Abstract
Aldehyde dehydrogenase 1B1 (ALDH1B1) is a mitochondrial enzyme sharing 65% and 72% sequence identity with ALDH1A1 and ALDH2 proteins, respectively. Compared to the latter two ALDH isozymes, little is known about the physiological functions of ALDH1B1. Studies in humans indicate that ALDH1B1 may be associated with alcohol sensitivity and stem cells. Our recent in vitro studies using human ALDH1B1 showed that it metabolizes acetaldehyde and retinaldehyde. To investigate the in vivo role of ALDH1B1, we generated and characterized a global Aldh1b1 knockout mouse line. These knockout (KO) mice are fertile and show overtly good health. However, ethanol pharmacokinetic analysis revealed ~40% increase in blood acetaldehyde levels in KO mice. Interestingly, the KO mice exhibited higher fasting blood glucose levels. Collectively, we show for the first time the functional in vivo role of ALDH1B1 in acetaldehyde metabolism and in maintaining glucose homeostasis. This mouse model is a valuable tool to investigate the mechanism by which alcohol may promote the development of diabetes.
Keywords: ALDH1B1, ethanol, acetaldehyde, glucose, pancreas, colon
Introduction
The superfamily of aldehyde dehydrogenases (ALDHs) are involved in the metabolism of a wide range of endogenous and exogenous aldehydes [1]. ALDH enzymes play diverse but physiologically important roles, as evidenced by the variety of human diseases associated with mutations in ALDH genes [2]. This gene encodes a mitochondrial protein (ALDH1B1), previously known as ALDHX or ALDH5 [3], which is 72% and 65% identical to mitochondrial ALDH2 and cytosolic ALDH1A1 proteins, respectively. ALDH1B1 is abundantly expressed in the liver, small intestine and testes, and to a lesser extent in other tissues, including the pancreas and colon [4].
To date, the physiological function of ALDH1B1 is largely unknown. We have previously reported that human ALDH1B1 is the second most efficient enzyme (Km = 55 μM) at oxidizing acetaldehyde after ALDH2 (Km = 3.4 μM) [4]. This biochemical feature of ALDH1B1 is suggestive of a potential role in ethanol metabolism. In line with this notion, human studies have identified ALDH1B1 polymorphisms to be associated with symptoms of acetaldehyde toxicity including ethanol hypersensitivity, hypertension and ethanol aversion in Caucasian populations [5, 6], where the well-studied ALDH2*2 variant is nearly absent [7]. In addition to its acetaldehyde metabolic effects, ALDH1B1 has the catalytic capacity for oxidation of retinaldehyde [8], which is supportive of ALDH1B1 playing a role in the differentiation and development of normal and cancer stem cells [9]. In this context, we have observed that ALDH1B1 is expressed specifically in the stem cell compartment in the normal colon and is drastically induced in human colon cancerous tissues [10]. In another study, we have found that ALDH1B1 is strongly expressed in the early pancreatic buds in developing mice [11]. With further development and differentiation, strong ALDH1B1 expression remains confined exclusively to tips and the trunk of the pancreatic epithelium and persists only in centroacinar-like cells by the time of birth [11]. In adult mice, ALDH1B1-expressing cells expand dramatically in the pancreas following acute experimental ablation of acinar or β cell populations [11]. Taken together, these findings indicate a role for ALDH1B1 in pancreatic development and regeneration. As such, ALDH1B1 may influence the functional integrity of pancreas tissue, and thereby impact glucose homeostasis.
In this current study, we generated a mouse line with global disruption of the Aldh1b1 gene through gene targeting. Utilizing this knockout (KO) mouse model, we explored the in vivo role of ALDH1B1 in ethanol metabolism and glucose homeostasis.
Materials and methods
Chemicals
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Preparation of targeting construct and generation of Aldh1b1 (−/−) KO mice
Details about the targeting construct and targeting procedures can be found in the Supplementary Materials. The Aldh1b1(-) allele has been backcrossed into the C57BL/6J background for >10 generations. All studies were carried out in accordance with the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee (IACUC).
