Abstract
Purpose
The poor performance of current tests for predicting the onset, progression and treatment response of diabetic nephropathy has engendered a search for more sensitive and specific urinary biomarkers. Our goal was to develop a new method for protein biomarker discovery in urine from these patients.
Experimental Design
We analyzed urine from normal subjects and patients with early and advanced nephropathy. Proteins were separated using a novel analysis process including immunodepletion of high abundance proteins followed by two stage LC fractionation of low abundance proteins. The proteins in the fractions were sequenced using MS/MS.
Results
Immunodepletion of selected high abundance proteins followed by two stage LC produced approximately 700 fractions, each less complex and more amenable to analysis than the mixture and requiring minimal processing for MS identification. Comparison of fractions between normal and diabetic nephropathy subjects revealed several low abundance proteins that reproducibly distinguished low glomerular filtration rate (GFR) from both high GFR diabetic and normal subjects, including uteroglobin, a protein previously associated with renal scarring.
Conclusions and clinical relevance
We developed a novel method to identify low abundance urinary proteins that enables the discovery of potential biomarkers to improve the diagnosis and management of patients with diabetic nephropathy.
Keywords: diabetic nephropathy, immunodepletion, liquid chromatography, proteomic methods, urinary biomarkers
1 Introduction
Diabetes mellitus is the leading cause of progressive chronic kidney disease and often requires renal replacement therapy with dialysis or transplantation. Approximately one third of patients with diabetes mellitus develop kidney damage characterized by proteinuria, hypertension and progressive diabetic nephropathy (DN) despite aggressive therapeutic intervention [1]. Diabetic nephropathy is asymptomatic in its early stages and current clinical biomarkers, such as serum creatinine and the urine albumin-creatinine ratio, lack the sensitivity and specificity for early detection of the disease, for monitoring its progression and for assessing response to pharmacological intervention. New urinary biomarkers for the diagnosis and management of diabetic nephropathy are urgently needed.
The identification of biomarkers in urine is complicated by the complexity of the urine proteome, low relative abundances of candidate biomarker proteins and the abundant presence of water. Above all, the total protein concentration varies widely in urine from kidney patients and may be more than 100 times normal, with albumin being the most abundant protein – a hallmark and prognostic indicator of diabetic nephropathy [2]. General approaches to overcoming these challenges include preprocessing steps to concentrate urine proteins, followed by fractionation of the complex proteome and then identification of individual proteins using mass spectrometry [3-5]. The method most frequently applied, and heretofore the technique of choice for urine proteome mapping, has been two-dimensional gel electrophoresis (2-DE) [6]. This method has yielded chromatograms with 1400 distinct spots, demonstrating both the complexity of the urine protein mixture and the ability of 2-DE to separate it [7]. Some disadvantages of the 2-DE technique are that it is time and labor intensive, it is not easily automated and gel spots can not be directly introduced into a mass spectrometer. Further, the reproducibility of the separation makes it difficult to compare samples separated on different gels. The latter problem can be overcome using two-dimensional difference gel electrophoresis, a technique whereby two protein mixtures are separately tagged with different fluorescent dyes and then separated simultaneously on one gel [8]. This technique allows for quantitative comparisons of two protein mixtures and has been used, for example, to compare the proteomes of normal subjects to patients with diabetic nephropathy [9, 10]. But problems with separation of proteins with gels remain, including the separation of low molecular weight, highly basic and hydrophobic proteins.
A second widely-applied method for urine protein separation and identification is capillary electrophoresis-mass spectrometry [11]. Coupling the capillary electrophoresis column to an electrospray ionization mass spectrometer allows direct separation and identification of urine proteins and has been used for investigation of many kidney-related diseases [12]. Advantages of this method include rapid separation and identification of complex mixtures; disadvantages are limitations to small sample sizes and lower molecular weight proteins.
A third well-known group of analysis methods for protein mixtures includes liquid chromatography (LC) separations [13]. Individually or in tandem, many different LC methods have been used to separate complex protein mixtures prior to protein identification with mass spectrometry. One tandem LC pipeline is chromatofocusing (CF) followed by non-porous reversed phase (RP) separation, which first separates proteins into fractions according to their isoelectric points, then separates the proteins in each fraction by hydrophobicity [14]. This two-dimensional technique not only provides the extensive fractionation beneficial for analysis of complex protein mixtures, but also proteins are eluted in solution and ready for mass spectrometry with minimal further processing.
The two-dimensional CF-RP LC process has been applied to cell lysates, rat plasma and normal human urine [14-16]. A problem complicating the analysis of blood serum and plasma is the presence of a few proteins in high abundance, especially albumin and immunoglobulins. These blood proteins are largely retained by healthy kidneys but may be present in large amounts in urine from proteinuric patients where their presence interferes with the analysis for lower abundance proteins which are the most likely candidates for biomarkers of disease. Selected proteins can be removed by immunoaffinity subtraction chromatography (immunodepletion), and immunodepletion of abundant proteins prior to RP LC has been demonstrated on human serum and plasma [17, 18], and on urine prior to 2-DE [10]. Abundant protein depletion of rat plasma prior to sequential CF-RP chromatography was shown to improve signal-to-noise ratios in subsequent MS analyses [19], and albumin and IgG depletion prior to multi-dimensional LC enhance MS detection of low abundance proteins in normal urine [20]. However, the promising combination of abundant protein depletion followed by CF-RP multi-dimensional LC has not been applied to compare urine proteins from patients with diabetic nephropathy to those from normal subjects. Here we demonstrate the application of this method to compare proteins in urine of DN patients to those from the urine of normal subjects.
