Abstract
Clostridium sporogenes DSM 795 is the type strain of the species Clostridium sporogenes, first described by Metchnikoff in 1908. It is a Gram-positive, rod-shaped, anaerobic bacterium isolated from human faeces and belongs to the proteolytic branch of clostridia. C. sporogenes attracts special interest because of its potential use in a bacterial therapy for certain cancer types.
Genome sequencing and annotation revealed several gene clusters coding for proteins involved in anaerobic degradation of amino acids, such as glycine and betaine via Stickland reaction. Genome comparison showed that C. sporogenes is closely related to C. botulinum. The genome of C. sporogenes DSM 795 consists of a circular chromosome of 4.1 Mb with an overall GC content of 27.81 mol% harboring 3,744 protein-coding genes, and 80 RNAs.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0016-y) contains supplementary material, which is available to authorized users.
Keywords: C. sporogenes, Anaerobic, Butanol, C. botulinum, Gram-positive, Stickland reaction
Introduction
C. sporogenes was isolated from human faeces [1-3], but can also be found in soil and marine or fresh water sediments [4-7]. C. sporogenes strain DSM 795 [8] serves as type strain for this species and as a consequence was chosen for whole genome sequencing.
Because C. sporogenes is closely related to C. botulinum group I strains, it is used as a non-toxic surrogate for this common food-borne pathogen. 16S rDNA sequencing revealed a 99.7% sequence similarity to proteolytic C. botulinum strains of serotypes A, B, and F [9]. In this context, the genome of C. sporogenes strain PA 3679 was sequenced and a draft sequence published in 2012 [10]. C. sporogenes can be isolated from infections, but does not play a prominent role as a pathogen. Only few clinical cases are reported, in which this species was found to participate. These cases include epynema, soft tissue abscesses, septic arthritis, or gas gangrene [11-16].
Requiring an anaerobic habitat, C. sporogenes is known to specifically colonize hypoxic areas of solid tumors. In 1964, Möse and co-workers demonstrated tumor colonization resulting in tumor lysis after intravenous application of C. butyricum M-55 in mice carrying Ehrlich carcinomas [17]. The respective strain was subsequently reclassified as C. oncolyticum and finally as C. sporogenesATCC 13732. They also demonstrated that C. sporogenes spores are immunologically inert by injecting them into themselves [18]. As an excellent tumor colonizer, C. sporogenes bears promising therapeutic potential for cancer therapy [19]. With CFU numbers up to 2 × 108 per gram of tumor tissue, C. sporogenes outperforms saccharolytic clostridia such as C. beijerinckii and C. acetobutylicum by far, as the latter reach only CFU numbers of 105-106 per gram of tumor tissue [20-22].
Restricted to the inner core of the tumor, clostridia cannot lyse the well-oxygenated outer rim of tumor cells, which remains viable and unaffected. Therefore, treatment with clostridial spores alone is not sufficient for complete tumor eradication. Several attempts have been made to genetically modify C. sporogenes for production of therapeutic proteins or pro-drug converting enzymes. The latter catalyze the conversion of innocuous pro-drug molecules into their active, cytotoxic form. This reaction takes place directly in the tumor allowing systemic application of higher drug concentrations and reduction of side effects [20]. Among others, cytosine deaminase and nitroreductase were used for this purpose. A recombinant C. sporogenesDSM 795 mutant expressing cytosine deaminase induced growth delay of tumors in a mouse model after application of spores and the pro-drug 5-fluorouracil [20]. Also, C. sporogenes mutants heterologously expressing nitroreductase exhibited a significant antitumor efficacy in different in vivo tumor models [23-25].
Organism information
Classification and features
C. sporogenes has been subject of extensive studies since the 1930s. Characteristic features of C. sporogenesDSM 795 are listed in Table 1.
Table 1.
