Abstract
It is widely accepted that Retinoblastoma protein (pRb) phosphorylation plays a central role in mediating cell cycle G1/S stage transition, together with E2 promoter-binding factors (E2F). The binding of pRb to E2F is controlled by the sequential and cumulative phosphorylation of pRb at various amino acids. In addition to the well characterized roles for pRb as a tumor suppressor, pRb has more recently been implicated in osteoprogenitor and other types of stem cell maintenance, proliferation and differentiation, thereby influencing the morphogenesis of developing organs. In this study, we present data characterizing the expression of three phosphorylated pRb (ppRb) isoforms - ppRbS780, ppRbS795, and ppRbS807/811- in developing mouse molar and incisor tooth buds. Also, we analyzed the co-localization of pRb isoforms and histone H3 expression in incisor tooth buds. Our results reveal distinct developmental expression patterns for individual ppRb isoforms in differentiating dental epithelial and dental mesenchymal cells, suggesting discrete functions for each in tooth development.
Keywords: Phosphorylated Retinoblastoma protein, Tooth development, Epithelial-mesenchymal interactions, Cell differentiation
Introduction
Retinoblastoma protein (Rb, pRb, pRb1, pRB), a tumor suppressor, plays a crucial role in many types of tumors (Lee, To et al. 1988). It is widely accepted that loss of pRb results in cancer – for example, loss of pRb in the developing human retina results in retinoblastoma tumor formation (Dunn, Phillips et al. 1988). Accumulating evidence indicates that the cell cycle withdrawal function of pRb is essential for tumor suppression, and that mutations in pRb can result in uncontrolled cell proliferation (Weinberg 1995). Cell cycle progression is regulated by the differential phosphorylation of pRb, which fluctuates from unphosphorylated, hypo-phosphorylated and hyper-phosphorylated status during the cell cycle, and regulates its ability to bind to E2 promoter-binding factors (E2F) and other transcription factors (Mittnacht 1998). Active pRb isoforms include newly synthesized unphosphorylated pRb, and hypophosphorylated pRb present in early G1 phase cells, which binds to E2Fs and represses their transcriptional activation. Cyclin-dependent kinase (CDK) complexes then hyperphorphorylate and inactivate pRb, freeing E2F, and allowing E2F to direct cells into S phase (DeCaprio, Ludlow et al. 1989; Ezhevsky, Ho et al. 2001). To date, 16 potential pRb phosphorylation sites have been identified, spanning the entire pRb amino acid sequence. Accumulating data suggest that site specific pRb phosphorylation regulates specific pRb-protein interactions. pRb binding to E2F is thought to be mediated either by cumulative phosphorylation (Brown, Phillips et al. 1999), or via discrete C-terminal or pRb insert domain phosphorylation (Knudsen and Wang 1997). Site specific pRb phosphorylation may play distinct roles in development and disease (Thakur, Siedlak et al. 2008).
In this study, we have used antibodies specific for three pRb phosphorylation sites (ppRbS780, ppRbS795, and ppRbS807/811) to systematically evaluate pRb phosphorylation status in developing mouse teeth. We found discrete expression patterns for each ppRb isoform, suggesting distinct roles for each in dental epithelial and dental mesenchymal cell interactions and differentiation.
Materials and Methods
Animal husbandry and dental tissue harvesting and preparation
Wild type mouse teeth were analyzed at four developmental stages: embryonic 12.5 days (E12.5), E14.5, E16.5 and Postnatal day 1 (P1). Samples were fixed in 10% neutral-buffered formalin overnight, embedded in paraffin, and serial sectioned. Haematoxylin and Eosin (H&E) staining was used to identify tooth containing regions, which were then used for immunohistochemical analyses. First molars from all four developmental stages, and P1 incisors were analyzed.
Immunohistochemical (IHC) analyses
Selected tooth containing sections were analyzed by IHC using the following antihuman pRb antibodies: polyclonal phospho Rb Serine 780 (ppRbS780, #9307, Cell Signaling Technology, Danvers, MA, USA); polyclonal phospho Rb Serine 795 (ppRbS795, #9301, Cell Signaling Technology); and polyclonal phospho Rb Serine 807/811 (ppRbS807/811, #9308, Cell Signaling Technology); according to the manufacturer’s protocol (Cell Signaling Technology). Vectastain ABC staining kit (Vector Laboratories, Burlingame, CA, USA) was used as detection method, along with DAB substrate and Fast Green counterstain. Immunostained samples were analyzed and imaged using a Zeiss Axiophot Imager Microscope and Digital camera (Zeiss, Frankfurt, Germany).
