Abstract
To determine the hierarchy of transcriptional regulation within the in vivo vertebrate embryo, we examined whether developmental enhancers were influenced by Nodal signaling during early embryogenesis in Xenopus tropicalis. We find that developmental enhancers, defined by the active enhancer chromatin marks H3K4me1 and H3K27ac, are established as early as blastula stage and that Smad2/3 only strongly associates with these regions at gastrula stages. Significantly, when we perturb Nodal signaling using the drug SB431542, most enhancers remain marked, including at genes known to be sensitive to Nodal signaling. Overall, as enhancers are in an active conformation prior to Nodal signaling and are established independently of Nodal signaling, we suggest that many developmental enhancers are marked maternally, prior to exposure to extrinsic signals.
Keywords: H3K4me1, H3K4me3, H3K27ac, H3K27me3, Smad2, Nodal, Xenopus tropicalis, Gastrulation, Enhancer
Introduction
Enhancers are distinct genomic regions that control temporal and spatial transcription. Hundreds of thousands of enhancers are scattered throughout the genome of all organisms, but only a fraction of these are utilized at any given time or any given cell. As enhancer usage is complex – varying in both developmental stages and cell types – elucidating these regions in vivo has been difficult. Many cell type specific enhancers, particularly in model organisms, have been identified and characterized using promoter bashing, gel shift assays and fingerprinting methodologies. Only recently have enhancers been identified on a genome-wide scale using chromatin immunoprecipitation followed by sequencing (ChIP-Seq) (Andersson et al., 2014; Bonn et al., 2012; Heintzman et al., 2009). Collectively, this work has shown that enhancers are regions of the genome that are depleted for nucleosomes. Further, the nucleosomes that exist on either side of the open chromatin contain specific histone modifications which are highly correlated with either active, repressed or poised neighboring gene transcription (Creyghton et al., 2010; Rada-Iglesias et al., 2011). Therefore, examining these open chromatin regions and their associated histone variants can strongly predict how a regulatory region is being utilized. H3K4me1, when bound either alone or together with H3K27me3, is not associated with transcription of neighboring genes and therefore enhancers containing these signatures are considered primed or poised (Shlyueva et al., 2014). If, however, H3K4me1 is bound together with H3K27ac then this combination is strongly associated with neighboring gene transcription and these regions are considered active enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Shlyueva et al., 2014). The presence of a specific transcription factor at such regions provides additional support for the enhancer being active (Kim et al., 2011). Overall, genomic regions that contain H3K4me1, H3K27ac and a transcription factor are highly likely to be functioning as active enhancers within that cellular context.
In the vertebrate embryo, it is unknown when enhancer marks are established during gestation, and whether their establishment is primarily dependent on maternal or zygotic proteins. Understanding how developmental enhancers are established and whether they are guided by specific transcription factors is the key to mechanistically understanding the hierarchy of transcriptional networks during embryogenesis. There is ample precedence from work in Drosophila showing that enhancer marks precede transcription factor binding (Bonn et al., 2012; Negre et al., 2011), although some transcription factors, termed pioneers, are known to unwind chromatin and recruit histone variants (Zaret and Carroll, 2011). In the Xenopus embryo, the maternally supplied pioneer factor FoxH1/Fast1 is known to be critical for proper germ layer specification and patterning of the early embryo, suggesting that maternal factors can act in the establishment of early gene transcription (Kofron et al., 2004; Watanabe and Whitman, 1999). However, while some activities of maternal FoxH1 are Smad-independent, a primary activity of FoxH1 is to activate gene transcription by binding activin response elements together with Smads, which are not active in the nucleus until after zygotic transcription begins (Chen et al., 1996, 1997). The timing of FoxH1 binding, enhancer mark deposition, and Smad binding at enhancers is unknown. There is also evidence that chromatin marks are remodeled prior to zygotic transcription, as the promoter mark H3K4me3 is established at some key early developmental genes through the action of β-catenin and the arginine methyltransferase Prmt2 (Blythe et al., 2010). However, the global hierarchy of transcription factor binding events and chromatin mark establishment is unclear: it remains unknown whether the transcription factor recruits enhancer chromatin marks or whether these chromatin marks permit transcription factor binding.
With the sequencing of Xenopus tropicalis, this organism has become amenable to examine the hierarchy of events that underlies the formation of the vertebrate body plan at the level of enhancers. In this report, we examine the sequence of deposition of the enhancer marks H3K4me1 and H3K27ac, and of the Nodal transcription factor, Smad2/3, immediately following zygotic transcription and continuing through gastrulation. We find that regions marked by H3K4me1 and H3K27ac are associated with key developmental genes at the onset of zygotic transcription. However, these regions are not generally associated with either the transcription factor Smad2/3 or with active promoters until gastrulation. Further, using perturbations within the Xenopus embryo, we find that the presence of H3K4me1 and H3K27ac at these regions is independent of functional Nodal signaling. Overall, we suggest that, in X. tropicalis, many important developmental enhancers are marked at or prior to zygotic transcription, but are not dependent on zygotic signaling, particularly Nodal, for their establishment.
Results and discussion
Promoter identification at blastula and gastrula stages using H3K4me3 and H3K27me3 in X. tropicalis
In order to define promoter elements in X. tropicalis, we examined the occupancy of H3K4me3 and H3K27me3 using ChIP-Seq. Promoters are nucleosome depleted regions of the chromatin. If H3K4me3 is present on the nucleosomes flanking these open chromatin regions there is a strong correlation with active transcription (Rada-Iglesias et al., 2011; Shlyueva et al., 2014). Conversely, if H3K27me3 is present, there is a correlation with absence of transcription (reviewed in Shlyueva et al., 2014). Therefore, these two epigenetic marks are useful in characterizing active vs silent promoters within the genome. Our analysis focused on the early stages of embryonic differentiation, including midblastula embryos near the onset of zygotic transcription (stage 8, Nieuwkoop and Faber staging criteria) (Nieuwkoop and Faber, 1967), late blastula embryos undergoing germ layer specification (stage 9), and early gastrula embryos undergoing dorsal–ventral patterning and gastrulation morphogenesis (stage 10.5). For each stage and histone mark, 10 to 59 million reads were aligned to version xenTro2 using MACS2.08 at a stringent q value of 0.0001 (see the Experimental procedures section).