Southern blot and PCR analysis
Details on Southern blotting and PCR screening of successfully targeted ES clones can be found in the Supplementary Materials. Genotyping in offspring was performed by PCR analysis using tail genomic DNA. The Aldh1b1(+) wild-type allele was detected using forward primer 5’-ACACTGCAACAGGAGGACCAAGAA-3’) and reverse primer 5’-ACATGCCCAATGACCTCACCT-3’, generating a 429 bp product. The Aldh1b1(−) KO allele was detected using same forward primer as (+) allele and reverse primer 5’-TTAAACGCGGCCGCCAATTGT-3’, generating a 200 bp product.
Reverse transcription and quantitative real time PCR (qRT-PCR)
Total RNA from selected tissues was extracted using TRI reagent and further purified with an RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized using the Maxima First-Strand cDNA Synthesis kit (Fisher Scientific Inc., Waltham, MA). qRT-PCR reactions were carried out using the Power SYBR Green Master Mix (Applied Biosystems Inc., Foster city, CA). Expression of β-2 microglobulin (B2M) was used for normalization of C data according to the 2−ΔΔCT method [12]. The mRNA level of examined genes in WT mice was set as the control (= 1), and relative mRNA levels are expressed as fold of control. Data are reported as the mean ± S.E. from three to four animals. Details on primer sequences used in qRT-PCR reactions can be found in the Supplementary Materials.
Western blot analysis
Western immunoblotting was performed using tissue whole-cell homogenates and mitochondrial extractions as previously described [13]. Primary antibodies were used at a dilution of 1:5000 for rabbit polyclonal anti-ALDH1B1 [4]. Protein bands were visualized using a chemiluminescence imaging system (PerkinElmer Life and Analytical Sciences, Waltham, MA).
Histological and immunohistochemical (IHC) analysis
Tissues were fixed in 4% paraformaldehyde, dehydrated in graded ethanol solutions, and embedded in paraffin. Tissue sections (5 microns thick) were cut, deparaffinized, rehydrated, and processed for hematoxylin and eosin (HE), IHC and periodic acid-Schiff (PAS) staining. IHC staining was performed using the TSA Biotin System Kit (PerkinElmer, Waltham, MA). A dilution of 1:750 was used for all primary antibodies, including rabbit polyclonal anti-ALDH1B1 [4], mouse monoclonal anti-insulin and anti-glucagon (Abcam, Cambridge, MA), and mouse monoclonal anti-Ki-67 (Sigma-Aldrich, St. Louis, MO). HRP-conjugated secondary antibodies were used at a 1:500 dilution. Immunoreactive signals were visualized using a DAB or AEC substrate kit (Vector laboratories, Burlingame, CA). PAS staining was performed according to the standard protocol.
Glucose tolerance test (GTT)
Twelve-wk old WT and KO male mice (n = 6) were fasted overnight with water available ad libitum. Mice were anesthetized with isoflurane anesthesia before collecting blood from the tail vein. After administration of 15% sterile D-glucose (1.5 g/kg, i.p.), blood was sampled from a tail nick immediately (t = 0) and 15, 30, 60 and 120 min thereafter. Blood glucose levels were determined using a glucometer (OneTouch Ultra, One Touch). The area under the curve (AUC) in the glucose concentration-time curve was calculated using SigmaPlot software (SPSS Inc., Chicago, IL). Given that the fasting blood glucose levels were different in KO mice at the time of glucose administration, the AUC was calculated using the increase in glucose (relative to t=0 levels) at each time point.