Immunofluorescent (IF) histochemical analyses
Double staining immunofluorescence assays were performed on selected paraffin sections using two primary antibodies: anti-phospho pRb Specific antibodies; and anti-phospho-histone H3 Serine 28 (ab11946, Abcam, Cambridge, MA). Primary antibody stained sections were then treated with Fluorescein (FITC)-Donkey anti- Goat IgG (#705-095-003, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), and Texas Red-Donkey anti Rabbit IgG (711-075-152, Jackson ImmunoResearch Laboratories, Inc.). Stained sections were mounted with VECTASHIELD HardSet Mounting Medium with DAPI (H-1500, Vector Laboratories), and digitally imaged using an Axiophot Imager Zeiss Microscope and Digital camera (Zeiss, Frankfurt, Germany).
Results
Expression of phosphorylated pRb isoforms in developing mouse molars
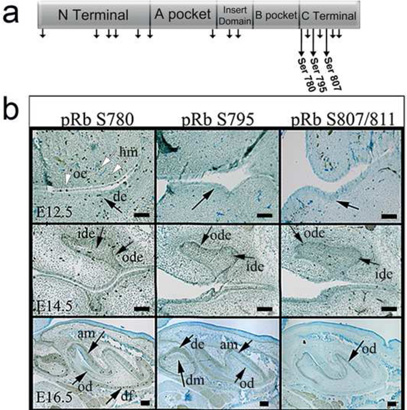
H&E staining was used to identify regions containing E12.5 bud stage, E14.5 cap stage, E16.5 bell stage, and P1 newborn mouse teeth of proper sagittal orientation, which we then used to examine the expression of ppRbS780, ppRbS795, and ppRbS807/811 (Figure 1a). E12.5, tooth buds are identifiable as a thickening of the dental epithelium (Figure 1b, E12.5, black arrows). No distinct tooth specific expression was observed at this time, although ppRb780 was detected in the nuclei of oral and dental epithelial (de) and head mesenchymal (hm) cells (Figure 1B, E12.5, white arrows). All three ppRb isoforms were detected in E14.5 stage teeth. ppRb780 exhibited punctuate expression in both the inner dental epithelium (ide) and outer dental epithelium (ode) of E14.5 stage teeth (Figure 1b, black arrows). ppRb795 was strongly expressed in the ide and weakly expressed in dental papilla, while ppRb807/811 exhibited punctate expression in the ide, ode, dental papilla, and dental follicle (df) cells (Figure 1b, black arrows). At E16.5, ppRbS780 was detected in differentiated ameloblasts (am), odontoblasts (od), and the dental follical (df) of M1 molar teeth (Figure 1b, black arrows). Strong ppRbS780 expression was also detected in the less developed M2 molar at this stage (see Figure 2e), indicating that ppRbS780 is expressed in both M1 and M2 dental epithelial and mesenchymal tissues. ppRb795 was detected in de and dm tissues of M2 molars, and in differentiated ameloblasts (am) and odontoblasts (od) of M1 molar teeth (Figure 1B, black arrows). In contrast, ppRb807/811 appeared restricted to differentiated odontoblasts (od) and undifferentiated ameloblasts, but was not detected in undifferentiated odontoblasts, or in differentiated ameloblasts at E16.5 (Figure 1b, black arrows).
Figure 1. Individual ppRb isoform expression in E12.5, E14.5 and E16.5 mouse tooth development.
a. Schematic of the pRb protein, with short arrows indicating identified phosphorylation sites throughout the pRb protein and with long arrows indicating the three ppRb isoforms examined in this study. b. Individual ppRb isoforms exhibit discrete expression in developmentally staged mouse teeth (arrows). Abbreviations: am, ameloblast; de, dental epithelium; dm, dental mesenchyme; df, dental follicle; ide, inner dental epithelium; od, odontoblast; ode, outer dental epithelium; oe, odontal epithelium; hm, head mesenchyme. Scale bars indicate 100µm.
