Preface
The human body contains several hundred cell types, all with the same genome. In metazoans, much of the regulatory code that drives cell type-specific gene expression resides in distal elements called enhancers. Enhancers are activated by proteins called transcription factors that bind specific DNA motifs and recruit co-regulators to ultimately activate transcription. While the human genome contains millions of potential enhancers, only a small subset of them is active in a given cell type. Densely spaced clusters of active enhancers, referred to as super-enhancers, are associated with the expression of genes that specify cell identity and function. On a genomic scale, the function of enhancers is influenced by, and in turn affects higher-order chromatin structure and nuclear organization.
Introduction
The molecular mechanisms that enable and mediate cell-specific transcriptional responses to intra- and extra-cellular cues remain poorly understood. Early experiments indicated that sequences far away from gene promoters are often required to regulate cell type-specific gene transcription1. Such genetic elements are termed enhancers, and were initially functionally defined as DNA sequences that have the potential to enhance basal transcription levels from gene promoters and transcriptional start sites (TSS)1, at distances ranging from hundreds of base pairs to megabases2. Recent genome-wide transcription factor-binding studies indicated that the majority of transcription factor binding sites are found in distal locations that frequently exhibit enhancer function3–9. This is consistent with the profound role that enhancers play in shaping signal-dependent transcriptional responses10–12.
When cell signaling induces an increase in the nuclear concentration and DNA binding of transcription factors, as occurs following the activation of steroid hormone receptors and NF-κB, the great majority of binding events typically occurs at genomic locations that already exhibit binding of other transcription factors and enhancer-like histone modifications5, 6. Because the complement of active cis-regulatory elements is different across cell types, these findings introduced the notion that pre-existing sets of enhancers are largely responsible for cell type-specific gene expression and responses to external stimuli13–15. The annotation of epigenetic features associated with enhancers in many different cell lines, primary cells and tissues by the ENCODE consortium provided evidence for the utilization of several hundreds of thousands of such elements in the human genome16, greatly exceeding the number of genes that encode mRNAs or long intergenic non-coding RNAs (lincRNAs). This raised the question of how the correct subsets of enhancers are selected from the large repertoire of potential enhancers in each particular cell type. Here, we review recent findings on the selection and function of enhancers that specify cell identity and underlie their distinctive responses to intra- and extracellular signals. We discuss the collaborative and hierarchical binding of transcription factors to DNA in the context of chromatin, which orchestrates enhancer selection and priming, and the transformation of chromatin from a silent, primed or poised state to one that actively supports transcription. We conclude with a discussion of the three-dimensional organization of enhancers in the nucleus and its importance for their function.
Enhancer characteristics
Genomic regions that function as transcriptional enhancers are enriched for closely spaced recognition motifs for sequence-specific transcription factors. Enhancer activation begins with the binding of transcription factors and local nucleosome remodeling. Recent genome-wide studies of nucleosome remodeling during differentiation of embryonic stem cells and induced pluripotent stem cells indicated that the majority of remodeling affects a single nucleosome, and that alterations in nucleosome occupancy are enriched at enhancers associated with pluripotency and differentiation17. Transcription factor binding leads to, and in some cases is facilitated by, the recruitment of co-regulators such as the histone acetyltransferase p30018, followed by the recruitment of RNA polymerase II (Pol II) and the transcription of enhancer-associated RNAs (eRNAs)19, 20. Co-regulator recruitment and transcription are accompanied by the covalent modification (methylation and acetylation, among others) of histone tails in enhancer-associated nucleosomes. In organisms whose DNA exhibits methylation in the context of CG dinucleotides (CpG methylation), these enhancers become demethylated upon their activation, concomitant with transcription factor binding21. Thus, epigenetic modification patterns can be used to distinguish between different enhancer activation states22 and have been used extensively to annotate putative enhancers in different cell types16.
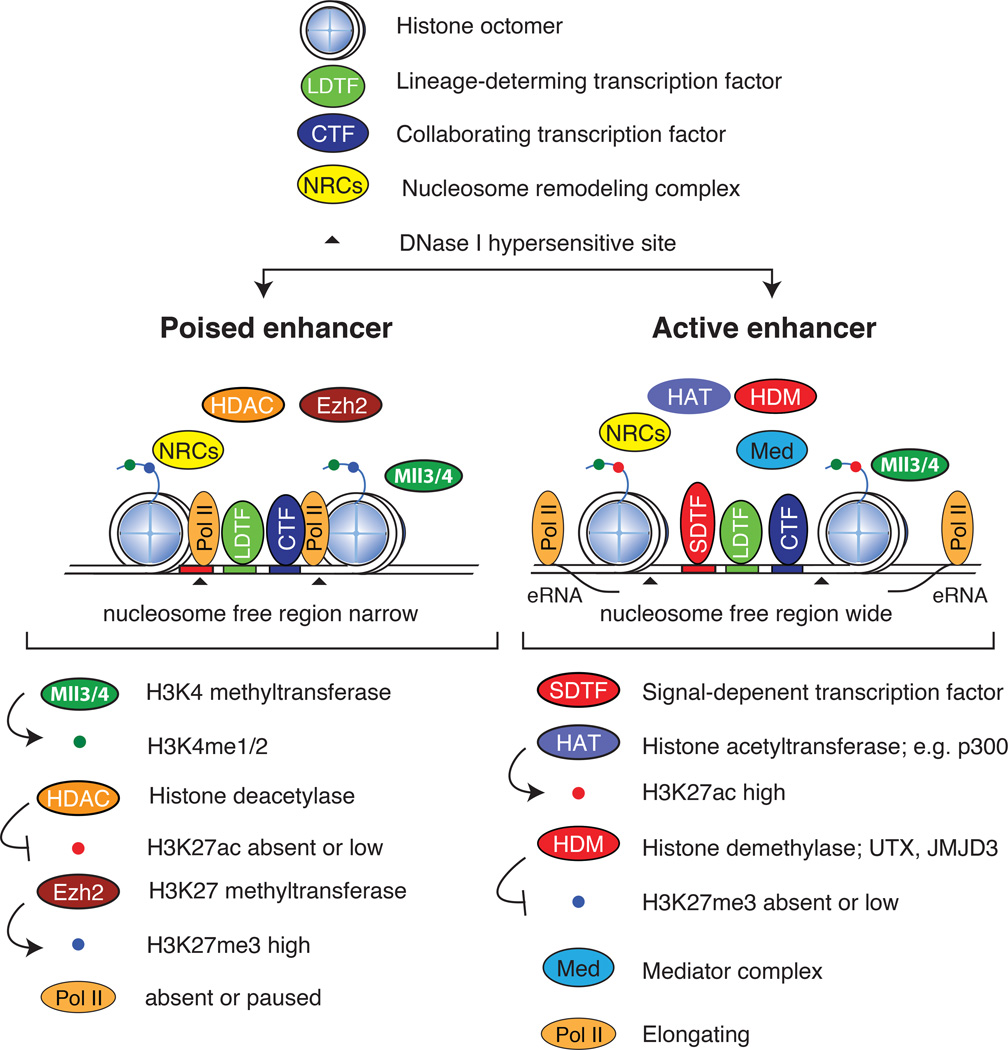
Enhancer states can broadly be classified as inactive, primed or poised, and active22. An inactive enhancer is essentially buried in compact chromatin and is devoid of transcription factor binding and histone modifications. Primed enhancers are characterized by closely bound, sequence-specific transcription factors that establish a DNase I-hypersensitive15, nucleosome-free23 region of open chromatin. However, they may require additional cues to accomplish their function, which may include signal-dependent activation, the recruitment of additional transcription factors, and the eventual recruitment of co-activators that lead to enhancer activation. Poised enhancers can be defined as primed enhancers that also contain repressive epigenetic chromatin marks (see below), a state that is most commonly found in embryonic stem cells. The characteristic features of poised and active enhancers are depicted in Figure 1.