Ethanol pharmacokinetics study
Eight-wk old WT and KO mice (n=6) were matched for body weight and administered with a single dose of 20% ethanol (5 g/kg, i.p.). Blood was collected at 0, 1, 3 and 24 h later by cardiac puncture and was immediately mixed with 2x volume of ice-cold 1.7N perchloric acid (PCA) solution to prevent the spontaneous formation of acetaldehyde. Following centrifugation at 15,000xg for 10 min at 4°C, de-proteinized supernatants were assayed immediately for ethanol and acetaldehyde concentrations by gas chromatography-mass spectrometry (GC-MS) as previously described [14, 15]. Details about the GC-MS procedures can be found in the Supplementary Materials. The concentrations of ethanol (mM) and acetaldehyde (μM) were calculated by plotting against respective standard curves. The total area under the curves (AUCs) in the concentration-time curves were calculated using SigmaPlot software (SPSS Inc., Chicago, IL).
Statistical analysis
All quantitative experiments were performed at least in triplicate. Data are expressed as means ± S.E. Statistical significance was determined using Student’s unpaired t-test using SigmaStat Statistical Analysis software (SPSS Inc., Chicago, IL). P < 0.05 was considered to be significant.
Results
Generation of global ALDH1B1 KO mice
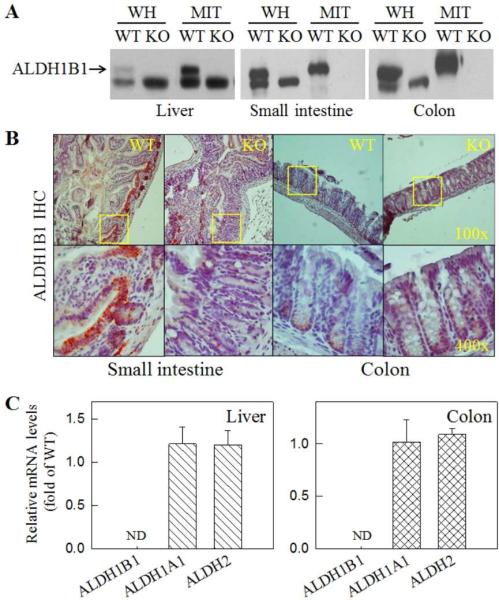
The targeting construct was designed to disrupt exon2 (2 Kb) of the Aldh1b1 gene, resulting in the removal of the complete coding region of the Aldh1b1 gene (Supplementary Fig. 1A). Independent ES clones harboring successful homologous recombination were confirmed by Southern blotting and PCR analysis (Supplementary Fig. 1B). Intercrossing of Aldh1b1(+/−) offspring generated from chimeric breeding produced Aldh1b1(+/+) (WT) and Aldh1b1(−/−) (KO) littermates (Supplementary Fig. 1C). As expected, Aldh1b1 KO mice do not express ALDH1B1 protein in the tissues that were examined, including liver, small intestine and colon,by Western immunoblotting (Fig. 1A) and IHC (Fig. 1B) analyses. The rabbit polyclonal anti-ALDH1B1 antibody [4] used in these analyses cross-reacted with a lower molecular weight peptide, which was unrelated to ALDH1B1 and appeared to exist in the mitochondria of the liver but not of the other two tissues (Fig. 1A). The identity of this peptide is unknown. In WT mice, ALDH1B1 was detected by immunohistochemistry in the bottom of the crypts of the small intestine and colon, but was lacking in KO mice (Fig. 1B). This pattern of ALDH1B1 expression in intestinal tissues is consistent with what we reported in human tissues [10]. Loss of ALDH1B1 expression in the liver and colon did not cause any compensatory changes in the messenger levels of Aldh1a1 and Aldh2 (Fig. 1C).
Fig. 1. Loss of ALDH1B1 expression in ALDH1B1 KO mice.
(A) Western blotting detection of ALDH1B1 protein in whole-cell homogenates (WH) and mitochondrial extractions (MIT) from liver (10 g), small intestine (10 g) and colon tissues (40 g). The rabbit polyclonal anti-ALDH1B1 antibody appeared to cross-react with an unknown lower molecular weight peptide. (B) Immunohistochemical (IHC) detection of ALDH1B1 in mouse small intestine and colon tissue sections. Cells at the crypt base showed ALDH1B1 immunoreactivity in WT but not in KO mice. Representative images are presented at two magnifications, with the square field in the top panel (100x) enlarged in the bottom panel (400x). (C) qRT-PCR detection of mRNAs of ALDH1B1, ALDH1A1 and ALDH2 in liver and colon tissues. Relative mRNA levels were expressed as fold of control (WT = 1) after normalization with β-2 microglobulin (B2M). Data are presented as mean + SE from 4 mice. ND, not detectable.
