Abstract
Background
Carotenoid compounds play essential roles in plants such as protecting the photosynthetic apparatus and in hormone signalling. Coloured carotenoids provide yellow, orange and red colour to plant tissues, as well as offering nutritional benefit to humans and animals. The enzyme phytoene synthase (PSY) catalyses the first committed step of the carotenoid biosynthetic pathway and has been associated with control of pathway flux. We characterised four PSY genes found in the apple genome to further understand their involvement in fruit carotenoid accumulation.
Results
The apple PSY gene family, containing six members, was predicted to have three functional members, PSY1, PSY2, and PSY4, based on translation of the predicted gene sequences and/or corresponding cDNAs. However, only PSY1 and PSY2 showed activity in a complementation assay. Protein localisation experiments revealed differential localization of the PSY proteins in chloroplasts; PSY1 and PSY2 localized to the thylakoid membranes, while PSY4 localized to plastoglobuli. Transcript levels in ‘Granny Smith’ and ‘Royal Gala’ apple cultivars showed PSY2 was most highly expressed in fruit and other vegetative tissues. We tested the transient activation of the apple PSY1 and PSY2 promoters and identified potential and differential regulation by AP2/ERF transcription factors, which suggested that the PSY genes are controlled by different transcriptional mechanisms.
Conclusion
The first committed carotenoid pathway step in apple is controlled by MdPSY1 and MdPSY2, while MdPSY4 play little or no role in this respect. This has implications for apple breeding programmes where carotenoid enhancement is a target and would allow co-segregation with phenotypes to be tested during the development of new cultivars.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0573-7) contains supplementary material, which is available to authorized users.
Keywords: Apple, Carotenoids, Fruit skin, Fruit flesh, Phytoene, Phytoene synthase, Promoter, Transient activation
Background
Carotenoid compounds have important roles in biochemical processes in plants such as light harvesting during photosynthesis and protecting the photosynthetic apparatus against damage. As secondary metabolites, these compounds accumulate in plant tissues to give attractive colours, which facilitate pollination and seed dispersal. In fruit and other plant tissues, colour is of high consumer and commercial value [1]. Carotenoids have potential health benefits in reducing the risk of diseases [2, 3]. In food crops, carotenogenesis contributes to nutritional quality through accumulation of alpha- and beta-carotene, which are major sources of pro-vitamin A [4–7]. Carotenoids serve as substrates for the biosynthesis of apocarotenoids such as abscisic acid and strigolactone which mediate stress and developmental signalling responses [8, 9].
Phytoene synthase (PSY) plays a pivotal role in the carotenoid pathway as the first committed step and acts to control flux through the pathway [10, 11]. The number of PSY genes differs between species, a result of duplication events, which have significance for function and modulation of carotenogenesis. Arabidopsis has a single PSY gene while two PSY genes have been reported for carrot [12, 13]. Tomato, cassava and members of the grass family, such as maize, rice and sorghum have three paralogs [14–17]. Since PSY plays such an important role in the carotenoid biosynthetic pathway, the implication of this gene duplication is not insignificant and may be related to producing carotenoids for diverse roles as suggested earlier [18, 19]. The presence of multiple PSYs in plants has resulted in distinct roles acquired by individual members [6]. In maize, ZmPSY1 is important for carotenoid accumulation in endosperm as well as stress-induced carotenogenesis in green tissues. ZmPSY2, which is upregulated by light, is associated with photosynthesis [20]. ZmPSY3 is stress-induced and specifically expressed in roots [21]. Similarly in rice, the multiple PSYs have distinct as well as overlapping roles. Rice OsPSY1 and OsPSY2 are involved in carotenoid biosynthesis under phytochrome control in green tissues, and as found in maize, OsPSY3 was also up-regulated during stress treatments [22]. In tomato, virus-induced gene silencing (VIGS) of SlPSY1 resulted in a yellow flesh phenotype, with complete disappearance of linear carotenoids and only trace amounts of other carotenoid compounds [15, 23]. On the other hand, silencing of SlPSY2 and SlPSY3 did not result in any obvious fruit phenotype, suggesting tomato PSY1 has a dominant role in the fruit. This is probably because tomato SlPSY2 functions mainly in chloroplast-containing tissues [24].
Apples are well known for their metabolites such as flavonoids and vitamin C, which are beneficial health compounds. Fruit flavour and colour are major apple breeding objectives because of their importance in determining consumer preferences [25–29]. In general, volatile apocarotenoids such as β-ionone and geranylacetone contribute to flavour while the accumulation of coloured carotenoid compounds in the fruit, together with chlorophyll and anthocyanins are responsible for fruit colour [30–32]. In a typical red skinned apple fruit, such as ‘Royal Gala’, the anthocyanin and carotenoid concentrations increase at maturity, while the chlorophyll concentration decreases [33]. We previously characterized the apple carotenoid pathway and found strong correlation between carotenoid accumulation in apple fruit and transcript levels of three genes, zeta-carotene isomerase (Z-ISO), carotenoid isomerase (CRTISO) and lycopene epsilon cyclase (LCYE) [34]. However, given the indispensable role PSY genes play in the carotenoid pathway as the first committed step, we have now also characterised the apple PSYs by assessing any overlapping or specialized roles they might have in fruit, from enzymatic function, protein localization and transcript levels. We also examined the transcriptional activation of the PSY promoters by APETALA2 domain/ethylene response transcription factors (AP2/ERF).
Methods
Sequence analysis
Apple PSY genes were identified in the phytozome database (http://phytozome.jgi.doe.gov). Multiple amino acid sequence alignments were performed with ClustalW [35] using default parameters and were manually adjusted in Geneious (www.geneious.com). Transit targeting peptides of full length PSYs were predicted using the ChloroP bioinformatic tool [36]. Phylogenetic analysis was conducted using MEGA6 [37]. Evolutionary relationships were inferred using the Neighbor-joining method, with 1000 Bootstrap re-sampling strategy. The database accession numbers of sequences used are: AtPSY (AAA32836), EjPSY1 (KF922363), EjPSY2A (KF922364), EjPSY3 (KF922367), MdPSY1 (KT189149 corresponds to MDP0000177623), MdPSY2 (KT189150 corresponds to MDP0000237124), MdPSY3 (KT189151 corresponds to MDP0000151924), MdPSY4 (KT189152 corresponds to MDP0000288336), MdSQS1 (AGS78117), MdSQS2 (AGS78118), MePSY1 (ACY42666), MePSY2 (ACY42670), MePSY3 (cassava4.1_033101m available at http://phytozome.jgi.doe.gov), OsPSY1 (AAS18307), OsPSY2 (AK073290), OsPSY3 (DQ356431), SbPSY1 (AY705389), SbPSY3 (AAW28997), SlPSY1 (ABM45873), SlPSY2 (ABU40771), SlPSY3 (Solyc01g005940), ZmPSY1 (Zea mays) AAX13806, ZmPSY2 (AAQ91837), ZmPSY3 (DQ356430).