Figure 1.
The anatomies of poised and active enhancers. The characteristic features of poised and active enhancers are shown, including the binding of lineage-determining transcription factors (LDTFs) and collaborating transcription factors (CTFs) to closely spaced recognition motifs (blue and green sites, respectively) on the DNA. The binding of these factors in concert with nucleosome remodeling complexes (NRCs) initiates nucleosome displacement to form narrow nucleosome free regions at poised enhancers (top). At poised enhancers, the redundant histone methyltransferases (HMTs) myeloid/lymphoid or mixed-lineage leukemia protein 3 (MLL3) and MLL4 deposit the active H3K4me1 and H3K4me2 marks, whereas EZH2 (a component of the polycomb complex) deposits repressive H3K27me3 and histone deacetylase (HDAC)-containing complexes maintain histones in a repressed deacetylated state. Pol II is either absent or low at poised enhancers. In response to various cues, signal-dependent transcription factors (SDTFs) associate with recognition motifs in close association with LDTFs, which results in additional nucleosome displacement (bottom), as observed by widening of the DNase I-hypersensitive sites. SDTFs recruit co-activator complexes containing histone demethylase (HDM) complexes that remove H3K27me3 marks, histone acetyltransferase (HAT) that deposit H2K27ac, and the mediator complex. The transformation to elongating Pol II results in bidirectional transcription — a hallmark of active enhancers — and the generation of enhancer RNAs (eRNA), which is closely coupled to enhancer activity.
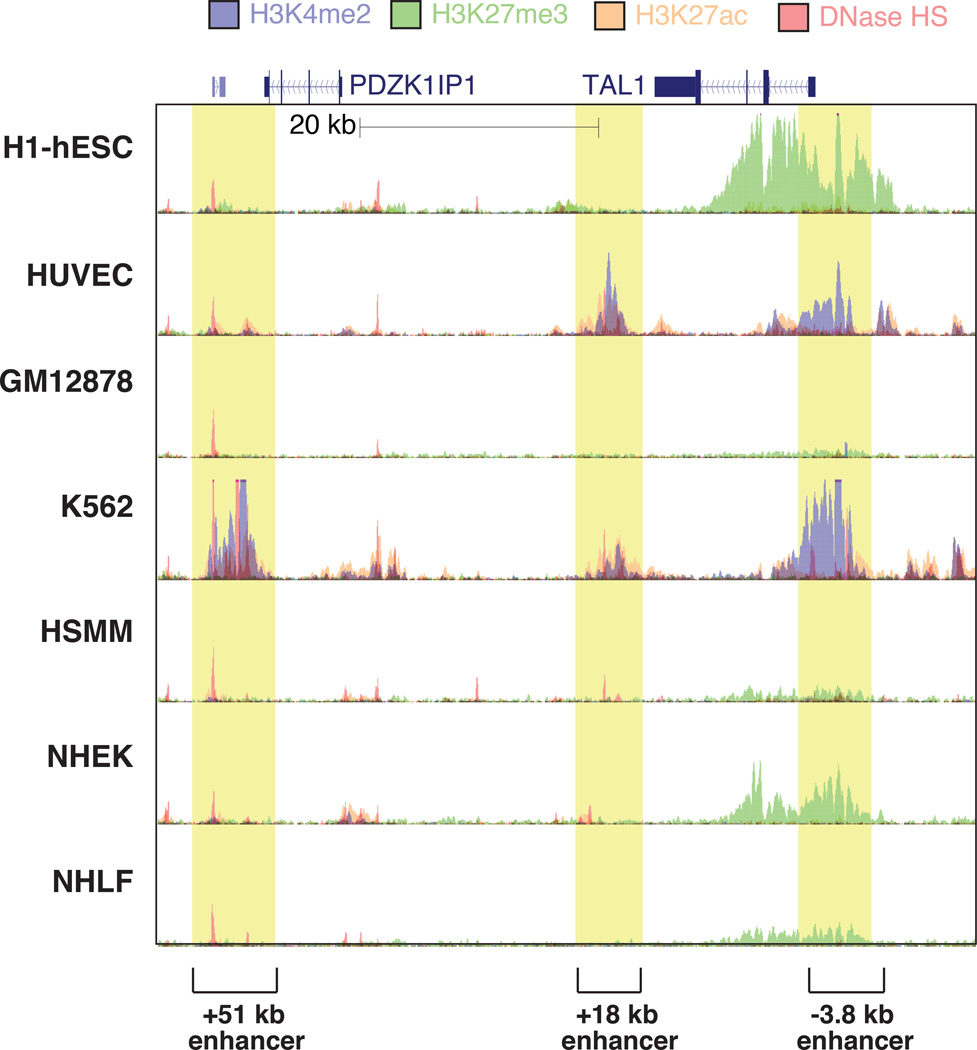
An insight important for the identification of potential enhancers was the understanding that specific histone methylation signatures mark enhancer-like regions. In particular, enhancers display enrichment for histone H3 Lys 4 mono- or dimethylation (H3K4me1 and H3K4me2, respectively) and depletion of H3K4me3, compared to promoters3. Whereas genomic regions exhibiting these features are not necessarily functional enhancers, it appears that the vast majority of regions that do function as enhancers exhibit these characteristics3, 7, 24. Specifically, primed enhancer-like regions are marked with H3K4me1 and H3K4me2 and lack histone acetylation, and enhancers marked by the trimethylation of histone H3 at lysine 27 (H3K27me3, a repressive mark) are considered to be poised24–26 (for a review, see27)(Fig. 1). Features associated with active enhancers include histone H3 Lys 27 acetylation (H3K27ac)25 and the presence of actively transcribing Pol II19. Examples of these features in the vicinity of the TAL1 locus in the genomes of 7 human cell lines, evaluated by the ENCODE consortium, are illustrated in Figure 2. Several developmental enhancers have been characterized for this locus: the −3.8 kb (upstream) and +19 kb (downstream) enhancers drive TAL1 transcription in endothelial cells (HUVEC) and hematopoietic stem and progenitor cells28, 29, whereas the +51 kb enhancer is required for TAL1 expression in erythroid cells (K562)30. On the whole, DNase I hypersensitivity at this locus corresponds with overall transcription factor binding, and the presence of the “active” epigenetic marks H3K4me2 and H3K27ac is correlated with cell type-specific enhancer activity. Conversely, in TAL1 non-expressing cells such hESC and NHEK, the +19 kb enhancer, promoter and gene body are devoid of DNase I-hypersensitive sites, and the −3.8 kb region and the gene body exhibit the repressive mark H3K27me3.
Figure 2.
Cell type-specific enhancers are marked by specific epigenomic features and chromatin accessibility. Genomic features of a ~60 kb region of human chromosome 1 centered around the TAL1 gene ENCODE consortium data of DNase-I hypersensitive (DNase HS) regions and ChIP-Seq for the marks H3K4me2, H3K27me3 and H3K27ac in 7 cell lines. Enhancers known to be responsible for TAL1 transcription in endothelial cells (the −3.8 kb and +19 kb enhancers, relative to the TAL1 promoter, in HUVEC cells) and erythroid cells (the +51 kb enhancer in K562 cells) exhibit cell type-specific DNase HS, H3K4me2 and H3K27ac signals. In cell types where TAL1 is not expressed, the promoter and gene body are devoid of DNase HS and histone modifications indicative of enhancer activation (H3K4me2, H3K27ac), and exhibit variable levels of the repressive H3K27me3 mark. Shaded boxes indicate cell-restricted or cell-specific enhancers regions.
