Abstract
Oligonucleotide array comparative genomic hybridization, karyotype and fluorescence in situ hybridization analyses were employed to delineate the cytogenetic abnormalities in a case of pediatric Acute Megakaryoblastic Leukemia. Here we present a unique genetic profile that includes bi-allelic deletions within 13q14, where the retinoblastoma tumor suppressor gene (RB1) resides, as well as isolated trisomy 21 without a concomitant mutation in the hematopoietic transcription factor GATA1s and translocation (17;22), that does not involve the megakaryoblastic leukemia 1 (MKL1) gene located on chromosome 22. Alteration of the RB1 gene is most likely the critical leukemogenic event in this patient.
Keywords: AML, Megakaryocytopoiesis, Pediatric Hematology/Oncology, Cancer Biology, Cytogenetics
Introduction
Acute Megakaryoblastic Leukemia (AMKL), a morphologically and cytogenetically heterogeneous disease characterized by the halted differentiation and pathologic proliferation of megakaryoblasts, occurs most commonly in children less than two years old [1]. The incidence of AMKL in children with Down Syndrome (AMKL-DS) is 600 times greater than in the general pediatric population [2]. A somatic mutation resulting in truncation of the hematopoietic transcription factor GATA1 on the X chromosome, occurs concomitantly with trisomy 21 [3]. Infant AMKL is commonly associated with translocation of the RNA Binding Motif 15 (RBM15) gene on chromosome 1 upstream of the Megakaryoblastic Leukemia 1 (MKL1) gene on chromosome 22, resulting in the fusion of RBM15 to MKL1 (AMKL-t(1;22)) [4,5]. During normal megakaryopoiesis, RBM15 expression is highest in hematopoietic stem cells and decreases with maturation, while MKL1 expression increases with megakaryocytic differentiation [6,7].
Here we report a pediatric AMKL patient with novel cytogenetic findings including transient trisomy 21 without GATA1 mutation, translocation between 17q and 22q without involvement of MKL1 and bi-allelic deletions within 13q14 with hemizygous loss of the RB1 gene.
Case Report
A 17-month-old female presented with irritability and bruising. Physical examination revealed pallor, petechiae and ecchymoses on her upper and lower extremities, a systolic ejection murmur, mild hepatosplenomegaly, and inguinal lymphadenopathy. Laboratory evaluation revealed leukocytosis (60 × 109/L) with 53% circulating blasts, anemia (6.1 g/dL) and thrombocytopenia (12 × 109/L), as well as a prolonged prothrombin time and elevated INR. Her lactate dehydrogenase was elevated at 3550, although her uric acid was normal.
Bone marrow (BM) aspiration demonstrated an infiltrative blast population with high nuclear to cytoplasmic ratio, basophilic cytoplasm, dense nuclear chromatin and indistinct nucleoli (Supplemental Figure 1). Flow cytometry of peripheral blood and BM was consistent with AMKL. Two immunophenotypically distinct myeloblast populations were identified, the predominant being CD41+ CD61+ CD42 + megakaryoblasts. Table I summarizes the immunophenotype and cytogenetic data collected from diagnosis to recurrence prior to hematopoietic stem cell transplant.
Table I.
