Summary
Several terminal uridyltransferases (TUTases) are known to modulate small RNA biogenesis and/or function via diverse mechanisms. Here, we demonstrate that Drosophila splicing-derived pre-miRNAs (mirtrons) are efficiently modified by the previously uncharacterized TUTase, Tailor. Tailor is necessary and sufficient for mirtron hairpin uridylation, and this modification inhibits mirtron biogenesis. Genomewide analyses demonstrate that mirtrons are dominant Tailor substrates, and three features contribute to substrate specificity. First, reprogramming experiments show Tailor preferentially identifies splicing-derived miRNAs. Second, in vitro tests indicate Tailor prefers substrate hairpins over mature miRNAs. Third, Tailor exhibit sequence preference for 3′-terminal AG, a defining mirtron characteristic. Our work supports that Tailor preferentially suppresses biogenesis of mirtrons, an evolutionarily adventitious pre-miRNA substrate class. Moreover, we detect preferential activity of Tailor on 3′-G canonical pre-miRNAs, and specific depletion of such loci from the pool of conserved miRNAs. Thus, Tailor activity may have had collateral impact on shaping populations of canonical miRNAs.
Graphical Abstract
Introduction
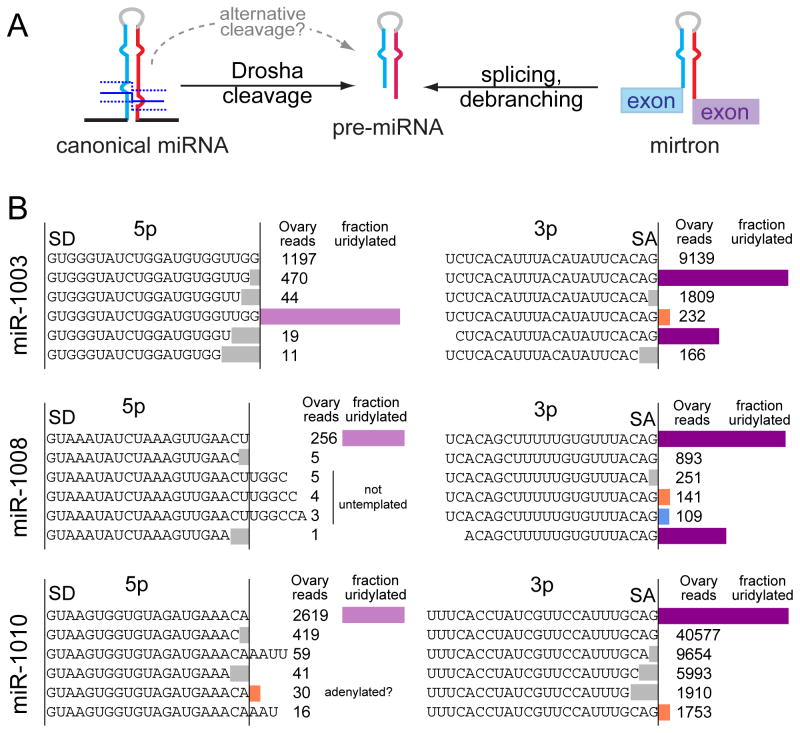
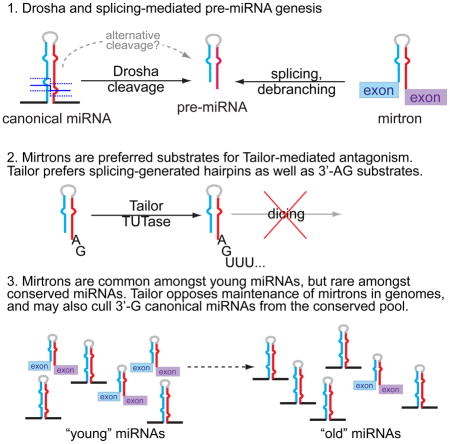
microRNA (miRNA) genes generate a major class of Argonaute-associate short RNA (sRNA), and are minimally defined by hairpin-bearing transcripts that are hewn into relatively specific regulatory sRNAs (Axtell et al., 2011). In animals, cleavage of a pri-miRNA transcript by the nuclear RNase III Drosha yields a pre-miRNA hairpin, which is then cleaved by the cytoplasmic RNase III Dicer. While most miRNAs are made by this canonical pathway, an alternative pathway was established with mirtrons, short hairpin introns that use splicing to bypass Drosha cleavage (Okamura et al., 2007; Ruby et al., 2007) (Figure 1A). The study of alternative miRNA biogenesis pathways now comprises both Drosha-independent and Dicer-independent strategies (Maurin et al., 2012; Yang and Lai, 2011), and non-canonical mechanisms continue to be identified (Okamura et al., 2013; Xie et al., 2013).
Figure 1.
Preferred uridylation of hairpins from an alternative miRNA biogenesis pathway. (A) Pre-miRNA hairpins can be generated by Drosha-mediated cleavage, or by splicing of a short hairpin intron (mirtron). (B) In general, most miRNA reads do not carry untemplated additions. However, many mirtron-3p species exhibit high rates of terminal uridylation, in some cases comprising the dominant species. Shown are three mirtrons used as models in this study. The fraction of uridylated species are calculated with respect to the splicing-derived species. Their corresponding mirtron-5p species essentially lack untemplated modifications, reflecting that uridylation likely occurred at the hairpin stage.
Curiously, while most well-conserved miRNAs are canonical, recently-emerged miRNAs in flies, worm, mouse and human are strongly represented by mirtrons (Berezikov et al., 2010; Chung et al., 2011; Ladewig et al., 2012). Therefore, mirtrons exhibit increased evolutionary flux relative to canonical miRNAs (Berezikov et al., 2010; Mohammed et al., 2013). In this light, mirtrons might conceivably contribute preferentially to species-specific regulation. However, notions of their overall regulatory impact are tempered by their typically modest accumulation, as is the case for most species-specific miRNAs. An alternate consideration is that newly-evolved miRNAs may be more likely to incur detrimental than beneficial effects (Chen and Rajewsky, 2007). In this view, if splicing fortuitously spawns many functional sRNAs, then a strategy to counteract the evolutionary emergence of mirtrons might be desirable.
While summary diagrams of sRNA biogenesis imply inexorable processes, biological pathways are usually regulated. Indeed, diverse modifications of sRNA pathway factors and substrates are documented to affect sRNA biogenesis and/or function (Ha and Kim, 2014). In particular, diverse terminal modifications have been detected for pre-miRNAs (Li et al., 2013; Newman et al., 2011) and mature small RNAs (Burroughs et al., 2010; Wyman et al., 2011), and terminal uridyltransferases (TUTases) with sRNA specificity exist in plants, worms, and mammals. For example, TUT4 and TUT7 can act with the Lin28 RBP to uridylate and degrade pre-let-7 (Hagan et al., 2009; Heo et al., 2008; Heo et al., 2009; Thornton et al., 2012; Viswanathan et al., 2008). This mechanism involves oligouridylation of pre-let-7 hairpins by Lin28/TUT4 complex, which recruits the Dis3L2 nuclease for substrate degradation (Chang et al., 2013). Interestingly, in the absence of Lin28, several enzymes (TUT4/7 and TUT2) monouridylate “group II” pre-let-7 family members that contain a single-nt 3′ overhang following Drosha cleavage, thereby facilitating Dicer processing (Heo et al., 2012). TUT4/7 were recently described to play broader roles in quality control during pre-miRNA biogenesis, by modifying defective substrates and recruiting the RNA exosome to degrade them (Liu et al., 2014).