Classification and general features of Clostridium sporogenes DSM 795 T
| MIGS ID | Property | Term | Evidence code a |
|---|---|---|---|
| Classification | Domain Bacteria | [26] | |
| Phylum Firmicutes | [27-29] | ||
| Class Clostridia | [30,31] | ||
| Order Clostridiales | [32,33] | ||
| Family Clostridiaceae | [32,34] | ||
| Genus Clostridium | [32,35,36] | ||
| Species Clostridium sporogenes | [8,32,37,38] | ||
| Type strain DSM 795 T | [8] | ||
| Gram stain | positive | IDA | |
| Cell shape | rod-shaped | IDA | |
| Motility | motile | [39] | |
| Sporulation | sporulating | IDA | |
| Temperature range | mesophilic, 25–45°C | [39] | |
| Optimum temperature | 30-40°C | IDA, [39] | |
| pH range; Optimum | 5.7-8.5; 7 | [39], IDA | |
| Carbon source | amino acids | [40-43] | |
| MIGS-6 | Habitat | human and animal gut, soil, marine and fresh water sediments | [4-7,44,45] |
| MIGS-6.3 | Salinity | growth in 2YT medium | IDA |
| MIGS-22 | Oxygen requirement | anaerobic | IDA |
| MIGS-15 | Biotic relationship | free living | IDA |
| MIGS-14 | Pathogenicity | low | [39] |
| MIGS-4 | Geographic location | Not reported | |
| MIGS-5 | Sample collection | Not reported | |
| MIGS-4.1 | Latitude | Not reported | |
| MIGS-4.2 | Longitude | Not reproted | |
| MIGS-4.4 | Altitude | Not reproted |
aEvidence codes - IDA: Inferred from Direct Assay. Evidence codes from the Gene Ontology project [46].
C. sporogenes belongs to the proteolytic branch of clostridia capable of amino acid fermentation. No carbohydrates are required for growth, although addition can have a stimulating effect [47]. Amino acids are degraded via the Stickland reaction for energy conservation [40-43]. Required media composition and further nutritional demands have already been elucidated [48-55]. Growth can be obtained anaerobically in complex medium, but also several minimal media supplemented with amino acids are described [52,53].
The Gram-positive nature of C. sporogenes was confirmed by Gram staining (Figure 1). Cell size can vary between 0.3-1.4 × 1.3-16.0 μm [39].
Figure 1.
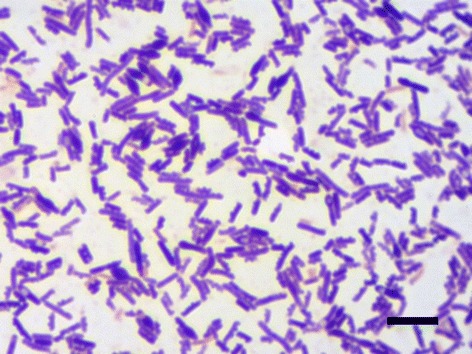
Gram staining of Gram-positive C. sporogenes DSM 795. Scale bar represents 5 μm.
A scanning electron microscopy image of C. sporogenesDSM 795 cell culture is shown in Figure 2. Several cell stages are depicted: vegetative dividing and sporulating cells and a mature spore (upper left part of the image).
Figure 2.
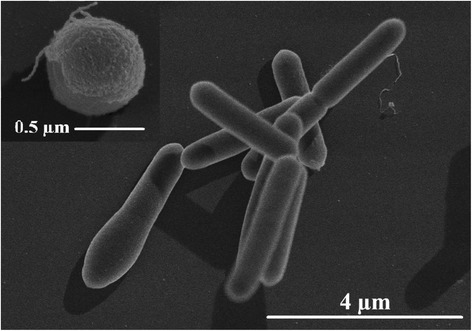
Scanning electron microscopy image of C. sporogenes DSM 795.
Transmission electron microscopy images (Figure 3) reveal membrane organizations and cell compartments of a dividing cell (Figure 3A), sporulating cells (Figure 3B and C), and a spore (Figure 3D).
Figure 3.
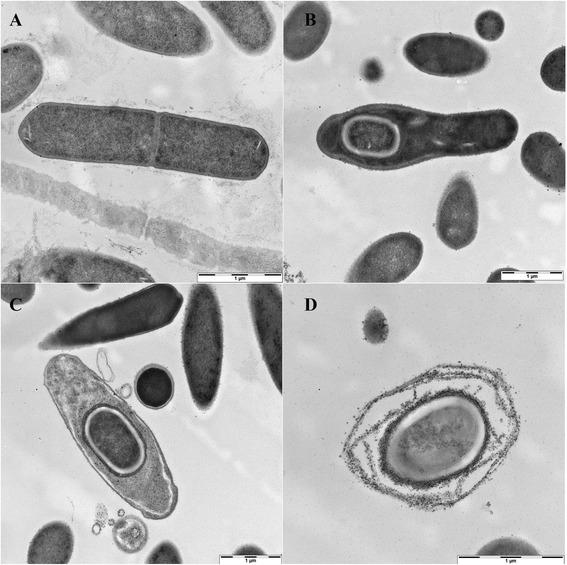
Transmission electron microscopy image of C. sporogenes DSM 795; A: dividing cell; B, C: sporulating cells; D: spore; scale bars represent 1 μm.
