Abstract
Background
Trichoderma is one of the most common fungi in soil. However, little information is available concerning the diversity of Trichoderma in soil with no previous history of cultivation. This study was conducted to investigate the most common species and the level of genetic relatedness of Trichoderma species from uncultivated soil in relation to cultivated soil and potting media.
Results
A total of 24, 15 and 13 Trichoderma isolates were recovered from 84 potting media samples, 45 cultivated soil samples and 65 uncultivated soil samples, respectively. Analysis based on the internal transcribed spacer region of the ribosomal RNA (rRNA) and the translation elongation factor gene (EF1) indicated the presence of 9 Trichoderma species: T. harzianum (16 isolates), T. asperellum (13), T. citrinoviride (9), T. orientalis (3), T. ghanense (3), T. hamatum (3), T. longibrachiatum (2), T. atroviride (2), and T. viride (1). All species were found to occur in potting media samples, while five Trichoderma species were recovered from the cultivated soils and four from the uncultivated soils. AFLP analysis of the 52 Trichoderma isolates produced 52 genotypes and 993 polymorphic loci. Low to moderate levels of genetic diversity were found within populations of Trichoderma species (H = 0.0780 to 0.2208). Analysis of Molecular Variance indicated the presence of very low levels of genetic differentiation (Fst = 0.0002 to 0.0139) among populations of the same Trichoderma species obtained from the potting media, cultivated soil and uncultivated soil.
Conclusion
The study provides evidence for occurrence of Trichoderma isolates in soil with no previous history of cultivation. The lack of genetic differentiation among Trichoderma populations from potting media, cultivated soil and uncultivated soil suggests that some factors could have been responsible for moving Trichoderma propagules among the three substrates. The study reports for the first time the presence of 4 Trichoderma species in Oman: T. asperellum, T. ghanense, T. longibrachiatum and T. orientalis.
Keywords: Biological control, Fallow soil, Phylogenetic analysis
Background
Trichoderma species are free living fungi that are commonly found in soil and woody materials. They are symbionts on plants and play an important role as antagonistic fungi towards pathogenic fungi [1, 2]. Trichoderma species are generally characterized by their rapid growth and ability to survive in variable environmental conditions. They are commonly found in agricultural lands, forests, and deserts. They are important contributors in the decomposition of plant materials. Some species of Trichoderma are economically important because of their ability to produce industrial enzymes and antibiotics [3, 4].
Soil-based system is the preferable choice for production of vegetables in the Arabian Peninsula and in different parts of the world. However, due to poor soil characteristics [5–7], many growers tend to use potting media and organic fertilizers in the production system of vegetables. In addition, they also replace farm soil with uncultivated soil (fallow soil) in order to reduce populations of soilborn fungal pathogens [7]. Trichoderma species are known to be very common in cultivated soils and potting media [8–11]. However, no information is available concerning diversity of Trichoderma in uncultivated soil and their relationship to Trichoderma species from cultivated soil and potting media. This makes it difficult for growers to know the impact of soil replacement on lowering Trichoderma diversity in farm soil.
Populations of fungi can vary in their levels of genetic diversity from one species to the other [12], one geographical region to the other [13] and between different cultivation systems [14]. As a result, several molecular markers have been developed and used to characterize the level of genetic diversity within and among fungal populations. These include the use of isozyme variation, Restriction Fragment Length Polymorphism (RFLP), Amplified Fragment Length Polymorphism (AFLP) and Random Amplified Polymorphic DNA (RAPD). AFLP has been found to be powerful in the characterization of genetic diversity among and within populations of different fungi [15, 16].
This study was conducted to characterize the genetic diversity of Trichoderma species in uncultivated soils and their genetic relationship with Trichoderma species isolated from cultivated soils and potting media. Results will provide a basis for future studies on Trichoderma in soil and potting media.
Results
Analysis of Trichoderma species in soil and potting media samples
Phylogenetic analysis based on the ITS rDNA and EF1 sequences separated Trichoderma isolates into clusters representing 9 species: T. asperellum (13 isolates), T. atroviride (2), T. citrinoviride (9), T. ghanense (3), T. hamatum (3), T. harzianum (16), T. longibrachiatum (2), T. orientalis (3) and T. viride (1) (Figs. 1, 2, 3).
Fig. 1.
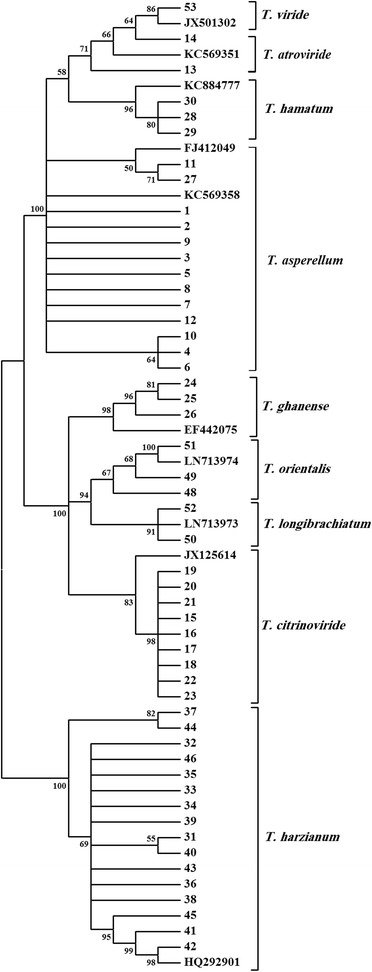
Phylogram representing the relationship of 52 Trichoderma isolates to sequences of 9 reference isolates. The analysis is based on the ITS rDNA sequences inferred by a neighbor-joining method search. Numbers within the tree represent the bootstrap values (values above 50 % are indicated; 1000 replications)
Fig. 2.