We earlier analyzed 3′ uridylation and adenylation of Drosophila miRNAs (Berezikov et al., 2011). Adenylated reads are not biased to either hairpin arm, and the non-canonical poly-A polymerase (PAP) Wispy adenylates maternal mature miRNAs, marking them for degradation during the maternal-to-zygotic transition (Lee et al., 2014). In contrast, greater aggregate uridylation of miRNA-3p vs. miRNA-5p species suggests a pre-miRNA preference (Berezikov et al., 2011). Curiously, the most highly uridylated miRNAs in flies were dominated by mirtron-3p reads, and preferred mirtron uridylation is conserved in other animals (Westholm et al., 2012).
In this study, we identify CG1091 (“Tailor”) as the Drosophila mirtron uridylase. Loss- and gain-of-function assays show Tailor is necessary and sufficient to induce mirtron tailing, an inhibitory modification. We use structure-function analyses and genomewide profiling to demonstrate that mirtrons are preferred substrates of Tailor. Nevertheless, canonical pre-miRNAs can be Tailor substrates, especially if they end in guanine. In vitro tests demonstrate Tailor prefers hairpins over mature miRNAs and also prefers terminal substrate 3′-G, thus fulfilling structural and sequence characteristics of mirtrons. Finally, we detect a corollary impact of uridylation of canonical 3′-G pre-miRNAs, as such loci are specifically depleted amongst well-conserved (but not recently-emerged) canonical miRNAs. Altogether, these data unveil a miRNA modification enzyme with biogenesis-dependent substrate specificity, which is tailored to suppress the evolutionarily adventitious mirtron pathway.
Results
Functional screening identifies CG1091 as the Drosophila mirtron uridylase
Mirtron-3p species dominate the catalog of highly uridylated Drosophila small RNAs (Westholm et al., 2012). In some cases, uridylated reads are more abundant than “AG” splice-terminal reads (Figure 1B), and the lack of modified mirtron-5p species (Figure 1B) suggests that mirtron hairpins incur tailing.
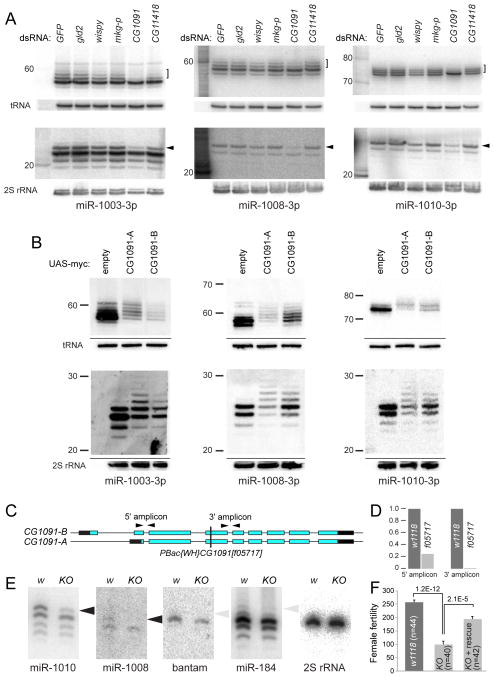
We performed Northern blotting using sequencing gels to provide single-nt resolution of hairpins. As pre-miRNAs accumulate modestly, we sensitized these tests by transfecting S2 cells with mirtron constructs. Assays of mirtrons mir-1003, mir-1008 and mir-1010 revealed a series of pre-miRNA bands (Figure S1A). To demonstrate this as tailing, we sought to reduce such modifications via RNAi screening of candidate nucleotidyltransferases. Having validated effective knockdowns (Figure S1B,C), we observed that loss of CG1091 depleted larger mirtron hairpin and mature mirtron-3p species (Figure 2A). Quantifications of replicate tests are shown in Figure S1D,E. Co-depletion assays showed only treatments including CG1091 dsRNA affected mirtron tailing, and to an extent comparable to single CG1091 knockdown (Figure S1B, F).
Figure 2.
Functional screening identifies CG1091 (Tailor) as the Drosophila mirtron uridylase. (A) Knockdown screening of a panel of terminal nucleotidyltransferases in S2 cells transfected with various mirtron plasmids; shown are Northern blots probed for the indicated small RNA species. Depletion of CG1091 selectively caused loss of tailed mirtron hairpins (brackets) and tailed mirtron-3p miRNAs (arrowheads). (B) Co-expression of CG1091-A or CG1091-B with the indicated mirtron constructs strongly enhanced accumulation of tailed hairpin and mature 3p species. (C) A transposon allele that disrupts the common open reading frame of CG1091 isoforms. (D) qPCR analyses show loss of CG1091 transcripts, especially distal to the insertion, confirming it as a knockout (KO). (E) Northern analysis of mature small RNAs from wildtype and KO ovaries shows loss of normally abundant tailed mirtron-3p miRNAs. In contrast, there are only modest CG1091-dependent species for the canonical miRNA-3p species of bantam and miR-184. (F) Loss of CG1091 reduces numbers of progeny per female fly, and is rescued by a genomic transgene. Error bars represent SEM and student’s t-tests were used to calculate significance. See also Figures S1 and S2.
We performed reciprocal tests by overexpressing CG1091. Two annotated isoforms differ at their N-termini but exhibit similar functional domains, including a single PAP/TUTase domain. When we expressed myc-tagged CG1091-A and CG1091-B with mirtron constructs, we observed robust polyuridylation of their pre-miRNAs (Figure 2B). This correlated with tailing of mature mirtron-3p species (Figure 2B), but not of partner mirtron-5p species (Figure S2A,C). Quantification of replicate tests indicated greater tailing of hairpins than mature species (Figure S2B,D). Furthermore, ectopic CG1091 proteins were active in cells depleted of endogenous CG1091 (Figure S2E).
To assess if CG1091 executes a similar function in the animal, we characterized the piggyBac insertion [f05717] (Figure 2C). We did not detect CG1091 transcripts 3′ of its insertion site (Figure 2D), suggesting it is a null allele. Northern analysis of RNAs from wildtype and CG1091[f05717] ovaries showed loss of mature, modified species of miR-1010 and miR-1008 (Figure 2E). We also note a minor degree of CG1091-dependent tailing of the canonical miRNAs miR-184 and bantam (Figure 2E). However, the amounts of modified canonical miRNAs were very modest compared to mirtrons.
