenotypes indicate that they should not be considered merely in terms of discrete presence-or-absence states, but as phenotypes that vary in both population level morphology (Fig. 2) and visual prominence (Fig. 4).
Fig. 4.
Temporal kin discrimination phenotype-emergence patterns for sample populations. (A) Kin discrimination phenotypes between evolved populations and their ancestor (KD-A) were classified into four qualitative categories: freely merging (green), inconsistent (gray), incompatible (light red), and strongly incompatible (red). White boxes indicate that the cycle could not be analyzed. (B) Ancestral colonies (Left) encountering P10 or P35 samples (Right) from different evolutionary time points.
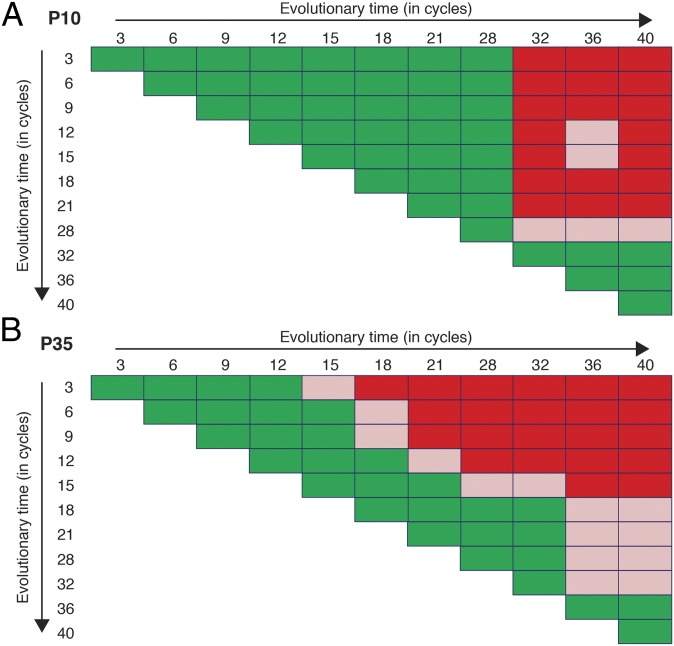
Within any given population over time, a KD-A incompatibility might be caused by a single mutation and then remain relatively constant over time or might rather be strengthened over time by multiple successive mutations. To distinguish between these possibilities, for a subset of populations we paired temporal samples from all evolutionary time points in all possible combinations and screened for kin discrimination between those samples (“KD-T” for kin discrimination among time-point samples). As observed for KD-A, distinct temporal patterns of KD-T were strikingly evident. In roughly half of the populations, an abrupt evolutionary transition occurred such that all temporal samples from after the transition were incompatible with all samples stored before the transition (Fig. 5A and SI Appendix, Fig. S3). In other populations, KD-T phenotypes evolved gradually such that their presence and strength among intermediate time-point samples correlated with the temporal distance between samples (and thus, presumably, genetic distance also; Fig. 5B and SI Appendix, Fig. S3). These contrasting temporal patterns suggest that kin discrimination in M. xanthus can evolve both in the form of all-or-nothing “kind discrimination” that is determined by distinct allelic states at a single locus and in more graded forms in which the degree of social incompatibility scales more gradually with overall genomic relatedness (5–7).
Fig. 5.
Kin discrimination phenotypes evolved both abruptly and gradually. Kin discrimination between temporal samples of the same population (KD-T) evolved abruptly in P10 (A) but gradually in P35 (B). Colony-encounter phenotype classifications for all possible pairs of time points within each evolved population are shown.
Kin Discrimination Evolved by Diverse Genetic Mechanisms.