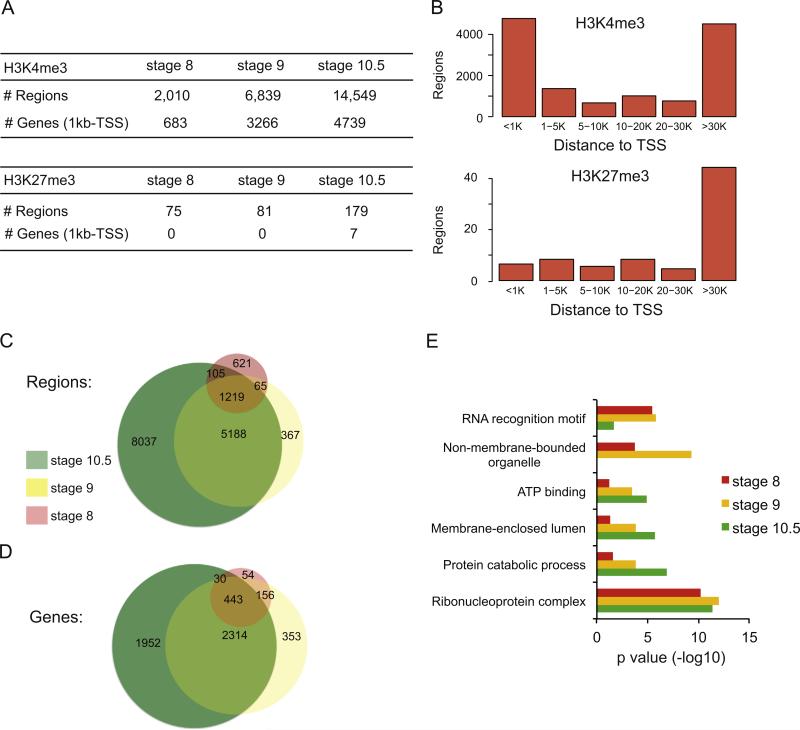
For the active promoter mark, H3K4me3, we identified 2,010, 6,839 and 14,549 peaks at stages 8, 9 and 10.5, respectively (Fig. 1A). At each stage these regions are predominantly located either within 1 kb of a transcription start site (TSS) or within intergenic regions greater than 30 kb from a TSS (Fig. 1B). Further, when we compare all regions that contain a H3K4me3 mark between all embryonic stages, we find significant overlap, with most of the marks present at stage 8 and 9 being represented at stage 10.5 (Fig. 1C).
Fig. 1.
Occupancy of H3K4me3 and H3K27me3 in X. tropicalis at stage 8, 9 and 10.5. (A) Table showing the breakdown of numbers from the ChIP-Seq datasets for H3K4me3 (top) and H3K27me3 (bottom), including the number of regions identified and the genes that could be associated to the regions. Each category is depicted for stages 8, 9 and 10.5. (B) Histograms showing where the regions bound by either H3K4me3 (top) or H3K27me3 (bottom) exist with respect to annotated TSS regions at stage 10.5. The number of bound regions is plotted on the Y axis, with the distance from nearest TSS along the X axis. (C) Venn diagram showing how the regions bound to H3K4me3 compare between stage 8, 9 and 10.5. (D) Venn diagram showing how the genes associated with H3K4me3 compare between stage 8, 9 and 10.5. (E) DAVID analysis for genes associated with H3K4me3 at stage 8, 9 and 10.5 (red, yellow and green, respectively).
Next we identified the genes that are associated with a H3K4me3 marked region within 1 kb of a transcription start site (TSS) using HOMER software (Heinz et al., 2010) (see the Experimental procedures section). We find 683, 3266, and 4739 genes at stages 8, 9 and 10.5, respectively. We next compared the overlap of these genes between each stage (Fig. 1D). The majority of genes with a promoter containing H3K4me3 at stage 8 remain marked at both stage 9 and stage 10.5, and most promoters that acquire a mark at stage 9 retain it at stage 10.5 (2757/3266). There is a gradual accumulation of blastula- or gastrula-specific active promoters that are used as development proceeds (353/3266 at stage 9 and 1952/4739 at stage 10.5). Overall, H3K4me3 bound promoters at stage 8 and stage 9 strongly predict the continued presence of this mark at stage 10.5—suggesting that active promoters remain stable as early development progresses.
We then examined the function of the genes associated with active promoters at all stages using the gene ontology analysis tool DAVID (Huang et al., 2009a, 2009b). We find significant enrichment for the terms “Ribonuclear protein complex” (stage 8, p=6.2 × 10−11; stage 9, p=1.0 × 10−12; stage 10.5, p=4.4 × 10−12) and “RNA recognition motif” (stage 8, p=3.3 × 10 −6; stage 9, 1.6 × 10 −6; stage 10.5, 0.022) (Fig. 1E) (Supplemental Table S1), likely reflecting functions involved in initiating zygotic transcription. The functional categories also include terms associated with housekeeping functions like “Protein catabolic process” (stage 9; p=1.5 × 10 −4; stage 10; p=1.3 × 10 −7), “ATP binding” (stage 9, p=3.3 × 10 −4; stage 10.5, p=1.4 × 10 −5), “Intracellular non-membrane organelle” (stage 8, p=1.7 × 10 −4; stage 9, p=4.7 × 10 −10), “Membrane enclosed lumen” (stage 9, p=1.6 × 10 −4; stage 10.5, p=1.9 × 10 −6). Overall, this suggests that most active promoters during early embryonic development are strongly enriched for basic metabolic functions. We do not see enrichment for terms associated with early embryonic development. Certainly, some early developmental regulators are marked by H3K4me3, but among the thousands of H3K4me3 promoters we identified, basic transcriptional and metabolic functions predominate at all three stages.
We conducted a similar analysis at each of the three stages with the repressive promoter mark, H3K27me3, and find very few instances of binding genome-wide, including only 75 regions at stage 8 and 179 at stage 10.5 (Fig. 1A). Of the regions identified at each time point, most are found within intergenic regions more than 30 kb from an annotated TSS (Fig. 1B). These results are consistent with what has previously been described in X. tropicalis (Akkers et al., 2009; van Heeringen et al., 2013), and support the notion that Polycomb Complex activity is minimal during early embryonic development in X. tropicalis (van Heeringen et al., 2013). Like other researchers, we conclude that promoter poising through bivalent H3K4me3/H3K27me3 marks is not a common mechanism for regulating gene expression in the early Xenopus embryo.
Putative enhancers are associated with developmental genes at early blastula stages
We next sought to elucidate enhancers during blastula and gastrula stages genome wide in X. tropicalis. While it is thought that enhancers are stable in adult organ systems, research in embryonic stem cells and in invertebrate model organisms suggests that enhancers are utilized transiently during development (Bonn et al., 2012; Calo and Wysocka, 2013; Lindeman et al., 2011; Negre et al., 2011; Rada-Iglesias et al., 2011; Rada-Iglesias and Wysocka, 2011). Therefore, understanding enhancers and how they vary throughout embryogenesis will provide both the means of targeting these regions and an appreciation for how they are used within a temporal context. To identify embryonic enhancers marked during blastula and gastrula stages in X. tropicalis, we performed ChIP-Seq for H3K27ac and H3K4me1 (Bonn et al., 2012; Creyghton et al., 2010; Shlyueva et al., 2014; Zentner et al., 2011). We find thousands of genomic regions containing each mark, increasing in number at each subsequent developmental stage (see Fig. 2A).