Plasmids and functional complementation
The pACCAR25ΔcrtB (ΔcrtB) plasmid has all the genes needed to produce zeaxanthin diglucoside, except for a gene encoding PSY [38]. The pACCRTE plasmid, carrying the bacterial crtE gene to produce geranylgeranyl pyrophosphate (GGPP), was constructed by removing the crtY, crtI, crtB genes from pAC-BETA [39] by digesting with SalI followed by religation of the vector. The pAC-PHYT vector for producing phytoene in bacteria was used as a positive control [40]. To test functional complementation, fruit cDNA fragments of MdPSY1, MdPSY2, MdPSY4, without predicted transit peptides, were amplified and cloned into the pET200/D TOPO vector (Life Technologies, Carlsbad, California, USA) to give pETPSY1ΔTP, pETPSY2ΔTP and pETPSY4ΔTP constructs respectively. Competent cells of DH5α, carrying either the ΔcrtB or pACCRTE, were co-transformed with the PSY constructs. Transformed cells were selected on LB plates with 34 mg/L chloramphenicol and 50 mg/L kanamycin at 37 °C overnight. Fifty mL of Luria-Bertani (LB) broth was inoculated with one mL of overnight bacterial culture, supplemented with antibiotics and grown at 37 °C for 8 h before induction with 10 mM IPTG. This was followed by incubation at 28 °C in the dark, with shaking at 200 rpm for 48 h and then an additional 48 h without shaking. Cultures were centrifuged for 15 min in the dark at 4000 x g, resuspended with five mL of methanol containing 1 % butylated hydroxytoluene (BHT) and sonicated twice with 30s pulses on ice, at 50 % of output power using a Microson Ultrasonic Cell Disruptor XL2005 (Heat Systems, Farmingdale, New York) equipped with a tapered 3 mm microtip. Samples were centrifuged at 4000 x g to pellet disrupted cells and the supernatant flushed to dryness with nitrogen. The dried extract was resuspended with acetone for high-performance liquid chromatography (HPLC) analysis.
Protein localization and fluorescent confocal microscopy
Full-length ‘Royal Gala’ apple fruit cDNAs were amplified by polymerase chain reaction (PCR) and cloned into a Gateway destination vector, pGWB441 [41, 42], in-frame with enhanced-yellow fluorescent protein under the Cauliflower Mosaic Virus 35S promoter. Transient expression of fluorescent fusion proteins in maize etiolated leaf protoplasts was visualized with a DMI6000B inverted confocal microscope with TCS SP5 system (Leica Microsystems CMS) as described previously [43]. Images were obtained by combining several confocal Z-planes.
Carotenoid and chlorophyll extraction
A 50 mg dry weight (DW) sample of powdered freeze-dried material from each sample was moistened with water (approx 100 μl) and first extracted overnight in 2 mL of acetone:methanol (7:3) with 200 mg mL−1 CaCO3. Extracts were kept at room temperature and covered with foil to exclude light. The extract was centrifuged for 5 min at 21,000 x g, the supernatant removed and re-extracted with an additional 1 mL of acetone:methanol (7:3). This process was repeated 3 times. The combined supernatants for each sample were partitioned with equal volumes of diethyl ether and water, and the diethyl ether fraction removed. This process was repeated until the acetone aqueous phase was colourless. The combined diethyl ether fractions were dried under O2-free N2 and the carotenoids dissolved in 1 mL of 0.8 % BHT/acetone as previously described [44] and then analysed by HPLC.
HPLC analysis
HPLC analysis was performed on a Dionex Ultimate 3000 solvent delivery system (Thermo Scientific, Sunnyvale, California) fitted with a YMC RP C30 column (5 μm, 250 x 4.6 mm), coupled to a 20 x 4.6 C30 guard column (YMC Inc. Wilmington, North Carolina) (column temperature 25 °C) and a Dionex 3000 PDA detector as previously published [34]. Phytoene was monitored at 280 nm and phytofluene at 350 nm. Coloured carotenoids and chlorophyll b were detected at 450 nm, while chlorophyll a and other chlorophyll derivatives were monitored at 430 nm. Carotenoid concentrations were determined as β-carotene equivalents /g DW of tissue. Chlorophyll b was determined using a chlorophyll b standard curve derived from a spinach extract [45]. Chlorophyll a and other chlorophyll derivatives were determined as chlorophyll a equivalents/g DW of tissue, again derived from a standard curve using the spinach extract and monitoring absorbance at 430 nm. β-carotene, and lutein were identified in the extracts by comparison of retention times and on-line spectral data with standards. All trans-β-carotene and lutein were purchased from Sigma Chemicals (St Louis, Missouri, U.S.A.). Other carotenoids were putatively identified by comparison with reported retention times and spectral data [46–50] and by comparison with carotenoids present in a spinach sample. Total carotenoid and chlorophyll content of the fruit tissue was also estimated using methods as previously described [51].
RNA extraction and cDNA synthesis
Total RNA was extracted by tissue homogenisation in CTAB buffer using a modified method from one previously described [34, 52]. cDNA was synthesised from total RNA (0.5-1 μg) using Superscript III reverse transcriptase (Life Technologies, Carlsbad, California, USA) following the manufacturer’s protocol. Reaction components included 50 μM oligo dT (12) primer, 500 μM dNTPs, 1X reverse transcription buffer, 5 mM MgCl2, 10 mM DTT, 40 units of RNaseOUT and 200 units of reverse transcriptase. The reactions were incubated at 50 °C for 50 min.
Quantitative real-time PCR analysis
Primers were designed using PRIMER3 software [53] to a stringent set of criteria. RT-qPCR was performed under conditions described previously [34, 54] (Additional file 1). First strand cDNA products were diluted 1:25 and used as templates for the PCR reaction. PCR analysis was performed using the LightCycler 1.5 system and the SYBR Green master mix (Roche, Penzberg, Germany), following the manufacturer’s protocol. Each reaction sample was analysed from biological replicates, with a negative control using water as template. PCR conditions were as follows: pre-incubation at 95 °C for 5 min followed by 40 cycles each consisting of 10 s at 95 °C, 10 s at 60 °C and 20 s at 72 °C. Amplification was followed by a melting curve analysis with continuous fluorescence measurement during the 65–95 °C melt. The relative expression was calculated using LightCycler software version 4 and the expression of each gene was normalised to apple Actin and Elongation factor1-α gene, whose expression are considered stable in these tissues [34, 55].
Cloning of apple PSY promoters and apple AP2/ERF transcription factors
MdPSY1 and MdPSY2 promoter fragments (1.5 kb) were amplified from ‘Royal Gala’ genomic DNA using primer pairs PSYprom1F (ATTCACTTTCAGGGAGGCGAAC) and PSY1prom1R (GGTTTTGGGTCTTGAGTGTGAG), and PSY2promF (CAGTATCGCGAATTTTTCGT) and PSY2promR (GAGGGTGTGAGTATGTGAGCTG) respectively. PCR fragments were cloned into pGEM-T Easy vector (Promega, Madison, USA) and then sub-cloned as Not I fragment into pGreen II 0800-LUC vector, upstream of the Luciferase reporter [56].
The apple AP2/ERF transcription factors were cloned as previously described [56, 57]. cDNAs from expressed sequenced apple libraries [58] were cloned into pART27 binary vector using restriction enzymes or pHEX2 using Gateway cloning.
Transient assay of promoter activation
Transient assays were performed as previously described [56, 57]. Agrobacterium tumefaciens strain GV3101 carrying a cloned PSY promoter construct or AP2/ERF construct were both resuspended in infiltration buffer (10 mM MgCl2, 0.5 μM acetosyringone) and infiltrated into the abaxial side of Nicotiana benthamiana leaves. The plants were left to grow for 2 days before 2 mm leaf discs were taken from infiltrated leaves and assayed with Victor 3 Multi-label Microplate Reader (Perkin Elmer, Waltham, Massachusetts, USA). Luciferase expression under PSY promoters relative to Renilla luciferase signals under the Cauliflower Mosaic Virus 35S promoter was measured.
Results
PSY sequence characterisation
Twelve apple PSY gene models were identified in the Phytozome sequence database (Additional file 1). Sequence analysis showed these gene models map to six positions on four chromosomes (3, 9, 11 and 17), suggesting six PSY genes are present in the apple genome [59]. We amplified four of these genes (MdPSY1-4) in fruit for analysis, while two of them, "MdPSY5" and "MdPSY6", which are present as additional genes on chromosomes 9 and 11 respectively, did not have transcripts available in the publicly available apple EST libraries [58] so were not further analysed.