Phylogram representing the relationship of 52 Trichoderma isolates to sequences of nine reference isolates. The analysis is based EF sequences inferred by a neighbor-joining method search. Numbers within the tree represent the bootstrap values (values above 50 % are indicated; 1000 replications)
Fig. 3.
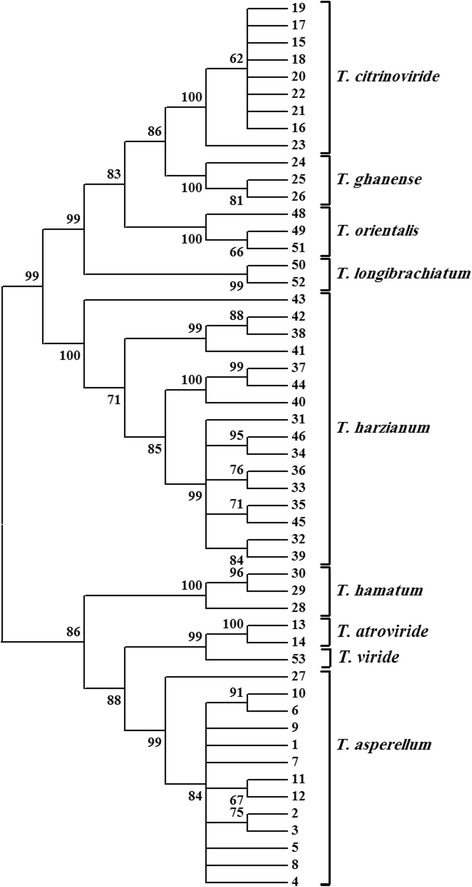
Phylogram representing clustering of 52 Trichoderma isolates based on combined analysis using ITS rDNA and EF sequences inferred by a neighbor-joining method search. Numbers within the tree represent the bootstrap values (values above 50 % are indicated; 1000 replications)
All species were found to occur in potting media samples (24 isolates obtained from 84 samples), with T. citrinoviride being the most common (8), followed by T. hamatum (3), T. harzianum (3), T. asperellum (3), T. atroviride (2), T. ghanense (2), T. longibrachiatum (1), T. orientalis (1), and T. viride (1).
Isolations from cultivated soil (15 isolates obtained from 45 samples) revealed the presence of T. asperellum (7), T. harzianum (5), T. citrinoviride (1), T. ghanense (1), and T. orientalis (1). On the other hand, isolations from the uncultivated soils (13 isolates obtained from 65 samples) showed that the most common species were T. harzianum (8), T. asperellum (3), T. orientalis (1) and T. longibrachiatum (1).
The ITS rRNA gene showed higher level of intraspecific variation compared to the EF1 gene, which gave higher resolution in separating Trichoderma species. The bootstrap support for separating species from each other was 99 % for the EF1 gene (Fig. 2). However, the bootstrap support for separating species based on the ITS rRNA gene showed that it ranges from 67 to 100 %, with one species (T. asperellum) not forming a tight cluster (Fig. 1). When the sequences of ITS rDNA and EF1 were used to produce a single tree, the resolution in separating Trichoderma species imporved to 99–100 % bootstrap support (Fig. 3).
No relationship was observed between clustering of the isolates based on ITS rDNA or EF1 sequences and clustering based on the substrates from which the isolates were obtained (Figs. 1, 2; Table 1). For example, T. harzianum isolates grouped into several sub-clusters. However, neither ITS-based sub-clustering, nor EF1-based sub-clustering correlated with the substrates from which the isolates were obtained. All ITS and EF1 sequences were deposited in the European Nucleotide Archive (Table 1).
Table 1.