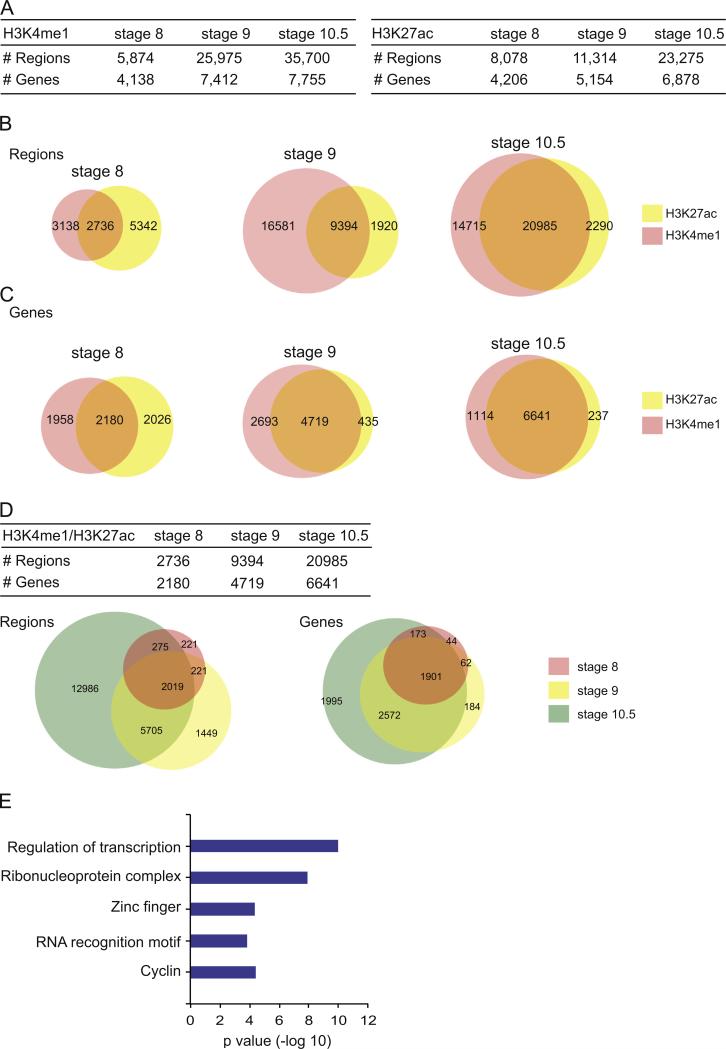
Fig. 2.
Occupancy of H3K27ac and H3K4me1 in X. tropicalis at stages 8, 9 and 10.5. (A) Table showing the breakdown of numbers from the ChIP-Seq datasets from H3K4me1 (left) and H3K27ac (right), including the number of regions identified and the genes that could be associated to the regions. Each category is depicted for stages 8, 9 and 10.5. (B) Venn diagrams showing overlap between regions bound by H3K27ac and H3K4me1 at stage 8, 9 and 10.5. (C) Venn diagrams showing overlap between the genes that can be associated with a region bound by H3K27ac and/or H3K4me1. (D) Table summarizing the number of regions bound by both H3K4me1 and H3K27ac and the associated genes at each stage. Venn diagrams depict the overlap of either the regions or associated genes. (E) DAVID analysis of genes associated to a region that is co-bound by H3K27ac and H3K4me1 at all stages (see intersection in Venn diagrams from (D)). X axis is the –log10 of p value for each term.
As regions containing both H3K4me1 and H3K27ac have been associated with active enhancers (Bonn et al., 2012; Creyghton et al., 2010; Rada-Iglesias et al., 2011; Shlyueva et al., 2014), we then identified regions genome-wide that contained overlapping H3K27ac and H3K4me1 at each stage (Fig. 2B). We find considerable overlap between the two marks at each stage genome-wide (Fig. 2B). Interestingly, by stage 10.5, 90% of regions that are marked by H3K27ac also contain H3K4me1. Given the body of literature linking these two variants to enhancer function, we designated regions marked by both H3K27ac and H3K4me1 as putative enhancers. Throughout the remaining text, we will refer to these as putative enhancers. Validation of known Nodal enhancer elements (see Fig. 4) suggests that many of these are active enhancers utilized during gastrulation in X. tropicalis.
Fig. 4.
Putative Nodal enhancers correspond to known functional enhancers in X. laevis and in mouse. (A) Genome browser shot of the gsc locus and its regulatory regions on xentro2 scaffold 185. Scale is shown at top. The gsc gene model is shown in purple. The ChIP-Seq read pile-up of Smad2/3 is shown in light green, with Nodal enhancer regions shown as black bars. ChIP-Seq read pileups are shown in orange for the putative enhancer histone marks H3K27ac (orange) and H3K4me1 (green), and the entire putative enhancer regions of H3K4me1/H3K27ac overlap for stage 10.5, stage 9, and SB431542-treated are shown as black lines. Note that the putative enhancer (H3K4me1/H3K27ac marked) regions are quite broad, while the region of Smad2/3 binding is narrow. The region corresponding to the published activin responsive element (ARE) for gsc in Xenopus laevis is shown in red, and the percent identity is indicated for the alignment of the X. laevis enhancer and orthologous X. tropicalis genomic coordinates. (B) Genome browser shot, putative enhancer regions, and known X. laevis enhancer position for eomesodermin on scaffold 26. ChIP-Seq data and X. laevis enhancer positions are marked as above. (C) Genome browser shot, putative enhancer regions, and known mouse enhancer position for pitx2 on scaffold 89. ChIP-Seq data and mouse enhancer positions are marked as above. (D) Genome browser shot, putative enhancer regions, and known mouse enhancer position for lefty on scaffold 719. (E) Genome browser shot, putative enhancer regions, and known mouse enhancer position for otx2 on scaffold 68.
We next examined whether putative enhancers change between embryonic stages (Fig. 2D). We find that, once marked, most of these elements remain throughout subsequent stages (Fig. 2D, left Venn). Of the 2736 putative enhancers identified at stage 8, 2019 (74%) remain marked at both stage 9 and stage 10.5. Less than 10% are found exclusively at stage 8 and only 15.5% are specific to stage 9. However, 62% of the putative enhancers are specific for stage 10.5, supporting the idea that as development becomes more complex there is a requirement for a greater repertoire of enhancers.
We next sought to identify genes associated with putative enhancers (Fig. 2C). We find 2180, 4719 and 6641 genes at stages 8, 9, 10.5, respectively (Fig. 2D). After comparing these stages, we find that 87% of genes associated with a putative enhancer at stage 8 stay associated through stage 10.5. We then performed DAVID analysis using the 1901 genes associated with a putative enhancer across all stages and found five enriched categories including “Regulation of transcription” (p=1.1 × 10 −10), “Ribonuclear protein complex” (p=1.2 × 10 −8), “Zinc Finger” (p=4.4 × 10 −5), “RNA recognition motif” (p=1.5 × 10 −4), and “Cyclin” (p= 3.8 × 10 −5) (Fig. 2E).