In order to understand the roles of the multiple apple PSYs in carotenogenesis, we analysed their gene sequences. Comparisons of PCR amplified ‘Royal Gala’ cDNA, and genomic DNA fragments of these genes revealed a strong nucleotide and amino acid sequence similarity between MdPSY1 on chromosome 17 and MdPSY2 on chromosome 9, and between MdPSY3 (on chromosome 3) and MdPSY4 (on chromosome 11). MdPSY1 has a predicted 400-amino acid protein while MdPSY2 has a predicted protein of 401 amino acids. MdPSY3 has a stop codon after residue 130, from the original ORF start site, resulting in a truncated protein. There is a potential methionine start site at residue 150, which could result in a 242-amino acid protein. However, it is possible this MdPSY3 transcript, sequenced from four fruit cDNA clones, is a result of mis-splicing; when the MdPSY3 transcript is compared with the MdPSY3 and MdPSY4 genomic DNA sequences, the stop codon appears to be the result of a 15 bp footprint sequence left on exon 3 during the splicing of intron 2. Thus, there could be other correctly spliced MdPSY3 transcripts present in apple without the internal stop codon. Such correctly spliced transcripts would result in a protein sequence identical to MdPSY4, which has an ORF of 1158 bp, encoding a predicted 386-amino acid protein.
Multiple sequence comparisons of the predicted proteins indicated that MdPSY1 has 94 % identity to MdPSY2, while MdPSY3 has 98 % identity to PSY4 (Figure 1). In contrast both MdPSY1 and MdPSY2 have 54 % identity to MdPSY4. Comparing genomic and cDNA sequences revealed different exon-intron boundaries for the four PSY genes (Additional file 2). MdPSY1 and MdPSY2 have five exons and four introns, similar to the reported gene structure of the closely related loquat EjPSY2A [60]. MdPSY1 and MdPSY2 have similar exon sizes but differently spliced introns. MdPSY4 has 6 exons and five introns, which is similar to MdPSY3 (both in size and position). Compared with the MdPSY4 protein sequence, the stop codon in MdPSY3 appears to be the result of a 15 bp footprint sequence left on exon 3 during the splicing of intron 2.
Fig. 1.
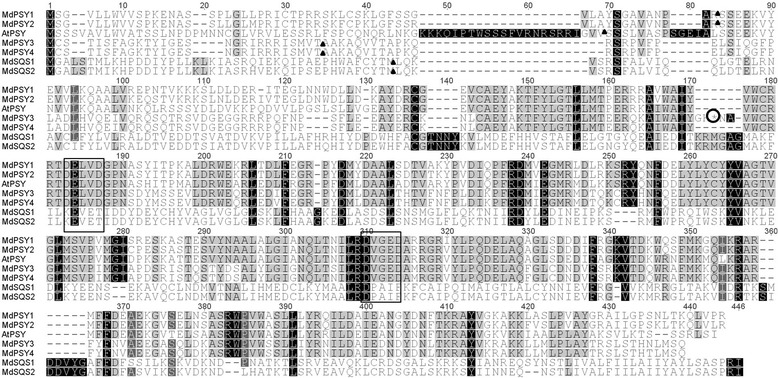
Alignment of the four apple Phytoene synthase (PSY) compared with Arabidopsis PSY and apple squalene synthase (MdSQS1 and MdSQS2). Multiple sequence alignment was conducted using ClustalW and manually adjusted in Geneious. The predicted chloroplast cleavage site by ChloroP is indicated by a black triangle. The stop codon present in PSY3 is indicated by a circled asterisk. Boxed sequence indicate the putative active site DXXXD [18]. Highlighting indicates similarity among residues ignoring the gaps in sequence; black, 100 %, dark grey >80 % and grey, >50 %
Phylogenetic analysis of predicted amino acid sequences (including transit peptides) classified PSY1, PSY2, PSY3 and PSY4 into two distinct clades, supporting the observation that these pairs arose from a single duplication event (Fig. 2). Apple MdPSY1 and MdPSY2 form a clade with maize ZmPSY2, rice OsPSY2 and loquat EjPSY2A. On the other hand, MdPSY3 and MdPSY4 grouped together with loquat EjPSY3, cassava MePSY3, and tomato SlPSY3.
Fig. 2.
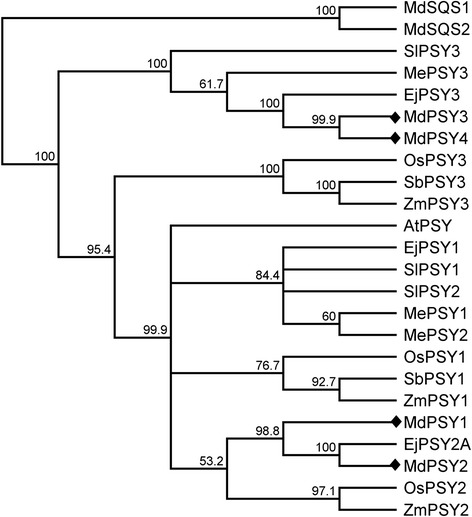
Phylogenetic tree was constructed using MEGA6 [37] from PSY and apple squalene synthase sequences retrieved from the GenBank database (except where noted): Arabidopsis, AtPSY; loquat, EjPSY1, EjPSY2A, EjPSY3; apple, MdPSY1, MdPSY2, MdPSY3, MdPSY4, MdSQS1, MdSQS2; cassava, MePSY1, MePSY2, MePSY3; rice, OsPSY1, OsPSY2, OsPSY3; sorghum, SbPSY1, SbPSY3; tomato, SlPSY1, SlPSY2, SlPSY3; maize, ZmPSY1, ZmPSY2, ZmPSY3. Evolutionary relationships were inferred using the Neighbor-joining method [85], with 1000 bootstrap re-sampling strategy. The four apple PSY sequences are indicated by diamond
Functional complementation
To test whether the apple PSY genes encode functional enzymes, we used a standard bacterial complementation method for assessing carotenoid pathway enzyme function [10, 61]. Escherichia coli test strains were produced by transforming them with either pACCRTE, which encodes the enzyme to produce GGPP in bacteria, or pACCAR25ΔcrtB (ΔcrtB) which requires PSY function to produce zeaxanthin and its glycosylated derivatives [38]. Next, vectors with cDNA fragments encoding the open reading frame of the apple PSYs with predicted transit peptides removed, were transformed into each of the test strains.
MdPSY1 and MdPSY2 constructs in cells with pACCRTE produced phytoene, with a peak whose retention time and spectral qualities were similar to the positive control (Fig. 3a). Similarly, ΔcrtB cells produced zeaxanthin diglucoside when transformed with MdPSY1 and MdPSY2 constructs (Fig. 3b). The extracts from these transformants had distinct yellow colouration and the peak retention times and fine spectral qualities were consistent with HPLC standards and previous publications (Fig. 3b) [22, 38]. Surprisingly, MdPSY4 in bacterial cells with pACCRTE or ΔcrtB did not result in the expected product peaks (Fig. 3). The same result was obtained with extended induction periods or using the full-length MdPSY4 including its transit peptide, suggesting MdPSY4 is not able to catalyse the conversion of GGPP to phytoene in bacteria.
Fig. 3.