Characteristics of Trichoderma species collected from cultivated and uncultivated soils and potting media
| Isolate No. | Identity | Accession # (ITS rRNA)a | Accession # (EF-1)a | Substrate | Cultivated speciesb | Countryb |
|---|---|---|---|---|---|---|
| 1 | T. asperellum | LN846676 | LN846728 | Uncultivated soil | - | Oman |
| 2 | T. asperellum | LN846677 | LN846729 | Cultivated soil | Cucumis sativus | Oman |
| 3 | T. asperellum | LN846678 | LN846730 | Cultivated soil | Cucumis sativus | Oman |
| 4 | T. asperellum | LN846679 | LN846731 | Cultivated soil | Cucumis sativus | Oman |
| 5 | T. asperellum | LN846680 | LN846732 | Cultivated soil | Cucumis sativus | Oman |
| 6 | T. asperellum | LN846681 | LN846733 | Cultivated soil | Phoenix dactylifera | Oman |
| 7 | T. asperellum | LN846682 | LN846734 | Uncultivated soil | - | Oman |
| 8 | T. asperellum | LN846683 | LN846735 | Cultivated soil | Phoenix dactylifera | Oman |
| 9 | T. asperellum | LN846684 | LN846736 | Potting media | - | Latvia |
| 10 | T. asperellum | LN846685 | LN846737 | Cultivated soil | Lycopersicum esculentum | Oman |
| 11 | T. asperellum | LN846686 | LN846738 | Uncultivated soil | - | Oman |
| 12 | T. asperellum | LN846687 | LN846739 | Potting media | - | Latvia |
| 13 | T. atroviride | LN846688 | LN846740 | Potting media | Estonia | Estonia |
| 14 | T. atroviride | LN846689 | LN846741 | Potting media | Germany | |
| 15 | T. citrinoviride | LN846690 | LN846742 | Cultivated soil | Cucumis sativus | Oman |
| 16 | T. citrinoviride | LN846691 | LN846743 | Potting media | - | Oman |
| 17 | T. citrinoviride | LN846692 | LN846744 | Potting media | - | Oman |
| 18 | T. citrinoviride | LN846693 | LN846745 | Potting media | - | Estonia |
| 19 | T. citrinoviride | LN846694 | LN846746 | Potting media | - | Estonia |
| 20 | T. citrinoviride | LN846695 | LN846747 | Potting media | - | Estonia |
| 21 | T. citrinoviride | LN846696 | LN846748 | Potting media | - | Finland |
| 22 | T. citrinoviride | LN846697 | LN846749 | Potting media | - | The Netherlands |
| 23 | T. citrinoviride | LN846698 | LN846750 | Potting media | - | UK |
| 24 | T. ghanense | LN846699 | LN846751 | Potting media | - | Estonia |
| 25 | T. ghanense | LN846700 | LN846752 | Potting media | - | Estonia |
| 26 | T. ghanense | LN846701 | LN846753 | Cultivated soil | Phoenix dactylifera | Oman |
| 27 | T. asperellum | LN846702 | LN846754 | Potting media | Estonia | Estonia |
| 28 | T. hamatum | LN846703 | LN846755 | Potting media | - | The Netherlands |
| 29 | T. hamatum | LN846704 | LN846756 | Potting media | - | Estonia |
| 30 | T. hamatum | LN846705 | LN846757 | Potting media | - | Estonia |
| 31 | T. harzianum | LN846706 | LN846758 | Potting media | - | Finland |
| 32 | T. harzianum | LN846707 | LN846759 | Potting media | - | The Netherlands |
| 33 | T. harzianum | LN846708 | LN846760 | Cultivated soil | Cucumis sativus | Oman |
| 34 | T. harzianum | LN846709 | LN846761 | Uncultivated soil | - | Oman |
| 35 | T. harzianum | LN846710 | LN846762 | Cultivated soil | Medicago sativa | Oman |
| 36 | T. harzianum | LN846711 | LN846763 | Cultivated soil | Phaseolus vulgaris | Oman |
| 37 | T. harzianum | LN846712 | LN846764 | Uncultivated soil | Dam soil | Oman |
| 38 | T. harzianum | LN846713 | LN846765 | Cultivated soil | Solanum tuberosum | Oman |
| 39 | T. harzianum | LN846714 | LN846766 | Uncultivated soil | Dam soil | Oman |
| 40 | T. harzianum | LN846715 | LN846767 | Uncultivated soil | Dam soil | Oman |
| 41 | T. harzianum | LN846716 | LN846768 | Uncultivated soil | Dam soil | Oman |
| 42 | T. harzianum | LN846717 | LN846769 | Cultivated soil | Phoenix dactylifera | Oman |
| 43 | T. harzianum | LN846718 | LN846770 | Uncultivated soil | Dam soil | Oman |
| 44 | T. harzianum | LN846719 | LN846771 | Uncultivated soil | Dam soil | Oman |
| 45 | T. harzianum | LN846720 | LN846772 | Potting media | - | Oman |
| 46 | T. harzianum | LN846721 | LN846773 | Uncultivated soil | - | Oman |
| 48 | T. orientalis | LN846722 | LN846774 | Potting media | - | Oman |
| 49 | T. orientalis | LN846723 | LN846775 | Uncultivated soil | - | |
| 50 | T. longibrachiatum | LN846724 | LN846776 | Uncultivated soil | Dam soil | Oman |
| 51 | T. orientalis | LN846725 | LN846777 | Cultivated soil | Capsicum annuum | Oman |
| 52 | T. longibrachiatum | LN846726 | LN846778 | Potting media | - | Germany |
| 53 | T. viride | LN846727 | LN846779 | Potting media | - | Germany |
aAccession number of sequences deposited in the European Nucleotide Archive
bThe sign (−) indicates that the soil was not cultivated with any plant species/no vegetation
Analysis of diversity within populations of Trichoderma species
Analysis of 52 Trichoderma isolates using 3 primer-pair combinations produced 993 polymorphic loci (100 % polymorphism), with the percentage of polymorphic loci ranging from 16 % to 80 % for the different Trichoderma species. The number of polymorphic loci produced by the three primer combinations were 277 (EcoRI-AGA/MseI-CAT), 389 (EcoRI-AGT/MseI-CAT) and 327 (EcoRI-AGT/MseI-CAA). The three primer pair combinations also resulted in moderate levels of Nei’s gene diversity, which were 0.1732, 0.2632 and 0.1810 respectively.