Examination of the 178 genes within the “Regulation of transcription” cluster reveals many important developmental transcription factors, including eomes, smad1, foxA1, foxA2, tcf3, sox17β.1, gata4, gata6, sox1, sox2, sox3, sox7, pax2, pax6, otx1, otx2, pitx1, pitx2, meis2, dlx1, dlx2, dlx3, irx2, irx3, pou3f4, pou3f1, six2, not1, not2, suz12, and twist1 (Supplemental Table S2). This is surprising as it suggests that, by early blastula stage, many key developmental loci already have H3K4me1 and H3K27ac marks, though these regions may not initiate expression until much later in development. Interestingly, most of these genes lack H3K4me3 at their promoters, suggesting that no transcription is occurring, consistent with the fact that most of these genes are not known to be expressed until gastrulation. We find that only smad1, sox2 and sox3 contain H3K4me3 marks near the TSS, suggesting that most of these developmental loci contain the signature of an active enhancer, but in the absence of transcription. Indeed, if we examine published RNA-Seq and microarray datasets for these transcripts, we find most (18/31) increase by over 2 fold from stage 8 to stage 12 (Yanai et al., 2011; Tan et al., 2013), strongly suggesting that while these developmental genes can be associated with enhancer marks at stage 8, they are not actively transcribed. Overall this suggests that, in Xenopus, the active enhancer marks H3K4me1 and H3K27ac precede gene expression by several hours and embryonic stages. This suggests that in Xenopus, enhancer marks are established before active transcription, in contrast to stem cells where enhancer marks are immediately associated with active transcription (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Shlyueva et al., 2014).
Identification of enhancers used by Nodal signaling in blastula and gastrula
To identify specifically those enhancers used by Nodal in the X. tropicalis embryo, we examined genomic regions occupied by Smad2/3. Nodal signaling is necessary and sufficient to establish both mesoderm and endoderm, but outside of the few characterized Nodal enhancers or Activin Response Elements (AREs) (Germain et al., 2000; Huang et al., 1995; Kimura et al., 1997; Rebbert and Dawid, 1997; Ryan et al., 2000; Saijoh et al., 1999; Shiratori et al., 2001; Watabe et al., 1995), it is unknown how and where Smad2/3 regulates transcription to drive Nodal responses. To this end, we performed ChIP-Seq (using an antibody that recognizes both Smad2 and Smad3) to examine Smad2/3 occupancy genome-wide at stage 8, 9 and 10.5. Although we were able to detect Smad2/3 binding in embryos at stage 8, we note that our Smad2 ChIP-Seq libraries for this stage were poor, with a high number of duplicate reads relative to our other libraries, and that we had to pool DNA from numerous ChIPs to generate enough material. We consider it likely that the small amount of DNA available in a stage 8 embryo, coupled with the small percentage of DNA that is bound by Smad2, contributed to poor library quality. Therefore, while we present our overall data for Smad2/3 binding at stage 8 as a potential resource, we acknowledge that more regions may be functionally bound by Smad2/3 at stage 8 than we were able to detect. Our libraries at stage 9 and stage 10.5, however, were very high quality, with high amounts of ChIP DNA and a low percentage of duplicated reads (see the Experimental procedures section).
Comparing stages, we find a small number of Smad2/3 occupied regions at stage 8 (207), which increases to 865 at stage 9 and 1024 at stage 10.5 (Fig. 3A). Smad2/3 occupied regions are not associated with more than one developmental stage (Fig. 3B, left). This is surprisingly distinct from what we observe with putative enhancers, which when marked at stage 8 remain through stage 10.5 (Fig. 2D).
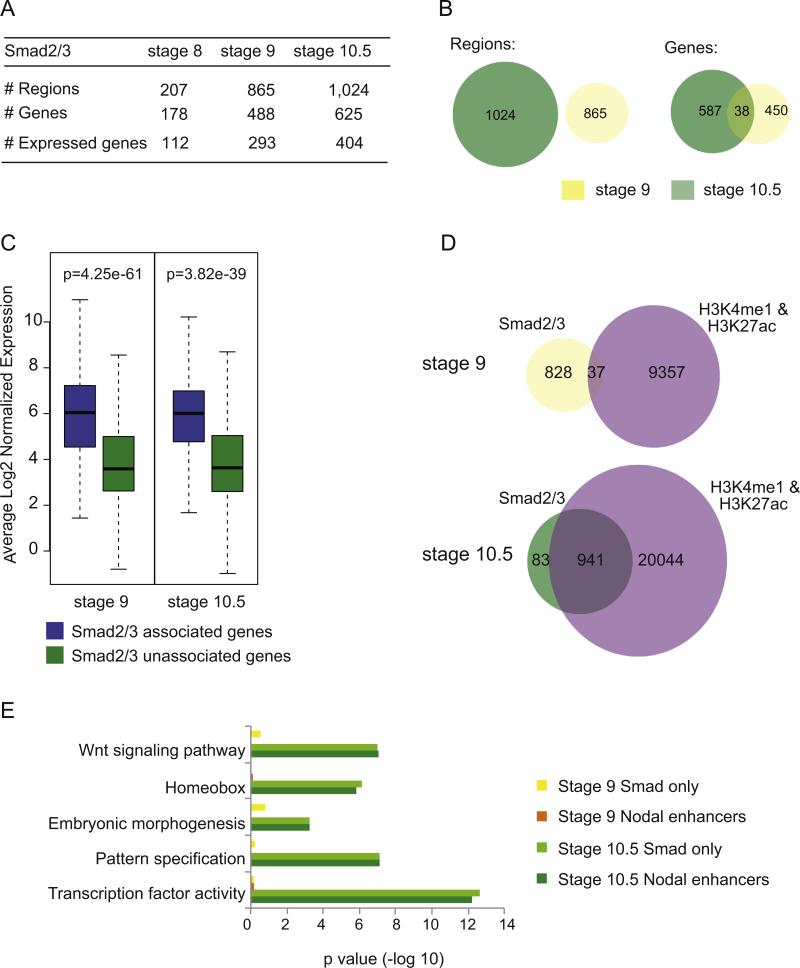
Fig. 3.
Identification of putative Nodal enhancers in blastula and gastrula. (A) Identification of regions bound by Smad2/3 and genes associated with these regions at stage 8, 9 and 10.5. The number of genes associated with Smad2/3 bound regions is further classified into those whose expression was detected by 3SEQ. (B) Venn diagrams comparing regions bound by Smad2/3 at stages 9 and 10.5 (left) and the genes that are associated to Smad2/3 binding at stages 9 and 10.5 (right). (C) Box plot representation of transcript abundance of genes associated with a Smad2/3 bound region (blue) and those that are not associated with a Smad2/3 bound region (green) at stage 9 (293 expressed genes associated with Smad2/3 binding vs. 6555 not associated) and stage 10.5 (404 genes vs. 6645 not associated). (D) Venn diagrams demonstrating the overlap between regions bound by Smad2/3 and regions bound by both H3K4me1 and H3K27ac at stages 9 and 10.5. (E) DAVID analysis of genes associated with Smad2/3, H3K4me1 and H3K27ac bound regions (Nodal enhancers) at stage 9 (orange), stage 10.5 (dark green) and genes associated only with Smad2/3 bound regions at stage 9 (yellow) and stage 10.5 (light green).