Phenotypically, Aldh1b1 KO mice display a growth curve that is no different from that of WT mice (Supplementary Fig. 2). Both KO male and female mice are fertile and show overtly good health. Histological analysis of tissues revealed no notable morphological abnormalities among any of the examined organs including liver (data not shown), small intestine, colon, rectum, lung, testes, and uterus (Supplementary Fig. 3).
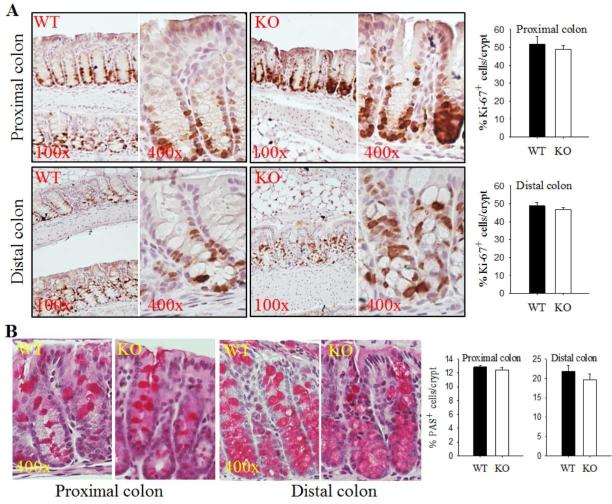
ALDH1B1 KO mice display normal cell proliferation and differentiation in the colon
We did not observe any notable histological abnormalities in the colon tissue of KO mice (Supplementary Fig. 3). Cellular proliferation and differentiation were examined by immunohistochemistry for Ki-67 and PAS, markers of cellular proliferation and secretory goblet cells, respectively. We did not find any differences in the intensity or extent of Ki-67 immunohistochemistry in KO mice relative to WT mice (Fig. 2A). Similarly, there were no differences in the number of goblet cells in colon crypts in WT mice relative to KO mice (Fig. 2B).
Fig. 2. Aldh1b1 KO mice show normal cell proliferation and differentiation in the colon.
(A) Cell proliferation in proximal and distal colon tissues in wild-type (WT) and Aldh1b1 KO (KO) mice was assessed by IHC staining for proliferation marker Ki-67. Representative images are presented at two magnifications, 100x and 400x. Ki-67+ cells were quantified in the proximal and distal colon and reported as % of total cells per crypt. (B) Cell differentiation in proximal colonic crypts was assessed using periodic acid-Schiff (PAS) staining for mucus-secreting goblet cells. Images are presented at 400x magnification. PAS+ cells were quantified in the proximal and distal colon and reported as % of total cells per crypt. Bars represent mean + SEM from 3 mice.