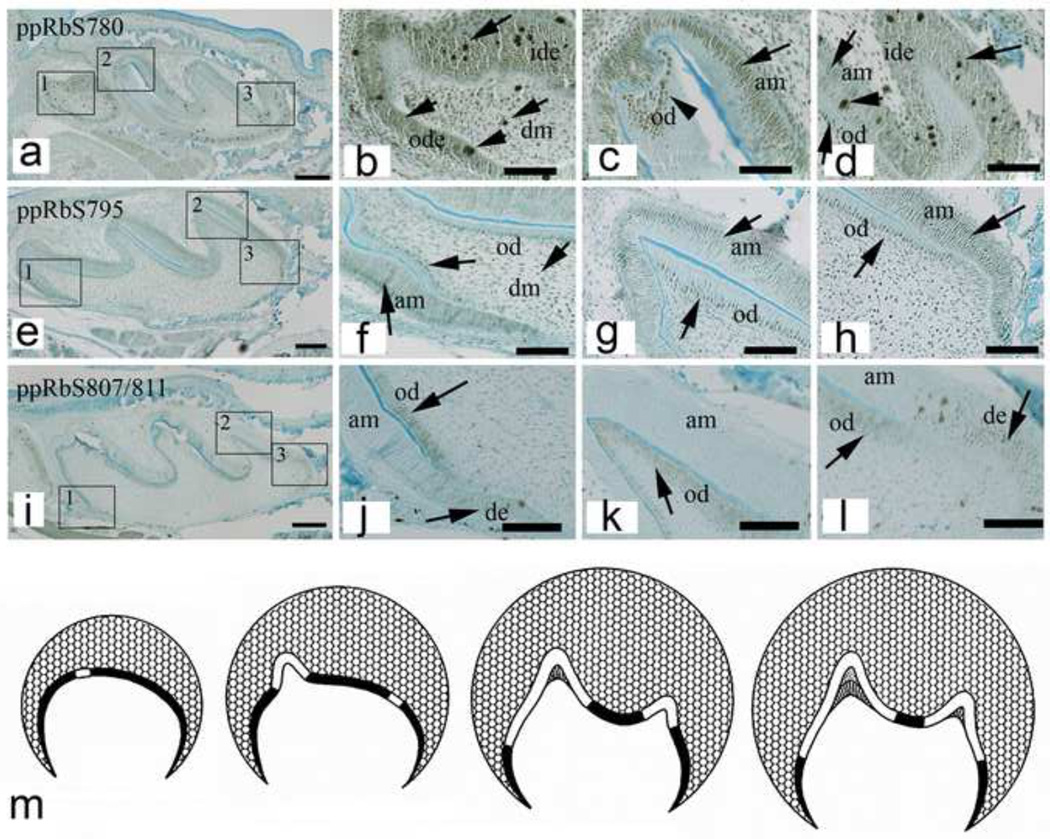
Figure 2. ppRb isoform expression in newborn (P1) mouse M1 molar teeth.
Individual ppRb isoforms exhibit discrete expression patterns in cusp and cervical loop regions of newborn M1 molar teeth (arrows). Boxes embedded in each left hand panels are shown enlarged in adjacent three right hand panels. The schematic molar tooth (m) indicates areas of dental epithelial cell proliferation (solid black areas), and non-proliferative zones (solid white areas). Abbreviations: am, ameloblast; de, dental epithelium; dm, dental mesenchyme; ide, inner dental epithelium; od, odontoblast; ode, outer dental epithelium. Scale bars indicate 100µm.
In newborn mice (P1) (Figure 2), ppRbS780 was detected in the ide, ode, and dm of M2 molars (Figure 2a), in differentiated ameloblasts (am) and odontoblasts (od) of M1 molars (Figure 2c,d, arrows), and in M1 molar undifferentiated ode (Figure 2d, arrows). ppRbS795 was detected fairly ubiquitously in all dental epithelial and mesenchymal cells, including both early progenitor and differentiated ameloblasts and odontoblasts (Figure 2e~h, arrows). ppRbS807/811 (Figure 2i) exhibited discrete expression restricted to undifferentiated dental epithelial cells, and differentiated odontoblasts (Figure 2i~l, arrows). Interestingly, ppRbS807/811 appeared restricted to proliferating dental epithelium, and was not detected in differentiated, non-proliferative dental epithelium (Figure 2i~l). And in contrast, ppRbS807/811 was not detected in proliferating dental mesenchymal cells, but was detected only in differentiated, non-proliferative dental mesenchyme derived odontoblasts, both in nuclei and cytoplasm (Figure 2i~l). The expression patterns of individual ppRb isoforms can be compared to the schematic shown in Figure 2m, which indicates proliferative dental epithelium in black, and non-proliferative, differentiated tissues in white.