As Smad2/3 is known to drive cell fate specification during blastula and gastrula stages, we sought to identify the target genes and their function. We identified 488 and 625 genes associated with Smad2/3 occupancy at stage 9 and 10.5, respectively (Fig. 3A). We found that only a few genes associated with a Smad2/3-bound region at stage 9 remain associated at stage 10.5 (38/488) (Fig. 3B, right). A similar pattern is found when comparing the data from stage 8 (data not shown). Although the role of Nodal signaling in cell fate specification at blastula stages is well established, our results suggest that Smad2/3 binding may be highly transient or weak during blastula stages and – while certainly it drives the expression of at least a handful of developmental genes – this binding falls below our detection thresholds. Lowering the false discovery cutoff did not significantly improve our ability to detect Smad2/3 occupancy at blastula stages. Smad2/3 is known to shuttle dynamically between the nucleus and cytoplasm, and only accumulates in the nucleus of Xenopus cells after the midblastula transition (Saka et al., 2007; Schmierer and Hill, 2005). Therefore, we consider it likely that by profiling Smad2/3 binding throughout the whole embryo at blastula stages, we are unable to capture transient events of Smad2/3 binding, which occur predominantly in only the prospective dorsal mesendoderm.
We then asked whether a Smad2/3 occupied region is predictive of neighboring gene transcription. To this end, we compared 3SEQ transcription levels from genes associated with a Smad2/3 occupied region to those genes not associated with a Smad2/3 occupied region. We found expression data for 293 and 404 genes at stages 9 and 10.5 respectively (see Fig. 3A) and compared the mean expression level of these gene sets with genes expressed at each stage but not occupied by Smad2/3 (6555 and 6645 genes, respectively). This analysis found a significant increase in transcription levels from genes associated with Smad2/3-occupied regions (Fig. 3C), strongly implying that Smad2/3 functions to drive transcription at these loci.
As Nodal signaling is known to drive many developmental processes, we next examined the overall function of genes that neighbor a Smad2/3 occupied region at each stage using DAVID. By stage 9, we identify weak enrichment in “HMG DNA binding process” (p=0.002), which is the top clustering term, driven by many sox and fox genes. By stage 10.5, DAVID analysis uncovers “Transcription factor activity” (p=2 10 −13) and many other developmental processes, including Wnt signaling (1.02 × 10 −7), homeobox (7.83 × 10 −7), pattern specification (7.63 × 10−8) and embryonic morphogenesis (5.9 × 10 −4) (Fig. 3E, light green). The genes represented in these categories include all the major known downstream effectors of Nodal signaling, including xbra, eomes, otx2, sox2, sox3, nodal, pitx1 and mix1, and many developmentally important genes, including Wnt, Tgfβ and Fgf pathway members (Supplemental Table S3). Overall, this captures the known role of Smad2/3 in driving critical developmental decisions during gastrulation and provides a wealth of enhancer information for this important transcriptional network.
Nodal enhancers are found predominantly during gastrula stage
We next sought to identify putative Nodal enhancers throughout the genome during blastula and gastrula formation in X. tropicalis. To this end, we defined which of the Smad2/3 occupied regions were also marked by both H3K27ac and H3K4me1. Surprisingly, at stage 9 only a small fraction of Smad2/3 occupied regions also contain both H3K4me1 and H3K27ac (37/865), but by stage 10.5, this has increased to 92% (941/1024) (Fig. 3D). Thus, by the gastrula stage, nearly all regions bound by Smad2/3 also contain both H3K4me1 and H3K27ac. To simplify nomenclature we refer to these regions as putative Nodal enhancers throughout the remaining text. Examples of 21 well-known Nodal target genes, showing screenshots of Smad2/3 and enhancer mark occupancy, are provided in Supplemental Fig. 1.
We next examined the function of genes associated with putative Nodal enhancers. At stage 10.5, DAVID analysis shows significant enrichment in transcription factor activity (p=6.18 × 10 −13), Homeobox (p=2.99 × 10 −6), Wnt signaling pathway (p=9.0 × 10 −8), embryonic morphogenesis (p=5.8 × 10 −4), and pattern specification process (p=7.3 × 10 −8) (Fig. 3E, dark green). However, no significant enrichment in biological processes – including developmental – was identified at stage 9 (or at stage 8). Overall, this suggests that, at stage 10.5, Nodal enhancers are robust, but that at earlier stages Smad2/3 must be weakly or transiently bound.
We next validated the putative Nodal enhancers, by examining whether the AREs previously identified in Xenopus laevis and mouse were found within this dataset (Kimura et al., 1997; Ryan et al., 2000; Saijoh et al., 1999; Shiratori et al., 2001; Watabe et al., 1995). Initially, we identified the orthologous regions of the X. tropicalis genome that correspond to the AREs in X. laevis and in mouse. We then compared the positions of the AREs with our putative Nodal enhancers at stage 10.5 (Fig. 4). X. laevis contains well-characterized AREs in the gsc and eomes promoters, which correspond perfectly to the positions of Smad2/3, H3K4me1 and H3K27ac occupancy in X. tropicalis (Fig. 4A and B). There is also extremely good evolutionary conservation of this region between the two species, with 93.2% and 84.9% nucleotide identity between X. tropicalis and X. laevis, respectively (Supplemental Fig. 2B and C). For pitx2, lefty, and otx2, we relied on functionally characterized AREs in the mouse. For these regions, we performed sequence alignments between the known mouse ARE region and the scaffold containing the X. tropicalis ortholog of each gene to identify the corresponding sequence in the X. tropicalis genome. We find that for each gene, the orthologous sequence in X. tropiclalis contains a predicted Nodal enhancer at the same position as the mouse ARE, and that predicted FoxH1/Fast1 and Smad2 sites are well conserved (Fig. 4C–E; Supplemental Fig. 2C– E). As Smad2/3 binding occurs predominantly (92%) in the presence of both H3K4me1 and H3K27ac at stage 10.5, we examined these genomic regions more closely for a broad distribution of H3K27ac and H3K4me1 surrounding narrow Smad2/3 bound regions. This distribution pattern has been associated with enhancer elements in stem cells, mouse adult tissues and Drosophila (Bonn et al., 2012; Creyghton et al., 2010; Rada-Iglesias et al., 2011; Zentner et al., 2011). At stage 9 and 10.5 we observe this trend clearly at many of the putative Nodal enhancers, including at the well-characterized gsc, eomes pitx2, lefty and otx2 AREs (Fig. 4). Overall we find that all 5 known AREs, from both X. laevis and mouse, are conserved in X. tropicalis. Importantly, these AREs were identified as putative Nodal enhancers in this study. This serves to validate the datasets and suggests we can effectively identify functional enhancers.