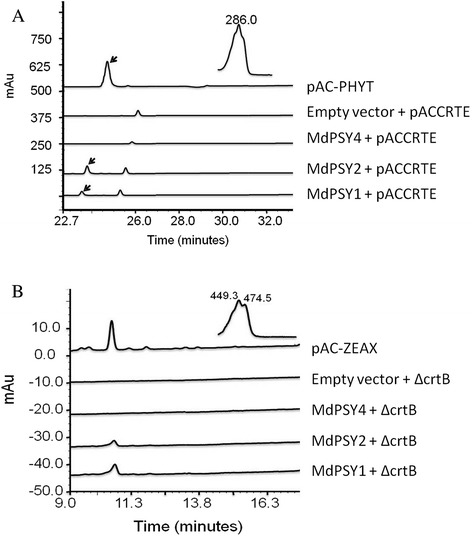
Functional complementation of apple PSY proteins. a. Escherichia coli cells harbouring the pACCRTE vector (which encodes CRTE, the enzyme catalyzing formation of geranyl geranyl pyrophosphate) were additionally transformed with apple PSY constructs or empty vector. Cells carrying the pAC-PHYT vector confer accumulation of phytoene [40] and were used as a positive control. HPLC chromatograms for the extracted pigments are shown. The peak representing phytoene (indicated by an arrow) was observed in cells with PSY1 and PSY2 constructs, but not with PSY4. The inset shows the absorption spectrum of the phytoene peak. b. E. coli cells harbouring pACCAR25ΔcrtB were transformed with the apple PSY constructs. Cells carrying the plasmid pAC-ZEAX [86] accumulating zeaxanthin were used as a positive control. The peak representing zeaxanthin diglucoside is shown. The inset shows the absorption spectra of the zeaxanthin diglucoside peak of pAC-ZEAX, which was similar to that from both PSY1 and PSY2 constructs with ΔcrtB
Protein localization
To ascertain the targeting of the apple PSYs, we fused PCR amplified fragments encoding apple PSYs including their predicted transit peptides to the enhanced yellow fluorescent protein (eYFP) [41, 42]. The fusion constructs were transiently expressed in maize etiolated leaf protoplasts and analysed using fluorescent confocal microscopy. Both MdPSY1 and MdPSY2 were co-localised with chlorophyll in the chloroplast confirming they are translocated to plastids (Fig. 4). MdPSY3 has a premature stop codon and was not further tested. MdPSY4 was localized to speckles associated with the chloroplasts (Fig. 4); these speckles are shown to be plastoglobuli using the maize plastoglobulin-2 as marker [18].
Fig. 4.
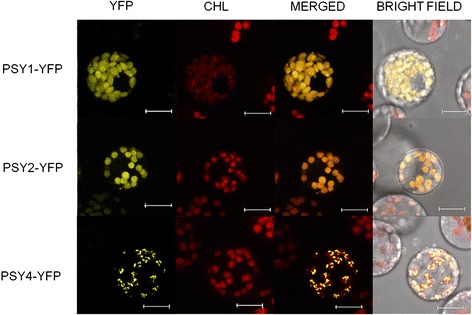
Transient expression of apple PSY-YFP fusion constructs in etiolated maize leaf protoplasts. PSY1 and PSY2 were localized throughout the plastids based on the fluorescent distribution pattern. PSY4 localized to speckles, suggesting localization to plastoglobuli [18]. CHL, chlorophyll autofluorescence; Bars = 10 μm
Carotenoid accumulation in fruit
To understand the role of PSYs in carotenogenesis during fruit development, we selected two commercial apple cultivars 'Granny Smith' and 'Royal Gala' based on the pigmentation of their fruit skin and flesh. ‘Granny Smith’ has a green skin and white flesh while 'Royal Gala’ has a red coloured skin with creamy flesh (Fig. 5a). Carotenoid and chlorophyll pigments were measured at different stages of fruit development. Total carotenoid concentration in ‘Granny Smith’ fruit skin was 2–5 fold greater than in ‘Royal Gala’ (Fig. 5b). In both cultivars, total carotenoid concentration in fruit skin appeared unchanged between 30 and 90 days after full bloom (DAFB) (~150 μg/g dry weight in ‘Granny Smith’ compared with ~75 μg/g dry weight for ‘Royal Gala’), followed by a significant decrease at 120 and 150 DAFB. Lutein and beta-carotene were the dominant compounds present in the analysed tissues and both compounds were up to 3-fold higher in ‘Granny Smith’ than ‘Royal Gala’ tissues (Additional file 3). The total carotenoid concentration in fruit flesh was about 7–10 fold lower than in skin and there was no clear pattern observed between the two cultivars. Total chlorophyll concentration (which includes breakdown compounds pheophytin a and b) in ‘Granny Smith’ tissues was 2–5 fold higher than in ‘Royal Gala’ (Additional file 3). A strong correlation was observed between total chlorophyll and carotenoid concentration (r = 0.97, p < 0.01) in these tissues, which suggested that most of the carotenoids present were associated with chloroplastic structures.
Fig. 5.
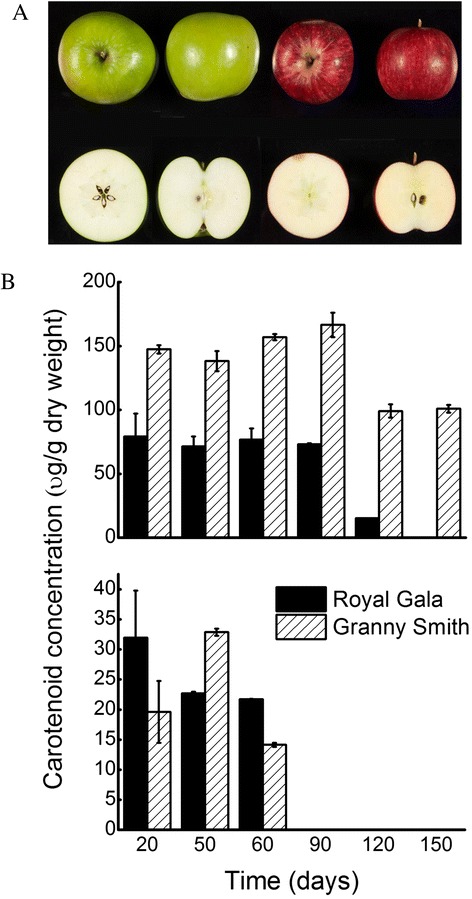
Carotenoid concentrations in fruit of apple cultivars selected based on the pigmentation of their skin and flesh. a. Ripe fruit (150 DAFB) of ‘Granny Smith’ (left) and ‘Royal Gala’ (right). b. Total carotenoid concentration as measured by HPLC in apple fruit skin (top panel) and flesh (lower panel). Fruit were harvested at different time points (days after full bloom) and separated into skin and flesh for carotenoid extraction and analysis. Error bars are standard errors from three biological replicates
Gene expression
We analysed the transcript levels of MdPSY1 and MdPSY2 in fruit skin and flesh tissues of the two cultivars. Both MdPSY1 and MdPSY2 were, in general, higher (1.1 to 12 fold) in fruit skin than in flesh, consistent with higher carotenoid concentrations (2–5 fold) in fruit skin compared to the flesh [34]. Both MdPSY1 and MdPSY2 had similar tissue transcript profiles in both cultivars, though the transcript level of MdPSY2 was higher than that of PSY1 (Fig. 6). The transcript levels were reduced in young fruit skin and the highest transcript level was observed at 60 days after full bloom (DAFB). After this stage, transcripts decreased, with the exception of that in the ‘Royal Gala’ 150 DAFB tissue. In the flesh, MdPSY1 and MdPSY2 transcript levels were similarly reduced at the early fruit stages (30 and 50 DAFB) and increased at 60 DAFB. After this stage, MdPSY1 transcripts reduced while MdPSY2 transcript levels increased in ‘Royal Gala’ flesh until 150 DAFB.
Fig. 6.