AFLP analysis of the 52 Trichoderma isolates produced 52 different AFLP genotypes (Table 2; Fig. 4). Each isolate representing a genotype differed from the others by at least 155 alleles. Trichoderma isolates showed a moderate level of genetic diversity (H = 0.2110). Trichoderma viride was excluded from population specific analysis of genotypic and genetic diversity because it consisted of one isolate. The seven populations differed in their level of gene diversity. The percent polymorphic loci ranged from 16 to 80 % and Nei’s gene diversity estimates ranged from 0.0780 to 0.2208 (Table 2).
Table 2.
Genotypic and genetic analysis of Trichoderma species obtained from potting media, uncultivated soils and cultivated soils in Oman
| Species | N | G | NPL | PPL | H |
|---|---|---|---|---|---|
| All | 52 | 52 | 993 | 100 | 0.2110 |
| T. asperellum | 13 | 13 | 799 | 80 | 0.2208 |
| T. atroviride | 2 | 2 | 225 | 23 | 0.1133 |
| T. citrinoviride | 9 | 9 | 571 | 58 | 0.1682 |
| T. ghanense | 3 | 3 | 260 | 26 | 0.1164 |
| T. hamatum | 3 | 3 | 363 | 37 | 0.1625 |
| T. harzianum | 16 | 16 | 767 | 77 | 0.2036 |
| T. orientalis | 3 | 3 | 323 | 33 | 0.1446 |
| T. longibrachiatum | 2 | 2 | 155 | 16 | 0.0780 |
N numbers of individuals, G genotypes, NPL numbers of polymorphic loci, PPL percentage of polymorphic loci (out of 993), H the gene diversity by Nei [30]
Fig. 4.

UPGMA dendrogram illustrating genetic similarity [31] among isolates and species of Trichoderma from potting media, cultivated soils and uncultivated soils based on AFLP fingerprinting analysis: a shows grouping based on species while b shows grouping based on isolates. The signs indicate that the isolates originate from cultivated soil (square sign), uncultivated soil (triangle sign) or potting media (circle sign)
Genetic similarity and cluster analysis
The level of genetic similarity among the seven populations was found to vary from 92 to 99 %. T. asperellum, T. harzianum and T. citrinoviride were found to share a high level of genetic similarity (Fig. 4a). Trichoderma atroviride shared the least level of genetic similarity with other Trichoderma species. Cluster analysis showed that T. asperellum, T. harzianum, T. citrinoviride, T. ghanense and T. longibrachiatum clustered together (Fig. 4a). Cluster analysis of the three most common species (T. asperellum, T. harzianum, and T. citrinoviride) showed that there was no relationship between clustering of isolate of the same species and the substrata from which they were isolated (Fig. 4b). In addition, there was no relationship between clustering of Trichoderma isolates and the species they belong to.
Partition of genetic variation
Analysis showed that the percent genetic variation is 1.39 % among populations of T. asperellum obtained from potting media, cultivated soil and uncultivated soil (F = 0.0139; P = 0.3059). The percent genetic variation among populations of T. harzianum obtained from potting media, cultivated soil and uncultivated soil was found to be 0.02 % (F = 0.0002; P = 0.4575). These values indicate that most of the genetic variation is within populations of the same species. They also indicate that the level of gene flow among populations of fungal isolates is high among potting media, cultivated soil and uncultivated soil.
Discussion
Trichoderma is a widespread genus of fungi. Previous studies in Oman provided evidence for occurrence of T. hamatum in the greenhouse soil [5] and T. harzianum, and T. parceramosum in desert crusts [17]. Our current investigation revealed presence of 9 Trichoderma species in potting media samples, 5 species in cultivated soil samples and 4 species in uncultivated soil samples. The higher level of diversity in Trichoderma species in potting media is related to the fact that these material consist of different organic and inorganic ingredients including peat moss, sphagnum, shredded bark, sawdust, vermiculite, perlite, clay and sand [18]. The Trichoderma species could have therefore been naturally occurring in these products or they could have been introduced after composting [18, 19].
Identification of Trichoderma to the species level based on reference sequences from the National Center for Biotechnology Information correlated with phylogenic analysis based on sequences of the ITS rRNA and EF1 genes. However, the limited intraspecific variation within Trichoderma species based on sequences of the EF1 gene helped gave better resolution in separating Trichoderma species when compared to sequences of the ITS region. The EF1 gene, especially when combined with ITS rDNA data, can give better resolution and is increasingly used in the identification of several fungal species [16, 20].