Summary of Immunophenotype and Cytogenetic Data
| Date | Immunophenotype | Cytogenetics |
|---|---|---|
|
02/09/07 Diagnosis |
CD45dim+, CD117+, CD33dim+, CD34−, CD4dim+, CD41−, CD61−, CD42−, HLA-DR+, CD11b−, CD13−, CD15−, TdT−, MPO− CD45dim+, CD117−, CD33dim+, CD34−, CD4dim+, CD41+, CD61+, CD42+, HLA-DR−, CD11b−, CD13−, CD15−, TdT−, MPO− |
BONE MARROW: 46–47,XX,del(13)(q14q14)x2,der(16)t(13;16)(?q31q33:p13.3),t(17;22)(?q25;q13),+21 Abnormal FISH - homozygous deletions of 13q14.3, heterozygous deletion of 13q14.2 (RB1), translocation 13q34;16, trisomy 21 and 17q;22q translocation |
| 02/19/07 |
PERIPHERAL BLOOD: 46,XX[2] Limited study Normal FISH - disomy 21 |
|
|
03/09/07 Induction I |
Normal | BONE MARROW: 46–47,XX,del(13)(q14)x2,der(16)t(13;16)(?q31q33:p13.3),t(17;22)(q25;q13),+21 |
|
04/17/07 Induction II |
Normal | BONE MARROW: 46,XX, Normal FISH |
|
05/16/07 Intensification I |
Normal | BONE MARROW: 46,XX, Normal FISH |
|
06/15/07 Intensification II |
Normal | BONE MARROW: 46,XX, Normal FISH |
|
08/10/07 Intensification III Recurrence |
CD45+ dim, CD117−, CD33dim+, CD34−, CD19−, CD41+, CD61+, CD42dim+, CD13dim+ |
BONE MARROW: 46,XX,del(13)(q12q14),del(13)(q14q14)[3]/46,XX Abnormal FISH - homozygous deletions of 13q14.3 and heterozygous deletion of 13q14.2(RB1) |
|
09/14/07 Re-induction I |
CD45+, CD33−, CD34−, CD10−, CD19−, glycophorin−, CD41+/−, CD61+/−, CD42 dim+ CD13− |
BONE MARROW: 46,XX,del(13)(q12q14),del(13)(q14q14),t(17;22)(q24;q13)[16]/46,XX[ Abnormal FISH - homozygous deletions of 13q14.3, heterozygous deletion of 13q14.2 (RB1), trisomy 21 and 17q;22q translocation |
|
10/01/07 Re-induction II |
CD45+, CD33−, CD10−, CD19−, glycophorin−, CD41+, CD61+, CD42+, HLADR+, CD11b−, CD13− |
BONE MARROW: 46,XX,del(13)(q12q14),del(13)(q14q14),t(17;22)(?q25;q13) Abnormal FISH - homozygous deletions of 13q14.3, heterozygous deletion of 13q14.2 (RB1) and 17q;22q translocation |
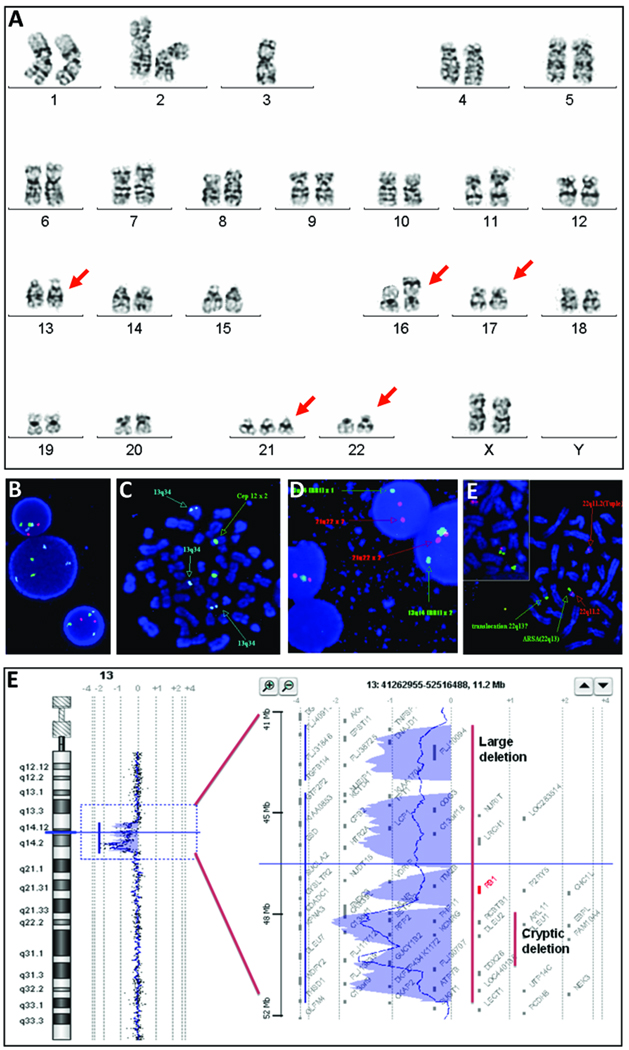
Chromosome analysis of cultured BM cells revealed an abnormal clone with multiple structural rearrangements in 13 of 17 metaphase cells. Consistent findings included a subtle deletion in the long arm of both copies of chromosome 13, a derivative 16 with 13q material added to its short arm, translocation(17q;22q), and trisomy 21. The karyotype was 47XX, del(13)(q12q14), der (16)t(13;16) (?q31q33;p13.3), t(17;22) (?q25;q13),+21 (Figure 1A).