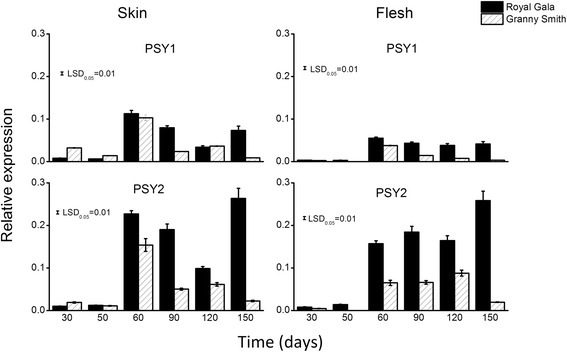
Gene expression profiles of PSY genes assessed in ‘Royal Gala’ and ‘Granny Smith’ apple fruit . PSY transcript levels in fruit skin and flesh picked at different time points (days after full bloom). The data were analysed using the target-reference ratios measured with LightCycler 480 software (Roche) using apple Actin and Elongation factor1-α (EF1-α) as reference genes. Data are analysed from biological replicates and presented as means ± SE (n = 4). Fisher's least significant difference (LSD) at P < 0.05 is shown
MdPSY transcript levels were next examined in various apple tissues (Fig. 7). MdPSY1 and MdPSY2 were present in varying levels in all tissues examined, including both photosynthetic and non-photosynthetic. In small and expanded leaves as well as in open and unopened flowers, MdPSY2 transcripts were at higher levels (3- to 5-fold) as compared to PSY1. Transcript levels for the two PSYs were similar in shoot tissues while in tissue-cultured roots, MdPSY2 levels were about 9-fold higher than for MdPSY1. Taken together, MdPSY2 represented the PSY gene with the most abundant transcripts. However, the PSY transcripts did not correlate with the total carotenoid levels in these tissues.
Fig. 7.
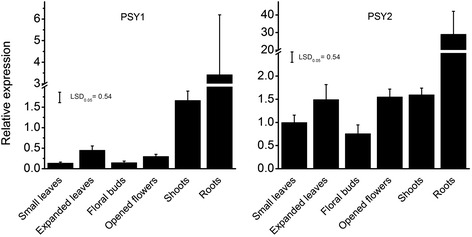
PSY transcript levels in different apple tissues from ‘Royal Gala’. Data were analyzed from biological replicates as described in Fig. 6 and presented as means ± SE (n = 4). Fisher’s least significant difference (LSD) at P < 0.05 is shown
Transient activation of PSY promoters
The transcript levels of MdPSY2 compared with MdPSY1 suggested these two paralogs are differentially regulated. To determine whether this differential regulation is related to diversity in the gene promoters, we amplified and sequenced 1.5 kb promoter fragments of MdPSY1 and MdPSY2 from ‘Royal Gala’ and found <30 % nucleotide similarity between these two gene promoters. Motif analysis by MatInspector [62] identified RAP2.2 (an APETALA2/ethylene response factor-type transcription factor) binding motifs in the PSY promoter sequences. Three RAP2.2 motifs were present in the PSY1 promoter, all within 500 bp upstream of the ORF, while only two motifs were found in the PSY2 promoter (Fig. 8a). In order to test functionality of the RAP2.2 binding motifs, both promoters were tested for transactivation with 36 apple AP2/ERFs transcription factors (TFs) [57] (Additional file 4, Additional file 6) using Agroinfiltration into young leaves of Nicotiana benthamiana [56, 63, 64]. Agrobacterium strains were separately transformed with constructs carrying PSY promoters cloned upstream of a Luciferase reporter or a gene encoding a AP2/ERF TF, which were together co-infiltrated into .Nicotiana benthamiana leaves. Transactivation was measured as an increase in luciferase levels compared to controls lacking a co-infiltrated transcription factor, normalised to 35S-Renilla [56]. The results showed differential trans-activation of the PSY promoters. For instance, the MdPSY2 promoter was trans-activated (~15-fold increase) by AP2D21, a homolog of AtERF3, compared to a ~5-fold increase of MdPSY1 promoter activity. Both PSY promoters were strongly activated by AP2D15 and AP2D26 homologs of AtRAP2.3 (47 % identity) and AtERF113 (49 % identity) respectively (Fig. 8b).
Fig. 8.
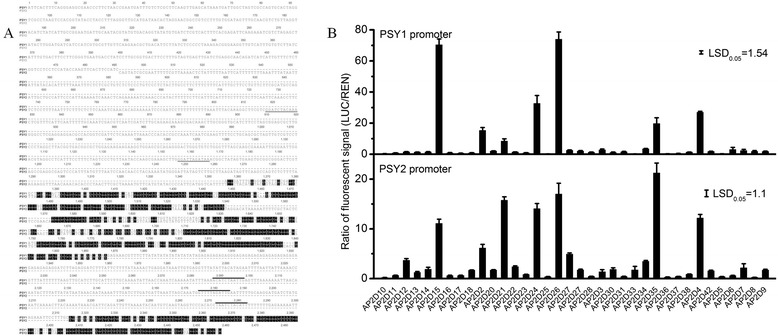
Transient activation of PSY promoters by apple AP2/ERF domain (AP2D) transcription factors. a. Alignment of ‘Royal Gala’ PSY1 and PSY2 promoters conducted using ClustalW in Geneious. Highlighting (black) indicate nucleotide similarity. RAP2.2 motifs are indicated by black (PSY1) and grey (PSY2) bars. b. Ratio of fluorescent signals measured from Nicotiana benthamiana leaves co-infiltrated with Agrobacterium constructs with AP2/ERF genes and a vector with firefly luciferase (LUC) under control of PSY promoter and Renilla luciferase (REN) under the control of CaMV 35S promoter. LUC/REN signal ratios were normalised to the basal promoter activity. Bars represent means ± SE (n = 4). Fisher's least significant difference (LSD) at P < 0.05 is shown
Physiologically relevant transactivation is predicated on overlapping in vivo expression of a candidate TF and the PSY target. Therefore, we analysed transcript levels of AP2D15 and AP2D26, in the skin and flesh tissues of the apple cultivars (Additional file 5). Both AP2D15 and AP2D26 transcript levels increased from early fruit stage (30 DAFB) through to ripe fruit stage (150 DAFB) in both apple cultivars. Co-expression measured as correlation between gene expression of transcription factors and PSYs showed strong positive correlation between AP2D26 and PSY2 in fruit skin (Table 1), further suggesting a potential regulatory relationship.
Table 1.
Pearson’s correlation between AP2D15, AP2D26 and apple PSY transcript levels in skin and flesh of ‘Royal Gala’ (RG) and ‘Granny Smith’ (GS). Bold values are statistically significant at P < 0.05
| Transcription factors | ||||||||
|---|---|---|---|---|---|---|---|---|
| AP2D15 | AP2D26 | |||||||
| RG | GS | RG | GS | |||||
| skin | flesh | skin | flesh | skin | flesh | skin | flesh | |
| PSY1 | 0.37 | 0.24 | 0.01 | −0.22 | 0.45 | 0.28 | 0.5 | −0.26 |
| PSY2 | 0.56 | 0.56 | 0.61 | −0.09 | 0.81 | 0.63 | 0.81 | −0.1 |
Discussion
Apples are consumed globally and are chosen to eat for their healthful metabolites and convenience. Consumers associate presence of nutritionally favourable compounds with fruit colour, partly contributed by carotenoids. Thus, carotenoid content has become an important apple breeding objective [25, 31, 34]. The phytoene synthase step is known to play a significant role in the carotenoid pathway because of its position as the first committed step, potentially controlling the flux downstream [65, 66]. Many plant species are known to have multiple PSY genes, including apple [34, 60].
The multiple PSY genes highlight the issue of functional diversity because of the potential to acquire a novel function, subfunctionalize or even lose the original function [67, 68]. The domesticated apple has 17 chromosomes, which may have been a result of both recent and older whole genome duplication (WGD) events [59]. The six apple PSYs are present on four chromosomes, which suggests that prior to the most recent genome duplication, at least two ancestral apple PSY genes on different chromosomes were present, resulting in the two homeologous pairs PSY1/PSY2 and PSY3/PSY4 described here.