To determine whether populations evolved KD-A by similar or distinct mechanisms, we staged colony encounters for 117 within-treatment pairs of terminal evolved populations (i.e., paired populations were descended independently from the same ancestor in the same environment). If the failure of independently evolved populations to each merge with their common ancestor is due to the same molecular mechanism, those populations are likely to be compatible with one another. Any failure of colonies of independently evolved populations to merge with each other (hereafter “KD-B” for between-population kin discrimination) implies that either they evolved distinct KD-A mechanisms that also cause the KD-B phenotype or they evolved distinct KD-A and KD-B mechanisms. More than half of all such population pairings (62 of 117, 53%) revealed clear KD-B phenotypes (SI Appendix, Table S3). From the pervasiveness of KD-B evolution across treatments, we infer that mere independence of the adaptive process is sufficient to generate diverse molecular forms of kin discrimination (SI Appendix, Table S3).
To document this molecular diversity at the genomic level, we sequenced clones from all 24 terminal populations from the CTT hard and soft agar treatments and analyzed mutation patterns (SI Appendix, Tables S4 and S5 and Fig. S4). Seventeen of these 24 populations (67%) evolved KD-A (SI Appendix, Table S2). Excluding the P29 clone (which is an apparent “mutator” carrying 435 mutations; ref. 26), evolved clones had accumulated an average of 13 mutations, a large majority of which altered amino acid sequences (SI Appendix, Table S4). No mutation was found in traA, a gene that encodes a protein implicated in outer membrane exchange (OME) among M. xanthus cells and has been hypothesized to have evolved as an adaptive greenbeard system for kin recognition in natural populations (27). However, the absence of traA mutations in our populations and the occurrence of colony-merger incompatibilities between natural isolates that carry the same traA allele (e.g., A47 and A96 in Fig. 1B, and ref. 27) together suggest that TraA functional incompatibilities evolved indirectly in evolutionary isolation after other social barriers were already established and do not represent a directly selected greenbeard system (27).
Fourteen genes were mutated in at least four populations and four genes were mutated in eight or more (SI Appendix, Table S5). Among the latter, only one locus, the CRISPR-associated gene cmr4, was mutated only in populations that had evolved KD-A (SI Appendix, Table S5). We thus sequenced cmr4 in all terminal populations and found two significant patterns. First, all cmr4 polymorphisms were identical (cmr4-P72H, see SI Appendix), and second, this mutation was present only in populations descended from four of the twelve ancestral subclones used to initiate our experiments (GJV1.1, 1.4, 1.5, and 1.6, SI Appendix, Table S2). Sequencing of cmr4 in ancestral subclones revealed that the mutation was already present in these four ancestors but not in any other. This result indicates that the source GJV1 culture from which the ancestral subclones were derived was polymorphic for this crm4 mutation. However, despite the fact that cmr4-P72H did not evolve via selection after the initiation of experimental evolution, populations carrying this mutation showed a statistically disproportionate tendency to evolve KD-A (binomial test, P = 0.002). Thus, although the cmr4-P72H mutation clearly does not directly generate KD-A itself, it may increase the probability of evolving KD-A. (See SI Appendix for additional discussion of the cmr4 mutation.)
As noted previously, the recurrent evolution of KD-B (i.e., multiple incompatibility allotypes among independent populations that share a common ancestor and evolved in the same selective conditions) implies that multiple distinct mechanisms of colony incompatibility evolved. Patterns of mutation among the 24 genome-sequenced populations are consistent with this implication. First, there is no single locus that was mutated in all 17 of the populations that evolved KD-A. Second, among seven genes that mutated independently in five or more populations after the initiation of experimental evolution (SI Appendix, Table S5), all of them were mutated in multiple populations that did not evolve KD-A as well as in populations that did. These mutation patterns further indicate that kin discrimination traits evolved due to mutations in different loci across populations.