C. sporogenes is considered as the non-toxic surrogate of neurotoxin producer Clostridium botulinum. Additional file 1: Table S1 provides an overview of all C. botulinum strains mentioned in this study. Generally, they are assigned to four groups (I-IV) based on their physiologic characteristics [56]. Strains belonging to group I are proteolytic [57]. They are further classified into serotypes A-F due to different types of the produced botulinum neurotoxin with several subtypes existing [56].
Phylogenetic relation of C. sporogenesDSM 795 to C. botulinum strains and other clostridia was investigated by calculation of a phylogenetic tree using 16S rDNA sequences (Figure 4). C. sporogenesDSM 795 positions itself in close relationship to proteolytic C. botulinum strains of types A, B, and F, confirming previous studies [9]. Clostridial 16S rRNA reference sequences were retrieved from GenBank (NCBI database). At first, these sequences were aligned with MAFFT version 7 using default settings except for “globalpair” in fast Fourier transform [58]. Then, based on the multiple sequence alignment, a phylogenetic tree was inferred with the program MrBayes 3.1.2 [59] using the default settings.
Figure 4.
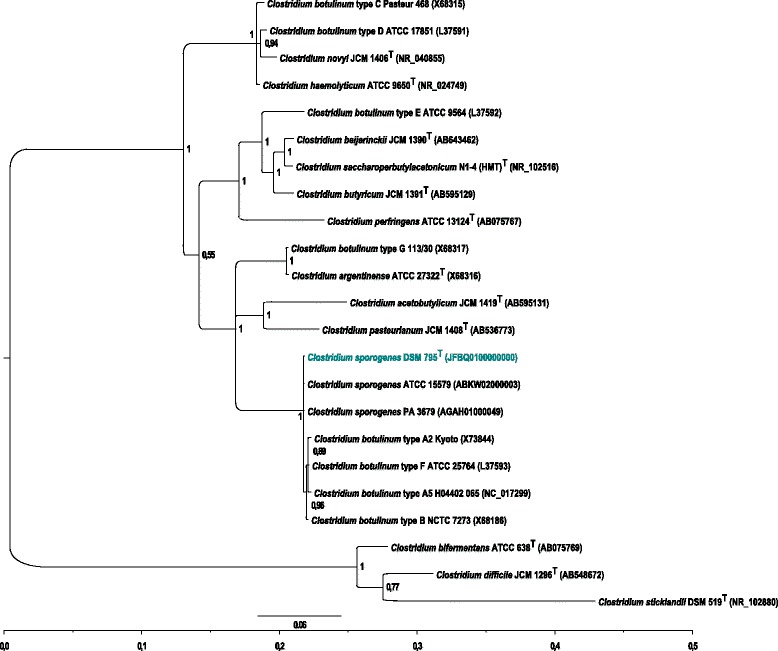
Phylogenetic tree of C. sporogenes DSM 795 based on 16S rDNA gene sequences. Estimation is based on Bayesian inference and MAFFT alignment.
C. sporogenesDSM 795 exhibits β-hemolysis on sheep and human erythrocytes (data not shown) due to production of clostridiolysin S [60]. Further enzymes produced are desoxyribonuclease, thiaminase, chitinase, kininase, L-methioninase, hyaluronate lyase, and superoxide dismutase [39].
In general, C. sporogenes is resistant to streptomycin, neomycin, kanamycin, tobramycin, and amikamycin and susceptible to penicillin G, metronidazole, tinidazole, chloramphenicol, tetracycline, and doxycycline [39]. C. sporogenes strain DSM 795 is additionally susceptible to thiamphenicol and erythromycin or clarithromycin in working concentrations of 15 μg/ml and 2.5 μg/ml, respectively.
Genome sequencing information
Genome project history
C. sporogenesDSM 795 was chosen for whole genome sequencing as it is the type strain of this species. Furthermore, it attracts special interest because of its potential use in tumor therapy and it is known as nontoxic surrogate of the food-borne and neurotoxin-producing pathogen C. botulinum. The sequencing of C. sporogenesDSM 795 genomic DNA delivered a high-quality draft genome sequence comprising 1 scaffold and 16 contigs. The sequence is deposited in GenBank database under the accession JFBQ00000000. A summary of the project information is listed in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Improved high-quality draft |
| MIGS-28 | Libraries used | Two genomic libraries: 454 pyrosequencing shotgun library, Illumina paired-end library (1 kb insert size) |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium, Illumina GAII and MiSeq |
| MIGS-31.2 | Fold coverage | 11.53 × 454, 51.25 × Illumina |
| MIGS-30 | Assemblers | Newbler 2.8, MIRA 3.4 |
| MIGS-32 | Gene calling method | YACOP, Glimmer |
| Locus Tag | CSPO | |
| Genbank ID | JFBQ00000000 | |
| GenBank Date of Release | 2014-05-06 | |
| GOLD ID | Gi0006347 | |
| BIOPROJCT | 239205 | |
| MIGS-13 | Source material identifier | DSM 795 |
| Project relevance | medical, butanol formation, amino acid degradation |
Growth conditions and genomic DNA preparation
C. sporogenesDSM 795 was cultivated in anaerobic 2YT medium containing 3% (w/v) tryptone, 2% (w/v) yeast extract, and 8.7 mM sodium thioglycolate ([61], mod.).