Figure 1.
Karyotype and FISH analysis on cultured unstimulated bone marrow cells. A: Chromosomal spread reveals a clonal abnormality detected at initial diagnosis including a 13q deletion, der (16), t(17;22) and acquired trisomy 21. B: Fish analysis on interphase cells reveals loss of 13q14.3 (red). C: FISH analysis on metaphase spread demonstrates homozygous loss of 13q14 (red) and the presence of 13q34 (aqua) on chromosome 16. D: Fish analysis on interphase cells using probes for RB1 gene and 21q22.13 (red) reveals only one copy of the RB1 gene in two cells. E: Fish analysis on metaphase spread identifies a translocation between 17q and 22q with the presence of the 22q13 signal (green) in the der(17) and the 22q11.2 signal (red) in the der(22). F: oaCGH analysis performed at time of first relapse defines a 1.72 Mb deletion of 13q14.2-q14.3 and a 9.34 Mb deletion of 13q14.11-q14.3.
Of 300 interphase cells analyzed by fluorescence in situ hybridization (FISH) using tri-colored probes for 13q14.3, 13q34 control locus, and 12cen, 38% were normal and 62% demonstrated three signals for the 13q34 probe and no signal for the 13q14.3 probe. Metaphase spreads revealed absence of 13q14.3 signals on both copies of chromosome 13, and presence of the 13q34 probe on 13q, as expected, and on the derivative 16p indicating an abnormal clone with homozygous deletions in the 13q14 region, and a translocated 13q34 segment on the derivative 16p (Figure 1B).
FISH for the RB1 gene at 13q14.2 and the 21q22 locus revealed that 89% of 200 interphase nuclei had deletion of RB1 (Figure 1C) and 57.5% had three signals for chromosome 21 (data not shown). Thus, one chromosome 13 had a large deletion involving the RB1 gene and the 13q14.3 locus, and the other had a submicroscopic deletion involving only the 13q14.3 locus. FISH for the TUPLE1 (HIRA) gene at 22q11.2 and the ARSA gene at 22q13 indicated a translocation between 17q and 22q (Figure 1D). FISH for the MKL1 gene at 22q13.1 revealed that 100% of 300 interphase cells had two adjacent signals consistent with absence of MKL1 gene rearrangements. Thus, the breakpoint in 22q is distal to TUPLE1 (17.7 Mb, 22q11.2) and proximal to MKL1 (39.1 Mb, 22q13.1), and the entire MKL1 gene is translocated onto the 17q. Additional analyses revealed one fusion signal on chromosome 22 and another fusion signal on derivative chromosome 17, confirming the 17;22 translocation (Figure 1D). There were no mutations in exon 2 of the GATA1 gene determined by direct sequencing of PCR amplified genomic fragments.
Chromosomal analysis of culture stimulated peripheral lymphocytes revealed a normal female karyotype. FISH for the RB1 gene and the 21q22.13 locus revealed that all 200 nuclei examined had a normal two-signal pattern for both probes, thus excluding constitutional or mosaic trisomy 21 and homozygous RB1 gene deletion.
The patient began Induction chemotherapy per COG protocol AAML0531 with cytarabine, daunorubicin and etoposide. Subsequent BM evaluation revealed blasts consistent with age appropriate hematopoietic recovery. Of 45 metaphase BM cells analyzed, an abnormal karyotype with the previously detected clonal abnormalities was identified in one cell.