Enhancer selection
The vast number of potential cis-regulatory elements in the genome and the cell-type selectivity with which they are utilized raises the question as to the series of events whereby unique enhancer repertoires are selected. Many lines of evidence indicate that enhancer selection is initially driven by so-called pioneer factors, exemplified by FOXA1, that are able to bind to their recognition motifs within the context of compacted chromatin31. By opening the conformation of the chromatin and initiating the process of enhancer selection, such pioneering factors can function as key cell lineage-determining transcription factors (LDTFs) to drive lineage-specific transcription programs. However, most sequence-specific transcription factors, including those that function as pioneer factors, recognize relatively short DNA sequences (of about 6 to 12 base pairs), and their typical DNA recognition motifs exhibit varying levels of degeneracy. This means that most sequence-specific transcription factors have millions of potential binding sites in the mammalian genome. Yet, chromatin immunoprecipitation followed by sequencing (ChIP-Seq) experiments have indicated that they bind only a small subset of all potential sites, and that a large fraction of the observed binding is associated with cell type-specific enhancers32. Cell type-specific binding sites often harbor motifs for additional pioneer factors, and experimental data strongly suggest that pioneer factors act in concert to jointly displace nucleosomes33, 34. Here, we review evidence supporting a model in which pioneer factors, or LDTFs, prime cell type-specific enhancers through collaborative interactions7, 23, 35–40,35–40.
The role of lineage-determining transcription factors
Experiments modulating the expression of LDTFs have demonstrated their ability to initiate the transition of enhancer elements from closed chromatin to a ‘primed’ or ’poised’ state, where transcription factors have gained access to the DNA and established nucleosome-free regions7, 10 (Figure 1). An example is provided by the ETS domain factor PU.1, a LDTF required for the development of macrophages and B cells. Importantly, PU.1 influences the establishment of distinct gene expression programs in each cell type41. The vast majority of PU.1 binding sites are located >500 bp from promoters and largely occupy different genomic locations in macrophages and B cells7. Macrophage-specific binding of PU.1 was observed at genomic locations that contained PU.1 binding sites in close proximity to binding sites of other macrophage LDTFs, such as the C/EBPs and AP-1 factors. Conversely, B cell-specific binding of PU.1 was observed in close proximity to other B cell LDTFs, including motifs recognized by EBF, E2A and Oct factors. The corresponding motifs were generally situated less than 100 bp from the PU.1 motif, but mostly not at a close (5–20 bp), invariable distance that would be indicative of direct ternary protein-protein-DNA interactions42. Notably, macrophage-specific PU.1-bound regions were depleted of B-cell LDTF motifs and vice versa, relative to neighbouring genomic regions. This, together with the finding that in a given cell type, non-bound PU.1 motifs that lie in transcriptionally inactive genomic regions are generally depleted of motifs of the LDTFs expressed in the cell32, suggests that (LDTF) motif composition may be one of the contributing factors to forming transcriptionally inactive and active genomic compartments (see below). Gain- and loss-of-function experiments revealed an interdependence of PU.1 with other LDTFs for effective DNA binding, suggesting that their collaborative interactions are necessary to compete with nucleosome for binding to DNA. By considering natural genetic variation between inbred strains of mice as mutations, LDTF binding site mutations not only impaired binding of the respective LDTF but also that of closely bound LDTFs43, 44, consistent with a model in which enhancer selection is a collaborative effort of multiple DNA binding factors. Other examples of LDTF co-operativity in establishing specific LDTF binding patterns have been observed in developmental systems such as zebrafish hematopoiesis45 and Drosophila melanogaster embryogenesis46.
The use of computational methods to identify binding motifs that are enriched in genomic regions marked by H3K4me1 resulted in the identification of LDTF motifs of the corresponding cell type. For example, binding sites for transcription factors that are capable of reprogramming fibroblasts into induced pluripotent stem cells are highly enriched in the H3K4me1-marked regions of the genome in embryonic stem (ES) cells7. Conversely, ChIP-Seq studies of LDTFs commonly revealed that they occupy large fractions of the enhancers within the cells in which they exert lineage-determining functions7, 10, 16, 36, 47, 48. Thus, whereas most cells express hundreds of transcription factors, the selection of a large fraction of cell type-specific regulatory elements may be driven by relatively simple combinations of LDTFs that interact with each other and with other factors. Collectively, these findings may facilitate computational efforts to predict the selection of cell type-specific enhancer elements based on the local organization of binding motifs and the combinations of expressed transcription factors.
The role of signal-dependent transcription factors
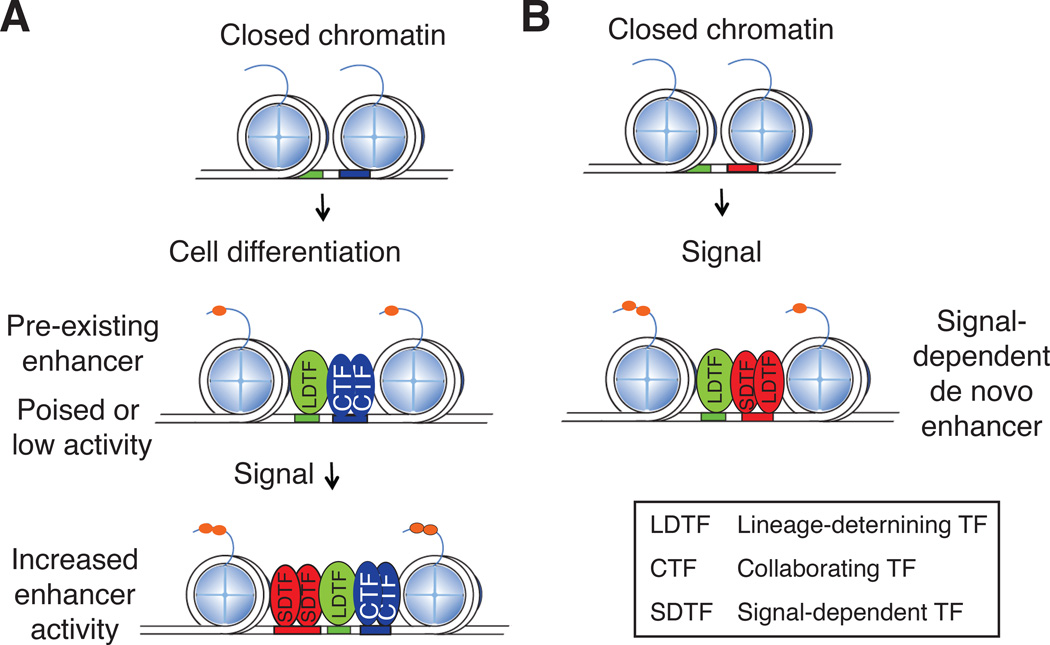
Whereas LDTF binding may be sufficient for activating some enhancers, additional signals will be required for other enhancers to be fully activated. Many of the cellular responses to internal and external signals are dependent on the function of widely expressed, signal-dependent transcription factors (SDTFs). Examples of SDTFs are members of the nuclear receptor and NF-κB families. These factors frequently activate common sets of genes in different cell types, but can also regulate gene expression in a cell type-specific manner. ChIP-Seq studies of SDTFs in different cell types found both common and cell type-specific binding sites37, 38, 48. Two types of mechanisms were suggested to account for cell type-specific binding of SDTFs. In one, such SDTF binding occurs at genomic locations that exhibit features of pre-selected enhancers7, 36, 45 (Fig. 3A). In these cases, there is a hierarchical relationship between SDTFs and LDTFs, which are the pioneer factors responsible for the initial enhancer selection through interactions with additional, collaborating transcription factors (CTFs). In many cases loss of function of the LDTF results in a failure of both the LDTF and the SDTF to bind the enhancer, but not vice versa7, 48–51. Alternatively, SDTFs could contribute directly to latent or de novo enhancer selection37, 50, 52 (Fig. 3B). This has been shown to involve collaborative interactions with LDTFs, which, owing to their restricted cell type-specific expression patterns, imposes cell type-specific enhancer selection at genomic locations that have the appropriate combination of motifs. Although mechanisms underlying collaborative DNA binding by transcription factors remain poorly understood, studies of the glucocorticoid receptor suggest that transcription factor binding can be highly dynamic and that even two factors that interact with the same recognition motif in the same cell can facilitate each other’s binding through a proposed assisted loading mechanism53.