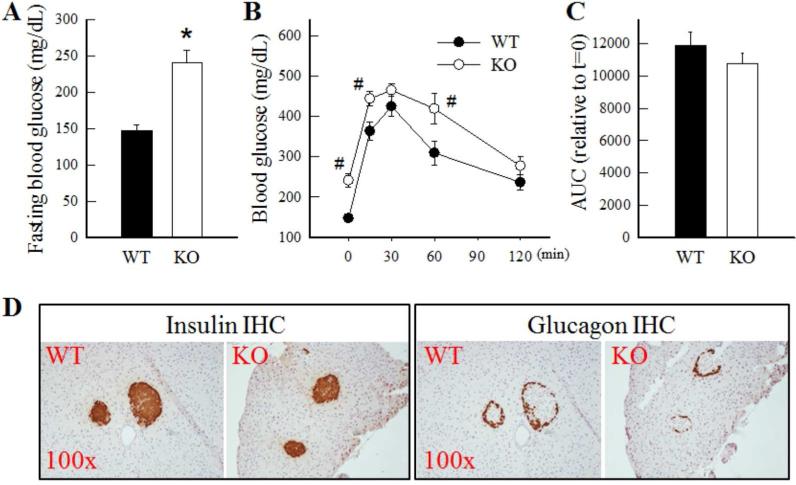
ALDH1B1 KO mice show disrupted glucose homeostasis
Histological analysis revealed the pancreas of KO and WT mice to be similar (data not shown). Interestingly, KO mice exhibited a 60% increase in the fasting blood glucose levels (Fig. 3A). However, when challenged with GTT, KO mice exhibited normal glucose tolerance (Fig. 3B), as indicated by no differences in the area under the glucose concentration-time curve (AUC) between KO and WT mice after normalization with respective glucose levels at t=0 (Fig. 3C). These results are suggestive of a disruption in glucose homeostasis in KO mice during fasting only. We then examined pancreatic islets architecture and number by IHC staining for insulin and glucagon. When compared with WT mice, we did not observe any difference in the intensity of insulin or glucagon immunohistochemistry (Fig. 3D) or in the numbers of insulin-/glucagon-positive cells in the pancreatic islets of KO mice (data not shown).
Fig. 3. ALDH1B1 KO mice show higher fasting blood glucose levels and decreased glucose tolerance.
Blood glucose levels were determined in overnight fasted WT and KO mice under basal conditions (A) and at various times after administration of glucose (1.5 g/kg body weight, i.p.) (B). *P < 0.05, compared to WT. #P < 0.05, compared to WT levels at the same time-point. (C) Area under the curve (AUC) was calculated after normalization relative to the levels of glucose at t = 0 for each genotype group.. Data represent mean ± SE from 6 mice. (D) Immunohistochemical (IHC) staining for insulin and glucagon in pancreas tissue sections. Representative images are presented at 100x magnification.
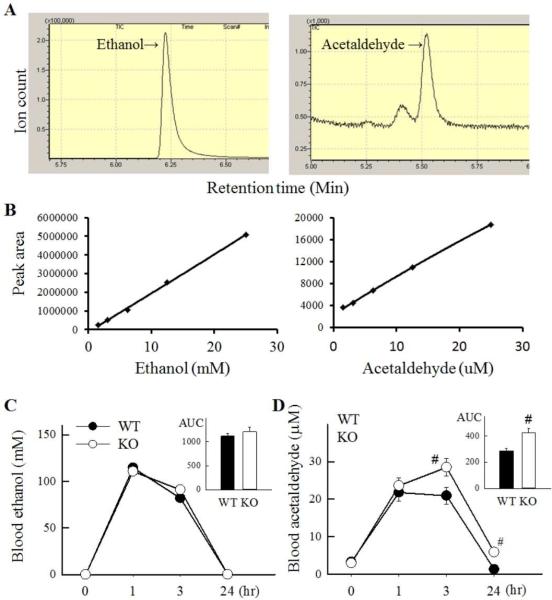
ALDH1B1 KO mice exhibit decreased clearance of blood acetaldehyde
We explored the impact of ALDH1B1 ablation on ethanol metabolism using an ethanol pharmacokinetics assay (Fig. 4). Following a single intra-peritoneal dose of ethanol (5 g/kg), ethanol and acetaldehyde concentrations were determined in blood samples obtained 0, 1, 3 and 24 h thereafter. Representative chromatograms of ethanol and acetaldehyde are shown in Fig. 4A. A best fit second order polynomial function was used to generate calibration curves for ethanol and acetaldehyde (Fig. 4B). KO and WT mice exhibited similar levels of ethanol following the ethanol administration (Fig. 4C). However, blood acetaldehyde levels were higher in KO mice than WT mice at 3 and 24 h following ethanol administration; this resulted in a ~40% increase in the AUC of the acetaldehyde concentration-time curve (Fig. 4D).