Enhancers in blastula predict where Smad2/3 will bind during gastrulation
We next examined whether putative enhancers preceded Smad2/3 occupancy during embryonic development. While much progress has been made in identifying enhancers in many organisms, how these enhancers are built over time to drive cell type specific transcription is an open question. Therefore, we pulled out the 865 regions bound by Smad2/3 at stage 9 and examined whether these regions had previously been marked at stage 8 by H3K4me3, H3K27ac, H3K4me1 or H3K27me3. For this purpose, first we extracted the tag densities for each mark at stage 8 surrounding the 5kb region bound by Smad2/3 at stage 9. Next, we visualized the patterning of these marks using TreeView (Saldanha, 2004). We represented these results as a heatmap with each Smad2/3 occupied region as a point on the Y axis (865) and the genomic coordinates surrounding each region on the X axis (2500 bp on either side of Smad2/3 occupancy). We observe very little correlation of any histone mark at stage 8 with regions occupied by Smad2/3 at stage 9 (Fig. 5A). To further define the overlap between putative enhancers at stage 8 and Smad2/3 at stage 9, we compared the occupancy data as a Venn diagram (Fig. 5C, left). This again reveals that very few putative enhancers at stage 8 are occupied by Smad2/3 at stage 9. Overall, stage 8 putative enhancers do not predict Smad2/3 binding at stage 9.
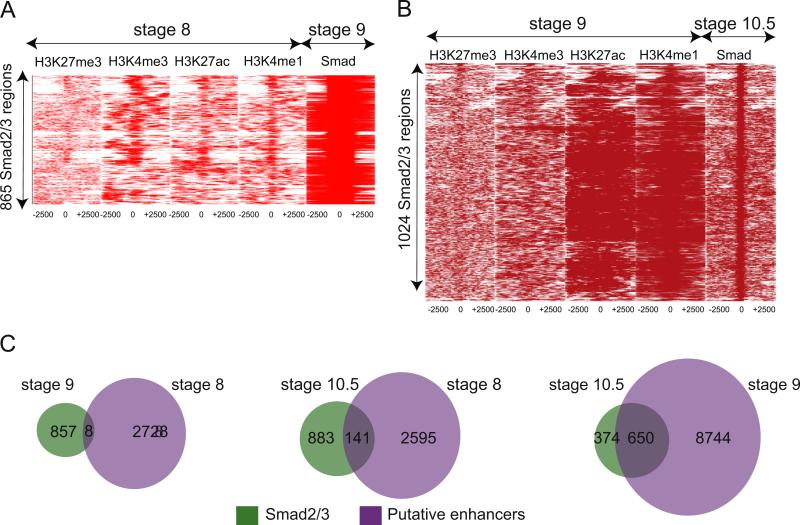
Fig. 5.
Deposition of enhancer marks precedes Smad2/3 binding. (A) Heat maps depicting binding intensities of H3K27me3, H3K4me3, H3K27ac, H3K4me1 at stage 8 (first four panels) at 5 kb region surrounding where Smad2/3 will bind at stage 9 (last panel). At stage 9, Smad2/3 occupies 865 regions, which is the Y axis. All regions are centered around Smad2/3 binding, with 2.5 kb on either side (X axis). (B) Heat maps depicting binding intensities of H3K27me3, H3K4me3, H3K27ac, and H3K4me1 at stage 9 (first four panels) at 5 kb region surrounding where Smad2/3 will bind at stage 10.5 (last panel). At stage 10.5, Smad2/3 occupies 1024 regions (Y axis). All regions are centered around Smad2/3 occupancy, with 2.5 kb on either side (X axis). (C) Venn diagrams depicting how H3K4me1 and H3K27ac present at stage 8 and stage 9 predict Smad2/3 association at stage 10.5. Left: comparison of stage 9 Smad2/3 bound regions with stage 8 putative enhancers. Middle: comparison of stage 10.5 Smad2/3 bound regions with stage 8 putative enhancers. Right: comparison of stage 10.5 Smad2/3 bound regions with stage 9 putative enhancers.
We next examined whether putative enhancers at blastula predict eventual Smad2/3 occupancy at gastrulation. To this end, we performed the same analysis as above, but compared histone marks at stage 9 with Smad2/3 occupancy at stage 10.5. Again, tag densities were represented as a heatmap with each Smad2/3 occupied region at stage 10.5 as a point on the Y axis (1024) and the genomic coordinates surrounding each region on the X axis (2500 bp on either side of Smad2/3 occupancy). We find that putative enhancer marks at stage 9 are present in regions that will eventually be bound by Smad2/3. We also show this association with Venn diagrams that depict the overlap between regions marked by H3K4me1 and H3K27ac at stage 9 and those bound by Smad2/3 at stage 10.5. This analysis shows that most Smad2/3 occupied regions at stage 10.5 contain a putative enhancer at stage 9 (650/1024, 63%; Fig. 5C, right). We next determined whether these putative enhancers were marked even earlier at stage 8. A Venn diagram comparing putative enhancers at stage 8 with regions bound by Smad2/3 at stage 10.5 show that, surprisingly, many putative enhancers used by Smad2/3 can be identified at early blastula stages (141/1024, 14%; Fig. 5D, middle). Interestingly, we find these 141 regions are associated with many known Nodal target genes, including xbra, lhx1, eomes, otx2, fgf8, sox2, sox3, pitx1, pitx2, fzd1, not1, tsg101, bambi, bmp4, sprout, hhex, irx3, foxc2, sox17b, gata3, gata4 and gata6, driving the enrichment for “Developmental protein” (8.2 × 10 −6) using DAVID. Overall this strongly suggests that H3K27ac and H3K4me1 precede or prime particular genomic regions at blastula stage prior to stable Smad2/3 association during gastrulation.