To demonstrate genetically divergent evolution of kin discrimination with finer temporal resolution, multiple clones from each of three populations showing KD-A (P10, P33, and P34) were isolated independently from both the generational time point at which KD-A first appeared (cycles 32, 9, and 9, respectively, Fig. 4) and the prior time point. [For P34, cycle 9 was the earliest cycle at which some individual sampled clones exhibited clear KD-A, although the KD-A phenotype of whole-population samples was not fully consistent until cycle 15 (Fig. 4A).] These clones were then tested for the presence or absence of all mutations detected in the terminal clone from the respective populations. Two mutations each first appeared at the first KD-A time points in P33 and P34, whereas four did so in P10 (SI Appendix, Table S6). Importantly, none of the eight mutations that are candidates for direct causation of KD-A in these three populations occurred in the same gene, further indicating that KD-A evolved by diverse molecular mechanisms across independent populations (SI Appendix, Table S6). As the genetic bases of kin-discriminatory behaviors become defined in an increasing number of social species (28–30), it will be of interest to determine how frequently kin discrimination is caused by a high level of allelic richness at only a single locus or variation at multiple loci.
Evolutionary Causation.
The many KD-B phenotypes documented in this study are indirect byproducts of evolutionary processes that occurred independently within fully isolated populations. Whether by direct selection, hitchhiking, or genetic drift, distinct social-gene alleles that rose to high frequency in completely allopatric populations reduce social compatibility of those populations upon secondary contact. In this regard, KD-B incompatibilities are conceptually analogous to Bateson–Dobzhansky–Muller (BDM) allele incompatibilities that evolve indirectly and reduce reproductive compatibility between divergent populations of sexual eukaryotes upon secondary contact (31, 32).
Just as BDM incompatibilities strengthen incipient species boundaries in sexual organisms, early barriers to social merger and cooperation in M. xanthus may constitute first steps in a long-term process of increasing social incompatibility. This view is supported by those populations in our experiments in which the likelihood of compatibility between time-point samples was found to be a function of evolutionary distance (e.g., P35, Fig. 5). Although BDM reproductive incompatibilities can result from differential selective forces, they can also result from populations following distinct genetic pathways of adaptation to an identical environment (33) just as indirectly generated forms of KD-B arose in our populations.
Similarly, incompatibilities among distinct lineages of other biological and cultural systems are proposed to have originated indirectly, including hybrid necrosis in plants (34), somatic cell incompatibilities in animals (16), and linguistic incompatibilities in humans (35). The commonality in our analogy between the KD-B incompatibilities that evolved in experimental populations of an asexual prokaryote and BDM incompatibilities in sexual eukaryotes or incompatibilities in many other complex systems (e.g., human language) lies in the indirectness of their evolution (36).
The rapid, pervasive and indirect evolution of KD-B phenotypes documented here among experimental populations of M. xanthus may help explain the great diversity of social allotypes found in natural populations (20). Crozier proposed that some “extrinsic” factor other than direct selection on kin-discriminatory interactions is necessary for the maintenance of diverse social allotypes because positive frequency-dependent selection on cooperative traits is expected to favor the most common social allotype (11, 16, 37, 38). Our results suggest that, at least in some biological systems, stochastic variation in adaptive trajectories may generate novel allotypes more rapidly than the most common allotype in a local population can increase toward fixation.
Direct selection favoring biological incompatibilities per se can also occur and adaptive explanations for interaction incompatibilities among other noncognitive organisms have been proposed, including allorecognition specificity among colonial invertebrates (15, 39) and vegetative fusion incompatibilities in fungi (40–43). In sexual systems, reproductive barriers can be directly reinforced by selection when hybridization upon secondary contact is maladaptive (44, 45).
In our experiments, it is possible that some mutations causing KD-A might have been adaptive in the social environment in which they first arose specifically because of kin-discriminatory effects. In this scenario, a KD-A mutation would be adaptive because it both (i) generated a fitness advantage requiring preferential interaction among clone-mates sharing the mutation and (ii) itself mechanistically caused such preferential interaction (46). Due to the large number of independent KD-A origins (n = 55) and the diversity of their molecular mechanisms, we consider it unlikely a priori that all 55 initial KD-A mutations rose to high frequency specifically by selection on kin-discriminatory effects of those mutations, although this hypothesis may apply in some cases.