Expression of phosphorylated pRb isoforms in mouse incisors
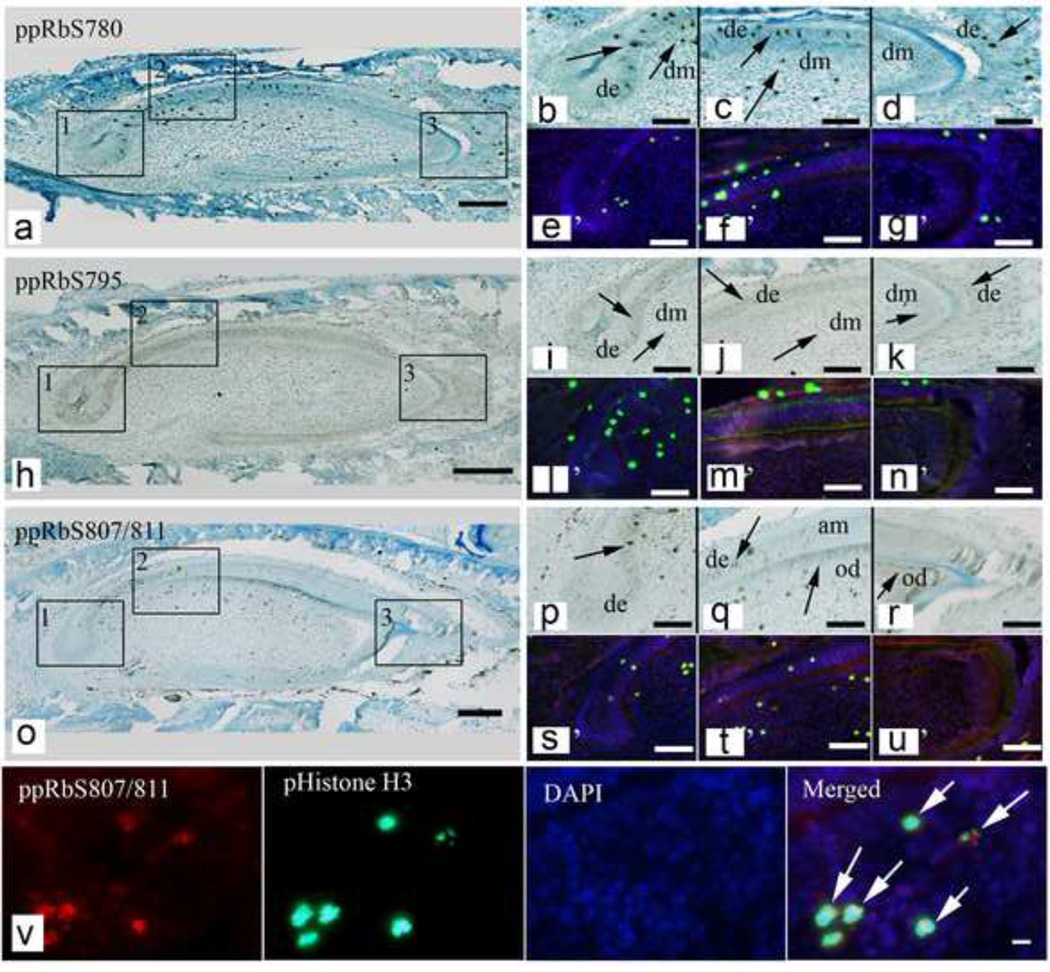
We next characterized ppRb isoform expression in P1 mouse incisors, taking advantage of the fact that the continuously erupting incisor model provides the opportunity to simultaneously analyze ppRb isoform expression in undifferentiated, differentiating, and fully differentiated tissues (Figure 3). We detected punctate ppRbS780 expression in dental epithelium including both the non-terminally differentiated labial cervical loop (LaCL), and the terminally differentiated lingual cervical loop (LiCL) (Figure 3a,b), and in the dental mesenchyme (Figure 3c). ppRbS780 was detected in dental epithelium at the tooth cusp (Figure 3d, arrow), but not in differentiated ameloblasts or odontoblasts. ppRbS795 exhibited fairly ubiquitous expression in both proliferating and differentiated dental epithelium and dental mesenchyme (Figure 3h~k). We detected strong ppRbS807/811 expression in the nuclei of dental epithelial progenitor cells located close to the cervical loop (Figure 3o, p), and detected no expression in differentiated ameloblasts (Figure o, q). In contrast, ppRbS807/811 was detected in both the cytoplasm and nuclei of differentiated dental mesenchymal cell derived odontoblasts located at the tooth cusps, and was not detected in less differentiated odontoblasts close to the cervical loop areas (Figure o, q, r). We next performed double IF staining for individual ppRb isoforms and phospho-histone H3, to correlate ppRb isoform expression with cell cycle progression. We found that all three ppRb isoforms were expressed in dividing dental mesenchymal and dental epithelial cells along with phospho-histone H3, (Figure 3, e~g, l~n, s~u). Enlarged images (Figure 3v) show an example of co-localized ppRbS807/811 and phospho-Histone H3 expression. Individual ppRb isoforms were also detected in non-dividing, Histone H3 negative cells (Figure 3). Schematic representations of color-coded ppRb isoform expression patterns in newborn (P1) M1 molars and incisors is shown (Figure 4).