For genomic DNA preparation via the hot phenol method, an overnight culture incubated at 37°C was used. The procedure was carried out as described previously [62]. Redistilled and in TE buffer equilibrated phenol (pH 7.5-8) was used for the extraction.
Electron microscopic images were taken from an overnight and a sporulating culture (10 d, 30 °C) of C. sporogenes. SEM and TEM cell samples were washed 3 times with PBS and fixed with 1 vol 5% (v/v) glutaraldehyde in 0.2 M phosphate buffer pH 7.3 containing 2% (w/v) sucrose. Further treatment and visualization were conducted by the Central Facility for Electron Microscopy, University of Ulm.
Genome sequencing and assembly
Whole-genome sequencing of C. sporogenes was performed with a combined approach using the 454 GS-FLX Titanium XL system (Titanium GS70 chemistry, Roche Life Science, Mannheim, Germany), the Genome Analyzer II, and the MiSeq (Illumina, San Diego, CA). Shotgun libraries were prepared according to the manufacturer’s protocols, resulting in 126,343 reads for 454 shotgun sequencing (11.53 × coverage) and 1,445,024 112-bp and 5,654,920 150-bp paired-end Illumina reads (263.72 × coverage). For the initial hybrid de novo assembly with MIRA 3.4 [63] and Newbler 2.8 (Roche Life Science, Mannheim, Germany), we used all of the 454 shotgun reads, 1,445,024 112-bp and 554,976 150-bp paired-end Illumina reads. The final assembly was composed of 298 contigs with an average coverage of 62.78. For scaffolding we used the Move Contigs tool of the Mauve Genome Alignment Software [64]. Additionally, contigs that could not be ordered with Mauve were examined via Gene Ortholog Neighborhoods based on bidirectional best hits implemented at the IMG-ER (Integrated Microbial Genomes-Expert Review) system [65,66]. For contig ordering tasks, the genomes of C. sporogenesATCC 15579 (ABKW00000000), C. botulinumATCC 3502 (AM412317, AM412318), and C. botulinum BoNT/B1 Okra (CP000939) were used as references. Sequence gaps were closed in the Gap4 (v.4.11) software of the Staden Package [67] by PCR-based techniques and primer walking with conventional Sanger sequencing, using BigDye 3.0 chemistry on an ABI3730XL capillary sequencer (Applied Biosystems, Life Technologies GmbH, Darmstadt, Germany). The resulting draft genome is composed of 16 contigs in 1 scaffold.
Genome annotation
The software tools YACOP and Glimmer [68] were used for automatic gene prediction, while identification of rRNA and tRNA genes was performed with RNAmmer and tRNAscan, respectively [69,70]. Automatic annotation was carried out with the IMG-ER (Integrated Microbial Genomes-Expert Review) system [65,66], but annotation was afterwards manually curated by employing BLASTP and the Swiss-Prot, TrEMBL, and InterPro databases [71].
Genome properties
The draft genome of C. sporogenesDSM 795 consists of one scaffold containing 16 contigs representing one circular chromosome with a size of 4.1 Mb and with an overall GC content of 27.81 mol%. 3,832 genes are encoded, from which 3,744 were putative protein coding, 8 were pseudo and 80 RNAs (10 rRNA and 70 tRNA genes). 77.51% of encoding genes could be assigned to a putative function while the remaining 843 genes were annotated as hypothetical proteins. The genome harbors 6 different selenocysteine-containing proteins, even SelD, the selenide water dikinase (CSPO_9c05010), necessary for incorporation of selenocysteine into proteins, contains one selenocysteine. 4 of the remaining gaps represent rRNA gene clusters and there are some indications in the draft genome that at least 4 of these clusters are double, triple, or even fivefold clusters. Statistics and genome properties are listed in Table 3.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,106,655 | 100.00% |
| DNA coding (bp) | 3,416,102 | 83.18% |
| DNA G + C (bp) | 1,142,131 | 27.81% |
| Number of scaffolds | 16 | |
| Total genes | 3,832 | 100.00% |
| Protein coding genes | 3,752 | 97.91% |
| RNA genes | 80 | 2.09% |
| Pseudo genes | 8a | |
| Genes in internal clusters | 2,960 | 76.77% |
| Genes with function prediction | 2,942 | 77.51% |
| Genes assigned to COGs | 2,456 | 64.09% |
| Genes with Pfam domains | 3,139 | 81.92% |
| Genes with signal peptides | 176 | 4.59% |
| Genes with transmembrane helices | 1,059 | 27.64% |
| CRISPR repeats | 0 |
aPseudo genes may also be counted as protein coding or RNA genes, so are not additive under total gene count.