Importantly, we note that initially latent colony-incompatibility traits that do not first evolve because they are specifically favored by selection may later be subject to direct selection in newly encountered social contexts. Some indirectly evolved kin discrimination traits may represent exaptations (47) that were initially nonadaptive (with respect to their potential to cause kin discrimination) but later proved adaptive during interactions with genotypes that are more competitive in mixed groups than in segregated groups. Parsing out temporally variable evolutionary forces acting on a given kin-discrimination trait will often be exceedingly difficult. However, given both the ease with which latent kin discrimination traits can arise indirectly (Fig. 2) and the strong effects that such traits can exert on the spatio-genetic distribution of social interactions among local neighbors (Figs. 1–4), we expect that such temporal variation in the forces maintaining kin discrimination in natural populations is common.
Studies examining interactions between conspecific natural isolates of social microbes under laboratory conditions are increasingly common (6, 20, 48, 49) and adaptive hypotheses for the origin of interaction phenotypes are often proposed, with varying degrees of support. Our results empirically demonstrate that kin discrimination phenotypes that appear to be consistent with the hypothesis of a directly adaptive origin mediated by kin selection can instead readily originate indirectly, simply as a result of differential independent adaptation to a common environment. In a similar vein, quantitatively striking examples of social cheating have previously been shown to evolve as indirect byproducts of adaptation to an unstructured habitat rather than as a result of selection for social cheating per se (50). Distinct natural strains of social microbes that are experimentally forced to compete during a focal social process (e.g., fruiting body development in Myxococcus bacteria or Dictyostelium amoebae) often exhibit unequal fitness (20, 48, 51, 52). Our results suggest that such fitness inequalities may often be indirect byproducts of alternative evolutionary forces rather than the result of the winning strains having undergone selection for increased within-group competitiveness during the focal social process.
Difficulties inherent to inferring what evolutionary forces have shaped naturally evolved traits in general (53) apply with equal force to social traits specifically. Our results highlight that the hypothesis that kin-discriminatory behaviors or other social interaction phenotypes originated by indirect processes should not be excluded without compelling positive evidence that they evolved primarily as directly selected social adaptations. Many extant social interaction traits are likely to have been shaped by complex combinations of indirect causes and direct selection.
Methods
To assess whether evolved populations discriminate between themselves and their ancestor (KD-A), samples from other evolutionary time-points from the same population (KD-T), or other evolved populations from the same evolutionary environment (KD-B), we staged colony-encounter assays as follows. One day before each assay, cells were inoculated in liquid media and plates were prepared by pouring 20 mL of CTT soft agar (0.5% agar) into 9-cm-diameter Petri dishes unless otherwise specified. To start each assay, 10 µL of each culture (previously adjusted to ∼5 × 109 cells per mL) were spotted 1 cm apart from each other. In experimental assays testing for KD-A, one spot on a plate contained the ancestor and the other contained an evolved population. In experimental assays testing for KD-B or KD-T, the two spots on a plate were from distinct evolved populations or different evolutionary time points within the same population, respectively. In control assays, two spots of the same genotype (or population sample) were tested for colony merger. After spotting, culture samples were allowed to dry in a laminar flow hood and plates were then incubated for three days, after which colonies were examined for the presence or absence of a clearly discernable line of demarcation between swarms.
For more detailed methods, see SI Appendix.
Supplementary Material
Acknowledgments
We thank the Genomic Diversity Center (GDC) at ETH Zürich and especially Jean-Claude Walser for his help in whole-genome sequence analysis. This work was supported in part by National Institutes of Health Grant GM079690 (to G.J.V.) and an EMBO Long-Term Fellowship (to O.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502251112/-/DCSupplemental.