Chromatin enhancer marks are independent of Nodal signaling
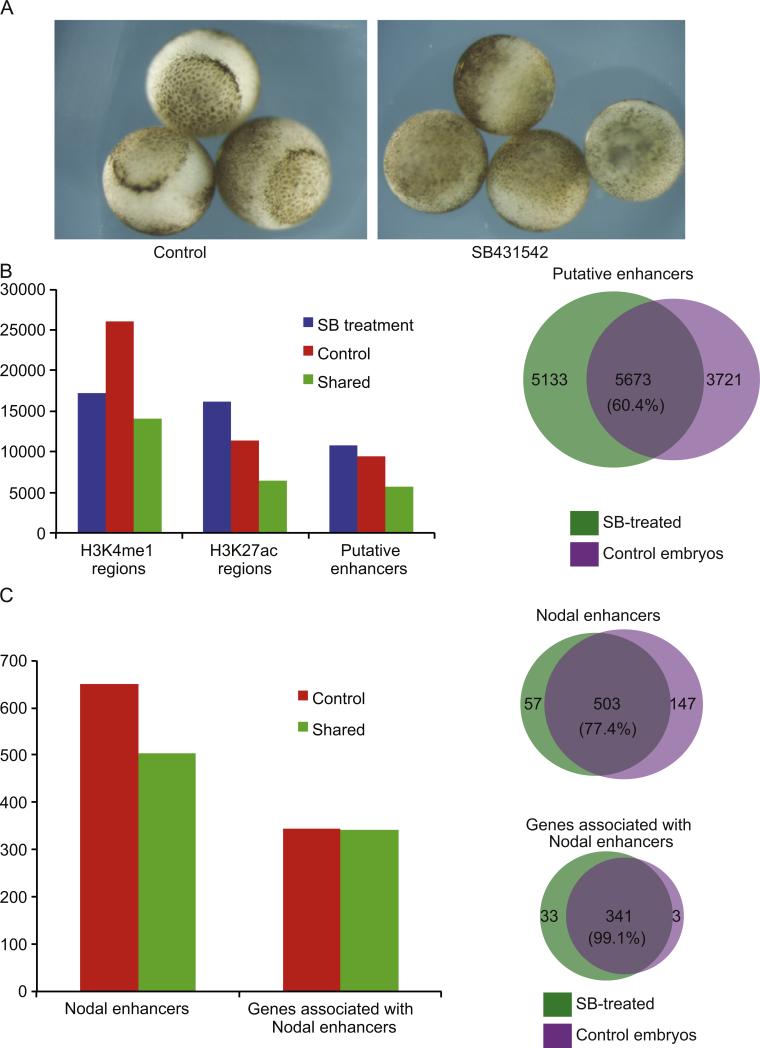
As our data suggests that Smad2/3 is recruited to enhancers that were marked much earlier in development, we next examined whether these marks are dependent upon Nodal signaling. To this end, we ablated Nodal signaling by treating embryos with the drug SB431542 at the 4 cell stage, which blocks the activity of Alk4/5 receptors and effectively inhibits all downstream Nodal signaling in Xenopus embryos (Luxardi et al., 2010; Skirkanich et al., 2011). Embryos treated with SB431542 fail to gastrulate as would be expected for inhibition of Nodal signaling (see Fig. 6A). The Nodal ablated embryos were harvested at stage 9, and examined by ChIP-Seq using antibodies against both H3K4me1 and H3K27ac to identify putative enhancers. We identified 10,806 in the SB431542-treated embryos and 9394 in the controls. Next, we compared the putative enhancers between the two conditions and found that 5673 were shared. Thus 60.4% (5673/9394) of putative enhancers are shared between SB431542-treated and control embryos at stage 9 (Fig. 6B).
Fig. 6.
Blocking Nodal signaling does not compromise H3K4me1/H3K27ac positioning. (A) Control embryos (left) or SB431542-treated embryos (right) showing the failure of blastopore formation after inhibition of Nodal signaling at stage 10.5. (B) Left: bar graph representing the number of regions marked by H3K4me1, H3K27ac, or both marks (putative enhancers) at stage 9, in either control embryos, SB431542-treated embryos, or both conditions (“Shared”); right: Venn diagram depicting the overlap of putative enhancers in Control and SB431542-treated embryos at stage 9. Percentage of enhancers that are shared is shown in the center. (C) Left: bar graph representing the number of putative enhancers at stage 9 that will acquire Smad2/3 binding at stage 10.5 (Nodal enhancers) in control embryos alone (“Control”) or both control and SB431542 treated embryos (“Shared”). Nodal enhancer regions and associated genes are both shown; right: Venn diagram showing the overlap of Nodal enhancers (top) and associated genes (bottom) in control and SB431542 treated embryos. The percentage of shared enhancers and genes is shown in the center of each diagram.
We next sought to determine how putative Nodal enhancers are affected after SB431542-treatment. Previously, we identified 650 putative Nodal enhancers that were marked at stage 9 and then occupied stably by Smad2/3 only at stage 10.5 (Fig. 5B and C). We then examined whether these 650 marks were maintained after SB431542-treatment. We find that 503 of the 650 putative Nodal enhancers maintain H3K4me1 and H3K27ac marks in the absence of Nodal signaling (77.4%; see Fig. 6C). When we associate these 503 maintained regions to their nearest neighboring genes, we find that 99.1% of genes retain a putative enhancer following Nodal inhibition (341/374 genes), including gsc, xbra, eomes, otx2, fgf8, sox2, sox3 nodal1, vegT, sox17b, gata4, and gata6. In Fig. 4 we highlight that putative enhancers at gsc, eomes, pitx2, lefty and otx2 are maintained after SB431542 treatment (black lines demarcating position of putative enhancers at stage 9 in control and SB431542-treated embryos). Thus, putative enhancers for critical developmental genes are marked normally in the absence of Smad2/3, importantly suggesting that their establishment is independent of zygotic Nodal signaling.
In this paper, we suggest that Nodal enhancers at important developmental loci are established early during embryogenesis—at least at blastula stages and possibly maternally. Further, we show that these enhancers are not dependent upon Nodal signaling for their establishment, strongly suggesting that a maternal protein is important for the identification of these regions and the subsequent recruitment of histone variants to particular nucleosomes. FoxH1 is known to function as a pioneer protein, which can recruit and establish open chromatin configurations (Serandour et al., 2011; Zaret and Carroll, 2011). As FoxH1 is also found maternally in the frog oocyte and embryo and is a well-studied Smad2/3 co-factor (Kofron et al., 2004), we propose that maternal FoxH1 may be responsible for the establishment of eventual Nodal enhancers within the embryo. It will be interesting to examine the role of FoxH1 as a potential maternal pioneer factor in establishing developmental enhancers. Overall, we suggest that Nodal enhancers are marked either maternally or very soon after zygotic transcription and are used transiently by Smad2/3 until gastrulation when they become stably bound. As it is an open question how developmental enhancers are established during embryogenesis in the vertebrate, this paper provides some of the first evidence showing that the chromatin is marked prior to zygotic signals and that enhancer marks are established independent of Nodal signaling.
Experimental procedures
X. tropicalis embryo isolation and culture
X. tropicalis embryos were generated by in vitro fertilization according to published methods (Khokha et al., 2002). Embryos were staged according to Nieuwkoop and Faber morphological criteria. We made an extra effort to raise embryos at consistent temperatures, so that embryos would reach a given developmental stage at a similar time following fertilization. For Nodal inhibition, embryos were cultured in SB431542 beginning at the 4 cell stage. For each cohort of treated embryos, a few embryos were raised to stage 10.5 and to stage 20, to confirm loss of blastopore and of dorsal anterior development.