2 Materials and Methods
2.1 Subjects and Samples
To demonstrate the method we used 24-hour urine samples from two male subjects with normal kidney function and six male subjects with type 2 diabetes and nephropathy. The diabetic subjects provided samples upon their enrollment into a double-blind, placebo-controlled clinical trial, had massive proteinuria (2.1 to 7.8 g/24 hr) and could be separated into two groups, three with stage 1 nephropathy and three with stages 3-5 nephropathy [21] (Table 1). Glomerular filtration rates were 95.1, 139.2 and 201.9 mL/min for the group with early stage disease and 11.1, 17.6 and 32.3 mL/min for the group with late stage disease; hyperfiltration in the high GFR group is consistent with early stage nephropathy. The proteins in these samples were concentrated, separated and identified using the analysis scheme detailed in Figure 1 and described below. Both the clinical trial and this study were approved by the UT Southwestern Medical Center Institutional Review Board and all subjects gave informed, written consent for the study.
Table 1. Subject Characteristics.
Results are presented as Mean (SD) or Median [range] and/or individual values.
| High GFR (n=3) | Low GFR (n=3) | Normal (n=2) | |
|---|---|---|---|
| Male | 3 | 3 | 2 |
| Black | 1 | 0 | 0 |
| Hispanic | 2 | 3 | 0 |
| Age (yr) | 58.7 (9.2) | 50.7 (2.1) | 57, 60 |
| Body mass index (kg/m2) | 33.1 (12.6) | 33.1 (10.0) | 20.4, 24 |
| Duration of diabetes (yr) | 9.0 (7.8) | 20.7 (6.7) | NA |
| Urinary albumin/creatinine ratio (mg/g) | 1714 [1038-2311] | 4838 [3917-5544] | |
| Serum creatinine (mg/dL) | 1.6 (0.9) | 3.2 (1.4) | 1.0, 1.04 |
| Blood urea nitrogen (mg/dL) | 35.7 (19.1) | 65.0 (13.9) | 11, 14 |
| Creatinine clearance (mL/min) | 97.7 (79.0) | 29.7 (19.2) | |
| GFR (mL/min) | 95.1, 139.2, 201.9 145.4 (53.7) | 11.1, 17.6, 32.3 20.3 (10.9) | |
| Proteinuria (g/day) | 3.3 (1.2) | 6.5 (1.8) |
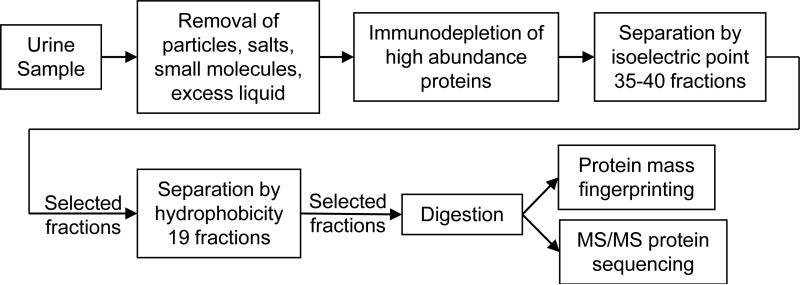
Figure 1.
Process for separation and identification of urine proteins.
2.2 Protein Concentration by Ultrafiltration
Urine samples were divided into portions after collection and stored at -80 °C. Individual portions were thawed on ice just before analysis, then spun at 4000 × g for 20 min at 10 °C to remove particles. The supernatant was decanted into a Millipore Amicon Ultra-15 3 kDa nominal molecular weight limit ultrafilter and spun at 4000 × g at 10 °C to a volume of 2 mL. Salts and small molecule metabolites and proteolysis products were washed from the concentrated protein by filling the ultrafilter with water and spinning again to a volume of 1 mL. Particles and precipitates were then removed by passing the protein through an Agilent 0.22 μm spin filter. Ultrafiltration of 160-180 mL of urine from normal subjects and 10-20 mL of urine from proteinuric patients was required to obtain sufficient protein for subsequent steps; larger volumes were required from the normal subjects because the protein concentration in their urine was much less than that of the patients with nephropathy. Typically 20 mg of protein was obtained for high protein samples and 5 mg for normal samples.
2.3 Immunodepletion of Abundant Proteins
Six selected proteins, here termed HAPs as they are high abundance proteins in blood fractions, were separated from the protein mixture using a Beckman System Gold LC system and an Agilent Hu-6HC Multiple Affinity Removal System column which retains albumin, IgG, IgA, transferrin, alpha-1-antitrypsin and haptoglobin. For each separation, 1.4 mg of the concentrated, desalted and filtered protein was diluted with Agilent Buffer A to 2 mL total volume and then loaded onto the column, where the six HAPs were retained while the remaining lower abundance proteins, termed the LAPs, passed through. After elution of the LAPs, the HAPs were cleared from the column by elution with Agilent Buffer B. Proteins exiting the column were detected by their absorption at 280 nm and were manually collected on ice into LAP and HAP fractions. Replicate separations (4-7) were required to obtain sufficient LAPs for subsequent steps. Each separation diluted the LAPs with buffer solution, so the replicate LAP fractions were pooled and reconcentrated using a 3 kDa ultrafilter. After removal of the HAPs, equal amounts of LAPs from the normal and diseased subjects were carried on for further analysis, thus compensating for their differing initial protein concentrations. Each pool typically contained 1-2 mg of low abundance protein.
2.4 Fractionation by Two-Dimensional Liquid Chromatography
The immunodepleted protein fractions were further separated by 2-D LC using a Beckman Coulter ProteomeLab PF 2D Protein Fractionation System consisting of tandem first and second dimension LC modules with an intermediate fraction collector/injector (FC/I) [15, 16, 19]. An Eprogen ProteoSep chromatofocusing column was used in the first dimension module. For each trial, 1 mg of protein was diluted to 5 mL with pH 8.5 Eprogen Start Buffer and injected into a 5 mL sample loop. Eprogen Eluent Buffer with pH adjusted to 4 was then passed through the column, establishing a moving pH gradient which separated and eluted proteins by pI. Fractions were collected into 96 well plates at 7.5 min intervals when the pH was constant, then in 0.3 unit pH intervals as it decreased from 8.5 to 4, resulting in the collection of 35-40 fractions in the FC/I. Protein elution was monitored by absorbance at 280 nm.