Fig. 4. Pharmacokinetics of ethanol and acetaldehyde in ALDH1B1 KO mice.
(A) Representative total ion chromatogram and selected ion monitoring profile of the blood samples for ethanol and acetaldehyde. Ion structure was set at 45 and 46 m/z for ethanol and 29, and 43 m/z for acetaldehyde. (B) Standard curves for ethanol and acetaldehyde measurement. (C) Blood ethanol and (D) acetaldehyde levels at 0, 1, 3 and 24 hr following a single ethanol dose (5 g/kg body weight, i.p). The area under the curve (AUC) was estimated for ethanol and acetaldehyde and presented as an inset, #P < 0.05, compared to WT levels at the same time-point.
Discussion
In this study, through gene targeting, we have successfully generated a global knockout mouse strain for ALDH1B1. The Aldh1b1 KO mice show normal growth and fertility and are in overtly good health, indicating ALDH1B1 as being dispensable for development and survival. In addition, we observed no compensatory changes in the expression of the highly related Aldh2 or Aldh1a1 genes in the liver and colon (and likely other tissues) from Aldh1b1 KO mice.
Several studies have linked ALDH1B1 with normal and cancer stem cells of the colon. Specifically, ALDH1B1 expression has been found to localize in the stem cell compartment of the normal colon and ALDH1B1 has been postulated to be a biomarker for human colon cancer [10] . In more recent studies, high ALDH1B1 expression has been found to correlate with poor prognosis in colorectal cancer patients [16, 17]. The Aldh1b1 KO mice exhibit normal histomorphology of the colon and show no dysregulated cell proliferation and/or differentiation in the colon. These results suggest that ALDH1B1 is not an essential cellular regulator in this tissue under physiological conditions. In contrast, our recent study has provided experimental evidence in support of an important role of ALDH1B1 in colon tumorigenesis [18]. In that study, ALDH1B1 knockdown in colon cancer cells using short hairpin RNA (shRNA) inhibited spheroid formation in culture and xenograft growth in mice [18]. Thus, the Aldh1b1 KO mouse model may serve as a valuable tool to examine the in vivo role of ALDH1B1 in colon tumorigenesis as well as understanding the molecular mechanisms involved in the process.
High ALDH activity has been used to isolate putative progenitor cells from the centroacinar compartment of the adult mouse pancreas. These cells are capable of generating endocrine and exocrine cells in culture [19]; however, the specific ALDH isozyme(s) contributing to this activity have not been identified due to the lack of specificity of the Aldefluor® assay [20]. Recently, we have found that (i) ALDH1B1 expression is enriched in progenitor pools for the acinar and endocrine cells in both embryo and adult pancreas [11], and (ii) ALDH1B1 is abundantly expressed in human pancreatic cancer and it contributes to proliferation in these tumor cells [21]. Collectively, our studies suggest that ALDH1B1 may be a key regulator of stem cell/progenitor status during pancreas development, regeneration and likely carcinogenesis. Given the important role played by the pancreas in regulating glucose homeostasis, we investigated whether genetic ablation of ALDH1B1 may affect blood glucose. In the present study, Aldh1b1 KO mice showed disrupted glucose homeostasis, characterized by elevations in fasting blood glucose and a normal response to exogenously-applied glucose. In addition, the pancreatic islets in the Aldh1b1 KO mice were indistinguishable from those of WT mice in displaying normal histomorphology and comparable numbers of insulin- and glucagon- positive cells. Circulating levels of insulin and glucagon and insulin sensitivity in Aldh1b1 KO mice were not investigated in the present study. Hence, the cause of the elevated fasting glucose levels in KO mice have not be identified. Certainly, the precise molecular mechanisms underlying the disrupted glucose homeostasis due to the loss of ALDH1B1 warrants further investigation.