Figure 3.
Cell type-specific enhancer selection and activation. A. Collaborative interactions between lineage-determining transcription factors (LDTFs) and collaborating transcription factors (CTFs) select enhancers for binding and activation by signal-dependent transcription factors (SDTFs). Prior to signal-dependent activation, such regions may be ‘poised’ enhancers or exhibit basal enhancer activity (‘pre-existing’ enhancers) that is further induced by the binding of a SDTF. The resulting transcription is cell type-specific because the enhancers are selected by the cell type-specific LDTFs. B. SDTFs can direct the selection of latent or de novo enhancers. In these cases, the SDTF functions as an essential collaborative transcription factor to LDTFs to enable concurrent binding of all factors involved. The transcriptional output is cell type-specific because of the requirement for cell type-specific LDTFs for enhancer priming.
The extent to which SDTFs operate on poised enhancers or participate in de novo enhancer selection appears to vary depending on the factor, cell type and signal in question. FoxP3, a SDTF required for the acquisition of the Th2 phenotype of CD4-positive T cells, was found to bind almost exclusively to a poised enhancers upon their activation36. In contrast, the receptor for the steroid hormone ecdysone, a member of the nuclear receptor family mediating transcriptional responses to ecdysone in insects, primarily binds newly selected enhancer elements in combination with cell type-specific transcription factors37. Both mechanisms of enhancer selection (Fig. 3) can occur simultaneously in the same cell type. For example, following macrophage activation by lipopolysaccharides, approximately 90% of the binding of the p65 subunit of NF-κB occurs at already primed enhancers, whereas the remainder is associated with the de novo selection of latent enhancers in collaboration with LDTFs such as PU.1 and C/EBPα43, 50. Recent studies of macrophage populations from different tissues demonstrated that local environmental signals regulate the expression and activities of TFs that can function to selectively activate enhancers that are primed in common between macrophage subsets or drive the priming and activation of enhancers that are specific to macrophage subsets or54, 55. Of note, the histone methylation signature of latent enhancers persists after the cessation of cell stimulation and is associated with more rapid and diverse transcriptional responses to subsequent stimulation52. These observations provide evidence that the writing of the H3K4me1 signature in enhancers provides a molecular memory of prior activation.
Enhancer activation
While transcription factor binding is a requirement enhancer activity, not all promoter-distal transcription factor binding sites appear to function as enhancers, as judged by H3K4me1 and H3K4me2, and not all regions of the genome enriched with H3K4me1 and H3K4me2 exhibit marks of active enhancers, such as H3K27ac. This raises the question of what determines whether transcription factor binding will result in an active enhancer. Many different enhancer states can be defined based on particular combinations of histone posttranslational modifications22 (Figure 1), which are deposited by transcription co-regulators that are recruited to enhancers and promoters by transcription factors. Transcription co-regulators include histone methyltransferases (HMT), including the MLL proteins56, histone acetyltransferases (HAT) such as p300 and CBP57, histone deacetylases (HDAC) which are components of co-repressors such as NCoR and SMART5, as well as chromatin remodelers such as the BRG1- or hBRM-associated factor (BAF) complexes (also known as the mammalian SWI/SNF complexes)58, 59 and the mediator complex60. Recruitment of co-regulators to enhancers is the more frequent the more transcription factors are co-bound to a given enhancer7, 61. Co-regulators are large proteins with multiple distinct interaction sites for transcription factors18, 62, 63, and likely serve as both facilitators and integrators of transcription factor binding and intracellular signals at enhancers, similar to their known roles at promoters64.
Enhancer transcription
The epigenetic marks deposited by co-regulator complexes serve as binding sites for chromatin readers such as TFIID65 and Brd4—P-TEFb66, which function in transcription pre-initiation complex assembly and in transcription elongation, respectively.
The presence of transcription pre-initiation complex and elongation factors at enhancers67, 68 is in line with the finding that Pol II is found at enhancers. More than 20 years ago Pol II was observed to generate non-coding RNAs at locus control regions69, but it was only recently appreciated that mammalian enhancers are broadly transcribed and generate enhancer RNAs (eRNAs)19, 20, 70–72. Pol II recruitment to enhancers and signaling-dependent changes in eRNA expression are highly correlated with changes in the expression of nearby genes, suggesting a functional link between eRNA and gene expression73, 50, 74, 75. The distinguishing features of eRNAs are that most are short (< 1 kb), are not subjected to polyadenylation or splicing19, 20, and are rapidly degraded by the exosome71. Similarly to what has been shown for short promoter antisense transcripts76, these characteristics are likely caused by the lack of a 5’ splice donor proximal to eRNA TSS71, 72, which is a prerequisite for splicing and promotes elongation77, packaging into messenger ribonucleoprotein particles (mRNP), polyadenylation and nuclear export78, all of which contribute to the stability of transcripts. As a side note, the fact that enhancers resemble promoters in almost every aspect, except for lacking proximal splice donors71 and H3K4me3 marks79, suggests that stable mRNAs or lincRNAs could be created by simply introducing a splice donor downstream of an eRNA TSS72. This would be in line with the ability of intronic enhancers to serve as alternative promoters80, and the fact that 98% of all lincRNAs have only two exons (that is, a single splice donor downstream of a promoter), compared with merely 6% of coding genes81.