The second dimension module used an Eprogen ProteoSep reversed phase column operated at 50 °C. First, a solution of 0.1% TFA in water was passed through the column and the FC/I injected 200 μL of sample from one first dimension fraction. A linear concentration gradient established by adding 0.08% TFA in ACN separated and eluted proteins by their hydrophobicity. Fractions were collected into 96 well plates in an eight place Gilson FC204 fraction collector at 1 min time intervals resulting in 19 second dimension fractions collected for each first dimension fraction. The protein elution profile was monitored by absorbance at 214 nm. After one first dimension fraction had cleared the column, initial solvent conditions were reestablished and the FC/I continued by injecting 200 μL of sample from the next first dimension fraction.
2.5 Protein Identification by Mass Spectrometry
In general, second dimension PF 2D fractions with absorbance peaks that increased (or decreased) in intensity from normal subjects through early stage to late stage disease were chosen for identification by MALDI-MS and MALDI-MS/MS sequencing. Each fraction was dried in a centrifugal evaporator, then resuspended in 25 μL 25 mM ammonium bicarbonate at pH 8.0. Fifteen ng trypsin was added and the solution incubated at 37 °C for 18 hours. One microliter of the tryptic digests was spotted directly onto the MALDI target plate and allowed to dry. A stock matrix solution (5 mg/mL CHCA in 50% (v/v) ACN) was diluted 1:1 with 50% ACN and 1 μL of the working matrix was then applied on the sample spot and allowed to dry.
Data were acquired with an Applied Biosystems 4700 MALDI-TOF/TOF Proteomics Analyzer (Life Technologies-Applied Biosystems, Carlsbad CA). The instrument was operated in positive ion reflectron mode, the mass range was 850-3000 Da and the focus mass was set at 1500 Da. For the MALDI-MS data acquisition, 2000 laser shots were acquired and averaged from each sample spot. Automatic external calibration was performed using known autolytic fragments of trypsin as internal standards. Following MALDI-MS analysis, MALDI-MS/MS was performed on several (5-8) abundant ions from each sample spot. A 1 kV positive ion MS/MS method was used to acquire data under postsource decay conditions. The instrument precursor selection window was ±3 Da and 4000 laser shots were acquired and averaged for each sample spot.
Applied Biosystems GPS Explorer (v. 3.0) software was used in conjunction with the MASCOT search engine (www.matrixscience.com) to search the Homo sapiens NCBI nr protein database using both the peptide mass fingerprint in the first MS and the MS/MS data from several precursor ions. MS peak filtering included the following: mass range, 800 to 4000 Da; minimum signal-to-noise filter, 10; mass exclusion list tolerance, 0.5 Da. Other search parameters were: maximum missed cleavages were set to unity; fixed modifications included cysteine carbamidomethylation; and variable modifications included methionine oxidation, N-terminal acetylation and pyroglutamylation of N28 terminal glutamine and glutamate residues. Precursor tolerance was set at 0.2 Da, MS/MS fragment tolerance was set at 0.3 Da and peptide mass values were monoisotopic. The significance of a protein match, based on both the peptide mass fingerprint in the first MS and the MS/MS data from several precursor ions, is based on expectation values. The expectation value is the number of matches with equal or better scores that are expected to occur by chance alone. Protein scores are calculated from the expectation value of the protein identification; the higher the protein score the more significant the match. The default significance threshold is p<0.05, so an expectation value of 0.05, or a protein score of 67, is considered to be on this threshold [22].
3 Results
The analysis scheme in Figure 1 meets two key requirements: removal of six selected high abundance blood proteins when present at either high or low concentrations and extensive fractionation of the remaining lower abundance proteins. The process first removes particles, salts, small molecules and excess liquid and then separates the HAPs from the remaining proteins. Using 2-D LC, the LAP fractions are first separated by pI and then by hydrophobicity into fractions each much less complex than the initial mixture. Finally, mass spectrometry is used to identify the proteins in individual fractions.
3.1 Protein Concentration by Ultrafiltration
The ultrafiltration process reduces the large and variable amounts of water in both normal and high protein urine samples, concentrating high protein urine about 10-20 times and normal urine about 160-180 times. It also separates urine proteins from small molecule metabolites (urea, urate, creatinine, citrate, and many others), low molecular weight proteolysis products and soluble salts. Some proteins smaller than 3 kDa may be retained by the ultrafilter because they are bound to larger proteins [23], but this association is disrupted in the immunodepletion step when the proteins are mixed with the denaturing start buffer. These small molecules are then released from the large proteins and pass through the chromatographic column with the unretained proteins during the immunodepletion.
We evaluated the benefit of the ultrafiltration by comparing immunodepletion chromatograms of urine without ultrafiltration and of both the retentate and filtrate fractions from the ultrafilter. (Details in Supporting Information.) These chromatograms show that the small molecule metabolites and low molecular weight proteolysis products constitute a highly absorbing component of the mixture. Without the ultrafiltration they co-elute with the low abundance proteins thereby passing through the immunodepletion step. In the subsequent CF separation their absorption will complicate the identification of fractions containing desirable proteins.