Figure 3. ppRb isoform expression in newborn mouse incisor teeth.
Discrete ppRb isoform expression was detected in P1 mouse incisor teeth (a, h, o). Boxed areas embedded in each bright field panel are shown enlarged to the right. Primed panels are double immunofluorescent images of phospho-Histone H3 and each corresponding ppRb isoform expression. Panel v. IF images of ppRbS807/811 demonstrate overlapping ppRbS807/811 and phospho-Histone H3 expression (arrows). Abbreviations: am, ameloblast; de, dental epithelium; dm, dental mesenchyme; LaCL, labial cervical loop; LiCL, lingual cervical loop; od, odontoblast. Scale bars indicate 100µm (A-C) or 10 µm (D).
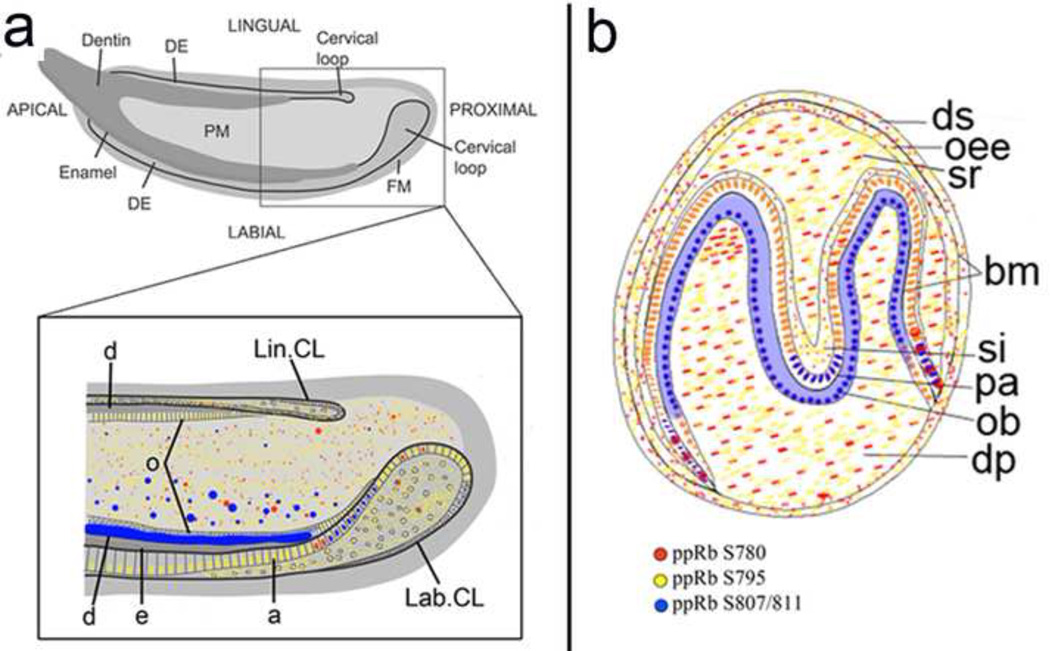
Figure 4. Schematic of ppRb isoform expression in mouse molar and incisor teeth.