After a second course of Induction chemotherapy with cytarabine, daunorubicin and etoposide, morphologic, cytometric, cytogenetic and FISH analyses of the BM were consistent with disease remission. The patient received Intensification I chemotherapy with high dose cytarabine and etoposide; BM evaluation demonstrated continued remission. After administration of Intensification II chemotherapy with high dose cytarabine and mitoxantrone, BM aspiration confirmed normal trilineage hematopoiesis.
Six weeks after completing a final course of Intensification III chemotherapy with high dose cytarabine and L-asparaginase, the patient developed thrombocytopenia, and BM aspiration revealed recurrent disease with approximately 30% megakaryoblasts. Chromosomal and FISH analyses revealed homozygous deletion of the 13q14.3 region in 12% of cells, a two signal pattern for 21q22 in only 25.5% of cells, and a single signal for the RB1 gene, consistent with relapse of the previously detected abnormal clone.
The patient tolerated re-induction chemotherapy with G-CSF stimulation, fludarabine, cytarabine and idarubicin, but recovered with persistent disease. Flow cytometry of peripheral blood and BM revealed megakaryoblasts with essentially the same abnormal immunophenotype. Cytogenetic analysis revealed persistence of the abnormal clone harboring deletions within the 13q14 region and a recurrence of the translocation between 17q24 and 22q13. FISH revealed bi-allelic deletions in the 13q14 region and hemizygous loss of RB1 with a subclone having trisomy 21.
A second attempt at re-induction of remission using cytarabine and clofarabine was unsuccessful as BM evaluation again revealed megakaryoblasts with similar cytogenetic abnormalities. Oligonucleotide array comparative genomic hybridization (oaCGH) analysis detected a 1.72 Mb deletion (Chr13:48,811,812-50,535,035) in 13q14.2q14.3 including known genes CAB39L, SETDB2, PHF11, RCBTB1, ARL11, EBPL, KPNA3, C13orf1, TRIM13, KCNRG, DLEU7, RNASEH2B, and GUCY1B2, and a 9.34 Mb deletion (Chr13:42,053,349-51,626,277) in the 13q14.11q14.3 containing approximately 58 genes (from TNFSF11,.., RB1,.., to NEK3 genes) (Figure 1E) with no other genomic gains or losses.
The patient underwent a BC mismatched unrelated umbilical cord blood transplantation. Less than four months post-transplant, the patient developed thrombocytopenia and BM evaluation revealed AMKL with additional cytogenetic aberrations including chromosome 1p duplication and 3q deletion. After a trial of decitabine and gemtuzumab, the patient received palliative chemotherapy with oral etoposide and died shortly thereafter.
Discussion
Initial cytogenetic findings suggested that this patient’s AMKL was related to the acquired trisomy 21 or a variant 22q13 translocation involving the MKL1 gene. The acquired trisomy 21 gradually diminished and disappeared from the later relapse clones indicating that it may have contributed to the initial tumor induction, but was not required for its maintenance and progression. FISH analysis ruled out a direct MKL1 rearrangement, however, the possibility of position effect on expression of the translocated MKL1 in the der(17) cannot be excluded. A pediatric non DS-AMKL case with a t(17;22)(q21;q13) and suspected involvement of the BRCA1 and WT-4 genes in the 17q21 and the MKL1 gene in the 22q13 regions has been reported, but no experiments were performed to detect these breakpoints [8]. The unexpected compound deletions at 13q14 on initial diagnosis, and the persistence of this abnormality upon multiple relapses, implicates this as the critical leukemogenic defect. oaCGH delineated a 1.72 Mb microdeletion, and the chromosomally observed del(13)(q12q14) as a 9.34 Mb deletion from 13q14.11 to 13q14.3.
Structural abnormalities within the 13q14 region, which includes the RB1 gene, occur in both lymphoid leukemia, which can harbor homozygous deletions [9], and myeloid leukemia, in which heterozygous deletions have been observed. Although the 13q14 deletion has not been reported in association with childhood AMKL, oaCGH analysis found it to be the most common chromosomal loss in AMKL cell lines [10]. The significance of the 13q14 deletion in pediatric AMKL remains unclear; however, reduced Rb protein expression is associated with poor outcomes in AML [11–14]. Given the refractory nature of this disease in this patient, deletion of the 13q14 region should be considered a poor prognosis indicator.