3.2 Immunodepletion Separation of Low- and High-Abundance Proteins
The kidneys of DN patients allow passage of blood proteins into the urine, thus high concentration blood proteins also tend to be high concentration in patient urine. For this reason a protein immunodepletion column for removal of high concentration proteins from blood serum and plasma, such as the Agilent Hu-6HC, is appropriate for urine from DN patients. The depletion efficiencies of the six proteins removed by this column have been measured by Brand et al. [24]. In plasma, and over the specified 200 regeneration cycle lifetime of the column, the depletion rates for albumin, transferrin, alpha-1-antitrypsin and haptoglobin were >99.9% for each target protein. The depletion rates for IgG and IgA were >99.9% for a new column, gradually falling to 99.1% for IgG and 99.7% for IgA after 200 cycles. Figure 2A shows that the immunodepletion step effectively separates the six HAPs from the LAPs in both normal and high protein samples. Similar results for immunodepletion of selected proteins have been observed for serum [17], plasma [18, 19] and urine [25] using various affinity removal systems.
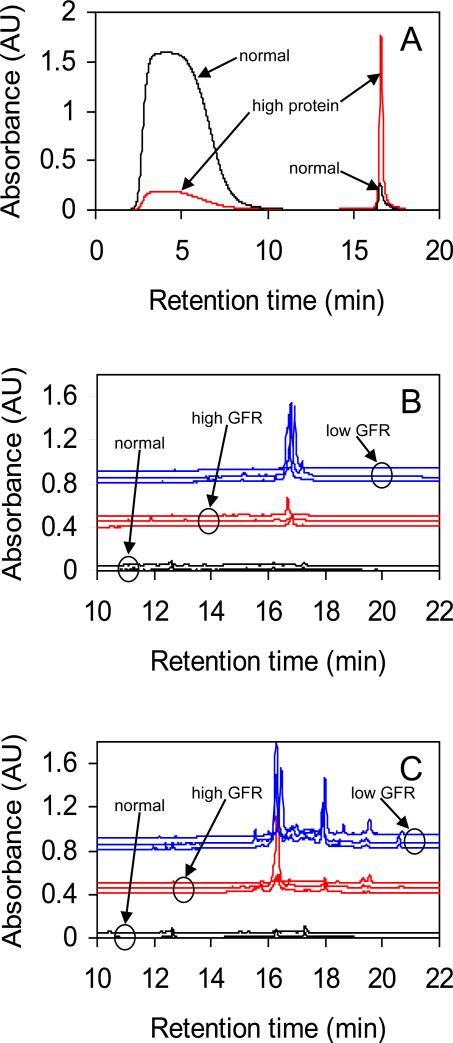
Figure 2.
A. Immunodepletion chromatograms for normal and high protein urine. Peaks at lower retention times are low abundance proteins (LAPs) and peaks at higher retention times are high abundance proteins (HAPs). A small baseline has been subtracted from each trace. B. and C. Second dimension (RP) chromatograms of first dimension (CF) fractions. Traces are offset for clarity and grouped into normal, high GFR and low GFR categories. First dimension pI ranges are: panel B - 6.1 to 5.8, panel C - 5.2 to 4.9.
This figure also shows that chromatograms from proteinuric patients have a large peak corresponding to the six targeted proteins, while chromatograms from normal subjects show a small peak because a much larger fraction of the urinary proteome of diseased patients is these six proteins. This is why the immunodepletion step is an important part of our analysis process: the abundance of these proteins in the urine of diseased patients complicates the comparison between the low abundance proteins in urine from diseased patients and normal subjects.
We evaluated the benefit of the immunodepletion step by comparing CF chromatograms of high protein urine without immunodepletion and of both the LAP and HAP fractions from the immunodepletion. (Details in Supporting Information.) The CF chromatogram of the HAP fraction exhibits several strong peaks in the pH range 4-7 which correspond more or less closely to the theoretical pIs of albumin, transferrin, alpha 1-antitrypsin and haptoglobin (http://www.uniprot.org). The polyclonal IgG and IgA elute over a broad pH range and contribute to an elevation of the baseline in the pH range 4-7. The CF chromatogram of the undepleted protein is a composite of the LAP and HAP chromatograms, weighted toward the HAPs because of their higher abundance. Without the immunodepletion step, the identification of CF fractions containing LAPs, the most likely source of potential biomarkers, is complicated because the HAP and LAP peaks overlap and because the presence of the HAPs reduces the concentration of LAPs.
3.3 Chromatofocusing and Reversed-Phase Liquid Chromatography
The low abundance proteins from each sample were separated by pI into 35-40 fractions using the CF column. Typically 25 fractions spanned the region of the first dimension chromatogram that included the pH change and these fractions contained many of the peaks in the first dimension chromatogram. Each of these 25 fractions was further separated using the RP column into 19 fractions resulting in 475 second dimension fractions for each LAP sample. Prominent peaks in the second dimension chromatograms indicated fractions containing the most abundant of the LAP proteins, though possibly only a small fraction of their total number. Fractions with smaller peaks contain the less abundant LAP proteins. Fractions without peaks contain proteins present at low concentrations or with low absorption coefficients, or contain no protein at all. Second dimension chromatograms of the LAPs from each subject's urine sample were matched by pI range and the positions and sizes of the peaks were compared between subjects. Panels B and C of Figure 2 illustrate pI-matched comparisons of second dimension traces for the 2 normal subjects, 3 patients with DN and high GFR and 3 patients with DN and low GFR. Using these comparisons, fractions were identified that exhibited the most changes in their peak intensities between the low GFR, high GFR and normal subject groups. We selected 105 fractions for MALDI-MS and MALDI-MS/MS protein identification.