Although the original CG1091[f05717] mutant stock was subviable, its vigor increased following outcrossing (see Methods). Nevertheless, quantification showed that loss of CG1091 compromises female fertility, and this was rescued by a CG1091 genomic transgene (Figure 2F). Therefore, this TUTase has molecular and physiological impact on the ovary. Altogether, we show that CG1091 is the major mirtron uridylase in cells and intact Drosophila, that it is necessary and sufficient for this process, and that both its isoforms share this functional capacity. Moreover, these data imply that it prefers mirtrons over canonical pre-miRNAs as substrates, consistent with known distributions of uridylated small RNAs (Westholm et al., 2012). We refer to CG1091 as “Tailor”, to reflect that it preferentially tails a specific sRNA population.
In vitro characterization of Tailor as a terminal uridyltransferase
We assessed activities of Tailor using in vitro assays. Initially, we incubated radiolabeled miR-1010-3p with lysates from S2 cells treated with GFP or Tailor dsRNA, supplemented with individual ribonucleotides. We specifically observed tailing activity in control GFP knockdown lysate that was supplemented with uridine, and this activity was lost in lysates depleted of Tailor (Figure S3A).
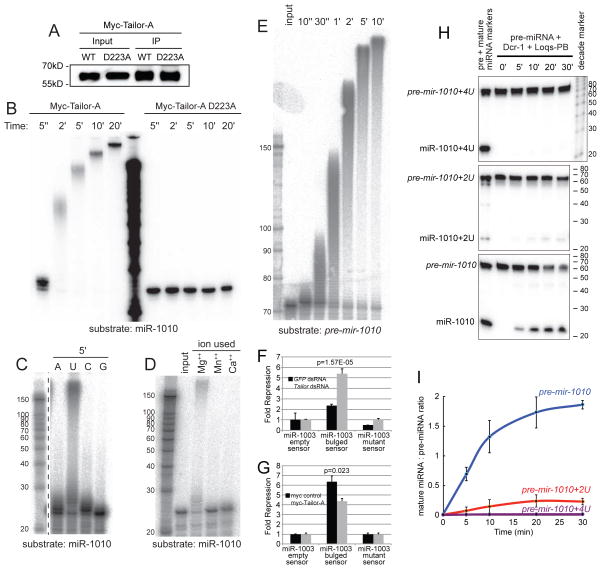
We performed direct tests using myc-Tailor-A immunopurified from transfected S2 cells (Figure 3A). Such material could uridylate miR-1010-3p within seconds, and generated tails of hundreds of nucleotides within minutes (Figures 3B and S3B). When less IP material was used, less substrate was modified, but these products still received long tails (Figure S3B). By contrast, a variant Tailor protein bearing a single mutation in the polβ-type catalytic site completely lacked tailing activity (Figure 3A,B). Bacterially-expressed Tailor also exhibited tailing activity (Figure S3C). Thus, Tailor is a bona fide terminal uridyltransferase.
Figure 3.
Tailor is a uridyltransferase that inhibits mirtron biogenesis and activity. (A) Immunopurified wildtype and catalytically inactive myc-Tailor from S2 cells, detected with anti-myc, used for subsequent tailing assays. (B) Timecourse assay of myc-Tailor incubated with radiolabeled mature miR-1010 in the presence of uridine. Its robust tailing activity is dependent on its catalytic site. (C) Tailor only generates extensively tailed products using uridine; 5′ reaction time. (D) Tailor is dependent on Mg++ ions. (E) Tailor is highly efficient on a mirtron hairpin (pre-mir-1010) substrate. (F, G) Reporter assays in S2 cells transiently expressing ub-Gal4, UAS-mir-1003 (mirtron) and the indicated luciferase sensors. Tailor knockdown enhances mirtron activity (F) while Ectopic Tailor antagonizes mirtron activity (G). Error bars represent SD and unpaired T-tests were used to calculate significance. (H) In vitro dicing of unmodified and uridylated pre-mir-1010 hairpins by recombinant Dcr-1/Loqs-PB. Tailing inhibits (for 2U) or abolishes (for 4U) the accumulation of mature species. (I) Quantification of three dicing assays; standard deviations are shown. See also Figure S3.
We compared tailing efficacies in the presence of different ribonucleotides. Uridine was the only one that supported formation of long tails, adenine was used much less efficiently, and guanine was not utilized (Figure 3C). Curiously, a majority of the substrate was modified when cytidine was present, but most of the product received only a single addition (Figure 3C). Assays of divalent cations shows that addition of Mg++, but neither Mn++ nor Ca++, supported efficient tailing with uridine (Figure 3D). Finally, given our presumption that Tailor modifies mirtrons as hairpin substrates, we examined its capacity to do so in vitro. We synthesized the full pre-mir-1010 hairpin, and observed this was very efficiently extended with uridine tails (Figure 3E). Overall, these tests provide biochemical validation of Tailor as a Mg++-dependent terminal uridyltransferase that acts on mirtron hairpins.
Uridylation inhibits mirtron activity
Although uridylation does not eliminate mirtron processing (Figure 1), we assayed its potential impact on mirtron function. To test the influence of endogenous uridylation, we treated S2 cells with GFP or Tailor dsRNA, then transfected them with mir-1003 expression construct and a luciferase sensor bearing four bulged miR-1003 target sites. We observed that depletion of Tailor increased target repression exerted by miR-1003 (Figure 3F). In reciprocal tests, we co-expressed myc-Tailor-A and mir-1003 expression constructs. Compared to co-transfection of empty myc vector, ectopic Tailor reduced the efficacy of mirtron-mediated target repression (Figure 3G). Therefore, Tailor-mediated modification antagonizes mirtron activity.
We considered that mirtron polyuridylation might inhibit dicing, as shown for pre-let-7 bearing long (~20 nt) tails (Heo et al., 2008). However, it was later shown that metazoan Dicers measure substrate cleavage relative to the 5′ end (Park et al., 2011), in which case shorter tails might be accommodated. To test the consequences of limited mirtron tails, we compared in vitro dicing efficiency of synthetic pre-mir-1010 bearing the 2-nt 3′ overhang expected from debranching of the spliced intron, relative to pre-mir-1010 bearing 2 or 4 terminal uridines (pre-mir-1010+2U or +4U). We radiolabeled these purified hairpins and incubated them with recombinant Dicer-1/Loqs-PB complex. Relative to dicing of the unmodified substrate, processing of the mirtron bearing the 2-nt uridine tail was less efficient, and that of the 4-nt uridine tail nearly abrogated (Figure 3H,I). Therefore, short 3′ tails, in the range of ones observed endogenously, inhibit mirtron processing.