The frequency of isolation of Trichoderma from cultivated soil (33 %) was higher than that from uncultivated soil (20 %). Since none of the sampled farms add commercial products of Trichoderma to soil, Trichoderma isolates could have been naturally occurring in the cultivated soil. Another possible source of Trichoderma into farms is the use of potting media. Many farmers in Oman use potting media products for germination of vegetable crops or in soil [7, 11]. Since findings from this study and from previous studies indicated that potting media act as a source of Trichoderma [10], it is possible that potting media contributed to moving Trichoderma isolates to cultivated soils. This is supported by AMOVA analysis which indicated the presence of low levels of genetic differentiation and high levels of gene flow among Trichoderma populations obtained from cultivated soil and potting media (Fst = 0. 01389 for T. asperellum and 0.00016 for T. harzianum).
Our study showed the occurrence of four Trichoderma species in soils with no previous history of cultivation. Wind driven sand, which is common is Oman, could have contributed to moving some Trichoderma propagules to areas without vegetation [7]. The lack of relationship between AFLP based clustering of Trichoderma isolates and their origin as well as the insignificantly (P > 0.05) very low levels of genetic differentiation among populations of the same species obtained from cultivated and uncultivated soils may explain the hypothesis of movement of Trichoderma isolates between cultivated and uncultivated soils. In addition, some of the Trichoderma isolates were recovered from soil trapped behind dams. During rainy periods in Oman, which are very limited in this part of the world, the flowing water from mountainous areas is usually trapped behind dams. Since it is common to find Acacia spp. and some other plants in the way of the flowing water, it is possible that Trichoderma propagules could have been moved by the flowing water and then deposited in the dam soil. A study is in progress to characterize naturally occurring fungal species in the path of flowing water and from the rhizosphere of wild plants.
Farmers in Oman usually replace farm soils with uncultivated soils imported from soils trapped behind dams or non-cultivated lands. Besides reducing pathogen inoculum in farms [7], this practice could help introduce beneficial Trichoderma isolates, especially T. asperellum and T. harzianum, into farms.
Several species of Trichoderma have been long used as biological control agents to manage diseases of vegetable and other crops [10, 21]. Trichoderma harzianum and T. asperellum are the two most common species and were isolated from potting media, cultivated soil and uncultivated soil. T. harzianum is a common species in soil and potting media and it is commercially used as a biological control agent [10, 21]. T. asperellum has been used for induction of resistance to different diseases [22, 23]. The relatively high level of occurrence of T. asperellum and T. harzianum in Oman soils (82 % of the total isolates) might contribute to improving soil health through disease suppression in different crop systems
The overall level of genetic diversity of Trichoderma isolates was found to be moderate (H = 0.2110). The level of genetic diversity for the different species varied from 0.1133 to 0.2208. However, clustering based on AFLP data did not correlate with sources of the isolates, nor with clustering based on the ITS rRNA and the EF1 genes. Variation in the level of genetic diversity and sequences among the different species could be related to several reasons. Multiple introductions of Trichoderma isolates into Oman could have played a major role in augmenting the genetic diversity of Trichoderma populations [15, 16, 24]. The hypothesis of multiple introductions is supported by the formation of several AFLP genotypes and the lack of relationship between AFLP data and the source of the isolates. It is also supported by the lack of relationship between ITS/EF1 based clustering and the sources of Trichoderma isolates. This may indicate that the isolates were introduced at different times and from different sources. Other factors which could have affected the level of genetic diversity is the differences in the reproduction modes and the level of sexual recombination between the different species [15, 16].
Conclusion
This study provided evidence that the diversity in Trichoderma species varies from one substrate to the other. This is the first report showing T. asperellum, T. ghanense, T. longibrachiatum and T. orientalis in Oman. AFLP analysis suggested possible introductions of Trichoderma isolates from potting media into cultivated soil and also movement of Trichoderma between cultivated and uncultivated soils. The relatively high level of diversity in Trichoderma species and isolates may give an indication about the health of soil and its ability to contribute in the suppression of soilborn diseases that affect different crops. In addition, the high diversity may make it possible to find and select Trichoderma isolates with high antagonistic properties. Studies are in progress to characterize the biological properties of some of the Trichoderma isolates obtained in this study. Future studies are required to focus on characterizing Trichoderma isolates and other biocontrol agents from extreme environments in order to come up with isolates that can tolerate the harsh conditions of arid countries.
Methods
Collection of Trichoderma
Trichoderma isolates were obtained from potting media, cultivated soil and uncultivated soil. Twenty four Trichoderma isolates were obtained from 84 potting media products originating from Oman, The Netherlands, Estonia, Germany, Finland, Latvia, or UK (trade names are kept anonymous). Isolations from these products were done using direct plating technique as previously described by Al-Sadi et al. [11].
Fifteen Trichoderma isolates were obtained from 45 samples of cultivated (farm) soils planted with date palm, cucumber, tomato, bean, alfalfa, pepper or potato. This was done by collecting approximately 50 g soil sample from the top 15 cm of the rhizosphere of each plant species. The samples were collected from Barka, to the North-West of Muscat and from Seeb during 2013.