3.4 Protein Identification
Proteins identified in selected fractions from each subject group are summarized in Table 2 (Details in Supporting Information Table 1). Each category in the table is defined by patient status - low GFR (late stage disease), high GFR (early stage disease) or normal kidney function - and by the pI of the fraction. The proteins observed in each disease stage-pI category are listed, along with the number of subjects for which they were found. Thus, for example, hemopexin was found for 3 of 3 subjects in the category low GFR and pI fraction 6.1 to 5.8. In general, more different kinds of proteins are observed in the urine of the more diseased patients, suggesting that decreasing GFR is indicative of increasing glomerular damage and hence greater protein leakage, or that increased amounts of potential biomarker proteins arise from the kidney due to more severe kidney damage.
Table 2. Low Abundance Proteins identified for subject categories low GFR, high GFR and normal kidney function.
(m/n) - found in m of n samples in a category; bold and underlined - found in all samples in a disease stage-pI category. The confidence interval for protein identification was ≥98% at a p-value of <0.05 for each protein listed.
| pI fraction | Low GFR (n=3) | High GFR (n=3) | Normal (n=2) | |||
|---|---|---|---|---|---|---|
| Protein | (m/n) | Protein | (m/n) | Protein | (m/n) | |
| 6.1 to 5.8 | Hemonexin | (3/3) | Zn-alpha-2-glycoprotein | (1/3) | NA | |
| Transthyretin | (1/3) | |||||
| 5.5 to 5.2 | Alpha-1-microglobulin | (3/3) | Alpha-1-beta-glycoprotein | (1/3) | MAP19 | (2/2) |
| Apolipoprotein A-I | (2/3) | Transthyretin | (1/3) | Zn-alpha-2-glycoprotein | (1/2) | |
| Hemonexin | (3/3) | Zn-alpha-2-glycoprotein | (2/3) | |||
| Transthyretin | (1/3) | |||||
| Vitamin D-binding Protein | (2/3) | |||||
| Zn-alpha-2-glycoprotein | (3/3) | |||||
| 5.2 to 4.9 | Alpha-1-microglobulin | (3/3) | Alpha-1-beta-glycoprotein | (1/3) | Zn-alpha-2-glycoprotein | (1/2) |
| Apolipoprotein A-I | (1/3) | Apolipoprotein A-I | (1/3) | |||
| Retinol-binding Protein 4 | (2/3) | Kininogen-1 | (1/3) | |||
| Uteroglobin | (3/3) | Vitamin D-binding Protein | (1/3) | |||
| Vitamin D-binding Protein | (3/3) | Zn-alpha-2-glycoprotein | (3/3) | |||
| Zn-alpha-2-glycoprotein | (3/3) | |||||
| 4.9 to 4.7 | Alpha-1-beta-glycoprotein | (3/3) | Alpha-1-beta-glycoprotein | (2/3) | Alpha-1-microglobulin | (2/2) |
| Alpha-1-microglobulin | (3/3) | Kininogen-1 | (1/3) | CD59 | (1/2) | |
| Kininogen-1 | (3/3) | Zn-alpha-2-glycoprotein | (3/3) | ITIH4 | (1/2) | |
| Vitamin D-binding Protein | (3/3) | Osteopontin | (1/2) | |||
| Zn-alpha-2-glycoprotein | 100 (3/3) | Zn-alpha-2-glycoprotein | 100 (2/2) | |||
The fractions chosen for MALDI-MS and MALDI-MS/MS analysis were those where peak intensities increased from normal to high GFR to low GFR groups, thus more proteins were identified in the disease groups. These fractions typically arose from first dimension fractions containing proteins with intermediate pI, less than 8.5 and greater than 4.0. In contrast, first dimension fractions containing proteins with pI outside the 8.5 to 4.0 range appear by comparison of peak intensities to contain more proteins in the normal subjects than the diseased patients. Since equal amounts of protein were loaded onto the column from all samples, a compensating amount of protein must be in these unanalyzed fractions.
The full-length molecular weights of proteins identified in the three subject categories (see Supporting Information Table 3 for a full list) are not simply correlated to the disease stage for several reasons. First, these proteins are only those from the LAP fractions and, since we carry forward equal amounts of protein from the immunodepletion process for both normal and diseased subjects, even a small leakage of high molecular weight proteins not targeted by the immunodepletion might be observable in the LAPs from normal subjects. Second, proteins of all molecular weights might originate within or downstream of the kidneys. Also, proteins and peptides in the urine may be subject to proteolysis by natural processes. Finally, MS analyses were performed using tryptic digests of protein fractions and the molecular weights of the full-length proteins are inferred.
3.5 Interpretation of Protein Identifications
The proteins most often found in low GFR samples include hemopexin, vitamin D-binding protein, kininogen and uteroglobin (Table 2). Apolipoprotein A-I, retinol-binding protein 4 and transthyretin are less frequently observed. The large sizes of hemopexin, vitamin D-binding protein and kininogen protect them from glomerular filtration by normal kidneys. The damaged glomerular basement membrane in patients with diabetic nephropathy, however, may allow these species to pass into the urine.
We observed other smaller proteins in the urine of diseased patients that are protected from filtration in normal subjects by their strong association with larger species. Both retinol-binding protein 4 and the very small retinol molecule circulate in plasma in a complex with the larger transthyretin molecule, whose size prevents glomerular filtration of the two smaller species [26, 27]. Apolipoprotein A-I (Apo-AI) and its fragments are bound to the large haptoglobin molecule and are therefore not excreted in normal subjects [28]. It is possible that Apo-AI and its carrier protein haptoglobin are excreted in the urine of diseased patients and the association between the two proteins disrupted by the denaturing starting buffer used in the immunodepletion column. Thus free Apo-AI is eluted with the LAPs even as haptoglobin is retained with the HAPs.