Biogenesis-selective pre-miRNA substrate preference of Tailor
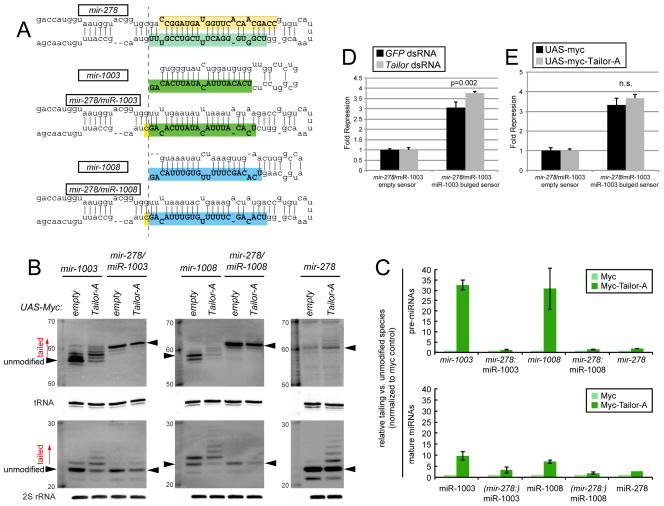
The fact of abundant uridylation of all mirtron-3p species but not of bulk canonical miRNA species (Westholm et al., 2012) suggests their seemingly similar hairpin precursors are somehow distinguished within cells. We tested whether biogenesis strategy influences the efficiency of pre-miRNA modification by swapping canonical and mirtron backbones. To do so, we reprogrammed mature miR-1003-3p and miR-1008-3p into the canonical miRNA backbone of mir-278 to generate mir-278/miR-1003 and mir-278/miR-1008 hybrid constructs (Figure 4A).
Figure 4.
Impact of biogenesis strategy on substrate recognition by Tailor. (A) The mature-3p species of mirtrons mir-1003 and mir-1008 were reprogrammed into the canonical mir-278 backbone. (B) Northern analysis of natural and reprogrammed mirtron constructs transfected into S2 cells, with or without Tailor expression construct. Robust tailing of mirtron hairpins and mature products is abrogated when their sequences are directed through the Drosha pathway. (C) Quantification of replicate experiments; SD is shown. The ratios of tailed to unmodified species (marked with arrowheads in B) in the presence of ectopic Tailor were normalized to parallel myc control conditions. The analysis shows mirtrons are preferred Tailor substrates, and their hairpins accumulate more tailed species than their mature miRNAs. (D, E) Reporter assays in S2 cells transiently expressing ub-Gal4, UAS-miRNA and the indicated luciferase sensors. (D) Activity of the reprogrammed mir-278/miR-1003 construct is less enhanced in Tailor-depleted cells than wildtype mir-1003 (Figure 3F). (E) Reprogrammed mir-278/miR-1003 construct is not inhibited by Tailor overexpression, as is wildtype mir-1003 (Figure 3G). Error bars represent SD and unpaired T-tests were used to calculate significance.
Analysis of these constructs yielded a clear picture. As shown earlier, mirtron hairpins of mir-1003 and mir-1008 were strongly uridylated in a Tailor-dependent fashion when expressed from their endogenous splicing context (Figure 4B). In contrast, their tailing decreased substantially when expressed as canonical pre-miRNAs. This was true within both pre-miRNA and mature-3p populations (Figure 4B). Analysis of mir-278 showed that its pre-miRNA was also only modestly tailed (Figure 4B). Quantification of substrate tailing in replicate experiments emphasized the strong majority of mirtron hairpins and resultant miRNA-3p species are elongated, whereas tailing incurred to canonical pre-miRNAs and miRNAs comprised only a minor fraction (Figure 4C). Moreover, the fraction of tailed mirtron-derived products was smaller in the mature pool than the hairpin pool (Figure 4B,C), in keeping with the notion that tailing inhibits dicing. Consistent with these data, the activity of mir-1003 reprogrammed into a canonical miRNA backbone was less sensitive to both Tailor-knockdown and overexpression (Figure 4D,E), relative to the endogenous mirtron context (Figure 3F,G). Altogether, these data define the first TUTase that exhibits miRNA substrate preference according to biogenesis class.
Genomewide analysis confirms mirtrons as dominant Tailor substrates
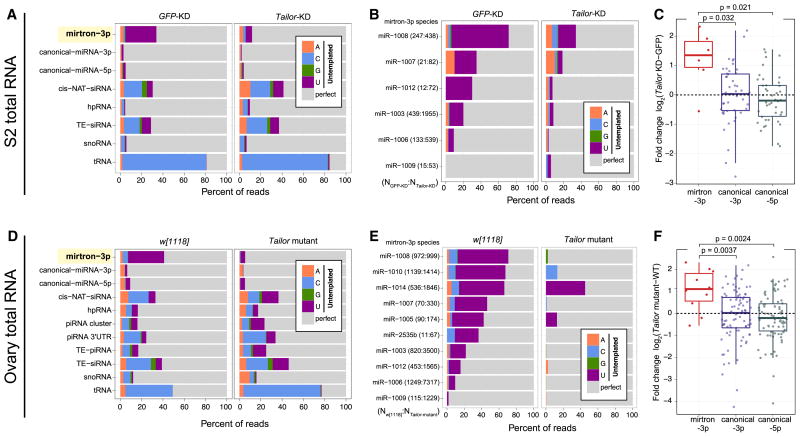
We extended analyses of Tailor substrate preferences genomewide. We began by comparing small RNA data from S2R+ cells with those depleted of Tailor (Table S1). Although various classes of sRNAs had detectable untemplated products, mirtron-3p species were by far the dominant uridylated species (Figure 5A). (Note tRNA fragments exhibit untemplated cytidine in this analysis, due to CCA addition). Moreover, mirtron-3p species were modified by Tailor, since its depletion largely eliminated their uridylation whereas other uridylated populations were little affected. The individual mirtrons that were expressed in S2 cells behaved similarly (Figure 5B). Notably, Tailor knockdown increased most individual mirtron-3p species, whereas there was no directional change in the levels of canonical miRNA-3p or miRNA-5p species (Figures 5C and S4A).
Figure 5.
Genomewide analysis of Tailor substrates in cultured cells and ovaries. Genome-matching reads are represented in gray while different untemplated nucleotides are color-coded. (A) Comparison of control and Tailor-depleted S2R+ cells demonstrates that mirtron-3p species are heavily uridylated, that this is nearly completely dependent on Tailor, and that other substrates are little affected by Tailor. (B) Analysis of individual mirtron-3p species shows their uridylation depends on Tailor. The number of mirtron reads in each library is shown in parentheses. (C) Expression levels of miRNAs in the presence and absence of Tailor. Most mirtron-3p species increase in Tailor-knockdown cells, whereas canonical miRNAs are not directionally affected. (D) Comparisons of w[1118] and Tailor-KO ovaries similarly shows that mirtron-3p species are the dominant class of Tailor-dependent, uridylated substrate. (E) Analysis of individual mirtron-3p species shows their Tailor-dependent modification. (F) Most mirtron-3p species increase in Tailor-knockout ovaries, whereas canonical miRNA species are not directionally affected. For analyses in A and D, all reads mapped to each sRNA class were pooled. For analyses in B–C and E–F, we used only mirtrons with ≥10 3p reads in both datasets under comparison. The box plots in C and F follow Tukey’s standard convention; significance was measured by t-test. See also Figure S4.