References
- 1.Sherman PW, Reeve HK, Pfennig DW. Recognition systems. In: Krebs JR, Davies NB, editors. Behavioural Ecology: An Evolutionary Approach. 4th Ed Blackwell Scientific Publications; Oxford, UK: 1997. [Google Scholar]
- 2.Wu HM, Holmes WG, Medina SR, Sackett GP. Kin preference in infant Macaca nemestrina. Nature. 1980;285(5762):225–227. doi: 10.1038/285225a0. [DOI] [PubMed] [Google Scholar]
- 3.Buss LW, Grosberg RK. Morphogenetic basis for phenotypic differences in hydroid competitive behavior. Nature. 1990;343(6253):63–66. [Google Scholar]
- 4.Keller L, Ross KG. Selfish genes: A green beard in the red fire ant. Nature. 1998;394(6693):573–575. [Google Scholar]
- 5.Kuehne HA, Murphy HA, Francis CA, Sniegowski PD. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr Biol. 2007;17(5):407–411. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- 6.Ostrowski EA, Katoh M, Shaulsky G, Queller DC, Strassmann JE. Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 2008;6(11):e287. doi: 10.1371/journal.pbio.0060287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 8.Karban R, Shiojiri K, Ishizaki S, Wetzel WC, Evans RY. Kin recognition affects plant communication and defence. P R Soc B. 2013;280:20123062. doi: 10.1098/rspb.2012.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusini J, Robin C, Franc A. To fuse or not to fuse? An evolutionary view of self-recognition systems. J Phylogen Evolution Biol. 2013;1:103. [Google Scholar]
- 10.Cadavid LF. Self/non-self discrimination in basal metazoa: Genetics of allorecognition in the hydroid Hydractinia. Integr Comp Biol. 2005;45(4):623–630. doi: 10.1093/icb/45.4.623. [DOI] [PubMed] [Google Scholar]
- 11.Rousset F, Roze D. Constraints on the origin and maintenance of genetic kin recognition. Evolution. 2007;61(10):2320–2330. doi: 10.1111/j.1558-5646.2007.00191.x. [DOI] [PubMed] [Google Scholar]
- 12.Axelrod R, Hammond RA, Grafen A. Altruism via kin-selection strategies that rely on arbitrary tags with which they coevolve. Evolution. 2004;58(8):1833–1838. doi: 10.1111/j.0014-3820.2004.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7(1):1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 14.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4(8):597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 15.Grosberg RK. The evolution of allorecognition specificity in clonal invertebrates. Q Rev Biol. 1988;63(4):377–412. [Google Scholar]
- 16.Crozier RH. Genetic clonal recognition abilities in marine invertebrates must be maintained by selection for something else. Evolution. 1986;40(5):1100–1101. doi: 10.1111/j.1558-5646.1986.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 17.Grant V. Selective origin of incompatibility barriers in plant genus Gilia. Am Nat. 1966;100(911):99–104. [Google Scholar]
- 18.Pfennig DW, Collins JP, Ziemba RE. A test of alternative hypotheses for kin recognition in cannibalistic tiger salamanders. Behav Ecol. 1999;10:436–443. [Google Scholar]
- 19.Smith J, Van Dyken JD, Velicer GJ. Nonadaptive processes can create the appearance of facultative cheating in microbes. Evolution. 2014;68(3):816–826. doi: 10.1111/evo.12306. [DOI] [PubMed] [Google Scholar]
- 20.Vos M, Velicer GJ. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr Biol. 2009;19(20):1763–1767. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dienes L. Reproductive processes in Proteus cultures. Proc Soc Exp Biol Med. 1946;63(2):265–270. doi: 10.3181/00379727-63-15570. [DOI] [PubMed] [Google Scholar]
- 22.Munson EL, Pfaller MA, Doern GV. Modification of dienes mutual inhibition test for epidemiological characterization of Pseudomonas aeruginosa isolates. J Clin Microbiol. 2002;40(11):4285–4288. doi: 10.1128/JCM.40.11.4285-4288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho HI, Hirose S, Kuspa A, Shaulsky G. Kin recognition protects cooperators against cheaters. Curr Biol. 2013;23(16):1590–1595. doi: 10.1016/j.cub.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan R, Sarwar Z, Garza AG, Kroos L. Developmental Gene Regulation. In: Yang Z, Higgs P, editors. Myxobacteria: Genomics, Cellular and Molecular Biology. Caister Academic Press; 2013. [Google Scholar]