Insights from the genome sequence
Of all protein coding genes 2,456 (64.09%) could be assigned to at least one COG category [72]; Table 4 shows the distribution into these functional groups. The two most abundant categories were “general function prediction” and “transcription”, to which 11.75% and 10.55%, respectively, could be assigned to, followed by “amino acid transport and metabolism”, “function unknown”, “signal transduction and mechanisms”, and “energy production and conversion” with 9.42%, 8.51%, 7.05% and 6.07%, respectively, of all protein coding genes.
Table 4.
Number of genes associated with the general COG functional categories
| Code | Value | % Age | Description |
|---|---|---|---|
| J | 167 | 6.07 | Translation, ribosomal structure and biogenesis |
| A | n. a. | n. a. | RNA processing and modification |
| K | 290 | 10.55 | Transcription |
| L | 110 | 4.00 | Replication, recombination and repair |
| B | n. a. | n. a. | Chromatin structure and dynamics |
| D | 28 | 1.02 | Cell cycle control, cell division, chromosome partitioning |
| V | 99 | 3.60 | Defense mechanisms |
| T | 194 | 7.05 | Signal transduction mechanisms |
| M | 130 | 4.73 | Cell wall/membrane biogenesis |
| N | 72 | 2.62 | Cell motility |
| U | 34 | 1.24 | Intracellular trafficking and secretion, and vesicular transport |
| O | 81 | 2.95 | Posttranslational modification, protein turnover, chaperones |
| C | 156 | 5.67 | Energy production and conversion |
| G | 131 | 4.76 | Carbohydrate transport and metabolism |
| E | 259 | 9.42 | Amino acid transport and metabolism |
| F | 78 | 2.84 | Nucleotide transport and metabolism |
| H | 124 | 4.51 | Coenzyme transport and metabolism |
| I | 49 | 1.78 | Lipid transport and metabolism |
| P | 164 | 5.96 | Inorganic ion transport and metabolism |
| Q | 27 | 0.98 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 323 | 11.75 | General function prediction only |
| S | 234 | 8.51 | Function unknown |
| - | 1,376 | 35.91 | Not in COGs |
9.42% of all protein coding genes were assigned to the COG category “amino acid transport and metabolism”, which indicates that utilization of amino acids plays an important role in the metabolism of C. sporogenes. Supporting this assumption, we identified several clusters coding for proteins involved in anaerobic amino acid degradation by the Stickland reaction [40-43]. One example is a gene cluster coding for several subunits of a proline reductase (CSPO_9c00030-CSPO_9c00180). This cluster contains two selenocysteine-containing proteins, the gamma subunit PrdB (CSPO_9c00070), and PrdC, a protein with high sequence homology to the C-subunit of Rnf-complex (CSPO_9c00100). The cluster shows a similar organization as in C. sticklandii [73,74], except for the additional presence of a transposase (CSPO_9c00150), a second copy of PrdH2 (CSPO_9c00160) and a non-selenocysteine-containing version of PrdC (CSPO_9c00170). Furthermore, we identified gene clusters for the degradation of glycine, another one for its derivative betaine (N,N,N-trimethylglycine), and a cluster involved in selenocysteine incorporation into proteins (Figure 5).
Figure 5.
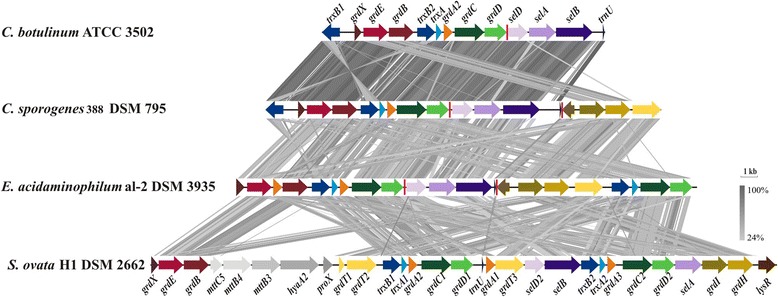
Tblastx comparison of glycine-, betaine-reductase and the selABC gene cluster of C. sporogenes DSM 795 with C. botulinum ATCC 3502, E. acidaminophilum al-2 DSM 3953 and S. ovata H1 DSM 2662: An E-value cutoff of 1e-10 was used and visualization was done with the program Easyfig [75]. The analyzed gene clusters are localized in different regions in the genomes of C. sporogenes DSM 795, C. botulinum ATCC 3502 as well as E. acidaminophilum al-2 DSM 3953. In S. ovata one genomic region includes all three gene clusters. Borders between the glycine-reductase (CSPO_4c08160-CSPO_4c08230), betaine-reductase (CSPO_4c10340-CSPO_4c10360) and selABCtrnU (CSPO_9c04980-CSPO_9c05010) gene cluster are indicated with red vertical lines.