Uteroglobin is especially interesting because it has been previously associated with renal scarring. In normal subjects, the excretion of uteroglobin is inhibited by its strong association with fibronectin. Uteroglobin knockout mice demonstrate that disruption of the uteroglobin gene results in abnormal deposits of fibronectin and collagen in the glomeruli, suggesting that uteroglobin plays a crucial role in preventing such deposits; the formation of uteroglobin-fibronectin heterodimers counteracts the fibronectin-fibronectin and fibronectin-collagen interactions required for abnormal tissue deposition [29]. Diabetic nephropathy is characterized by abnormal deposits of fibronectin and collagen in the glomeruli and such deposition is induced by high glucose levels through a downstream mediator chain that stimulates fibronectin matrix synthesis [30]. We speculate that excretion of uteroglobin in the urine of DN patients with low GFR and heavy proteinuria is associated with renal scarring, possibly by increased expression of uteroglobin in the kidney in response to fibronectin deposition. Although preliminary, the detection of this protein only in patients with low GFR suggests it may be a marker of intrarenal injury associated with more advanced diabetic nephropathy.
4 Discussion
The principal result of this study is demonstration of a method to compare low abundance urine proteins between patients despite widely varying degrees of proteinuria. Urine from patients with kidney disease often contains large amounts of several proteins that are not likely biomarker candidates. Immunodepletion chromatography removes these high abundance proteins so that those remaining may be more readily separated and identified. Thus, the low abundance proteins from proteinuric patients may be compared to proteins from subjects with normal protein levels.
A further observation is consistently differing amounts of particular proteins between the normal, high GFR and low GFR patient groups, suggesting that this method can detect variations in proteins as the disease progresses. And, we show that in many instances the amount of protein increases from the normal to low to high GFR subject groups, which may reflect the general loss of kidney function with disease progression (Figure 2 B and C). Thus this method may be useful to identify candidate biomarkers of disease state and to follow progression of disease in addition to measuring and predicting response to therapy.
This method has several advantages over alternative techniques for identifying potential urinary biomarkers in patients with diabetes and nephropathy. First, removal of the highest abundance proteins makes it easier to compare the remaining proteins between normal subjects and proteinuric patients. Second, the coupled 2-D CF-RP fractionation provides extensive separation of the lower abundance proteins. Third, similar fractions can be matched between samples so that their protein contents can be compared. Also, there is no need to excise gel spots and perform in-gel digestions, thus eliminating the potential for contamination from handling along with concerns for full recovery of the protein from the spot. And, proteins are recovered in solution and ready for identification by mass spectrometry with a minimum of further processing.
In this study we identified fewer urine proteins than have been found in some previous analyses [7, 10, 20, 25, 31, 32]. One reason is that the identification method we chose, MALDI-MS, may be less sensitive for low concentration protein detection than competing technologies. We could improve our MALDI-MS detection by separating larger quantities of protein and by pooling fractions from replicate separations to increase the amounts of recovered protein. As an alternative to MALDI-MS, fractions from the final RP separation should lend themselves well to a subsequent CE-MS or LC-MS separation and identification, and these methods may be more suitable for peptides and small proteins. Another reason we identified fewer proteins is that we analyzed only selected fractions; after the CF separation we chose fractions spanning the region where the pH changed and after the RP separation we chose fractions with prominent peaks. Further, we did not identify peptides because we chose to digest before MALDI-MS analysis and infer full-length proteins from the resulting fragments. And, we may have lost peptides that passed through the 3 kDa ultrafiltration. Peptide identification would have been possible without digestion before MALDI-MS, or by choosing CE-MS or LC-MS methods. These potential improvements to the analysis process are the subjects of further investigations.
A limitation of this process is that it is not quantitative, however the extensive fractionation provided by the method followed by protein sequencing using mass spectrometry allows identification of potential biomarkers. Quantitative methods, such as ELISA, must then be used to verify the performance of individual candidates and then larger-scale tests conducted to validate them. A quantitative, high-throughput and preferably inexpensive test will then make the analysis of individual biomarkers clinically applicable.
The numbers of subjects in the groups in this demonstration are too small to draw clinically applicable conclusions about the identified proteins, however the results show that this analysis pipeline allows comparison of lower abundance proteins in spite of the initial presence of a few proteins with large and widely varying concentrations. Larger sample groups are required to assess and compensate for normal inter-subject biological variations and to allow valid conclusions about the progression of disease. One could also follow individual patients from onset of nephropathy to renal failure, however the progression of this disease is highly variable and commonly takes years to advance from early to late stages. And, many patients die before reaching end stage renal disease, frequently from cardiovascular complications.
Our interest in these protein separation methods is motivated by the lack of clinical biomarkers that currently exist to predict response to pharmacological intervention in DN patients. Previous studies have shown that candidate protein biomarkers, or protein patterns, can predict the response of IgA nephropathy patients to angiotensin converting enzyme inhibitor (ACEi) therapy and can distinguish between steroid-sensitive and steroid-resistant nephrotic syndrome [33-35]. In DN patients, drugs that block the renin-angiotensin-aldosterone system, including angiotensin converting enzyme inhibitors, angiotensin II type 1 receptor blockers (ARB) and mineralocorticoid antagonists (MRA), reduce proteinuria and sometimes slow progression of the disease [36-40]. And, some DN patients in our clinical trial who had persistent albuminuria despite treatment with an optimal dose of an ACEi exhibited a decreased rate of disease progression when aggressively treated with either an MRA or ARB [21]. The response to treatment was hypervariable, however, and no current clinical biomarkers exist that predict individual patient response to these multi-drug interventions. Such biomarkers would be valuable in clinical practice for diagnosis, triage and treatment of proteinuric patients with DN. To this end, we will use this new method to identify candidate biomarkers that reproducibly predict the antiproteinuric response to treatment with an MRA or ARB, drugs currently used to treat this disease.
Supplementary Material
Clinical Relevance.