MdPSY3/PSY4 cluster with maize and rice PSY3, which have had their function proven in bacteria. Others, such as cassava MePSY3, tomato SlPSY3and loquat EjPSY3, which also share high homology to the apple PSY4 (66 %, 72 % and 96 % identity respectively) have not been tested or have been found to be non-functional [15, 21, 22, 60]. The non-functionality of MdPSY4 in bacteria could be because of the acquisition of a new function or perhaps mutation of some active sites. However, sequence analysis showed it was closer to PSY sequences than squalene synthase [18, 69]. It must be noted however, that while heterologous expression of plant genes in bacteria is a widely used method, the plastid environment where these enzymes function is absent, which may affect catalytic activity due to improper membrane localization of the enzyme and/or protein complex formation [19]. The localization of apple MdPSY4 to the plastoglobuli, in contrast to MdPSY1 and MdPSY2 in the chloroplast, may be important here in the sense that the catalytic activity of MdPSY4 could be influenced by its protein location. The tomato SlPSY3, for instance, was recently shown through virus induced gene silencing to affect carotenoid accumulation [15]. It remains to be seen if MdPSY4 has acquired a different function or its catalytic activity is affected by protein complex formation.
MdPSY2 has a dominant expression pattern in apple
We examined MdPSY1 and MdPSY2 transcript levels because of the ability of the encoded proteins to catalyze the conversion of GGPP to phytoene. Between these two genes, there was no tissue specific expression among the wide ranging apple tissues we examined to suggest subfunctionalization. Subfunctionalization of duplicated genes can take the form of complementary gene expression patterns in different tissues or partitioning protein function between paralogs [70, 71]. The absence of gene expression partitioning among the apple PSYs contrasts with what is observed in plants, such as maize, where subfunctionalization is observed among the PSYs [20]. This lack of difference in tissue-specific expression between MdPSY1 and MdPSY2 could be because the gene duplication between them is a recent event.
One obvious difference between MdPSY1 and MdPSY2 was their unequal gene expression levels. Unequal gene expression between paralogs in duplicated genomes can be an immediate consequence of the polyploidization or a result of changes introduced over time [72, 73]. The variation in gene expression between these PSY paralogs could be related to gene dosage effects or may simply be immaterial [74]. However, the higher relative expression of MdPSY2 over MdPSY1 is consistent with previous study where MdPSY2 showed higher transcript levels (3–5 fold) over MdPSY1 in different apple cultivars [34]. This could mean MdPSY2 has a dominant role in apple and may be primarily responsible for this first carotenoid pathway step in apple tissues. However, both PSY transcripts do not correlate with total carotenoid levels, suggesting post-transcriptional processes may be important for determining flux through this enzymatic step. Recent knowledge on regulation of PSY in controlling carotenoid biosynthesis has shown that post-transcriptional mechanisms play a major role in the pathway [75]. With adequate expression of the PSY transcript, which we have shown is mainly PSY1 and PSY2 in apple fruit, it is then the stability of the protein which defines PSY as the rate limiting step in carotenogenesis [75]. In Arabidopsis this stability is modulated by the chaperone protein, AtOR [75]. Such knowledge has implications for apple breeding, where marker assisted selection is accelerating the breeding of better cultivars [76].
Transactivation of PSY promoters
The possibility of different transcriptional mechanisms controlling the different expression of apple MdPSY1 and MdPSY2 is supported by the significant sequence differences in their promoters as well as the varied responses to transient activation by the AP2/ERF transcription factors we tested. The AP2/ERFs are implicated in many plant development processes such as floral development and response to biotic and abiotic stress [77, 78]. Their role in the carotenoid pathway was highlighted in Arabidopsis, when AtRAP2.2 was implicated in binding of the PSY promoter [79]. The apple AP2/ERF family has been characterised with some members implicated in fruit development [57, 58]. The expression pattern of AP2D15 and AP2D26, with increased transcript levels as the fruit matured, suggests these genes have a role in fruit development. This, in addition to the transient activation of PSY promoters and the high correlation observed between their gene expression and PSYs point to a potential relationship where these AP2/ERFs are possibly regulating the PSYs. There is circumstantial evidence supporting AP2/ERFs in carotenogenesis in tomato, where they have been shown to regulate the carotenoid pathway negatively during fruit ripening. SlAP2a indirectly affects the carotenoid pathway through regulation of the ethylene pathway, as RNAi repression of this gene resulted in a concomitant increase in both ethylene and carotenoid compounds in fruit. Similarly, SlERF6 was shown to affect carotenoid accumulation, where reduced expression by RNAi knockout resulted in increased carotenoid concentration in fruit. While these examples suggest both wild type genes are negative regulators of the pathway [80,81], they do not show any direct regulation of the carotenoid biosynthetic genes by the AP2/ERFs. The PSY step in carotenogenesis is under complex transcriptional and posttranscriptional regulation. The tomato SlPSY1 and SlPSY2 are targets of the MADS-box transcription factor Ripening Inhibitor, and SlPSY1 gene expression and enzyme activity is inhibited by tomato Stay-green 1 [82, 83]. AtPSY is regulated by AtPIF1 through direct binding to the promoter to repress transcription and more recently, the Arabidopsis Orange (OR) and OR-like proteins are showed to be major posttranscriptional regulators of AtPSY, controlling AtPSY protein levels and carotenoid content [75, 84]. The further analysis of apple AP2/ERFs interactions with apple PSY promoters will increase our understanding of their role in the carotenoid pathway and maybe reveal how the apple PSYs are transcriptionally controlled.
Conclusion
The data presented here suggest that the first committed carotenoid pathway step in apple is encoded by two functional genes MdPSY1 and MdPSY2, with other apple PSYs playing little or no role in this respect. This has implications for apple breeding programmes that have fruit colour as a breeding target. Characterisation of the PSY gene family members increases our understanding of how the first carotenoid pathway step is controlled in apple and for instance, would allow co-segregation with fruit colour phenotypes to be tested during the development of new cultivars.
Acknowledgements
This work was supported by Plant & Food Research Core programme for Consumer Traits in pipfruit. The Wurtzel laboratory was supported through funding from the National Institutes of Health (grant GM081160). We are grateful to Tim Holmes for photography and illustrations, Steve Arathoon for HPLC measurements, Bradley Drayton who spent a summer studentship at PFR studying the apple PSYs, Robert Schaffer and Ross Atkinson for helpful comments. We are grateful to Andrew Gleave and the Genomic Technologies team, Karen Grafton and Karen Bolitho for EST retrieval and cloning of AP2/ERFs into Agrobacterium vectors. Thanks to Anne Gunson and the Science Publication Office (PFR) for editing the manuscript.
Abbreviations
- bp
Basepairs
- g
Gram
- dw
Dry weight
- DAFB
Days after full bloom
- GGPP
Geranylgeranyl pyrophosphate
- PSY
Phytoene synthase
Additional files
List of primers used and apple PSY gene models (DOCX 14 kb)
The structure of the apple PSY genes. The exons (black bars) and introns (black lines) of the apple PSY genes were mapped by cDNA and genomic DNA sequence comparisons and constructed using FancyGENE version 1.4 (http://bio.ieo.eu/fancygene/). The location of the 15 bp insertion in MdPSY3 is indicated by triangle. (JPEG 241 kb)
Table of carotenoid concentration (μg/g dry weight) measured by HPLC in skin and flesh tissues of ‘Royal Gala’ and ‘Granny Smith’ apple fruit at different stages of development. (DOCX 15 kb)
Phylogenetic tree of AP2/ERF amino acid sequences from apple and Arabidopsis using the Neighbor-joining method in MEGA6 and reliability of tree construction estimated by boostrap method based on 1000 replicates. Apple sequences used in this analysis are indicated with black triangles. The database accession numbers of the sequences used are presented in Additional file 6. (JPEG 60 kb)
Relative expression of AP2D15 and AP2D26 in ‘Royal Gala’ (RG) and ‘Granny Smith’ (GS) apple fruit as described in Fig. 6. Data are presented as means ± SE (n = 4). (JPEG 1012 kb)
List of genes and associated accession numbers used in the AP2/ERFs Phylogenetic tree (Additional file 4). (DOCX 13 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
CAD, RVE and ACA conceived the experimental design. CAD and ND provided analyses of the bioinformatic, transcriptomic and transactivation data. DL performed the HPLC analysis of fruit carotenoid content. MS and ETW provided analysis of protein localization. XC collected fruit samples and assisted in RNA extraction. CAD wrote the body of the paper; all authors reviewed and edited the manuscript.