Schematic colorized drawings of individual ppRb isoform expression in newborn mouse M1 molar and incisor teeth. ppRbS780 expression is indicated in red, ppRbS795 expression is shown in yellow, and ppRbS807/811 expression is indicated in blue. Overlapping expression of ppRbS780 and ppRbS795 is indicated in orange (Panel b, ameloblast, am). Abbreviations: am, ameloblast; bm, basement membrane; DE, dental epithelium; dp, dental papilla; ds, dental sac; Lab. CL, labial cervical loop; Lin. CL, lingual cervical loop; od, odontoblast; oee, outer enamel epithelium; pa, pre-ameloblasts; si, stellate reticulum; sr, stellate reticulum.
Discussion
pRb, one of the first identified tumor suppressor genes, participates in a variety of cellular functions including cell division, differentiation, senescence, and apoptosis (Poznic 2009). Recently, the mechanism of pRb phosphorylation in cell growth control has become an important area of study (Brugarolas, Moberg et al. 1999). To our knowledge, no report of pRb isoform expression in tooth development has been published to date. One report of pRb phosphorylation in an ameloblastoma, a benign tumor originating from odontogenic epithelium, confirmed that aberrant Rb expression was related to this type of oral carcinoma (Pande, Mathur et al. 1998), and demonstrated pRb and ppRb expression in the nuclei of inner enamel epithelium and stratum intermedium cells. Another study reported that pRb expression correlated with neoplastic odontogenic epithelial cell proliferation (Kumamoto and Ooya 2006).
In the results presented here, we show that the three ppRb isoforms analyzed in this study - ppRbS780, ppRbS795, and ppRbS807/811 - were detected in both dental epithelial and dental mesenchymal cells and derived tissues (Schematized in Figure 4). All three ppRb isoforms were co-expressed with Histone H3 in dividing cells, suggesting roles in progression to the cell division stage of the cell cycle (Figure 3). Importantly, all three ppRb isoforms were also expressed in non-dividing, phosphor-Histone H3 negative cells. Interestingly, ppRbS807/811 was detected only in immature dental epithelial cells and not in differentiated ameloblasts, and in differentiated dental mesenchymal cell derived odontoblasts, and not in immature progenitor dental mesenchymal cells. This distinct “switch” in ppRbS807/911 expression (see Figure 3o) suggests roles for ppRbS807/811 in mediating epithelial/mesenchymal cross-talk required for odontoblast and ameloblast differentiation.
pRb, together with p107 and p130, control the cycle via phosphorylation by CDK (Cobrinik 2005). pRB null mice died in utero by E14.5, and exhibited defects restricted to cells of the central nervous system and developing erythropoietic cells (Clarke, Maandag et al. 1992). p107/p130 double null mice showed impaired terminal differentiation of interfollicular epidermal keratinocytes, and delayed and abnormal hair and tooth development, including odontoblast hypoplasia. Study of pRb function in the epidermis revealed roles in guiding morphogenetic events leading to the formation of specialized ectodermal organs (Ruiz, Segrelles et al. 2003). Roles for pRb family members in tissue morphogenesis are supported by the finding that they can form functional complexes with transcription factors containing paired-like homeodomains (Wiggan, Taniguchi-Sidle et al. 1998). One such factor, Alx-4, is expressed only at sites of epithelial-mesenchymal interactions, including the developing tooth bud (Hudson, Taniguchi-Sidle et al. 1998).
Cdks are serine/threonine kinases that are activated through binding with a regulatory subunit, cyclin (Endicott, Noble et al. 1999). Although the functions of pRb in tooth development remain to be determined, cell cycle regulators such as cyclins and CDK inhibitors have been identified in tooth germs, suggesting that odontogenic cell fates can be influenced by the cell cycle (Kumamoto, Kimi et al. 2001). To date, at least four Cdks - Cdk1, Cdk2, Cdk4, and Cdk6 - have been shown to participate in cell cycle regulation in mammalian cells. Cdk4 and Cdk6 are expressed in a variety of tissues and cell culture lines, and have distinct physiological roles in vivo although they appear to be biochemically indistinguishable (Meyerson and Harlow 1994; Kitagawa, Higashi et al. 1996). A recent report showed that Cdk4 and Cdk6 phosphorylate pRb with distinct amino acid residue specificities in vitro (Takaki, Fukasawa et al. 2005). Our demonstration of distinct ppRb isoform expression in developing teeth suggests that Cdk4 and Cdk6 may also be similarly differentially expressed.