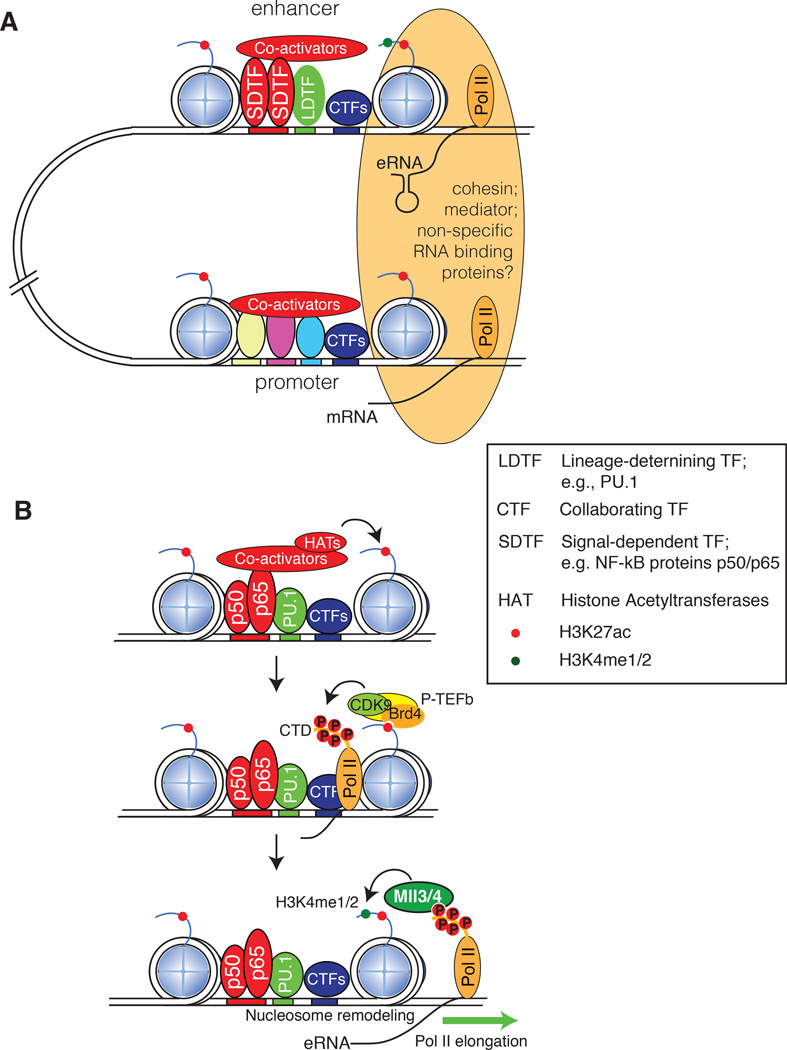
The occurrence of global enhancer transcription in mammalian cells raises the question of its functional significance. Recent studies provide evidence that eRNAs contribute to local enhancer activity, potentially by facilitating enhancer--promoter interactions through chromatin looping, recruitment of co-factors such as the mediator complex (Fig. 4A; reviewed in ref. 66) and release of negative elongation factor82. As of yet there is limited evidence for specific sequence features of eRNAs that could be necessary for their function, and not all eRNAs appear to contribute to enhancer function. To date, little attention has been directed at the possibility that the process of enhancer transcription itself (independent of the eRNA product) could influence enhancer activity. Pol II is a powerful nucleosome remodeling machine83, and transcription initiated from an enhancer sequence may contribute to maintaining an open chromatin configuration that enables access of sequence-specific transcription factors. In addition, enhancer transcription may play an important role in contributing to the deposition of H3K4me1 and H3K4me2 marks at enhancers (Fig. 4B). Genetic studies indicate that the D. melanogaster H3K4 methyltransferase trithorax-related (Trr) and its mammalian homologues MLL3 and MLL4 play important roles in the writing of these marks84, 85, but the mechanisms that recruit these enzymes and determine the overall distribution of histone methylation remain poorly understood. Studies of newly selected or de novo enhancers in activated macrophages provided evidence that the methylation of H3K4, but not the acetylation of H3K27, required enhancer transcription and the presence of MLL3 and MLL450.
Figure 4.
Enhancer activation and function. A. Interactions between enhancers and promoters involve structural connections (orange oval) that include cohesin and the mediator complex to promote pre-initiation complex formation, initiate transcription and/or overcome Pol II pausing. A potential role of enhancer RNAs (eRNAs) could be to promote transcription by facilitating chromatin looping, possibly by mediating interactions with cohesin. Another potential role could be to mediate interactions with protein complexes required for transcriptional elongation, such as the mediator complex. LDTFs, lineage-determining transcription factors; CTFs, collaborating transcription factors; SDTFs, signal-dependent transcription factors. B. Potential roles of enhancer transcription. In activated macrophages the NF-κB proteins p50 and p65 are signal-dependent transcription factors and PU.1 is a lineage-determining transcription factor that collaboratively select de novo enhancers. The subsequent recruitment of histone acetyltransferases (HAT) results in histone acetylation, which is bound by the Brd4 component of the P-TEFb complex, allowing its Cdk9 component to phosphorylate the C-terminal domain (CTD) of Pol II. Phosphorylated CTD acts as docking sites for the MLL3 and MLL4 histone H3K4 methyltransferases. MLL3 and MLL4 are proposed to deposit H3K4me1 and H3K4me2 during successive rounds of Pol II elongation.
A model of enhancer activation based on time-resolved studies of transcription factor binding, eRNA transcription, H3K4 methylation and H3K27 acetylation at de novo enhancers, and on results of gain- and loss-of-function experiments50, is illustrated in Fig. 4B: Signal-dependent activation of NF-κB (p50 and p65) results in its collaborative binding with PU.1 and the recruitment of co-activator complexes that contain histone acetyltransferases (HAT). These events result in nucleosome remodeling, histone acetylation and the recruitment of Pol II. The conversion of Pol II from a paused to an elongating form involves P-TEFb, which is recruited to at least some sites of transcription initiation by interactions between Brd4 and acetylated histone H4. Cyclin-dependent kinase 9 (CDK9), a component of P-TEFb, phosphorylates the C-terminal domain (CTD) of Pol II, providing docking sites for the histone methyltransferases complexes myeloid/lymphoid or mixed-lineage leukemia protein 3 (MLL3) and MLL4. MLL 3 and MLL4 progressively methylate H3K4 during successive rounds of transcription elongation. Consistent with this model is the distribution of H3K4me1 and H3K4me2, which was found to correlate with the extent of enhancer transcription, and to be dependent on transcription elongation50. The generality of this model with respect to the mechanisms by which H3K4 methylation marks are established at other classes of enhancers, such as those that are selected during cellular differentiation, remains to be determined. For example, in contrast to the activation of de novo enhancers in the context of extracellular signaling responses, studies of the distribution of H3K4me1 and H3K4me2 at cell type-specific enhancers selected during muscle and adipocyte differentiation suggested that MLL complexes can interact directly with LDTFs such as C/EBPβ and MyoD at cell type-specific enhancers, where MLL3 and MLL4 are also required for acetylation of H3K27, mediator and Pol II recruitment85.
The function of H3K4me1 and H3K4me2 marks remains an open question. As they are known to recruit histone remodeling complexes86, they could conceivably contribute to keeping previously bound and modified enhancers open and accessible, which would help explain the observation that previously activated latent enhancers are more rapidly re-activated by subsequent stimuli52.
Enhancer function
Promoter activation requires that many components of the transcriptional machinery come together in order to assemble the pre-initiation complex, initiate transcription, overcome Pol II pausing, and eventually lead to productive transcription elongation. Through looping of the intervening DNA, enhancers get into close proximity of promoters, and are thought to affect any or all of the aforementioned processes by increasing the local concentration of the factors that carry them out87 (Fig. 4A). These factors include co-activator complexes such as the mediator complex, which increases the loading of transcription factors on promoters and enhancers60; scaffold proteins such as cohesin, that mediate stable, often cell type-specific promoter-enhancer interactions60, 88; and factors involved in releasing paused Pol II and in the initiation of elongation, such as Brd489. A major challenge in deciphering cell-specific enhancer functions is connecting active enhancers to their target genes in vivo.
Super-enhancers
Based on their epigenetic features, and depending on the experimental methods used to define enhancers, ~10,000–50,000 putative enhancers can be identified in a given cell type13, 16, 90, implying there are more enhancers than expressed genes. Along the linear DNA molecule, enhancers are located non-uniformly in respect to genes, such that some genes reside in enhancer-rich regions of the genome, whereas others have few or no enhancers in their vicinity. Although a single enhancer is sufficient to activate the expression of a nearby gene37, high levels of cell type-specific and/or signal-dependent gene expression are most frequently observed for genes located in enhancer-rich regions of the genome, exemplified by the relationship of enhancer-rich locus control regions and the expression of the globin genes in erythroid cells69. Such enhancer-dense regions have recently been termed “super-enhancers”91–93.