Contributor Information
Charles Ampomah-Dwamena, Email: charles.dwamena@plantandfood.co.nz.
Nicky Driedonks, Email: n.driedonks@science.ru.nl.
David Lewis, Email: david.lewis@plantandfood.co.nz.
Maria Shumskaya, Email: mashum@gmail.com.
Xiuyin Chen, Email: xiuyin.chen@plantandfood.co.nz.
Eleanore T. Wurtzel, Email: eleanore.wurtzel@lehman.cuny.edu
Richard V. Espley, Email: richard.espley@plantandfood.co.nz
Andrew C. Allan, Email: andrew.allan@plantandfood.co.nz
References
- 1.Iglesias I, Echeverria G, Soria Y. Differences in fruit colour development, anthocyanin content, fruit quality and consumer acceptability of eight ‘Gala’ apple strains. Sci Hortic. 2008;119(1):32–40. doi: 10.1016/j.scienta.2008.07.004. [DOI] [Google Scholar]
- 2.Johnson EJ. The role of carotenoids in human health. Nutrition in clinical care : an official publication of Tufts University. 2002;5(2):56–65. doi: 10.1046/j.1523-5408.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 3.Krinsky NI, Johnson EJ. Carotenoid actions and their relation to health and disease. Mol Asp Med. 2005;26(6):459–516. doi: 10.1016/j.mam.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, Papacchioli V, et al. Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 2006;6. [DOI] [PMC free article] [PubMed]
- 5.Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, et al. Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol. 2000;18(6):666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- 6.Wurtzel ET, Cuttriss A, Vallabhaneni R. Maize provitamin A carotenoids, current resources, and future metabolic engineering challenges. Frontiers in Plant Science. 2012;3. [DOI] [PMC free article] [PubMed]
- 7.Moise AR, Al-Babili S, Wurtzel ET. Mechanistic Aspects of Carotenoid Biosynthesis. Chem Rev. 2014;114(1):164–193. doi: 10.1021/cr400106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, et al. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45(6):982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 9.Van Chien H, Antonio Leyva-Gonzalez M, Osakabe Y, Uyen Thi T, Nishiyama R, Watanabe Y, et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci U S A. 2014;111(2):851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher CE, Matthews PD, Li F, Wurtzel ET. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004;135(3):1776–1783. doi: 10.1104/pp.104.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welsch R, Arango J, Bar C, Salazar B, Al-Babili S, Beltran J, et al. Provitamin A Accumulation in Cassava (Manihot esculenta) Roots Driven by a Single Nucleotide Polymorphism in a Phytoene Synthase Gene. Plant Cell. 2010;22(10):3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E. Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot. 2008;59(13):3563–3573. doi: 10.1093/jxb/ern210. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Villalon A, Gas E, Rodriguez-Concepcion M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009;60(3):424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- 14.Dibari B, Murat F, Chosson A, Gautier V, Poncet C, Lecomte P, et al. Deciphering the genomic structure, function and evolution of carotenogenesis related phytoene synthases in grasses. BMC Genomics. 2012;13. [DOI] [PMC free article] [PubMed]
- 15.Fantini E, Falcone G, Frusciante S, Giliberto L, Giuliano G. Dissection of Tomato Lycopene Biosynthesis through Virus-Induced Gene Silencing. Plant Physiol. 2013;163(2):986–998. doi: 10.1104/pp.113.224733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arango J, Wust F, Beyer P, Welsch R. Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 2010;232(5):1251-1262. [DOI] [PubMed]
- 17.Chaudhary N, Nijhawan A, Khurana JP, Khurana P. Carotenoid biosynthesis genes in rice: structural analysis, genome-wide expression profiling and phylogenetic analysis. Mol Genet Genomics 2010;283(1):13-33. [DOI] [PubMed]
- 18.Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET. Plastid Localization of the Key Carotenoid Enzyme Phytoene Synthase Is Altered by Isozyme, Allelic Variation, and Activity. Plant Cell. 2012;24(9):3725–3741. doi: 10.1105/tpc.112.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shumskaya M, Wurtzel ET. The carotenoid biosynthetic pathway: Thinking in all dimensions. Plant Sci. 2013;208:58–63. doi: 10.1016/j.plantsci.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008;147(3):1334–1346. doi: 10.1104/pp.108.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008;146(3):1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsch R, Wust F, Bar C, Al-Babili S, Beyer P. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 2008;147(1):367–380. doi: 10.1104/pp.108.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fray RG, Grierson D. Identification and genetic-analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22(4):589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- 24.Fraser PD, Kiano JW, Truesdale MR, Schuch W, Bramley PM. Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid synthesis in ripening fruit. Plant Mol Biol. 1999;40(4):687–698. doi: 10.1023/A:1006256302570. [DOI] [PubMed] [Google Scholar]
- 25.Chagne D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, et al. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics. 2007;8. [DOI] [PMC free article] [PubMed]
- 26.Yuan K, Wang C, Wang J, Xin L, Zhou G, Li L, et al. Analysis of the MdMYB1 gene sequence and development of new molecular markers related to apple skin color and fruit-bearing traits. Mol Genet Genomics. 2014;289(6):1257–1265. doi: 10.1007/s00438-014-0886-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XJ, Wang LX, Chen XX, Liu YL, Meng R, Wang YJ, et al. A and MdMYB1 allele-specific markers controlling apple (Malus x domestica Borkh.) skin color and suitability for marker-assisted selection. Genet Mol Res. 2014;13(4):9103–9114. doi: 10.4238/2014.October.31.26. [DOI] [PubMed] [Google Scholar]
- 28.Dunemann F, Ulrich D, Malysheva-Otto L, Weber WE, Longhi S, Velasco R, et al. Functional allelic diversity of the apple alcohol acyl-transferase gene MdAAT1 associated with fruit ester volatile contents in apple cultivars. Mol Breed. 2012;29(3):609–625. doi: 10.1007/s11032-011-9577-7. [DOI] [Google Scholar]
- 29.Souleyre EJF, Chagne D, Chen X, Tomes S, Turner RM, Wang MY, et al. The AAT1 locus is critical for the biosynthesis of esters contributing to “ripe apple’ flavour in “Royal Gala’ and “Granny Smith’ apples. Plant J. 2014;78(6):903–915. doi: 10.1111/tpj.12518. [DOI] [PubMed] [Google Scholar]
- 30.Telias A, Bradeen JM, Luby JJ, Hoover EE, Allan AC. Regulation of anthocyanin accumulation in apple peel. Hortic Rev. 2011;38:357–391. [Google Scholar]
- 31.Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13(3):99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Simkin AJ, Schwartz SH, Auldridge M, Taylor MG, Klee HJ. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles beta-ionone, pseudoionone, and geranylacetone. Plant J. 2004;40(6):882–892. doi: 10.1111/j.1365-313X.2004.02263.x. [DOI] [PubMed] [Google Scholar]
- 33.Reay PF, Fletcher RH, Thomas VJG. Chlorophylls, carotenoids and anthocyanin concentrations in the skin of 'Gala' apples during maturation and the influence of foliar applications of nitrogen and magnesium. J Sci Food Agric. 1998;76(1):63–71. doi: 10.1002/(SICI)1097-0010(199801)76:1<63::AID-JSFA908>3.0.CO;2-K. [DOI] [Google Scholar]
- 34.Ampomah-Dwamena C, Dejnoprat S, Lewis D, Sutherland P, Volz RK, Allan AC. Metabolic and gene expression analysis of apple (Malus x domestica) carotenogenesis. J Exp Bot. 2012;63(12):4497–4511. doi: 10.1093/jxb/ers134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform. 2002;2:2.3.1-2.3.22 [DOI] [PubMed]