pRb phosphorylation is regulated by phosphate stoichiometry, and site specific combinations of pRb phosphorylation sites. A better understanding of functional roles for individual pRb isoforms can only be achieved by evaluating pRb phosphorylation status at each site. The effect of pRb phosphorylation on cell cycle progression was examined in vitro, where Western blot analysis showed that ppRbS780, ppRbS795, and ppRbS807/811 phosphorylation was associated with the G0/S phase transition (Boylan, Sharp et al. 1999). Previous reports suggested that hypo-phosphorylated pRb is tightly bound to the nucleus, while hyper-phosphorylated pRb loosely binds to nucleus (Mittnacht and Weinberg 1991). The fact that hypo-phosphorylated pRb exhibits nuclear localization, while ppRb isoforms can exhibit both cytoplasmic and nuclear localization, suggests that site specific Rb phosphorylation mediates specific functions for Rb (Mittnacht, Lees et al. 1994). Another report indicated that ppRb phosphorylation status correlated with nuclear versus cytoplasmic localization in encephalitic midfrontal cortex neurons (Jordan-Sciutto, Wang et al. 2000). Similarly, our results indicate that ppRb phosphorylation status fluctuates during odontoblast and ameloblast differentiation. In particular, the distinct expression patterns of ppRbS780 and ppRbS807/811 suggest important roles dental cell division and differentiation. Additional evidence suggests that ppRb and E2F regulate cell-to cell contacts and adherens junction formation. Lack of pRb during telencephalon development results in ectopic proliferation of neural precursor cells, and cell autonomous neuronal cell migration defects (Ferguson, McClellan et al. 2005).
The rodent incisor contains two cervical loops – the labial cervical loop is larger, non-terminally differentiated, and contains populations of resident dental stem cells (Harada, Kettunen et al. 1999), while the smaller lingual cervical loop is terminally differentiated, and lacks stem cells. The labial cervical loop serves as a niche for dental epithelial stem cells whose progeny proliferate to form a pool of transit amplifying cells, which subsequently differentiate into ameloblasts or root epithelium. The discrete expression of ppRb isoforms in the labial cervical loop suggests distinct roles in dental epithelial and dental mesenchymal stem cell maintenance and differentiation. Most dramatically, that fact that ppRbS807/811 expression is restricted to undifferentiated dental epithelial cells, and to differentiated dental mesenchymal cells, suggests important roles in mediating dental epithelial/dental mesenchymal cell interactions leading to proper tooth morphogenesis and development. A recent report found that eliminating pRb production in osteoblasts had profound consequences on osteoblast cell adhesion, including altered cadherin expression and disruption of adherens junctions, resulting in bone structure abnormalities (Gutierrez, Kong et al. 2008; Sosa-Garcia, Gunduz et al. 2010). These data provide direct evidence that pRb depletion can profoundly affect the capacity of cells to interact with one another, and suggest possible roles for pRb as a regulator of adherens junction formation and cell adhesion in tooth development.
In summary, here we demonstrate, for the first time, discrete expression of ppRb isoforms S785, S790, and S807/811 in developing molar and incisor teeth (Figure 4, schematic). Importantly, ppRb isoforms were detected in both dental epithelial and dental mesenchymal cells and tissues, and ppRbS807/811 was found to be differentially expressed in differentiated versus undifferentiated dental epithelial and mesenchymal cells. Our results suggest roles for ppRb isoforms in regulating proper cell-cell interactions required for dental cell differentiation. Future studies will determine the mechanism of differential pRb phosphorylation. One possibility is that multiprotein complexes may limit access of Cdks to pRb (Zarkowska and Mittnacht, JBC 1997). In addition, the combinatorial phosphorylation of pRb at specific sites may regulate the binding of specific pRb partners, and modulate pRb’s influence on cell cycle control and differentiation (Knudsen and Wang, JBC 1996; Mittnacht, Curr Opinion Gen Dev 1998). Future studies will focus on elucidating individual ppRb isoform function in dental stem cell maintenance and differentiation.
ACKNOWLEDGEMENTS
The authors would like to thank the Yelick and Hinds lab members for support and expert advice.
Footnotes
We acknowledge that there are no perceived or actual conflicts of interest to disclose, and support from ARRA funded NIH/NIDCR grant DE016962 (PCY).