The identification of novel biomarkers for kidney disease in general and for diabetic nephropathy in particular has high priority in clinical medicine because of the growing population of patients with progressive chronic kidney disease. Aggressive therapeutic intervention with drugs that block the renin-angiotensin-aldosterone system, including angiotensin converting enzyme inhibitors, angiotensin receptor blockers and mineralocorticoid antagonists, reduces proteinuria and sometimes slows progression of the disease. The response, however, is hypervariable among patients with diabetic nephropathy, and current clinical biomarkers, such as serum creatinine and the urine albumin-creatinine ratio, lack the sensitivity and specificity for predicting response to these pharmacological interventions. The goal of this study was to develop methods to analyze and compare proteins in the urine of normal subjects to those from patients with diabetic nephropathy, comparisons which are complicated by the presence of high and variable protein concentrations in proteinuric urine. We demonstrate that removal of high abundance urinary proteins and extensive fractionation of the remaining low abundance proteins allows these comparative analyses. We will use this method to test the hypothesis that urinary biomarkers can predict a renoprotective response to long-term administration of drugs that block the renin-angiotensin-aldosterone system in patients with diabetic nephropathy.
Acknowledgments
Support was provided by the American Diabetes Association (grant no. 1-08-CR-65) (R.D.T.), the University of Texas Southwestern Medical Center O'Brien Kidney Research Core (P30DK079328) (R.D.T.), the Welch Foundation Endowment in Chemistry and Related Science (grant no. L-AU-0002) (K.P.R.), the National Institutes of Health, National Heart, Lung and Blood Institute's Proteomics Initiative NO1-HV-28184 (K.P.R.), and by the Center for Clinical and Translational Sciences, which is funded by National Institutes of Health Clinical and Translational Award UL1 RR024148-05 from the National Center for Research Resources (K.P.R.). The NIH and Welch Foundation had no role in the study design, data collection and analysis, decision to publish or preparation of this manuscript.
Abbreviations
- ACEi
angiotensin converting enzyme inhibitor
- ARB
angiotensin II type 1 receptor blocker
- CF
chromatofocusing
- DN
diabetic nephropathy
- FC/I
fraction collector/injector
- GFR
glomerular filtration rate
- HAP
high abundance protein
- LAP
low abundance protein
- MRA
mineralocorticoid antagonist
Footnotes
All authors declare they have no financial/commercial conflicts of interest.
References
- 1.Brenner BM. Brenner and Rector's The Kidney. 8th Ed. Saunders; Philadelphia: 2007. [Google Scholar]
- 2.de Zeeuw D, Remuzzi G, Parving HH, Keane WF, et al. Proteinuria, a target for renoprotection in patients with type 2 diabetic nephropathy: lessons from RENAAL. Kidney Int. 2004;65:2309–2320. doi: 10.1111/j.1523-1755.2004.00653.x. [DOI] [PubMed] [Google Scholar]
- 3.Fliser D, Novak J, Thongboonkerd V, Argilés À, et al. Advances in urinary proteome analysis and biomarker discovery. J. Am. Soc. Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 4.Thongboonkerd V. Recent progress in urinary proteomics. Proteomics Clin. Appl. 2007;1:780–791. doi: 10.1002/prca.200700035. [DOI] [PubMed] [Google Scholar]
- 5.Mischak H, Julian BA, Novak J. High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics Clin. Appl. 2007;1:792–804. doi: 10.1002/prca.200700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candiano G, Santucci L, Petretto A, Bruschi M, et al. 2D-electrophoresis and the urine proteome map: where do we stand? J. Proteomics. 2010;73:829–844. doi: 10.1016/j.jprot.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, et al. Characterization of the human urinary proteome: a method for high resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct spots. Proteomics. 2004;4:1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 8.Ünlü M, Morgan ME, Mindan JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 9.Sharma K, Lee S, Han S, Lee S, et al. Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics. 2005;5:2648–2655. doi: 10.1002/pmic.200401288. [DOI] [PubMed] [Google Scholar]
- 10.Rao PV, Lu X, Standley M, Pattee P, et al. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care. 2007;30:629–637. doi: 10.2337/dc06-2056. [DOI] [PubMed] [Google Scholar]
- 11.Kolch W, Neusüß C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom. Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 12.Metzger J, Schanstra J, Mischak H. Capillary electrophoresis-mass spectrometry in urine proteome analysis: current applications and future developments. Anal. Bioanal. Chem. 2009;393:1431–1442. doi: 10.1007/s00216-008-2309-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Fang A, Riley CP, Wang M, et al. Multi-dimensional liquid chromatography in proteomics - a review. Anal. Chim. Acta. 2010;664:101–113. doi: 10.1016/j.aca.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong BE, Yan F, Lubman DM, Miller FR. Chromatofocusing non-porous reversed-phase high-performance liquid chromatography/electrospray ionization time-of-flight mass spectrometry of proteins from human breast cancer whole cell lysates: a novel two-dimensional liquid chromatography/mass spectrometry method. Rapid Commun. Mass Spectrom. 2001;15:291–296. doi: 10.1002/rcm.227. [DOI] [PubMed] [Google Scholar]
- 15.Soldi M, Sarto C, Valsecchi C, Magni F, et al. Proteome profile of human urine with two-dimensional liquid phase fractionation. Proteomics. 2005;5:2641–2647. doi: 10.1002/pmic.200401269. [DOI] [PubMed] [Google Scholar]