Progress in our understanding of cytogenetic abnormalities and their clinical relevance in pediatric malignancies prompts an integrated cytogenetic and molecular diagnostic approach. We proposed that a screen for constitutional or acquired trisomy 21, rearrangements involving 22q13, especially the t(1;22)(p13;p13), and submicroscopic 13q14 deletion potentially involving RB1, may improve risk stratification and ultimately aid in more effective targeted therapies. Mutational analysis of the GATA1 gene must be considered in patients with trisomy 21.
In summary, we have presented a case of pediatric AMKL with unique cytogenetic abnormalities involving transient acquired trisomy 21, t(17;22)(q23;q13), and compound deletions at 13q14. Possible candidate genes within this region and their role in megakaryocyte differentiation deserve further investigation.
Supplementary Material
Supplementary Figure 1. Bone marrow aspirate smear demonstrates a prominent population of blasts consistent with leukemic infiltration.
References
- 1.Ballerini P, Blaise A, Mercher T, et al. A novel real-time RT-PCR assay for quantification of OTT-MAL fusion transcript reliable for diagnosis of t(1;22) and minimal residual disease (MRD) detection. Leukemia. 2003;17(6):1193–1196. doi: 10.1038/sj.leu.2402914. [DOI] [PubMed] [Google Scholar]
- 2.Zipursky A, Thorner P, De Harven E, et al. Myelodysplasia and acute megakaryoblastic leukemia in Down's syndrome. Leuk Res. 1994;18(3):163–171. doi: 10.1016/0145-2126(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 3.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 4.Mercher T, Coniat MB, Monni R, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A. 2001;98(10):5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Z, Morris SW, Valentine V, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28(3):220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- 6.Raffel GD, Mercher T, Shigematsu H, et al. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci U S A. 2007;104(14):6001–6006. doi: 10.1073/pnas.0609041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X, Renda MJ, Wang L, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27(8):3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitlur MB, Bhambhani K, Mohamed AN, et al. Acute megakaryoblastic leukemia with t(17;22)(q21;q13) and liver dysfunction. Cancer Genet Cytogenet. 2004;154(2):167–168. doi: 10.1016/j.cancergencyto.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Hermanson M, Grander D, et al. 13q deletions in lymphoid malignancies. Blood. 1995;86(5):1911–1915. [PubMed] [Google Scholar]
- 10.Alvarez S, MacGrogan D, Calasanz MJ, et al. Frequent gain of chromosome 19 in megakaryoblastic leukemias detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;32(3):285–293. doi: 10.1002/gcc.1192. [DOI] [PubMed] [Google Scholar]
- 11.Kornblau SM, Andreeff M, Hu SX, et al. Low and maximally phosphorylated levels of the retinoblastoma protein confer poor prognosis in newly diagnosed acute myelogenous leukemia: a prospective study. Clin Cancer Res. 1998;4(8):1955–1963. [PubMed] [Google Scholar]
- 12.Kornblau SM, Xu HJ, del Giglio A, et al. Clinical implications of decreased retinoblastoma protein expression in acute myelogenous leukemia. Cancer Res. 1992;52(17):4587–4590. [PubMed] [Google Scholar]
- 13.Kornblau SM, Xu HJ, Zhang W, et al. Levels of retinoblastoma protein expression in newly diagnosed acute myelogenous leukemia. Blood. 1994;84(1):256–261. [PubMed] [Google Scholar]
- 14.Sauerbrey A, Stammler G, Zintl F, et al. Expression of the retinoblastoma tumor suppressor gene (RB-1) in acute leukemia. Leuk Lymphoma. 1998;28(3–4):275–283. doi: 10.3109/10428199809092683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Bone marrow aspirate smear demonstrates a prominent population of blasts consistent with leukemic infiltration.