In the mouse, ALDH1B1 is most highly expressed in the liver and parts of the small intestine, i.e., ileum and jejunum [4]. Such a pattern of expression is similar to that of ALDH2, the principal acetaldehyde-metabolizing enzyme [4, 22]. Despite having normal levels of ALDH2, Aldh1b1 KO mice exhibit a greater (~40%) accumulation of blood acetaldehyde following an acute dose of ethanol. This result corroborates our previous in vitro study showing that ALDH1B1 is second only to ALDH2 in oxidizing acetaldehyde, and indicates that ALDH1B1 plays a functionally significant role in acetaldehyde clearance in vivo. This finding may have clinical implications given that several functional polymorphisms of ALDH1B1 gene have been identified in the human population [8]. Alcohol hypersensitivity has been observed in carriers of the ALDH1B1*2 variant [5], which metabolizes acetaldehyde at a slower rate than wild-type ALDH1B1 [8].
Alcohol abuse is one of the major etiological factors for a variety of pancreatic disorders, including pancreatitis and diabetes [23, 24]. Numerous studies have demonstrated that ethanol and acetaldehyde exert deleterious effects on pancreatic exocrine and endocrine cells [25, 26]. The observations that ALDH1B1 metabolizes acetaldehyde in vivo and likely plays a role in maintaining glucose homeostasis provide for a potentially pivotal role of ALDH1B1 in the link between alcohol consumption and diabetes. Similarly, it is well established that chronic alcohol consumption is a strong risk factor for colon cancer [21]. Given that ALDH1B1 plays an active role in acetaldehyde metabolism and mediates oncogenic effects in colon cancer cells, it may be proposed that suppressed ALDH1B1 activity due to genetic variations would be associated with a high incidence of alcohol-related colon cancer. Thus, it is intriguing to speculate that carriers of certain human ALDH1B1 polymorphisms may represent a sub-population that is highly susceptible to alcohol-associated diseases. Future disease-association studies in the human population are warranted.
In summary, using our global Aldh1b1 KO mouse model, we show for the first time that ALDH1B1 participates in vivo in ethanol-derived acetaldehyde metabolism and glucose homeostasis. The Aldh1b1 KO mouse model therefore represents a useful and timely in vivo model for future studies aimed at understanding the functional roles of ALDH1B1 in human diseases, particularly those associated with alcohol abuse, including diabetes and cancer.
Supplementary Material
Highlights.
Establishment of Aldh1b1 knockout (KO) mice, which develop normally and show overtly good health.
Ethanol pharmacokinetic analysis revealed ~40% increase in blood acetaldehyde levels in KO mice.
KO mice exhibit higher fasting blood glucose levels and abnormal glucose tolerance.
Acknowledgements
This work was supported in part by NIH grants AA022057, AA021724 (VV). Hongbin Dong was supported by the NIAAA T32 AA007464.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert opinion on drug metabolism & toxicology. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart MJ, Malek K, Xiao Q, Dipple KM, Crabb DW. The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun. 1995;211:144–151. doi: 10.1006/bbrc.1995.1789. [DOI] [PubMed] [Google Scholar]
- 4.Stagos D, Chen Y, Brocker C, Donald E, Jackson BC, Orlicky DJ, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1: molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab Dispos. 2010;38:1679–1687. doi: 10.1124/dmd.110.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin. Exp. Allergy. 2010;40:123–130. doi: 10.1111/j.1365-2222.2009.03398.x. [DOI] [PubMed] [Google Scholar]
- 6.Husemoen LL, Fenger M, Friedrich N, Tolstrup JS, Beenfeldt Fredriksen S, Linneberg A. The association of ADH and ALDH gene variants with alcohol drinking habits and cardiovascular disease risk factors. Alcohol. Clin. Exp. Res. 2008;32:1984–1991. doi: 10.1111/j.1530-0277.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 7.Brennan P, Boffetta P. Mechanistic considerations in the molecular epidemiology of head and neck cancer. IARC Sci Publ. 2004:393–414. [PubMed] [Google Scholar]