In contrast to Sporomusa ovata [76], in which all above mentioned gene are organized in one large cluster, the clusters identified in the genomes of C. sporogenes, C.botulinumATCC 3502 and Eubacterium acidaminophilum al-2 DSM 3953 are localized in separate regions of the genomes. This is also true for the selenocysteine-incorporation genes (selABC [lilac tones]) as well as the Sec-specific tRNA (trnU [dark lilac]) as shown in Figure 5. The gene clusters of C. sporogenes show identical organization to those identified in C. botulinum and show only slightly differences to those found in E. acidaminophilum [77], whereas genes coding for the betaine-specific reductase is missing in the genome of C. botulinum. The glycine reductase cluster of C. sporogenes lacks the second copy of grdA [orange] in comparison to E. acidaminophilum, but contains two paralogs for thioredoxin reductases of the thioredoxin system [blue tones]. These two paralogs could also be identified in the cluster found in C. botulinum. In C. sporogenes the gene cluster coding for the betaine reductase (grdRIH [brown tones]) is much shorter than E. acidaminophilum’s cluster, as genes coding for thioredoxin (trxA [light blue]) and thioredoxin reductase (trxB [dark blue]) as well as the two genes coding for the C-subunit of the reductase (grdCD [green tones]) are not present. The 32.5 kb comprising cluster of S. ovata includes genes coding for the glycine-specific subunit (grdEB [red tones]), genes coding for the betaine-specific subunit (grdIH [brown tones]), two copies of genes coding for the substrate-unspecific subunit C (grdCD [green tones]), and two copies coding for the thioredoxin and a thioredoxin reductase (trxAB [blue tones]). All these genes show identical clustering as identified in the genome of C. sporogenes. The genes coding for proteins necessary for the selenocysteine-incorporation show a different arrangement as identified in C. sporogenes,C. botulinum and E. acidaminophilum, where these genes are organized in a selABCtrnU operon [lilac tones] [78]. It is also visible in Figure 5 that genes coding for glycine-specific subunit (grdBE [red tones]) show high sequence homology to genes coding for betaine-specific subunit (grdIH brown tones]).
C. sporogenes is able to produce solvents such as ethanol and butanol [79,80]. The genome of C. sporogenesDSM 795 harbors the complete set of genes necessary for glycolysis (phosphoglucomutase, glucose-6-phophate isomerase, 6-phopsphofructokinase, 1-phosphofructokinase, fructose-bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, aldehyde:ferredoxin oxidoreductase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, enolase, pyruvate kinase, pyruvate dehydrogenase) as well as aldehyde dehydrogenase and several bifunctional aldehyde-alcohol dehydrogenases, essential for ethanol production. Genes coding for key enzymes of butanol fermentation, such as butyryl-CoA dehydrogenase, acetyl-CoA acetyltransferase, 3-hydroxybutyryl-CoA dehydrogenase, 3-hydroxybutyryl-CoA dehydratase, several alcohol dehydrogenases, acetate kinase, phosphate acetyltransferase, two copies of butyrate kinase, two copies of phosphate butyryltransferase as well as three copies of formate acetyltransferase are also present (Additional file 2: Table S2). In contrast to other solventogenic clostridia, such as C. beijerinckii, C. saccharobutylicum, C. saccharoperbutylacetonicum, or C. acetobutylicum, C. sporogenes is not able to produce acetone. In solventogenic clostridia, CoA transferase and acetoacetate decarboxylase, key enzymes of acetone production, are organized in the sol operon [81-86]. In C. beijerinckii, C. saccharobutylicum, and C. saccharoperbutylacetonicum aldehyde dehydrogenase is also part of this operon, whereas in C. acetobutylicum this enzyme is replaced by alcohol/aldehyde dehydrogenase. We could not identify CoA transferase and acetoacetate decarboxylase in the genome of C. sporogenesDSM 795 and both, alcohol/aldehyde dehydrogenase and aldehyde dehydrogenase are present, but located in different regions of the genome.