- 25.Velicer GJ, et al. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc Natl Acad Sci USA. 2006;103(21):8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schofield MJ, Hsieh P. DNA mismatch repair: Molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 27.Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 2013;9(11):e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benabentos R, et al. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr Biol. 2009;19(7):567–572. doi: 10.1016/j.cub.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321(5886):256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirose S, Benabentos R, Ho HI, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science. 2011;333(6041):467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 32.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11(3):175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 33.Unckless RL, Orr HA. Dobzhansky-Muller incompatibilities and adaptation to a shared environment. Heredity (Edinb) 2009;102(3):214–217. doi: 10.1038/hdy.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5(9):e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagel M. Human language as a culturally transmitted replicator. Nat Rev Genet. 2009;10(6):405–415. doi: 10.1038/nrg2560. [DOI] [PubMed] [Google Scholar]
- 36.Wright KM, Lloyd D, Lowry DB, Macnair MR, Willis JH. Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biol. 2013;11(2):e1001497. doi: 10.1371/journal.pbio.1001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert OM. Histocompatibility as adaptive response to discriminatory within-organism conflict: A historical model. Am Nat. 2015;185(2):228–242. doi: 10.1086/679442. [DOI] [PubMed] [Google Scholar]
- 38.Holman L, van Zweden JS, Linksvayer TA, d’Ettorre P. Crozier’s paradox revisited: Maintenance of genetic recognition systems by disassortative mating. BMC Evol Biol. 2013;13:211. doi: 10.1186/1471-2148-13-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes RN, Manríquez PH, Morley S, Craig SF, Bishop JDD. Kin or self-recognition? Colonial fusibility of the bryozoan Celleporella hyalina. Evol Dev. 2004;6(6):431–437. doi: 10.1111/j.1525-142X.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 40.Bégueret J, Turcq B, Clavé C. Vegetative incompatibility in filamentous fungi: Het genes begin to talk. Trends Genet. 1994;10(12):441–446. doi: 10.1016/0168-9525(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 41.Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. Reproductive isolation and phylogenetic divergence in Neurospora: Comparing methods of species recognition in a model eukaryote. Evolution. 2003;57(12):2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. [DOI] [PubMed] [Google Scholar]
- 42.Glass NL, Kuldau GA. Mating type and vegetative incompatibility in filamentous ascomycetes. Annu Rev Phytopathol. 1992;30:201–224. doi: 10.1146/annurev.py.30.090192.001221. [DOI] [PubMed] [Google Scholar]
- 43.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64(3):489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noor MAF. Reinforcement and other consequences of sympatry. Heredity (Edinb) 1999;83(Pt 5):503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- 45.Turner E, Jacobson DJ, Taylor JW. Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet. 2011;7(8):e1002204. doi: 10.1371/journal.pgen.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackermann M, Chao L. Evolution of cooperation: Two for one? Curr Biol. 2004;14(2):R73–R74. doi: 10.1016/j.cub.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 47.Gould SJ, Vrba ES. Exaptation - a missing term in the science of form. Paleobiology. 1982;8(1):4–15. [Google Scholar]
- 48.Buttery NJ, Rozen DE, Wolf JB, Thompson CR. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Curr Biol. 2009;19(16):1373–1377. doi: 10.1016/j.cub.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 49.Kraemer SA, Velicer GJ. Social complementation and growth advantages promote socially defective bacterial isolates. P B Sci R Soc. 2014;281(1781):20140036. doi: 10.1098/rspb.2014.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404(6778):598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 51.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408(6815):965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 52.Rendueles O, Amherd M, Velicer GJ. Positively frequency-dependent interference competition maintains diversity and pervades a natural population of cooperative microbes. Curr Biol. 2015 doi: 10.1016/j.cub.2015.04.057. [DOI] [PubMed] [Google Scholar]
- 53.Williams GC. Adaptation and Natural Selection. A Critique of Some Current Evolutionary Thought. Princeton University Press; Princeton, NJ: 1966. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.