- 8.Jackson BC, Reigan P, Miller B, Thompson DC, V. V. Human ALDH1B1 polymorphisms may affect the metabolism of acetaldehyde and all-trans retinaldehyde – in vitro studies and computational modeling. Pharmaceutical Research, Under revision. 2014 doi: 10.1007/s11095-014-1564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405:173–179. doi: 10.1016/j.bbrc.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ioannou M, Serafimidis I, Arnes L, Sussel L, Singh S, Vasiliou V, Gavalas A. ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev Biol. 2013;374:153–163. doi: 10.1016/j.ydbio.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isse T, Oyama T, Kitagawa K, Matsuno K, Matsumoto A, Yoshida A, Nakayama K, Kawamoto T. Diminished alcohol preference in transgenic mice lacking aldehyde dehydrogenase activity. Pharmacogenetics. 2002;12:621–626. doi: 10.1097/00008571-200211000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson CJ, Mizoi Y, Fukunaga T. The determination of acetaldehyde in human blood by the perchloric acid precipitation method: the characterization and elimination of artefactual acetaldehyde formation. Anal Biochem. 1982;125:259–263. doi: 10.1016/0003-2697(82)90004-5. [DOI] [PubMed] [Google Scholar]
- 16.Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B, Chang LJ. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195:52–60. doi: 10.1016/j.cbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langan RC, Mullinax JE, Ray S, Raiji MT, Schaub N, Xin HW, Koizumi T, Steinberg SM, Anderson A, Wiegand G, Butcher D, Anver M, Bilchik AJ, Stojadinovic A, Rudloff U, Avital I. A Pilot Study Assessing the Potential Role of non-CD133 Colorectal Cancer Stem Cells as Biomarkers. Journal of Cancer. 2012;3:231–240. doi: 10.7150/jca.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, Vasiliou V. ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/β-catenin, notch and PI3K/Akt signaling pathways. PLoS One. doi: 10.1371/journal.pone.0121648. in press (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arcaroli JJ, Powell RW, Varella-Garcia M, McManus M, Tan AC, Quackenbush KS, Pitts TM, Gao D, Spreafico A, Dasari A, Touban BM, Messersmith WA. ALDH+ tumor-initiating cells exhibiting gain in NOTCH1 gene copy number have enhanced regrowth sensitivity to a gamma-secretase inhibitor and irinotecan in colorectal cancer. Mol Oncol. 2012;6:370–381. doi: 10.1016/j.molonc.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V. Acetaldehyde and retinaldehyde-metabolizing enzymes in colon and pancreatic cancers. Adv. Exp. Med. Biol. 2015;815:281–294. doi: 10.1007/978-3-319-09614-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V. Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug Metab. Dispos. 2005;33:649–656. doi: 10.1124/dmd.104.002964. [DOI] [PubMed] [Google Scholar]
- 23.Clemens DL, Wells MA, Schneider KJ, Singh S. Molecular mechanisms of alcohol associated pancreatitis. World J Gastrointest Pathophysiol. 2014;5:147–157. doi: 10.4291/wjgp.v5.i3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zasimowicz E, Wolszczak B, Zasimowicz B. [Influence of ethyl alcohol on diabetes pathogenesis type] Pol Merkur Lekarski. 2014;36:212–214. [PubMed] [Google Scholar]
- 25.Gonzalez-Reimers E, Santolaria-Fernandez F, Martin-Gonzalez MC, Fernandez-Rodriguez CM, Quintero-Platt G. Alcoholism: a systemic proinflammatory condition. World J Gastroenterol. 2014;20:14660–14671. doi: 10.3748/wjg.v20.i40.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JY, Lee DY, Lee YJ, Park KJ, Kim KH, Kim JW, Kim WH. Chronic alcohol consumption potentiates the development of diabetes through pancreatic beta-cell dysfunction. World J Biol Chem. 2015;6:1–15. doi: 10.4331/wjbc.v6.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.