Super-enhancers were initially defined as large (tens of kilobases-long) genomic loci with an unusually high density of enhancer-associated marks, such as binding of the mediator complex, relative to most other genomic loci91, 92. These regions can also be defined by high density91 and/or extended (> 3 kb)94 depositions of the histone mark H3K27ac. Using differences in the density of mediator complex-binding sites or of H3K27ac marks to distinguish super-enhancers from regular enhancers, most cell types are found to have between 300 and 500 super-enhancers91. A substantial fraction of super-enhancers and nearby genes are cell type-specific, and the gene sets that are associated with super-enhancers in a given cell type are highly enriched for the biological processes that define the identities of the cell types91,94. For example, many of the genes encoding factors required for pluripotency and self-renewal of ES cells reside near ES cell-specific super-enhancers91. In keeping with their tissue-specificity, super-enhancers active in certain cell types are enriched for disease-associated alleles relevant to that cell type91,94. Not surprisingly, the individual enhancers of cell type-specific super-enhancers are enriched for binding sites of the corresponding LDTFs92. Collectively, the specific set of super-enhancers within a particular cell type may provide a means of simplifying the problem of defining what are the quantitatively most important transcriptional programmes required for establishing cell identity, and to identifying disease-relevant, non-coding genetic variation.
Three-dimensional chromatin interactions
In the nucleus, the genome is organized and partitioned into functional compartments in the three-dimensional (3D) space95, and considerable effort is being directed at understanding enhancer function in the context of 3D chromatin interactions. One strategy is to identify the long-range looping interactions involving enhancer elements using a variety of chromosome conformation capture (3C)-based techniques96. Genome-wide applications of these techniques to define the chromatin interactomes of human and mouse cells confirmed that the genome is divided into active and inactive compartments96. These are further organized into sub-megabase-sized topologically associated domains (TADs) that correlate with regions of the genome that constrain the spread of heterochromatin and are relatively conserved across cell types97. Although the genome-wide resolution of such studies remains somewhat limited, the resulting chromatin connectivity maps suggest that only approximately 7% of the looping interactions are made between adjacent genes, indicating that linear genomic adjacency is not necessarily a good predictor for long-range interactions98. In addition, promoters and distal enhancer elements are frequently engaged in multiple long-range interactions and form active chromatin hubs98, 99 (Fig. 5). Whereas super-enhancers are identified along the linear DNA sequence by virtue of their high density of typical epigenomic features, it is clear that the enhancers within a super-enhancer form 3D interactions that are a feature of the folded genome in the nucleus93 (Fig 5). Interestingly, studies of tumor necrosis factor α (TNFα)-responsive enhancers in human fibroblasts indicated that they are already in contact with their target promoters before the activation of TNFα signaling100, suggesting that enhancer-promoter interactomes are already set up during development. This is in line with data from D. melanogaster showing that only 6% of spatial genome interactions change during early embryonal development101. It is not yet known when higher-order chromatin interactions are established during development, but it likely coincides with the occurrence of gap phases following the mid-blastula transition, which is accompanied by establishment of a non-random nuclear chromatin conformation and the transcriptional activation of chromatin domains102.
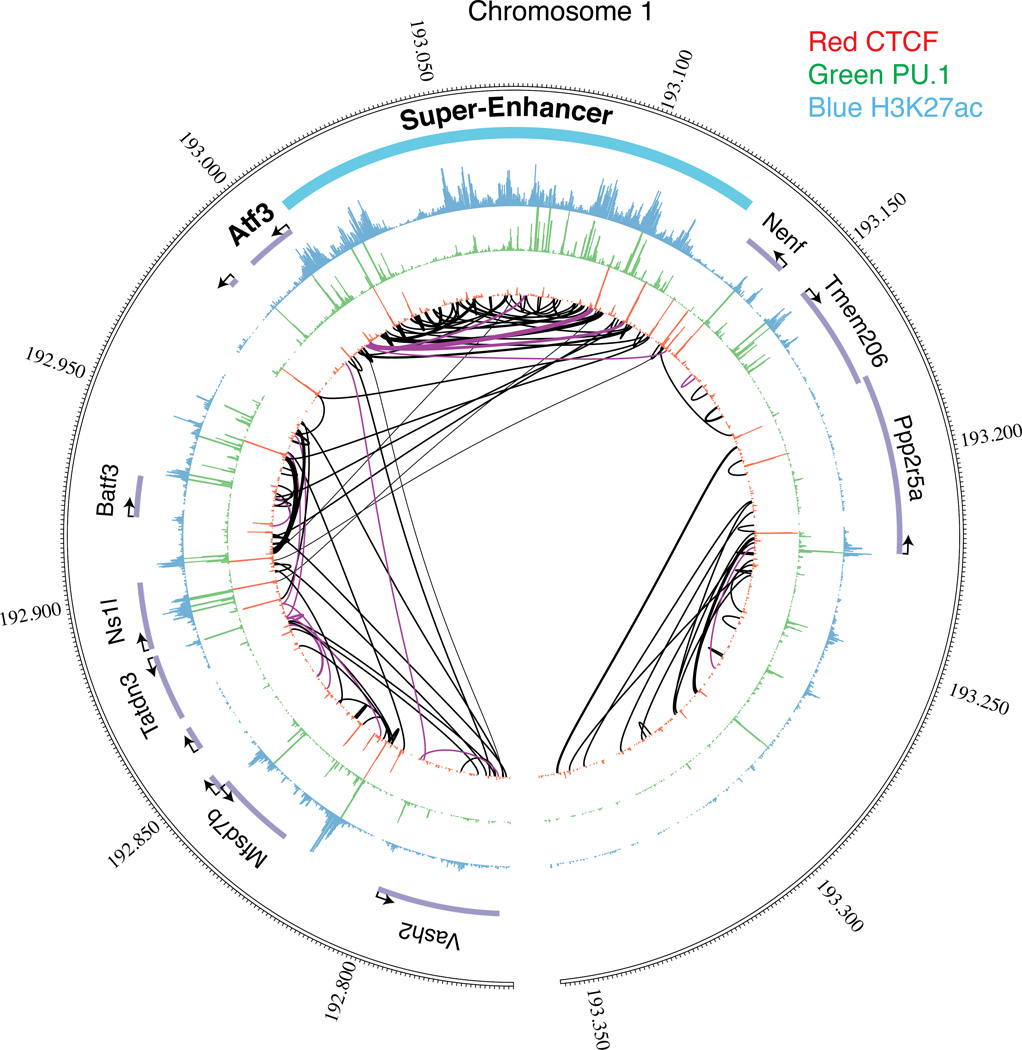
Figure 5.
The linear and the three-dimensional organization of enhancers in the nucleus. The outer circle represents the linear coordinates of a region of human chromosome 1 surrounding the Atf3 (activating transcription factor 3) gene in C57BL/6J mouse macrophages. The locations of individual genes are indicated by gene names and purple bars. The three successive concentric inner circles depict ChIP-Seq data of, respectively, histone H3 Lys 27 acetylation (H3K27ac), the transcription factor PU.1, and the transcription repressor CCCTC-binding factor (CTCF), which is enriched at boundaries of topological domains. A region of high density of H3K27ac in the vicinity of the Atf3 gene is designated as a super-enhancer. Purple and black lines in the center of the circle indicate physical contacts involving promoters and other genomic regions, respectively, as determined by statistically significant genome-wide chromatin connectivity measurements determined by tethered conformation capture130. This locus demonstrates the multitude of connections between the individual enhancers comprising the Atf3 super-enhancer, which essentially forms its own TAD, as well as the longer-range enhancer-enhancer and enhancer-promoter interactions outside of the TAD.