Genome comparison
C. sporogenes is renowned as a nontoxic surrogate for the proteolytic C. botulinum, an organism which produces the botulinum neurotoxin (BoNT). C. botulinum is classified into seven serotypes (A to G) according to the neurotoxin antigenic specificity [57,87]. Serotypes A, B, E, and F cause human botulism, C and D are mainly described in animal toxicity, and no botulism case has been reported for serotype G [88,89]. For genome comparisons, two C. sporogenes species (ATCC 15579 and PA3679) and available representatives of all serotypes of C. botulinum, except for serotype G, were chosen and retrieved from NCBI (Figure 6A). For this purpose and to prepare data for comparisons we used the scripts ncbi_ftp_download v0.2, cat_seq v0.1 and cds_extractor v0.6 [90]. Proteinortho v5.04 [91] was utilized to identify orthologs between the different organisms with an identity cutoff of 50% and an E-value of 1e-10. The core genome of all three C. sporogenes species consists of 2,920 CDS with a total pan genome of 4,754 CDS. C. sporogenesDSM 795 has 3,258 orthologous genes with C. sporogenes PA379 and 2,981 with C. sporogenesATCC 15579 (Figure 6B). C. sporogenesDSM 795 has the least number of genome specific proteins of the three C. sporogenes strains with 270 singletons, as C. sporogenesATCC 15579 contains 577 singletons and C. sporogenes PA3679 557. We identified 163, 178, and 189 paralogs in C. sporogenesDSM 795, ATCC 15579, and PA3679, respectively; but these genes were not included into analysis.
Figure 6.
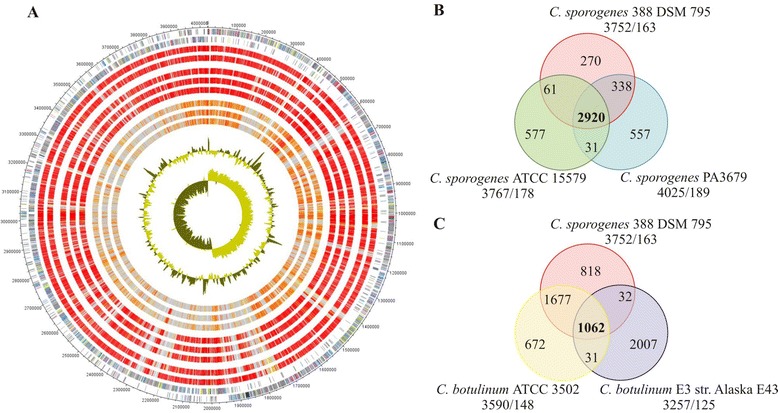
Genome comparison of C. sporogenes with different C. sporogenes and C. botulinum strains: A: Genes encoded by the leading and the lagging strand (circle 1 and 2) of C. sporogenes DSM 795 are marked in COG colors in the artificial chromosome map. The presence of orthologs (circle 3 to 10) is indicated for the genomes of C. sporogenes PA3679 (AGAH00000000), C. sporogenes ATCC 15579 (ABKW00000000) , C. botulinum ATCC 3502 (CP000727.1), C. botulinum B1 str. Okra (CP000939.1, CP000940.1), C. botulinum F str. Langeland (CP000728.1, CP000729.1), C. botulinum E3 str. Alaska E43 (CP001078.1), C. botulinum D str. 1873 (ACSJ01000001), C. botulinum C str. Eklund (ABDQ01000001) are illustrated in red to light yellow and singletons in grey (grey: >e−10-1; light yellow: <e−50- > e−10; gold: <e−50- > e−90; light orange: <e−90- > e−100; orange: <e−100- > e−120; red: <e−120-0). The two innermost plots represent the GC-content and the GC-skew. The artificial chromosome was built after scaffolding with Mauve alignment tool and concatenating the 16 contigs of the draft genome. Venn diagrams showing orthologs genes between the three sequenced C. sporogenes species (B) and between C. sporogenes DSM 795, the phylogenetic closely related C. botulinum ATCC 3502 and distantly related C. botulinum E3 str. Alaska E43 (C). Ortholog detection was done with the Proteinortho software (blastp) with an identity cutoff of 50% and an E-value of 1e-10. The total number of genes and paralogs, respectively, were depicted under the corresponding species name.