REFERENCES
- Boylan JF, Sharp DM, et al. Analysis of site-specific phosphorylation of the retinoblastoma protein during cell cycle progression. Exp Cell Res. 1999;248(1):110–114. doi: 10.1006/excr.1999.4389. [DOI] [PubMed] [Google Scholar]
- Brown VD, Phillips RA, et al. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol. 1999;19(5):3246–3256. doi: 10.1128/mcb.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Moberg K, et al. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after gamma-irradiation. Proc Natl Acad Sci U S A. 1999;96(3):1002–1007. doi: 10.1073/pnas.96.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, et al. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58(6):1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- Dunn JM, Phillips RA, et al. Identification of germline and somatic mutations affecting the retinoblastoma gene. Science. 1988;241(4874):1797–1800. doi: 10.1126/science.3175621. [DOI] [PubMed] [Google Scholar]
- Endicott JA, Noble ME, et al. Cyclin-dependent kinases: inhibition and substrate recognition. Curr Opin Struct Biol. 1999;9(6):738–744. doi: 10.1016/s0959-440x(99)00038-x. [DOI] [PubMed] [Google Scholar]
- Ezhevsky SA, Ho A, et al. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol Cell Biol. 2001;21(14):4773–4784. doi: 10.1128/MCB.21.14.4773-4784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KL, McClellan KA, et al. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 2005;24(24):4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez GM, Kong E, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci U S A. 2008;105(47):18402–18407. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147(1):105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R, Taniguchi-Sidle A, et al. Alx-4, a transcriptional activator whose expression is restricted to sites of epithelial-mesenchymal interactions. Dev Dyn. 1998;213(2):159–169. doi: 10.1002/(SICI)1097-0177(199810)213:2<159::AID-AJA1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Jordan-Sciutto KL, Wang G, et al. Induction of cell-cycle regulators in simian immunodeficiency virus encephalitis. Am J Pathol. 2000;157(2):497–507. doi: 10.1016/S0002-9440(10)64561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Higashi H, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15(24):7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17(10):5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto H, Kimi K, et al. Detection of cell cycle-related factors in ameloblastomas. J Oral Pathol Med. 2001;30(5):309–315. doi: 10.1034/j.1600-0714.2001.300509.x. [DOI] [PubMed] [Google Scholar]
- Lee EY, To H, et al. Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science. 1988;241(4862):218–221. doi: 10.1126/science.3388033. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14(3):2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8(1):21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- Mittnacht S, Lees JA, et al. Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 1994;13(1):118–127. doi: 10.1002/j.1460-2075.1994.tb06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnacht S, Weinberg RA. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65(3):381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- Pande P, Mathur M, et al. pRb and p16 protein alterations in human oral tumorigenesis. Oral Oncol. 1998;34(5):396–403. doi: 10.1016/s1368-8375(98)00024-4. [DOI] [PubMed] [Google Scholar]
- Poznic M. Retinoblastoma protein: a central processing unit. J Biosci. 2009;34(2):305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Segrelles C, et al. Abnormal epidermal differentiation and impaired epithelial-mesenchymal tissue interactions in mice lacking the retinoblastoma relatives p107 and p130. Development. 2003;130(11):2341–2353. doi: 10.1242/dev.00453. [DOI] [PubMed] [Google Scholar]
- Sosa-Garcia B, Gunduz V, et al. A role for the retinoblastoma protein as a regulator of mouse osteoblast cell adhesion: implications for osteogenesis and osteosarcoma formation. PLoS One. 2010;5(11):e13954. doi: 10.1371/journal.pone.0013954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki T, Fukasawa K, et al. Preferences for phosphorylation sites in the retinoblastoma protein of D-type cyclin-dependent kinases, Cdk4 and Cdk6, in vitro. J Biochem. 2005;137(3):381–386. doi: 10.1093/jb/mvi050. [DOI] [PubMed] [Google Scholar]
- Thakur A, Siedlak SL, et al. Retinoblastoma protein phosphorylation at multiple sites is associated with neurofibrillary pathology in Alzheimer disease. Int J Clin Exp Pathol. 2008;1(2):134–146. [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Wiggan O, Taniguchi-Sidle A, et al. Interaction of the pRB-family proteins with factors containing paired-like homeodomains. Oncogene. 1998;16(2):227–236. doi: 10.1038/sj.onc.1201534. [DOI] [PubMed] [Google Scholar]