Trichoderma isolates (13) were also obtained from 65 uncultivated soil samples with no known history of previous cultivation. Collection of soil samples from uncultivated soil was done by collecting 50 g soil from the top 15 cm of soil (Table 1). The sites from which the samples were collected were 5–17 Km away from the coastal area of Barka and Seeb districts. The sites have no previous history of cultivation with any crop and the temperature ranges from 15 °C in the winter to 49 °C in summer (avg. 32 °C). Collection of soil samples from cultivated and un-cultivated soil was during the period from April to November 2013.
Isolation and identification of Trichoderma
Isolation of Trichoderma from soil samples was achieved using direct plating as described by Al-Sadi et al. [11]. Approximately 100–150 mg of soil sample was spread on the surface of 2.5 % potato dextrose agar (PDA) amended with 50 mg l−1 Rose Bengal. Each soil was plated onto three replicate Petri dishes and the Petri dishes were incubated at 25 °C for 5–7 d. Growth of fungal isolates which was typical of Trichoderma species was excised and transferred to 2.5 % PDA amended with 10 mg l−1 rifampicin.
Fungal isolates with typical growth of Trichoderma species were confirmed to the species level using sequences of the internal transcribed spacer region of the ribosomal RNA gene (ITS rDNA). Mycelia were collected from 7–10 day old cultures of Trichoderma species. Mycelia were collected in Eppendorf tubes and kept at −80 °C overnight. This was followed by freeze drying of the collected mycelia.
DNA was extracted from 80 mg freeze dried mycelia following a modified protocol of Lee and Taylor [25]. The ITS rRNA gene region of the fungal isolates was amplified using the universal primers ITS1 and ITS4 [26]. The polymerase chain reaction mixture was carried out using PuReTaq™ Ready-To-Go PCR™ beads (HVD Life Sciences, Vienna, Austria), 0.4 μM ITS1, 0.4 μM ITS4, 25 ng DNA and made up to 25 μl with sterilized distilled water. Thermocycling was run with the following settings: heating at 95 °C (10 min); then 35 cycles of 95 °C (30 s), 55 °C (30 s) and 72 °C (90 s). The final extension was done at 72 °C for 10 min. Amplification of the ITS region was checked out by running 5 μl of the sample on 1.5 % agarose gel in 0.5× Tris-borate-EDTA buffer (TBE) at 120 V for 50 min.
PCR for EF1 gene was conducted using primers EF1-728 F and EF1-986R [20]. The PCR mixture was as described for ITS rDNA. PCR conditions consisted of denaturation at 94 °C for 4 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 60 s. This was followed by extension at 72 °C for 10 min. PCR products were checked by running 5 μl of a PCR product on 1.5 % agarose gel in 5× TBE at 120 V for 40 min.
PCR products were sequenced at Macrogen Inc. (Korea, Seoul) in both senses using the ITS1, ITS4, EF1-728 F and EF1-986R. The resulting forward and reverse ITS and EF1 sequences were aligned and edited using ChromasPro v. 1.41 (Technelysium Pty Ltd). Then, the obtained sequences for each isolate were compared to sequences available at the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) using BLAST search.
Nine reference ITS sequences representing 9 Trichoderma species were obtained from NCBI. The nine representative sequences were aligned with the sequences of the 52 isolates obtained in this study using Clustal W [27]. A neighbour joining tree was constructed using the Kimura 2 parameter evolutionary model (Mega 5) [28]. Consensus trees were generated using 1000 replications (55 % bootstrap criteria).
AFLP Fingerprinting
Amplified Fragment Length Polymorphism (AFLP) was used to assess genetic diversity within populations of Trichoderma species. AFLP fingerprinting was done as described by Al-Sadi et al. [16] using FAM-6-labelled EcoRI-AXX selective primers. The primer combinations used in this study were EcoRI-AGA/MseI-CAT, EcoRI-AGT/MseI-CAT, and EcoRI-AGT/MseI-CAA.
DNA restriction and ligation were performed as described by Al-Sadi et al. [16]. The pre-selective amplification reaction mixtures consisted of 0.65 μl of 10 μM each of EcoRI + A (5′-GACTGCGTACCAATTCA-'3) and MseI-C (5′-GATGAGTCCTGAGTAAC-′3) primers, 3.7 μl of diluted restriction/ligation mix, PuReTaqTM Ready-To-GoTM PCR beads and Milli-Q water made up to a volume of 25 μl. The cycling profile was conducted as detailed by Al-Sadi et al. [15].
The pre-selective amplification product was diluted by adding 210 μl of TE0.1 buffer (20 mM Tris–HCl, 0.1 mM EDTA, pH 8) to the remaining amount. The selective amplification reaction was as above except using 0.13 μl of 10 μM FAM-6 labeled EcoRI selective primer, 0.63 μl of 10 μM MseI selective primer, 6 μl of diluted pre-selective amplification and Milli-Q water to a volume of 25 μl. The cycling parameters for the selective amplification were as described by Al-Sadi et al. [16]. Fragment analysis of the PCR products from the selective amplification reactions was carried out at Macrogen Inc. (Seoul) using ABI 3730XL (Applied Biosystems, Carlsbad, CA).