- 16.Linke T, Ross AC, Harrison EH. Proteomic analysis of rat plasma by two-dimensional liquid chromatography and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Chromatogr. A. 2006;1123:160–169. doi: 10.1016/j.chroma.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 17.Martosella J, Zolotarjova N, Liu H, Nicol G, Boyes BE. Reversed-phase high-performance liquid chromatographic prefractionation of immunodepleted human serum proteins to enhance mass spectrometry identification of lower-abundant proteins. J. Proteome Res. 2005;4:1522–1537. doi: 10.1021/pr050088l. [DOI] [PubMed] [Google Scholar]
- 18.Zolotarjova N, Mrozinski P, Chen H, Martosella J. Combination of affinity depletion of abundant proteins and reversed-phase fractionation in proteomic analysis of human plasma/serum. J. Chromatogr. A. 2008;1189:332–338. doi: 10.1016/j.chroma.2007.11.082. [DOI] [PubMed] [Google Scholar]
- 19.Linke T, Doraiswamy S, Harrison EH. Rat plasma proteomics: effects of abundant protein depletion on proteomic analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;849:273–281. doi: 10.1016/j.jchromb.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y-T, Tsao C-Y, Li J-M, Tsai C-Y, et al. Large-scale protein identification of human urine proteome by multi-dimensional LC and MS/MS. Proteomics Clin. Appl. 2007;1:577–587. doi: 10.1002/prca.200600769. [DOI] [PubMed] [Google Scholar]
- 21.Mehdi UF, Adams-Huet B, Raskin P, Vega GL, Toto RD. Addition of angiotensin receptor blockade or mineralocorticoid antagonism to maximal angiotensin-converting enzyme inhibition in diabetic nephropathy. J. Am. Soc. Nephrol. 2009;20:2641–2650. doi: 10.1681/ASN.2009070737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pappin DJC, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 23.Lowenthal MS, Mehta AI, Frogale K, Bandle RW, et al. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin. Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- 24.Brand J, Haslberger T, Zolg W, Pestlin G, Palme S. Depletion efficiency and recovery of trace markers from a mutiparameter immunodepletion column. Proteomics. 2006;6:3236–3242. doi: 10.1002/pmic.200500864. [DOI] [PubMed] [Google Scholar]
- 25.Tyan Y-C, Guo H-R, Liu C-Y, Liao P-C. Proteomic profiling of human urinary proteome using nano-high performance liquid chromatography/electrospray ionization tandem mass spectrometry. Anal. Chim. Acta. 2006;570:158–176. doi: 10.1016/j.aca.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 26.Kanai M, Raz A, Goodman DS. Retinol binding protein: the transport protein for Vitamin A in human plasma. J. Clin. Invest. 1968;47:2025–2044. doi: 10.1172/JCI105889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naylor HM, Newcomer ME. The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry. 1999;38:2647–2653. doi: 10.1021/bi982291i. [DOI] [PubMed] [Google Scholar]
- 28.Candiano G, Bruschi M, Petretto A, Santucci L, et al. Proteins and protein fragments in nephrotic syndrome: clusters, specificity and mechanisms. Proteomics Clin. Appl. 2008;2:956–963. doi: 10.1002/prca.200780157. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee AB, Kundu GC, Mandal AK, Pattabiraman N, et al. Uteroglobin: physiological role in normal glomerular function uncovered by targeted disruption of the uteroglobin gene in mice. Am. J. Kidney Dis. 1998;32:1106–1120. doi: 10.1016/s0272-6386(98)70093-9. [DOI] [PubMed] [Google Scholar]
- 30.Weston BS, Wahab NA, Mason RM. CTGF mediates TGF-beta-induced fibronectin matrix deposition by upregulating active alpha5beta1 integrin in human mesangial cells. J. Am. Soc. Nephrol. 2003;14:601–610. doi: 10.1097/01.asn.0000051600.53134.b9. [DOI] [PubMed] [Google Scholar]
- 31.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castagna A, Cecconi D, Sennels L, Rappsilber J, et al. Exploring the hidden human urinary proteome via ligand library beads. J. Proteome Res. 2005;4:1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 33.Rocchetti MT, Centra M, Papale M, Bortone G, et al. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics. 2008;8:206–216. doi: 10.1002/pmic.200700492. [DOI] [PubMed] [Google Scholar]
- 34.Woroniecki RP, Orlova TN, Mendelev N, Shata IF, et al. Urinary proteome of steroid-sensitive and steroid-resistant idiopathic nephrotic syndrome of childhood. Am. J. Nephrol. 2006;26:258–267. doi: 10.1159/000093814. [DOI] [PubMed] [Google Scholar]
- 35.Khurana M, Traum AZ, Aivado M, Wells MP, et al. Urine proteomic profiling of pediatric nephrotic syndrome. Pediatr. Nephrol. 2006;21:1257–1265. doi: 10.1007/s00467-006-0165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis EJ, Hunsicker LG, Clarke WR, Berl T, et al. Renoprotective effects of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N. Engl. J. Med. 2001;345:851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- 37.Brenner BM, Cooper ME, de Zeeuw D, Keane WF, et al. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med. 2001;345:861–869. doi: 10.1056/NEJMoa011161. [DOI] [PubMed] [Google Scholar]
- 38.Parving H-H, Lehnert H, Bröchner-Mortensen J, Gomis R, et al. The effect of irbesartan on the development of diabetic nephropathy in patients with type 2 diabetes. N. Engl. J. Med. 2001;345:870–878. doi: 10.1056/NEJMoa011489. [DOI] [PubMed] [Google Scholar]
- 39.Ravid M, Savin H, Jutrin I, Bental T. Long-term stabilizing effect of angiotensin-converting enzyme inhibition on plasma creatinine and on proteinuria in type II diabetic patients. Ann. Intern. Med. 1993;118:577–581. doi: 10.7326/0003-4819-118-8-199304150-00001. [DOI] [PubMed] [Google Scholar]
- 40.Chan JCN, Ko GTC, Leung DHY, Cheung RCK, et al. Long-term effects of angiotensin-converting enzyme inhibition and metabolic control in hypertensive type 2 diabetic patients. Kidney Int. 2000;57:590–600. doi: 10.1046/j.1523-1755.2000.00879.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.