We confirmed these trends in the animal, by analyzing small RNA libraries from wildtype and Tailor-KO ovaries. Once again, mirtron-3p species were the dominant uridylated species in vivo, and comparison with Tailor mutant ovaries demonstrate that mirtron-3p species were its major in vivo substrate (Figure 5D). The diversity of mirtrons expressed in the ovary is greater than in S2R+ cells, and mirtron uridylation is overall more robust in ovaries than S2 cells (Westholm et al., 2012). Accordingly, the modification behavior of individual mirtrons presented a clearer picture of their nearly universal Tailor-made 3′ untemplated uridines (Figure 5E). Also consistent with S2R+ analysis, mutation of Tailor specifically increased the accumulation of most mirtron-3p miRNAs (Figure 5F and S4B). Overall, these genomewide data from cells and animals support that Tailor modifications to mirtrons are inhibitory to their biogenesis (Figures 2–4), and that mirtrons are preferred substrates of this enzyme.
Canonical miRNA-3p Tailor substrates are enriched for terminal G and AG
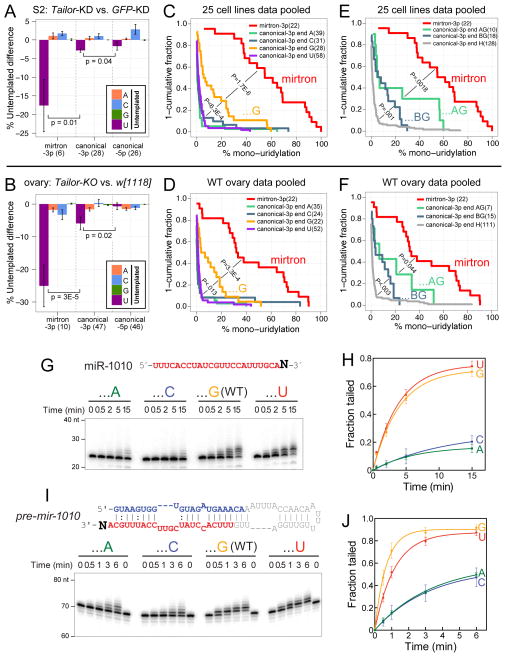
Closer inspection revealed a minor population of canonical miRNA-3p species in S2R+ cells and ovaries bore Tailor-dependent uridylation (Figure 6A,B). This trend was evident when considering loci that normally acquire ≥1% uridylation (detailed in Table S2). In aggregate, these modifications were greater for miRNA-3p than for 5p species (Figure 6A,B), reflecting canonical pre-miRNAs as Tailor substrates.
Figure 6.
Intrinsic activity of Tailor for 3′-G hairpin substrates. (A, B) Analysis of miRNAs with ≥10 reads and ≥1% untemplated uridylation in both paired datasets under comparison. In addition to the dominant uridylation of mirtron-3p reads, canonical miRNA-3p reads exhibit a greater degree of Tailor-dependent untemplated uridylation than do canonical miRNA-5p reads, in S2R+ cells (A) and ovaries (B). Wilcoxon rank-sum test (one-sided) was used to measure statistical significance; mean±SEM shown. (C–F) Analysis of miRNAs with ≥10 reads in the pooled cell line or ovary datasets (see Table S1). Wilcoxon rank-sum one-sided tests, incorporating multiple testing correction by Holm method, measured statistical significance of uridylation fraction enrichment between indicated small RNA classes. (C, D) Untemplated uridylation of mirtrons and canonical miRNA-3p species segregated by terminal nucleotide. While mirtrons exhibit far greater uridylation than canonical miRNAs, those canonical miRNA-3p species ending in G acquire greater uridylation than other miRNA subsets. (E, F) Terminal dinucleotide analyses of canonical miRNA-3p species and their uridylation. miRNAs ending in 3′-AG are more uridylated than other terminal G miRNAs (GG, CG, UG, i.e. “BG”), which in turn acquire greater uridylation than other terminal identities (A, U, C, i.e. “H”). This is true in both cell line (E) and ovary (F) data. (G–J) In vitro tailing assays of substrates bearing different 3′ nucleotides. To highlight substrate preferences, these assays utilize immunopurified Tailor-B material from stable cells, which is not as active as those in Figure 3. (G, H) Tailing assays on mature miR-1010 3′ variants. Tailor is most active on the normal 3′-G isoform, and also exhibits strong activity on the 3′-U variant. (I, J) Tailing assays on pre-mir-1010 3′ variants. Tailor maintains 3′-nt selectivity on hairpins, and tails them more effectively than mature miRNAs. See also Figure S5 and Tables S2 and S3.
We asked whether Tailor preferred particular canonical pre-miRNAs. In particular, we investigated potential impact of splice acceptor-like 3′ termini, which might mimic mirtron hairpins. We tested this by analyzing uridylation of canonical miRNA-3p species, segregated according to terminal nucleotides. Since tailing of canonical miRNAs is modest (Berezikov et al., 2011; Westholm et al., 2012), we enhanced our perspective by pooling large sets of ovary and cell line data (Wen et al., 2014) (Table S1).
As expected, mirtron-3p species were by far the dominant uridylated population in these aggregate data. Intriguingly, however, canonical miRNA-3p species ending in G were subject to greater untemplated uridylation than all other miRNA-3p species (ending in A/U/C, i.e. “H”). This was the case in cell lines (Figure 6C) and in ovaries (Figure 6D). We further segregated the 3′-G miRNA-3p species by terminal dinucleotides, and observed that 3′-AG canonical miRNAs collectively received greater uridylation than did 3′-BG miRNAs (where “B” represents U/C/G). Moreover, the trends of greater 3′-AG vs. 3′-BG uridylation, and of greater uridylation of 3′-BG vs. 3′-H miRNAs, was characteristic of both cell line (Figure 6E) and ovary (Figure 6F) datasets. Additional details of statistical comparisons between different classes of terminal-nt miRNAs are provided in Table S3. Overall, this is the first observation of differential modification of canonical miRNAs based on terminal nucleotides, and show that uridylation acts preferentially on terminal 3′-G and especially 3′-AG hairpins.
Intrinsic preference for terminal hairpin 3′-G substrates by Tailor
The above observations suggest that class-specific preference of Tailor for splicing-derived pre-miRNAs over Drosha-cleaved substrates may be partly driven by an intrinsic characteristic of the former, namely their invariant 3′ terminal nucleotide identities. We sought direct evidence for this scenario.
As the in vitro uridylation reactions were extremely efficient, we modified the Tailor assays to yield efficient, but less extensive tailing, of wildtype mature miR-1010 (see Methods). Using these conditions, we analyzed a panel of miR-1010 variants bearing the four different terminal nucleotides (Figure 6G). Timecourse assays demonstrated that normal miR-1010 bearing terminal G or U were the most favorable substrates. Quantification of multiple experiments shows that both the rate of modification and amount of input modified were greatest for the terminal G and U substrates (Figure 6H). These data are consistent with the notion that terminal G may be a preferred characteristic of de novo Tailor substrates, but as Tailor effectively generates extensive uridylation (Figure 3), we infer it should also have capacity to extend terminal U substrates effectively.
We performed similar experiments using pre-mir-1010 (Figure 6I). We observed qualitatively similar results as with miR-1010, except that the normal (terminal G) substrate was modified slightly more quickly than the terminal U variant (Figure 6J), consistent with the endogenous substrate. Notably, the modification of all hairpin substrates was enhanced relative to small RNA substrates. We observed greater yield of its optimal terminal G/U hairpin substrates in the 6 minute timecourse (Figure 6J) than in the 15 minute timecourse with corresponding small RNA substrates (Figure 6H).