It is unclear how the 3D organization of the genome is determined, however what is known is that cohesin and the mediator complex60, which are scaffold proteins of the replication machinery and the transcription machinery, respectively, are involved in the formation of high-order chromatin structures. Since cohesion appears to be recruited to enhancers through clusters of LDTFs103, it is likely that protein-protein interactions and the genomic sequence together shape 3D genomic conformations, although this hypothesis still awaits experimental confirmation.
Given that the conformation of the genome appears to be mostly fixed across developmental stages101, individual cells104, cell types97 and signaling states100, it is tempting to speculate how enhancers work in the 3D space: Promoters are known to also function as transcriptional enhancers with regard to the activation of promoters in their proximity105, and enhancers have sequence features identical to those found in promoters71, 72. Both are juxtaposed within TADs as part of linear super-enhancers92, as well being brought into proximity by higher-order structures74, leading to the co-regulation of promoters and enhancers within a domain74,106. Knocking out enhancers within a TAD shows that the loss of an enhancer often only leads to a graded reduction in expression107, 108,74 and in developmental dysregulation109 of the associated gene, suggesting that at least in some cases enhancers work in an additive manner. The distribution of gene regulation among a multitude of enhancers, some of which reside linearly within or beyond neighboring genes (in their “shadow”, hence the term “shadow enhancers”110) but are close in 3D space, is thought to increase robustness of the regulatory system to mutations111. Whereas the high-order chromatin structure of genomic regions >1 Mb is largely invariant, single loci can move between inert and active chromosomal compartments, depending on their activation status, leading respectively to stable repression or to a state poised for transcription112. In contrast, within these large-scale compartments, inside TADs, the chromatin structure of regions smaller than 100 kb does differ in a cell type-specific fashion100, implying that different regulatory regions within TADs can be dynamically juxtaposed in a stimulus-specific manner. This way, genome topology could contribute to cell type-specific transcription programs, meaning that mapping the genomic topology and elucidating the mechanisms that govern the 3D structure of the genome will be important steps toward understanding how the genome functions.
Conclusions and perspective
Although initially described more than 30 years ago, we still do not have a clear understanding of the mechanisms by which enhancers regulate gene expression. However, the development of a plethora of methods for genome-wide mapping of diverse enhancer features, their functional relationship with promoters, and their ultimate transcriptional outputs have resulted in a number of striking and unexpected discoveries, ranging from the identification of the great number of enhancers in metazoan genomes to the widespread production of enhancer-derived RNAs. The observation that more than 80% of disease-associated alleles identified by genome-wide association studies reside in non-coding regions of the genome113 implies they have yet unappreciated regulatory functions. Consistent with this, several studies have demonstrated an enrichment of disease-associated loci in cell type-specific regulatory regions, including in super-enhancers, of the corresponding disease-relevant cells types91, 114–117, and a number of studies are beginning to document the direct effects of common variation in enhancer elements on enhancer states118–120, gene expression117, 121, 122 and disease123–127.
Beyond the simple annotation of regulatory regions in the genome, it is important to understand how cells select the full complement of enhancers that is required for maintaining their identities and functions. In essence, we would like to be able to ‘read’ the genomic template and predict from the combination of active transcription factors the enhancers that will be functional in a cell type-specific manner. The principle of collaborative transcription factor-interactions at closely-spaced DNA recognition motifs provides a starting point for predicting genome-wide patterns of transcription factor binding required for enhancer selection. These predictions can be validated by mutating binding sites or by taking advantage of naturally-occurring genetic variations. However, transcription factor-binding maps are insufficient to predict enhancer activity. The discovery that enhancer transcription is highly correlated with nearby gene expression is likely to be an important clue in understanding how enhancers function. The evidence that eRNAs contribute to activities of at least some enhancers provide impetus for determining their mechanisms of action. In addition, the relative importance of enhancer transcription itself in maintaining enhancer accessibility and contributing to enhancer-related H3K4 methylation requires further study, as the functional roles of H3K4 methylation beyond providing a memory of prior enhancer activation, remain obscure52.
Defining functional enhancer-promoter interactions remains an important goal. Despite being informative, chromatin connectivity maps do not directly relate chromatin interactions to the regulation of gene expression. Definitive evidence that a specific enhancer-like region exerts a transcriptional regulatory function requires mutating that region, and encouragingly, site-specific mutagenesis should be greatly facilitated by recently developed genome editing methods128. Such tools will enable us to systematically delete enhancer elements and modify enhancer sequences to evaluate chromatin connectivity and gene expression. As a complementary approach, recent studies have demonstrated the utility of using natural genetic variation as a tool to study the relationships between transcription factor binding, enhancer selection and the regulation of gene expression43. Improving the understanding of the mechanisms underlying the selection and function of enhancers will likely not only enable predicting the consequences of genetic variation on gene expression and phenotype, but may also provide approaches to directly alter enhancer function for therapeutic purposes.
Acknowledgements
These studies were primarily supported by NIH grants DK091183, CA17390, and DK063491 and the San Diego Center for Systems Biology. C.E.R. was supported by the American Heart Association (12POST11760017) and the NIH Pathway to Independence Award (1K99HL12348).
Glossary
- DNase I-hypersensitive site
Genomic DNA is packaged into chromatin, which makes it less accessible to DNase I, and nucleases in general. Binding of proteins to regulatory regions makes these genomic sites more accessible, and hypersensitive to DNase digestion.
- Locus control region (LCR)
Confers tissue-specific expression to a linked transgene irrespective of the integration site of the transgene in the genome. Thus, LCRs display characteristics of both enhancers and insulators.
- Exosome
A multiprotein complex that is involved in surveillance, degradation and maturation of RNA transcripts in both nucleus and cytoplasm. In the nucleus, the exosome is involved in 3’ processing of rRNA and snoRNA, degrades many types of aberrant RNA transcripts, including pre-mRNAs and pre-tRNAs, as well as non-coding transcripts such as eRNAs and promoter upstream transcripts (PROMPTs), and. In the cytoplasm, mRNAs are the primary target of the exosome, where it contributes to mRNA turnover, nonsense-mediated decay of mRNAs with premature stop codons, mRNAs without a stop codon, and 5’ ends of mRNAs that have stalled in translation or have been cleaved by the RNAi pathway129.
- Mediator complex
A multisubunit protein complex that influences nearly all stages of transcription and in conjunction with cohesion contributes to the 3D organization of the genome. The mediator complex is recruited to the DNA by binding to sequence-specific transcription factors via its individual subunits, and integrates a wide gamut of intracellular signals to affect pre-initiation complex formation, transcription initiation and elongation.
- Chromosome conformation capture (3C)
A method to probe the higher order structure of genomic DNA in the nucleus. In living cells, DNA gets fixed to the chromatin in living cells by formaldehyde, digested with a restriction enzyme, and protein-DNA complexes get diluted and ligated. DNA that were spatially close in the nucleus are more likely to reside within the same protein-DNA complex. These are preferentially ligated together, and interaction frequencies of studied loci are quantified by quantitative PCR. This experimental strategy forms the basis for unbiased genome-wide methods such as Hi-C and tethered conformation capture (TCC), where the restriction sites are labeled with biotin prior to ligation, and either arm of the biotinylated, mixed ligation product is sequenced to reveal the genome-wide interaction frequencies of all genomic loci in a cell.
- Chromatin hub
A spatial arrangement of regulatory DNA elements (LCRs, enhancers, promoters) and genes into a domain that leads to correct gene expression of the associated genes. The smallest unit of a hub could be a topologically associated domain (TAD), and the largest could comprise an entire nuclear compartment (see TAD).