Analysis of AFLP data
Analysis of AFLP data was done to estimate gene diversity, genotypic diversity, genetic distance and genetic differentiation within and among different populations. AFLP alleles in the range of 50 to 500 base pairs (bp) were evaluated using Gene Mapper 4.0 with 0 for absence and 1 for presence of each amplified fragment. The binary data were analyzed using POPGENE version 1.32 [29] which was used to calculate the number of polymorphic loci, Nei’s gene diversity [30] and genetic distance and identity within and among the different populations of Trichoderma species. Genetic distance based on Nei’s [31] unbiased measurement of genetic distance was also determined between samples and populations of Trichoderma species using POPGENE. A dendrogram was constructed based on Nei’s unbiased measures of genetic distance using UPGMA (unweighted pair group method with arithmetic mean; NTSYSpc v. 2.21 m).
Analysis of molecular variance (AMOVA) was used to determine genetic differentiation among the different populations using Arlequin v. 3.1 [32]. AMOVA was conducted only for populations of T. asperellum and T. harzianum because each species included at least 2 isolates from each substratum.
Acknowledgements
We would like to acknowledge Sultan Qaboos University for financial support of the study through the strategic project SR/AGR/CROP/10/01 and IG/AGR/CROP/13/01 and Mohammed Al-Maamari and Issa Al-Mahmooli for technical help.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AMA, SE and FAA designed the experiments; FAA, HAA and AMF performed the laboratory experiments; AMA, SE and FAA prepared the manuscript. All of the authors examined and agreed with the final manuscript.
Contributor Information
Abdullah M. Al-Sadi, Phone: +96899424633, Email: alsadi@squ.edu.om
Fatma A. Al-Oweisi, Email: 12213700@live.harper.ac.uk
Simon G. Edwards, Email: sedward@harper-adams.ac.uk
Hamed Al-Nadabi, Email: m087182@student.squ.edu.om.
Ahmed M. Al-Fahdi, Email: m086014@student.squ.edu.om
References
- 1.Harman GE, Taylor AG, Stasz TE. Combining effective strains of Trichoderma harzianum and solid matrix priming to improve biological seed treatments. Plant Dis. 1989;73(8):631–7. doi: 10.1094/PD-73-0631. [DOI] [Google Scholar]
- 2.Gruber S, Omann M, Zeilinger S. Comparative analysis of the repertoire of G protein-coupled receptors of three species of the fungal genus Trichoderma. BMC Microbiol. 2013;13:108. doi: 10.1186/1471-2180-13-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yurnaliza R, Aryantha INP, Esyanti RR, Susanto A. Antagonistic activity assessment of fungal endophytes from oil palm tissues against ganoderma boninense pat. Plant Pathol J. 2014;13:257–67. doi: 10.3923/ppj.2014.257.267. [DOI] [Google Scholar]
- 4.Rahnama N, Foo HL1, Rahman NAA, Ariff A, Shah UKM. Saccharification of rice straw by cellulase from a local Trichoderma harzianum SNRS3 for biobutanol production. BMC Biotechnol. 2014;14:103. doi: 10.1186/s12896-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Sadi AM, Al-Said FA, Al-Kiyumi KS, Al-Mahrouqi RS, Al-Mahmooli IH, Deadman ML. Etiology and characterization of cucumber vine decline in Oman. Crop Prot. 2011;30:192–7. doi: 10.1016/j.cropro.2010.10.013. [DOI] [Google Scholar]
- 6.Al-Mawaali QS, Al-Sadi AM, Khan AJ, Al-Hasani H, Deadman ML. Response of cucurbit rootstocks to Pythium aphanidermatum. Crop Prot. 2012;42:64–8. doi: 10.1016/j.cropro.2012.07.017. [DOI] [Google Scholar]
- 7.Al-Sa’di AM, Drenth A, Deadman ML, Al-Said FA, Khan I, Aitken EAB. Potential sources of Pythium inoculum into greenhouse soils with no previous history of cultivation. J Phytopathol. 2008;156:502–5. doi: 10.1111/j.1439-0434.2008.01396.x. [DOI] [Google Scholar]
- 8.Migheli Q, Balmas V, Komoñ-Zelazowska M, Scherm B, Fiori S, Kopchinskiy A, Kubicek C, Druzhinina I. Soils of a Mediterranean hot spot of biodiversity and endemism (Sardinia, Tyrrhenian Islands) are inhabited by pan- European, invasive species of Hypocrea/Trichoderma. Environ Microbiol Rep. 2009;11:35–46. doi: 10.1111/j.1462-2920.2008.01736.x. [DOI] [PubMed] [Google Scholar]
- 9.Tsurumi Y, Inaba S, Susuki S, Kamijo S, Widyastuti Y, Hop D, Balijinova T, Sukarno N, Nakagiri A, Susuki K, et al. Distribution of Trichoderma species in four countries of Asia. 9. UK: International Mycological Congress Edinburgh; 2010. pp. 1–8. [Google Scholar]