We further analyzed hairpin variants with different penultimate nucleotide identities. For this purpose, we utilized pre-mir-1003, which is not as efficiently tailed as pre-mir-1010. We observed that the normal -AG mirtron was the best substrate, although this was only nominally better than the -GG variant; however, both of these were substantially better tailed than either -CG or -UG variants (Figure S5). Thus, Tailor exhibits structural and terminal dinucleotide substrate preferences that correlate with preferred recognition of mirtron hairpins.
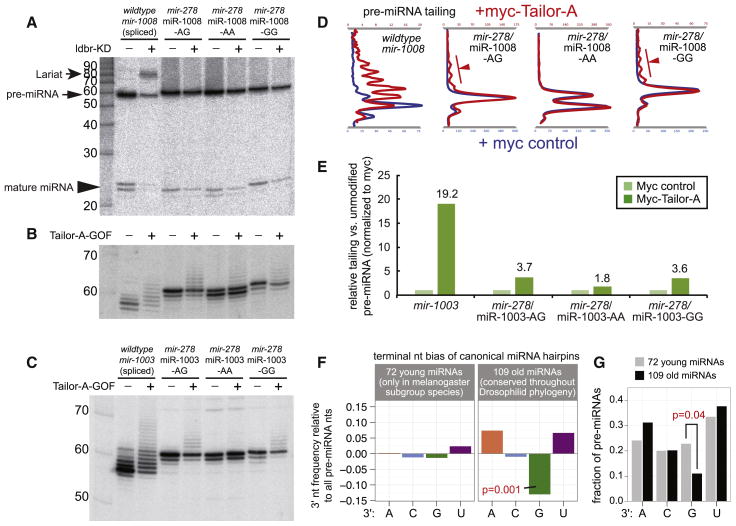
We complemented these in vitro assays by analyzing in vivo responses of ectopically expressed, reprogrammed, miRNA constructs. Since the mir-278/miR-1008 construct retains a potentially functional splice acceptor sequence, we assayed its behavior in cells depleted of lariat debranching enzyme (ldbr), whose activity is essential to linearize branched mirtron lariat intermediates so that they can adopt a hairpin conformation. Comparison of control and ldbr knockdown cells showed that transfection of mir-1008 constructs resulted in accumulation a lariat precursor and strong reduction of hairpin and mature miR-1008 (Figure 7A). In contrast, no lariats were detected when ldbr was knocked down in the presence of mir-278/miR-1008 constructs, with only minor reduction of processed forms (Figure 7A). Thus, mir-278/miR-1008 is not substantially spliced. We also constructed additional mir-278/miR-1008 variants in which the AG splice acceptor sequence was changed to AA or GG (Figure 7A).
Figure 7.
Terminal sequence-specific activity of Tailor and correlation with miRNA evolution. (A–C) Analysis of mirtrons and their hybrids in a canonical mir-278 backbone. These include a straight reprogramming of mirtron-3p sequences into mir-278 (including its 3′-AG splice acceptor), and variants where the splice site was altered to AA or GG. Expression constructs were transfected into S2 cells that were manipulated for ldbr or Tailor as indicated, and analyzed by Northern blotting. (A) Dependency of mir-1008 constructs on ldbr. Only the maturation of the unmodified mirtron is sensitive to ldbr depletion, resulting in lariat accumulation (arrow), reduction of pre-miRNA and loss of mature miR-1008. Note that ldbr knockdown cells are relatively poorly transfected, leading to generally decreased products from the canonical miRNA constructs. (B, C) Tailing response of different mir-1008 variants (B) and mir-1003 variants (C) to ectopic Tailor. (D, E) “Wiggle” plots (D) and bar plots (E) comparing relative levels of tailing of mirtron and reprogrammed canonical miRNA substrates. Splicing-derived pre-miRNAs are preferred Tailor substrates, but 3′-G also enhances Tailor modification within the canonical pre-miRNA context. (F) 3′-terminal bias of canonical pre-miRNAs. We compared frequencies of 3′-terminal nucleotides against background frequencies of all bases in canonical miRNA hairpins, separately for young and old loci. There is significant depletion of 3′-G specifically amongst well-conserved canonical miRNA hairpins, as determined by binomial test. (G) Direct comparison of 3′-terminal nucleotide frequency between young and old canonical pre-miRNAs. There is specific depletion of 3′-G amongst well-conserved miRNA hairpins, as determined by Fisher’s exact test. See also Figure S6 and Table S4.
We compared the capacity of Tailor to modify these constructs. Notably, the efficacy of Tailor-dependent tailing of miR-1008 sequences was reduced in all cases when they were introduced into the canonical miRNA backbone, relative to the endogenous splicing-dependent context (Figure 7A,B). Nevertheless, we observed a clear influence of terminal nucleotides on tailing, within the canonical miRNA context. That is, both mir-278/miR-1008 variants bearing terminal G were better-modified by Tailor than the terminal A variant (Figure 7A,B). To confirm these data, we generated similar terminal nucleotide variants of miR-1003-3p, reprogrammed into the mir-278 backbone. Again, we observe that 3′-G enhances the Tailor-dependent modification of canonical pre-miRNAs (Figure 7C). The separable impact of biogenesis strategy and 3′ nucleotide identity is emphasized by graphic visualization of tailing patterns (Figure 7D) and quantification of substrate tailing (Figure 7E).
Taken together, these data provide a mechanistic basis for the Tailor-dependent modification patterns observed on individual substrates and in genomewide surveys, and show how Tailor can preferentially modify mirtrons based on structural and terminal nucleotide features.
Conserved, but not newly-emerged, canonical miRNA hairpins are depleted for 3′-G
Mirtrons are frequent amongst newly-emerged Dicer substrates, but few mirtrons persist over evolution (Berezikov et al., 2010; Chung et al., 2011; Ladewig et al., 2012). We speculate that inhibitory Tailor-mediated modification of splicing-dependent miRNA biogenesis may reflect an evolutionary mechanism to suppress these adventitious miRNA substrates. If so, given that Tailor can modify canonical miRNAs, we might expect to observe differences in the distribution of 3′-terminal nucleotides amongst canonical miRNA hairpins.
We tested this scenario by segregating canonical miRNA loci according to their evolutionary age, as we earlier observed that “old” and “young” miRNAs exhibit distinct properties (Mohammed et al., 2013). We hypothesized if Tailor restricts miRNA evolutionary persistence, there might be depletion of well-conserved canonical miRNA-3p bearing terminal 3′-G. In principle, such differences might be less overt amongst recently-evolved miRNAs, which may not have been in existence for sufficient time for such culling action to operate.