- Active enhancers
In addition to marks of poised enhancers, active enhancers are marked with acetylated H3 lysine 27 (H3K27ac), produce eRNAs, are bound by the Mediator complex, and exert regulatory function to increase the transcription of target genes.
- Latent or de novo enhancers
An inactive enhancer locus whose selection requires the binding of a combination of transcription factors that includes SDTFs.
- Primed enhancers
Enhancers that have been selected by LDTFs and CTFs and are marked with histone modifications characteristic of enhancers, such as mono- and dimethylation of histone H3 lysine 4 (H3K4me1 and H3K4me2), but do not produce eRNA.
- Poised enhancers
Similar to primed enhancers, but distinguished by the presence of tri-methylation of H3 lysine 27, which must be removed for the transition to an active enhancer state.
- Topologically Associated Domains (TADs)
Genomic, largely self-interacting domains of sub-megabase sizes, further organized into multi-megabase-sized structures called nuclear compartments. Genes within TADs are co-regulated and their expression patterns are highly correlated. TADs are determined by chromosome conformation capture experiments and are largely conserved across cell types and throughout development.
Biographies
Sven Heinz is the Director of the Next Generation Sequencing Core at the Salk Institute for Biological Studies in La Jolla, California, USA. After receiving his Ph.D. in Chemistry from the University of Regensburg, Germany, he did postdoctoral work with Christopher Glass at the University of California, San Diego, California, USA. His research focuses on the transcription regulatory mechanisms that underlie genome function in different cell types and individuals during development, homeostasis and disease.
Casey E. Romanoski completed her Ph.D. in 2010 in Human Genetics under supervision of Aldons J. Lusis at the University of California, Los Angeles. There, she studied the genetics of endothelial cell expression and responsiveness to oxidized phospholipids. Since, she has worked as a postdoctoral fellow with Christopher K. Glass where she is interested in the molecular mechanisms of gene regulation and the consequences of genetic variation on expression networks and disease traits.
Christopher Benner is the Director of the Integrative Genomics and Bioinformatics Core at the Salk Institute. Dr. Benner's research focuses on how genomic sequences are organized to encode complex regulatory mechanisms. He develops innovative software tools for the analysis of large-scale sequencing-based genomics experiments, including ChIP-Seq, GRO-Seq and conformation capture assays. Dr. Benner is the developer of the HOMER software suite that is widely used by the genomics community.
Christopher K. Glass is Professor of Cellular and Molecular Medicine and Professor of Medicine at the University of California San Diego School of Medicine. He has had a long-standing interest in understanding how sequence-specific transcription factors, co-activators and co-repressors regulate macrophage gene expression. His current work employs genome-wide approaches to investigate mechanisms underlying the selection of enhancers that determine macrophage identity and function.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 3.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 4.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 5.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes & development. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. This paper shows that the LDTF PU.1 binds to different locations in different cell types, which is dependent on the complement of LDTFs expressed in a cell. The loci become marked by H3K4me1, serve as beacons for SDTFs and drive cell type-specific signal responses.
- 8.Lefterova MI, et al. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen R, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghisletti S, et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. This paper shows that PU.1 is an LDTF necessary for priming signal-dependent enhancers in macrophages that define their transcriptional response to inflammatory stimuli.
- 11.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 12.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS biology. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. This paper summarizes the work of the ENCODE consortium to annotate functional DNA elements within the human genome.
- 17.West JA, et al. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun. 2014;5:4719. doi: 10.1038/ncomms5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 19.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. The papers by De Santa et al., and Kim, et al., report widespread transcription at enhancers, which correlates with transcription of neighboring genes.
- 21.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 22.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottgens B, et al. The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5' bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol Cell Biol. 2004;24:1870–1883. doi: 10.1128/MCB.24.5.1870-1883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez M, et al. An SCL 3' enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development. 1999;126:3891–3904. doi: 10.1242/dev.126.17.3891. [DOI] [PubMed] [Google Scholar]
- 30.Delabesse E, et al. Transcriptional regulation of the SCL locus: identification of an enhancer that targets the primitive erythroid lineage in vivo. Mol Cell Biol. 2005;25:5215–5225. doi: 10.1128/MCB.25.12.5215-5225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaret KS, et al. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham TH, et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman BG, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. This paper and the paper by Mullen (reference 48) show that SDTFs bind to the open chromatin landscape that is defined by combinations of LDTFs.
- 37.Shlyueva D, et al. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Molecular cell. 2014;54:180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 38.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, et al. Combinatorial Assembly of Developmental Stage-Specific Enhancers Controls Gene Expression Programs during Human Erythropoiesis. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 42.Kazemian M, Pham H, Wolfe SA, Brodsky MH, Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefflova K, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 2012;22:2018–2030. doi: 10.1101/gr.132811.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercer EM, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–425. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. This paper and the paper by Samstein (reference 36) show that SDTFs bind to the open chromatin landscape that is defined by combinations of LDTFs.
- 49.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 50. Kaikkonen MU, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. This paper shows that the SDTF NF-κB contributes to the priming (and activation) of enhancers de novo and in conjunction with LDTFs, and demonstrates that transcription elongation is required for mono- and dimethylation of flanking histone H3K4 by MLL3 and MLL4.
- 51.Sullivan AL, et al. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Molecular and Cellular Biology. 2011;31:861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. This paper describes signal dependent selection of latent enhancers in macrophages and their persistence following signal termination as a molecular 'memory' of prior activation.
- 53.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Molecular cell. 2014;53:859–866. doi: 10.1016/j.molcel.2014.02.033. The papers by Gosselin, et al., Lavin, et al., demonstrate how specific tissue environments differentially influence the selection and function of enhancers within the corresponding resident macrophage populations.
- 57.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Euskirchen GM, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris SA, et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siersbaek R, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 62.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 63.Blazek E, Mittler G, Meisterernst M. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 64.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 66.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 68.Zhang W, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collis P, Antoniou M, Grosveld F. Definition of the minimal requirements within the human beta-globin gene and the dominant control region for high level expression. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 76.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 78.Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nature reviews. Genetics. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 79.Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 80.Kowalczyk MS, et al. Intragenic enhancers act as alternative promoters. Mol Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 81.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schaukowitch K, et al. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herz HM, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes & development. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee JE, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife (Cambridge) 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 87.Plank JL, Dean A. Enhancer Function: Mechanistic and Genome-Wide Insights Come Together. Molecular cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. The papers by Hnisz, et al., and Whyte, et al., describe regions of the genome that are highly enriched for marks of active enhancers and reside near genes that play essential roles in determining cellular identity and function.
- 93.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cremer T, et al. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 96.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. The papers by Dixon, et al., and Sanyal, et al, use global chromatin conformation capture assays to interrogate three dimensional organization of the functional elements within the genome.
- 99.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 100.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 102.Vassetzky Y, Hair A, Mechali M. Rearrangement of chromatin domains during development in Xenopus. Genes Dev. 2000;14:1541–1552. [PMC free article] [PubMed] [Google Scholar]
- 103.Yan J, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 104.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. This publication shows that promoters function as transcriptional enhancers.
- 106.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bender MA, et al. The hypersensitive sites of the murine beta-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012;119:3820–3827. doi: 10.1182/blood-2011-09-380485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sur IK, et al. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 109.Rosenbauer F, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nature genetics. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 110.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays. 2012;34:135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. The papers by Kasowski, et al., Kilpinen et al., and McVicker, et al., report on the effects of natural genetic variation in humans on the binding of transcription factors and histone modifications associated with enhancers and gene expression.
- 121.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gaffney DJ, et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 125.Cowper-Sal lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 130.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]