RNA isolation and 3SEQ library preparation
Total RNA was isolated by collecting cohorts of 100 embryos into Trizol (Invitrogen). 20 mg of starting total RNA was used for each library. mRNA was prepared from total RNA by two rounds of purification using oligo-DT Dynabeads (Invitrogen, cat #610.06). 400 ng of poly-A selected RNA was used for library preparation. Libraries for 3SEQ were prepared according to published methods (Beck et al., 2010), using HPLC or PAGE purified primers, 3% NuSieve GTG agarose for gel purification, and an optimized ratio of 0.75ul primer P7_oligodT (10 mM) per 200 ng polyA RNA for the initial heat shearing step. Primers used were:
P7_OligoDT:
5′ CAA GCA GAA GAC GGC ATA CGA GCT CTT CCG ATC TTT TTT TTT TTT TTT TTT TTT TTT TVN 3′
P5_LinkerA:
5′ CCG AGA TCT ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT 3′
P5_LinkerB:
/5PHOS/GAT CGG AAG AGC GTC GTG TAG GGA AAG AGT 3′ (5′ Phos required for linker ligation)
PCR_F:
5′ AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T 3′
PCR_R:
5′ CAA GCA GAA GAC GGC ATA CGA GCT CTT CTT CCG ATC 3′
ChIP and ChIP-Seq library preparation
Chromatin immunoprecipitation from whole embryo lysates was performed according to published methods (Blythe et al., 2009; Wills et al., 2013), using pools of 250 embryos. DNA yields from ChIP were pooled until 1ug of total DNA was generated; this 1ug of DNA was used to generate libraries for ChIP-Seq. Antibodies used for ChIP were as follows:
| Target | Catalog number | Company |
|---|---|---|
| H3K27ac | ab4729 | Abcam |
| H3K4me1 | ab8895 | Abcam |
| H3K4me3 | 39159 | Active Motif |
| H3K27me3 | 39535 | Active Motif |
| Smad2/3 (FL-425) | sc-8332 | Santa Cruz Biotechnology |
ChIP-Seq data analysis
ChIP-Seq reads are mapped to xenTro2 genome using the bowtie and bwa-sa tools (Langmead et al., 2009). GA II and HiSeq sequencing platforms gave different yields of alignable reads, with GA II generally yielding 10–20 aligned million reads and HiSeq 40–100 million aligned reads. Both platforms gave sequencing coverage that was more than adequate to our analyses. Prior to analysis, for each library and each biological replicate, all reads were filtered for quality, and duplicate and multimapped reads were discarded. After discarding these reads, the remaining reads from multiple biological replicate samples were pooled to provide greater analytical power. We pooled reads from two biological replicates for Smad2/3 Stage 9; H3K4me1 stage 8, 9, and 10; H3K4Me1 Stage 8, 9 and 10.5; H3K27ac stage 8, 9 and 10.5, and H3K4me3 stage 8, 9 and 10.5. These pooled reads were then aligned to the xentro2 (JGI4.1) X. tropicalis genome. For H3K4me3, we aligned 22,581,352; 10,828,491; and 10,227,842 reads to the genome at stages 8, 9 and 10.5, respectively (Fig. 1 data). For H3K27me3, we aligned 59,206,117; 24,305,339; and 37,175,470 reads to the genome at stages 8, 9 and 10.5, respectively. For H3K27ac, we aligned 17,998,987; 76,511,738; and 50,444,132 reads to the genome at stages 8, 9 and 10.5, respectively (Fig. 2A data). For H3K4me1, we aligned 26,879,020; 41,716,284; and 54,294,470 reads at stages 8, 9 and 10.5, respectively. For Smad2/3 we aligned 18,473,099; 27,673,586 and 7,611,088 reads to the genome at stages 8, 9, and 10.5, respectively (Fig. 3 data). Aligned ChIP-Seq files are used to call peaks with the MACS2.08 software (Feng et al., 2012; Zhang et al., 2008) at a stringent q value of 0.00001. For histone marks, “—broad option” of the software is used. Input datasets are also aligned to xenTro2 genome and are used as baseline for peak calling. Called peaks are annotated using HOMER software to the closest gene (Heinz et al., 2010). For promoter associated epigenetic marks H3k4me3 and H3K27me3, peaks are assigned to genes only if they are in the vicinity of a transcription start site, i.e., 1 kb upstream or downstream of TSS. To visualize peaks using UCSC Genome Browser, HOMER software (Heinz et al., 2010) was used to generate bigWig and bedGraph files. To intersect peaks of different marks, bedtools software (bedIntersect) was used (Quinlan and Hall, 2010). All datasets are available in GEO: accession number GSE56000.
3SEQ data analysis
Raw counts from 3SEQ datasets are calculated and annotated to the closest genes using the Unipeak software (Foley and Sidow, 2013). DESeq software is used to normalize the raw counts data and to find the differentially expressed genes (Anders and Huber, 2010). In-house Perl scripts are used to intersect expression data with gene lists derived from the ChIP-Seq data.
Clustering and gene ontology analysis
Tag density values around 2.5 kb up and downstream of called peaks are generated using the HOMER software. Extracted tag density values around peaks are visualized using the TreeView software (Saldanha, 2004).
Gene ontology analysis was performed using the “Functional Annotation Clustering” tool of DAVID software (Huang et al., 2009a, 2009b). Gene lists were submitted using the “Official_Gene_Symbol” preset and restricted to X. tropicalis IDs. Only categories with enrichment values greater than 1.3 (generally corresponding to p values at or near 0.01) were considered significant, according to the software's author recommendations.
Supplementary Material
Acknowledgements
We thank Christine Reid for critical comments on the manuscript, and members of the Baker lab for thoughtful and productive discussion during this project. This work was supported by grants from the National Institute of General Medical Sciences and the National Institute of Child Health and Human Development (NIH R01GM103787, NIH R01HD076839, and NIH R01GM095346). The authors declare no competing interests.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2014.08.034.
References
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AH, Weng Z, Witten DM, Zhu S, Foley JW, Lacroute P, Smith CL, Tibshirani R, van de Rijn M, Sidow A, West RB. 3′-End sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev. Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JW, Sidow A. Transcription-factor occupancy at HOT regions quantitatively predicts RNA polymerase recruitment in five human cell lines. BMC Genomics. 2013;14:720. doi: 10.1186/1471-2164-14-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang HC, Murtaugh LC, Vize PD, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, Papalopulu N, Smith JC, Zorn AM, Harland RM, Grammer TC. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I. Cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development. 1997;124:3929–3941. doi: 10.1242/dev.124.20.3929. [DOI] [PubMed] [Google Scholar]
- Kofron M, Puck H, Standley H, Wylie C, Old R, Whitman M, Heasman J. New roles for FoxH1 in patterning the early embryo. Development. 2004;131:5065–5078. doi: 10.1242/dev.01396. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P, Collas P. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev. Cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Luxardi G, Marchal L, Thome V, Kodjabachian L. Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development. 2010;137:417–426. doi: 10.1242/dev.039735. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba XS, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis. North Holland Publishing Company, Amsterdam. 1967 [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med. 2011;3:36. doi: 10.1186/gm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbert ML, Dawid IB. Transcriptional regulation of the Xlim-1 gene by activin is mediated by an element in intron I. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9717–9722. doi: 10.1073/pnas.94.18.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Garrett N, Bourillot P, Stennard F, Gurdon JB. The Xenopus eomesodermin promoter and its concentration-dependent response to activin. Mech. Dev. 2000;94:133–146. doi: 10.1016/s0925-4773(00)00300-2. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Mochida K, Ohishi S, Hirao A, Hamada H. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes Dev. 1999;13:259–269. doi: 10.1101/gad.13.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Hagemann AI, Piepenburg O, Smith JC. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol. Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev. Biol. 2011;357:478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, Veenstra GJ. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2013 doi: 10.1101/gr.159608.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho KW. Molecular mechanisms of Spemann's organizer formation: conserved growth factor synergy between Xenopus and mouse. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development. 1999;126:5621–5634. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- Wills AE, Gupta R, Chuong E, Baker JC. Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis. Methods. 2013 doi: 10.1016/j.ymeth.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev. Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.