This is indeed what we observed. Analysis of 181 canonical miRNA-3p loci showed 3′-G was the only nucleotide significantly depleted, compared to the nucleotide distributions across the genome or in pre-miRNA hairpins (Figure S6A). Notably, we observed distinct behaviors when we split these into 72 recently-emerged loci (i.e. present at most in the 5 melanogaster subgroup species) and 109 deeply-conserved loci (i.e. miRNAs present across all flies). Depletion of 3′-G was enhanced when restricting analysis to “old” canonical miRNAs (Figure 7F and S6B). Reciprocally, there was no bias in 3′-terminal nts amongst the “young” canonical miRNAs (Figure 7F).
We directly compared 3′ nucleotide properties of recently-evolved and well-conserved miRNAs. Using Fisher’s exact test, we observed specific depletion only of 3′-terminal G amongst the set of pan-Drosophilid miRNAs (Figure 7G, see also Figure S6C and Table S4). Finally, we did not observe depletion of 3′-terminal G amongst canonical miRNA-5p species, demonstrating specificity to the effect observed with 3p hairpin arms. In fact, guanine was enriched amongst conserved miRNA-5p species (Figure S6C).
Altogether, these data support the notion that Tailor-dependent modification of 3′-G terminal small RNA hairpins shapes miRNA evolutionary dynamics, both between substrate classes (e.g. Drosha- and splicing-mediated pathways) and within an individual substrate class (e.g. different terminal nucleotide hairpins produced by Drosha).
DISCUSSION
A Drosophila small RNA uridylase with substrate-selective activity
Many of the best-studied miRNA tailing pathways regulate specific substrates, such as pre-let-7 hairpins (Hagan et al., 2009; Heo et al., 2012; Heo et al., 2009; Thornton et al., 2012) or mature miR-122 species (Katoh et al., 2009). Tailing can also be deployed broadly, as seen with adenylation of maternal miRNAs (Lee et al., 2014). In plants, all miRNAs are generally protected from a tailing and degradation pathway by their acquisition of 3′ methylation (Rogers and Chen, 2013), and the lack of animal miRNA methylation underlies their susceptibility to tailing and degradation in the presence of artificial highly complementary targets (Ameres et al., 2010).
In this study, we define a new tailing mechanism that preferentially modifies and antagonizes miRNA substrates based on biogenesis strategy, namely splicing-derived miRNAs. In particular, we show that CG1091/Tailor prefers to uridylate the 3′ ends of mirtron hairpins, and to a lesser extent, canonical pre-miRNA hairpins. Its activity is reminiscent of vertebrate TUT4 and TUT7, which mediate terminal uridylation of let-7 pre-miRNAs (Norbury, 2013). On the other hand, that Tailor is a pre-miRNA-modifying factor was not obvious, since TUT4/TUT7 belong to a specialized TUTase class bearing duplicated polymerase domains, which do not exist in Drosophila.
A unique feature of Tailor is its capacity to distinguish substrates by terminal nucleotides. In vivo, in vitro, and bioinformatic analyses show that Tailor prefers 3′-G, and especially 3′-AG, substrates. While the in vitro preference of Tailor for endogenous pre-mir-1010 (3′-G) over the 3′-U variant is marginal (although 3′-A and -C are clearly disfavored, Figure 6I–J), we note Tailor exhibits greater 3′-G>U selectivity using pre-mir-1003 variants (Reimão-Pinto et al., 2015). In general, pre-mir-1010 is more efficiently tailed than pre-mir-1003 (compare Figure 6I–J with Figure S5), so the latter is more suited to highlight substrate discrimination. Nevertheless, as Tailor can extend long uridine tails, it must be able to add onto a 3′ substrate uridine, potentially in a processive mode. We speculate the initial recognition of Tailor hairpin substrates ending in 3′-AG may be preferred. Further studies of broader substrate panels and detailed kinetics, potentially using single-molecule assays, are needed to clarify these issues. As well, recombinant protein tests can determine if substrate structure/sequence-selective properties are truly intrinsic to Tailor, or are influenced by partners.
Biological and evolutionary significance of conserved mirtron-specific tailing
What is the utility of a pathway that suppresses splicing-derived miRNA biogenesis? Evolutionary considerations may provide context. While some general features of canonical pri-miRNAs are known (Auyeung et al., 2013; Han et al., 2006), it is still not possible to specifically identify bona fide miRNA loci genomewide without using small RNA or comparative data. However, it is possible to separate invertebrate mirtrons from amongst bulk introns (Chung et al., 2011). Considering the yield of invertebrate mirtrons from amongst hairpin introns of appropriate length (tens out of a few hundred) is greater than of canonical miRNAs from putative inverted repeats (100s out of 100,000s), a plausible inference is that de novo emergence of mirtrons is more prevalent than Drosha-processed miRNAs. This notion is supported by the fact that mirtrons contribute substantially to the pool of recently-evolved miRNAs in both insects (Berezikov et al., 2010) and vertebrates (Berezikov et al., 2007; Ladewig et al., 2012), even though few mirtrons are deeply-conserved. Notably, amongst hundreds of human and mouse mirtrons annotated via stringent evidence of small RNA duplexes, hardly any are shared between primates and rodents (Ladewig et al., 2012).
Newly-evolved miRNAs are proposed, on balance, as likely to mediate more detrimental regulatory interactions than beneficial ones (Bartel and Chen, 2004; Chen and Rajewsky, 2007). Therefore, only a minority of de novo miRNAs may eventually overcome the hurdles of incorporating into substantially useful regulatory networks and/or achieving efficient biogenesis. On this basis, we hypothesize that the facile emergence of mirtrons in diverse animal species may have necessitated implementation of a system that can preferentially identify and suppress mirtrons. This would create an additional evolutionary hurdle for mirtrons to overcome, in order to be incorporated into conserved networks. Ideally, such a system would antagonize, but not completely suppress miRNA biogenesis, due to potential collateral effects on canonical miRNA loci.
We provide molecular, genetic, biochemical, and genomewide data that Tailor is endowed with functional attributes that fulfill such a system. Tailor specifically uridylates mirtrons and inhibits their biogenesis, thereby imposing resistance to the general capacity for mirtrons to survive during evolution. A similar function for Tailor in mirtron regulation is reported in a companion study (Reimão-Pinto et al., 2015). This activity provides a molecular explanation for observed dynamics of accelerated turnover of mirtrons, relative to canonical miRNA loci. Moreover, this pathway appear to have had impact on reducing the representation of well-conserved, canonical, pre-miRNAs with mirtron-like 3′ termini. Thus, we uncover a molecular mechanism that imposes unsuspected evolutionary dynamics on different classes of miRNAs.
Experimental Procedures
Northern blotting
Probes and procedures for Northern blotting were described (Okamura et al., 2007). To obtain single-nucleotide resolution of 55–80 nt pre-miRNAs, 8 to 15 μg of total RNA was loaded on 12% polyacrylamide-urea sequencing gels (40cm long).
RNAi screen for terminal nucleotidyltransferases
We cloned ~500 bp TUTase amplicons (Table S5) into pLitmus (NEB), and used them as T7 templates for dsRNA production using MEGAscript (Ambion). We used a double-knockdown approach. 2–3x106 cells were suspended in 1ml serum-free Schneider medium supplemented with 15μg of dsRNA for 30–60 min. After addition of 1ml 20% FBS Schneider medium, cells were plated in 6-wells. After 4 days, cells were treated with another round of dsRNA and analyzed after 4 more days.