- 36.Emanuelsson O, Nielsen H, Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8(5):978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, et al. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990;172(12):6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham FX, Jr, Pogson B, Sun Z, McDonald KA, DellaPenna D, Gantt E. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell. 1996;8(9):1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham FX, Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6(8):1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, et al. Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in Transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71(8):2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 43.Quinlan RF, Shumskaya M, Bradbury LMT, Beltran J, Ma C, Kennelly EJ, et al. Synergistic Interactions between Carotene Ring Hydroxylases Drive Lutein Formation in Plant Carotenoid Biosynthesis. Plant Physiol. 2012;160(1):204–214. doi: 10.1104/pp.112.198556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ampomah-Dwamena C, McGhie T, Wibisono R, Montefiori M, Hellens RP, Allan AC. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. J Exp Bot. 2009;60(13):3765–3779. doi: 10.1093/jxb/erp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Albert NW, Zhang H, Arathoon S, Boase MR, Ngo H, et al. Temporal and spatial regulation of anthocyanin biosynthesis provide diverse flower colour intensities and patterning in Cymbidium orchid. Planta. 2014;240(5):983–1002. doi: 10.1007/s00425-014-2152-9. [DOI] [PubMed] [Google Scholar]
- 46.Burns J, Fraser PD, Bramley PM. Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry. 2003;62(6):939–947. doi: 10.1016/S0031-9422(02)00710-0. [DOI] [PubMed] [Google Scholar]
- 47.Fraser PD, Pinto MES, Holloway DE, Bramley PM. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 2000;24(4):551–558. doi: 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- 48.Kamffer Z, Bindon KA, Oberholster A. Optimization of a Method for the Extraction and Quantification of Carotenoids and Chlorophylls during Ripening in Grape Berries (Vitis vinifera cv. Merlot) J Agric Food Chem. 2010;58(11):6578–6586. doi: 10.1021/jf1004308. [DOI] [PubMed] [Google Scholar]
- 49.Lee MT, Chen BH. Separation of lycopene and its cis isomers by liquid chromatography. Chromatographia. 2001;54(9–10):613–617. doi: 10.1007/BF02492187. [DOI] [Google Scholar]
- 50.Xu C-J, Fraser PD, Wang W-J, Bramley PM. Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J Agric Food Chem. 2006;54(15):5474–5481. doi: 10.1021/jf060702t. [DOI] [PubMed] [Google Scholar]
- 51.Wellburn AR. The spectral determination of chlorophyll-a and chlorophhyll-b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol. 1994;144(3):307–313. doi: 10.1016/S0176-1617(11)81192-2. [DOI] [Google Scholar]
- 52.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plan Mol Biol Rep. 1993;116(5):113–6. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 53.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods mol biol (Clifton, NJ) 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 54.Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50–0. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nieuwenhuizen NJ, Green SA, Xiuyin C, Bailleul EJD, Matich AJ, Wang MY, et al. Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 2013;161(2):787–804. doi: 10.1104/pp.112.208249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1. [DOI] [PMC free article] [PubMed]
- 57.Tacken E, Ireland H, Gunaseelan K, Karunairetnam S, Wang D, Schultz K, et al. The Role of Ethylene and Cold Temperature in the Regulation of the Apple POLYGALACTURONASE1 Gene and Fruit Softening. Plant Physiol. 2010;153(1):294–305. doi: 10.1104/pp.109.151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newcomb RD, Crowhurst RN, Gleave AP, Rikkerink EHA, Allan AC, Beuning LL, et al. Analyses of expressed sequence tags from apple. Plant Physiol. 2006;141(1):147–166. doi: 10.1104/pp.105.076208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat Genet. 2010;42(10):833. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 60.Fu X, Feng C, Wang C, Yin X, Lu P, Grierson D, et al. Involvement of multiple phytoene synthase genes in tissue- and cultivar-specific accumulation of carotenoids in loquat. J Exp Bot. 2014;65(16):4679–4689. doi: 10.1093/jxb/eru257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song GH, Kim SH, Choi BH, Han SJ, Lee PC. Heterologous Carotenoid-Biosynthetic Enzymes: Functional Complementation and Effects on Carotenoid Profiles in Escherichia coli. Appl Environ Microbiol. 2013;79(2):610–618. doi: 10.1128/AEM.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. Matlnspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 63.Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, et al. Multiple Repeats of a Promoter Segment Causes Transcription Factor Autoregulation in Red Apples. Plant Cell. 2009;21(1):168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang YN, Li RG, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22(6):543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- 65.Giuliano G. Plant carotenoids: genomics meets multi-gene engineering. Curr Opin Plant Biol. 2014;19:111–117. doi: 10.1016/j.pbi.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez-Suarez C, Mellado-Ortega E, Hornero-Mendez D, Atienza SG. Increase in transcript accumulation of Psy1 and e-Lcy genes in grain development is associated with differences in seed carotenoid content between durum wheat and tritordeum. Plant Mol Biol. 2014;84(6):659–673. doi: 10.1007/s11103-013-0160-y. [DOI] [PubMed] [Google Scholar]
- 67.Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166(2):1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kellogg EA. What happens to genes in duplicated genomes. Proc Natl Acad Sci U S A. 2003;100(8):4369–4371. doi: 10.1073/pnas.0831050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Emparan A, Quezada-Martinez D, Zuniga-Bustos M, Cifuentes V, Iniguez-Luy F, Laura Federico M. Functional Analysis of the Brassica napus L. Phytoene Synthase (PSY) Gene Family. Plos One. 2014;9(12). [DOI] [PMC free article] [PubMed]
- 70.Dong S, Adams KL. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol. 2011;190(4):1045–1057. doi: 10.1111/j.1469-8137.2011.03650.x. [DOI] [PubMed] [Google Scholar]
- 71.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 72.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci U S A. 2003;100(8):4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Renny-Byfield S, Gallagher JP, Grover CE, Szadkowski E, Page JT, Udall JA, et al. Ancient Gene Duplicates in Gossypium (Cotton) Exhibit Near-Complete Expression Divergence. Genome Biol Evol. 2014;6(3):559–571. doi: 10.1093/gbe/evu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Veitia RA. Paralogs in polyploids: One for all and all for one? Plant Cell. 2005;17(1):4–11. doi: 10.1105/tpc.104.170130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, et al. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci. 2015;112(11):3558–3563. doi: 10.1073/pnas.1420831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Nocker S, Gardiner SE. Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Horticulture Research. 2014;1. [DOI] [PMC free article] [PubMed]
- 77.Okamuro JK, Caster B, Villarroel R, VanMontagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci U S A. 1997;94(13):7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379(6):633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 79.Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007;145(3):1073–1085. doi: 10.1104/pp.107.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JM, Joung J-G, McQuinn R, Chung M-Y, Fei Z, Tieman D, et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012;70(2):191–204. doi: 10.1111/j.1365-313X.2011.04863.x. [DOI] [PubMed] [Google Scholar]
- 81.Chung M-Y, Vrebalov J, Alba R, Lee J, McQuinn R, Chung J-D, et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010;64(6):936–947. doi: 10.1111/j.1365-313X.2010.04384.x. [DOI] [PubMed] [Google Scholar]
- 82.Fujisawa M, Nakano T, Shima Y, Ito Y. A Large-Scale Identification of Direct Targets of the Tomato MADS Box Transcription Factor RIPENING INHIBITOR Reveals the Regulation of Fruit Ripening. Plant Cell. 2013;25(2):371–386. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo Z, Zhang J, Li J, Yang C, Wang T, Ouyang B, et al. A STAY-GREEN protein SlSGR1 regulates lycopene and beta-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013;198(2):442–452. doi: 10.1111/nph.12175. [DOI] [PubMed] [Google Scholar]
- 84.Toledo-Ortiz G, Huq E, Rodriguez-Concepcion M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2010;107(25):11626–11631. doi: 10.1073/pnas.0914428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saitou N, Nei M. The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 86.Sun Z, Gantt E, Cunningham FX., Jr Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem. 1996;271(40):24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]