- 10.Blaya J, López-Mondéjar R, Lloret E, Pascual JA, Ros M. Changes induced by Trichoderma harzianum in suppressive compost controlling Fusarium wilt. Pestic Biochem Physiol. 2013;107:112–9. doi: 10.1016/j.pestbp.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Al-Sadi AM, Al-Said FA, Al-Jabri AH, Al-Mahmooli IH, Al-Hinai AH, de Cock AWAM. Occurrence and characterization of fungi and oomycetes transmitted via potting mixtures and organic manures. Crop Prot. 2011;30:38–44. doi: 10.1016/j.cropro.2010.09.015. [DOI] [Google Scholar]
- 12.Garzon CD, Geiser DM, Moorman GW. Diagnosis and population analysis of Pythium species using AFLP fingerprinting. Plant Dis. 2005;89(1):81–9. doi: 10.1094/PD-89-0081. [DOI] [PubMed] [Google Scholar]
- 13.Harvey PR, Butterworth PJ, Hawke BG, Pankhurst CE. Genetic variation among populations of Pythium irregulare in southern Australia. Plant Pathol. 2000;49(5):619–27. doi: 10.1046/j.1365-3059.2000.00493.x. [DOI] [Google Scholar]
- 14.Francis DM, St Clair DA Population genetics of Pythium ultimum. Phytopathology. 1997;87(4):454–61. doi: 10.1094/PHYTO.1997.87.4.454. [DOI] [PubMed] [Google Scholar]
- 15.Al-Sadi AM, Al-Ghaithi AG, Al-Balushi ZM, Al-Jabri AH. Analysis of diversity in Pythium aphanidermatum populations from a single greenhouse reveals phenotypic and genotypic changes over 2006 to 2011. Plant Dis. 2012;96:852–8. doi: 10.1094/PDIS-07-11-0624. [DOI] [PubMed] [Google Scholar]
- 16.Al-Sadi AM, AL-Wehaibi AN, Al-Shariqi RM, Al-Hammadi MS, Al-Hosni IA, Al-Mahmooli IH, Al-Ghaithi AG. Population genetic analysis reveals diversity in Lasiodiplodia species infecting date palm, Citrus, and mango in Oman and the UAE. Plant Dis. 2013;97:1363–9. doi: 10.1094/PDIS-03-13-0245-RE. [DOI] [PubMed] [Google Scholar]
- 17.Abed RMM, Al-Sadi AM, Al-Shihi MA, Al-Hinai S, Robinson MD. Diversity of free-living and lichenized fungal communities in biological desert crusts of the Sultanate of Oman and their role in improving soil properties. Soil Biol Biochem. 2013;57:695–705. doi: 10.1016/j.soilbio.2012.07.023. [DOI] [Google Scholar]
- 18.Kausar H, Ismail MR, Saud HM, Habib SH, Othman R, Bhuiyan MSH. Changes of physical and chemical characteristics during microbial composting of rice straw at various pH levels. Compost Sci Utilizat. 2014;22:153–63. doi: 10.1080/1065657X.2014.918864. [DOI] [Google Scholar]
- 19.Huang K, Li F, Wei Y, Chen X, Fu X. Changes of bacterial and fungal community compositions during vermicomposting of vegetable wastes by Eisenia foetida. BioresourTechnol. 2013;150:235–41. doi: 10.1016/j.biortech.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–6. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 21.Yu C, Fan L, Wu Q, Fu K, Gao S, Wang M, Gao J, Li Y, Chen J. Biological role of Trichoderma harzianum-derived Platelet-Activating Factor Acetylhydrolase (PAF-AH) on stress response and antagonism. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández E, Segarra G, Trillas MI. Physiological effects of the induction of resistance by compost or Trichoderma asperellum strain T34 against Botrytis cinerea in tomato. Biol Control. 2014;78:77–85. doi: 10.1016/j.biocontrol.2014.06.012. [DOI] [Google Scholar]
- 23.Guzmán-Valle P, Bravo-Luna L, Montes-Belmont R, Guigón-López C, Sepúlveda-Jiménez G. Induction of resistance to Sclerotium rolfsii in different varieties of onion by inoculation with Trichoderma asperellum. Eur J Plant Pathol. 2014;138:223–9. doi: 10.1007/s10658-013-0336-y. [DOI] [Google Scholar]
- 24.Al-Sadi AM. Phylogenetic and population genetic analysis of Ceratocystis radicicola infecting date palms. J Plant Pathol. 2013;95:47–55. [Google Scholar]
- 25.Lee SB, Taylor JW. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A Guide to Methods and Applications. New York, USA: Academic; 1990. pp. 282–7. [Google Scholar]
- 26.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A Guide to Methods and Applications. New York, USA: Academic; 1990. pp. 315–22. [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biol Evolut. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh RC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot. 1997;129:157. [Google Scholar]
- 30.Nei M. Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci. 1973;70:3321–3. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–90. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolut Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]