When introducing mirtron constructs to dsRNA-treated cells, we transfected them using Effectene (Qiagen) on day 6 of the procedure above (i.e., the day following the second round of dsRNA soaking), without further dilution of the cells, and analyzed samples 3 days later.
Tailor and mirtron expression constructs
We amplified Tailor-A and Tailor-B from cDNAs using primers in Table S5, and cloned them into pENTR-d-TOPO. We then transferred them into ptMW, a pUAS-6xMyc vector. To reprogram mirtrons into mir-278, we utilized PCR-sewing, using the scheme and primer sets in Table S5. Cloning details are in the Supplementary Methods.
Expression plasmids for mir-1003, mir-1008, and mir-1010 and the miR-1003 luciferase sensor were reported (Okamura et al., 2007). We assayed sensors in cells manipulated for mirtron and/or Tailor activity, as detailed in the Supplemental Methods.
Fertility tests
The PBac{WH}CG1091[f05717] insertion (i.e., Tailor-KO) carries the mini-w+ marker. To remove genetic background, we conducted five rounds of back-crossing of the original insertion to w[1118], and made an isogenic stock for further analysis. We rescued the mutant with a pBDP-Tailor genomic transgene inserted at attP2 (BestGene, Inc). We individually crossed 2 day old virgin w[1118], Tailor-KO or rescued females with three w[1118] males, and performed three serial transfers to new vials every third day. Total progeny from the four vials was recorded.
In vitro uridylation assays
We used synthetic mature miRNAs or pre-miRNAs that were assembled using ligation (see Supplementary Methods). Following radiolabeling, we gel-purified these substrates for in vitro assays. To assay uridylation using immunoprecipitated proteins, we adapted a published method (Heo et al., 2009) for use with myc-tagged Tailor isolated from transiently-transfected cells, as detailed in Supplementary Methods. We mixed 15μl of beads with 15μl of 2x reaction mix (6.4mM MgCl2, 2mM DTT, 0.5mM rNTP, 1000 cpm of radiolabeled RNA substrate) and incubated at 25°C for the indicated times.
To assay terminal nucleotide preferences, we utilized S2 cells stably expressing actin-Flag-Myc-Tailor-B. These Tailor immunoprecipitates were less active than those from transient transfections, and were better suited to observe substrate preferences. Details of Tailor immunopurification from stable cells are in the Supplementary Methods. We used conditions described for in vitro RNAi assays (Ameres et al., 2010) except the ATP-regeneration system and ATP were omitted, and UTP was added to a final concentration of 500 μM.
In vitro dicing assay
We radiolabeled pre-mir-1010, pre-mir-1010-2U and pre-mir-1010-4U as above. We prepared recombinant His-Dcr-1 and His-Loqs-PB from Sf9 cells infected with Baculovirus expression constructs (Jiang et al., 2005). Dicing reactions were carried out with 20nM His-Dcr-1 and His-Loqs-PB under conditions described (Park et al., 2011).
Small RNA datasets and analysis
We cloned small RNA libraries from S2 cells (Tailor-KD and GFP-KD), and ovaries (w[1118] and Tailor mutant) and processed them as described (Okamura et al., 2011). The data were submitted to NCBI (GSE67984). We also used publicly available D. melanogaster sRNA data, whose GEO or SRA IDs are summarized in Table S1. Details of sRNA analysis are provided in the Supplemental Methods.
Supplementary Material
Highlights.
Splicing-derived pre-miRNAs (mirtrons) are preferentially uridylated amongst sRNAs
Mirtron uridylation is mediated by the TUTase Tailor, which inhibits their biogenesis
Tailor recognizes spliced hairpins and 3′-AG hairpins characteristic of mirtrons
Tailor opposes adventitious mirtron emergence, and shapes the canonical miRNA pool
Acknowledgments
We thank the Bloomington Stock Center for the Tailor mutant, Qinghua Liu for baculovirus constructs, Erik Ladewig for initial tailing analyses, Jr-Shiuan Yang for TUTase dsRNAs, Sonali Majumdar for Northern analysis, and Jaaved Mohammed for discussion. Taya Feldman, Xuejun Jiang, Dhirendra Simanshu and Dinshaw Patel provided critical support for recombinant protein purification. Research in K.O.’s group was supported by the National Research Foundation (NRF2011NRF-NRFF001-042). Work in E.C.L.’s group was supported by the Burroughs Wellcome Foundation and R01-GM083300 and R01-NS083833.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond Secondary Structure: Primary-Sequence Determinants License Pri-miRNA Hairpins for Processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome biology. 2011;12:221–221. 213. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature genetics. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Molecular cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Liu N, Flynt AS, Hodges E, Rooks M, Hannon GJ, Lai EC. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nature genetics. 2010;42:6–9. doi: 10.1038/ng0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome research. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome research. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013 doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nature reviews Genetics. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome research. 2011;21:286–300. doi: 10.1101/gr.113050.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & development. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & development. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome research. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H, et al. Adenylation of maternally inherited microRNAs by Wispy. Molecular cell. 2014;56:696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, You X, Chen T, Mackowiak SD, Friedlander MR, Weigt M, Du H, Gogol-Doring A, Chang Z, Dieterich C, et al. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic acids research. 2013;41:3619–3634. doi: 10.1093/nar/gkt072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Q, Vrettos N, Maragkakis M, Alexiou P, Gregory BD, Mourelatos Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Molecular cell. 2014;55:868–879. doi: 10.1016/j.molcel.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin T, Cazalla D, Yang JS, Bortolamiol-Becet D, Lai EC. RNase III-independent microRNA biogenesis in mammalian cells. RNA. 2012;18:2166–2173. doi: 10.1261/rna.036194.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J, Flynt AS, Siepel A, Lai EC. The impact of age, biogenesis, and genomic clustering on Drosophila microRNA evolution. RNA. 2013;19:1295–1308. doi: 10.1261/rna.039248.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nature reviews Molecular cell biology. 2013;14:643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes & development. 2013;27:778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Robine N, Liu Y, Liu Q, Lai EC. R2D2 organizes small regulatory RNA pathways in Drosophila. Molecular and cellular biology. 2011;31:884–896. doi: 10.1128/MCB.01141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimão-Pinto M, Ignatova V, Burkard T, Hung J-H, Manzenreither R, Sowemimo I, Herzog V, Reichholf B, Fariña-Lopez S, Ameres SL. Uridylation of RNA-hairpins by Tailor confines the emergence of miRNAs in Drosophila. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.05.033. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. The Plant cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Mohammed J, Bortolamiol-Becet D, Tsai H, Robine N, Westholm JO, Ladewig E, Dai Q, Okamura K, Flynt AS, et al. Diversity of miRNAs, siRNAs and piRNAs across 25 Drosophila cell lines. Genome research. 2014;24:1236–1250. doi: 10.1101/gr.161554.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA. 2012;18:177–192. doi: 10.1261/rna.030627.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome research. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.