Phylogenetic analysis based on 16S rDNA revealed that C. sporogenes is closely related to serotypes A, B, and F C. botulinum strains, whereas it is distantly related to serotypes C, D, E, and G. These results were confirmed by gene content analyses as we identified 2,739 orthologous proteins between C. sporogenesDSM 795 and C. botulinumATCC 3502 (serotype A) (Figure 6C). In contrast, there are only 1,094 orthologous genes between C. sporogenes and C. botulinum E3 str. Alaska E43 (serotype E). This number is nearly identical to the quantity of orthologs (1,093) found between C. botulinumATCC 3502 and C. botulinum E3 str. Alaska E43. We identified 163, 148, and 125 paralogs in C. sporogenesDSM 795, C. botulinumATCC 3502, and C. botulinum E3 str. Alaska E43, respectively; but these genes were not included into analysis.
A genome comparison between C. sporogenesDSM 795 and C. botulinumATCC 3502 revealed that both organisms have 2,739 orthologs in common, with 818 singletons in C. sporogenesDSM 795 and 672 singletons in C. botulinumATCC 3502. The most important difference between both strains is the presence of the botulinum neurotoxin (BoNT/A) gene cluster in C. botulinumATCC 3502 and its absence in C. sporogenesDSM 795 (Figure 7).
Figure 7.
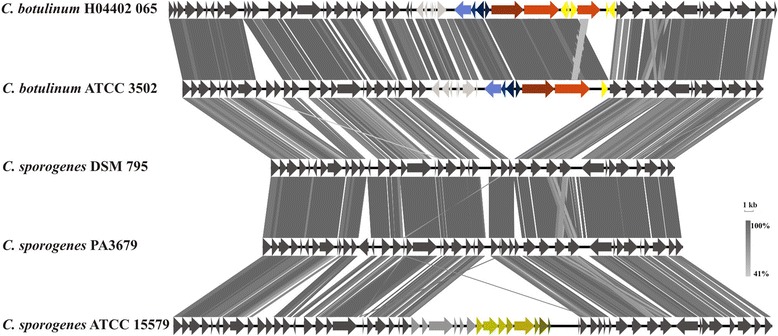
Tblastx comparison of the botulinum neurotoxin cluster (BoNT/A) and the flanking regions between C. botulinum and C. sporogenes strains. For the tblastx comparison an E-value cutoff of 1e-10 was set. Visualization was done with Easyfig. The neurotoxin cluster was marked in red tones, haemagglutinin components of the neurotoxin complex in blue tones, transposases in bright yellow, genes identified only in the C. botulinum strains in light grey, the CRISPR/cas system of C. sporogenes ATCC 15579 in olive tones, and singletons identified for the latter strain in grey. Core genes are anthracite-colored.
The region of the neurotoxin gene cluster is flanked by genes coding for several hypothetical proteins, components of different ABC transporters, as well as a ferrous iron transport system and several regulatory proteins (data not shown). As shown in Figure 7, these flanking genes are present in the C. botulinum strains as well as in all C. sporogenes strains. This region might be an area of high genome plasticity, as in C. sporogenesATCC 15579 a subtype I-B/TNEAP CRISPR/cas system [92] is inserted, which could not be identified in the other strains used for this comparative approach.
Conclusions
Members of the non-toxic species C. sporogenes are closely related to neurotoxin producer C. botulinum. This study presents an overview of physiological, morphological, and genomic characteristics of the type strain C. sporogenesDSM 795. Detailed insight into its proteolytic metabolism was gained on genomic level. Also, the ability of C. sporogenes to produce solvents such as ethanol and butanol was linked to a set of genes and compared to other solventogenic clostridia. Genome comparison of C. sporogenesDSM 795 with two other sequenced strains of this species revealed high similarity. C. sporogenesDSM 795 was also compared at the genomic level with two strains of the close relative C. botulinum.
Acknowledgements
This work was supported by a personal grant (LGFG) to K.R. by the state of Baden-Württemberg, Germany and carried out in frame of the Cooperative Research Training Group “Pharmaceutical Biotechnology” (Kooperatives Promotionskolleg Pharmazeutische Biotechnologie) of the University of Ulm and the Biberach University of Applied Sciences.
We thank the “Bundesministerium für Ernährung, Landwirtschaft und Verbraucherschutz (BMELV)” for support (project REACTIF within program ERA-IB 3). We would like to thank Paul Walther (Central Facility for Electron Microscopy, University of Ulm) for generating the electron microscopic images. We thank Frauke-Dorothee Meyer and Kathleen Gollnow for technical support.
Additional files
Overview of all C. botulinum strains mentioned in this study [56].
Overview of enzymes, gene tags and locus tags of C. sporogenes DSM 795.
Footnotes
Anja Poehlein and Karin Riegel contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AP and RD planned the genome sequencing, AP did the genome sequencing, AP did the genome annotations, AP and KR prepared the figures, KR generated all microscopic images and the phylogenetic tree. SMK carried out the genomic DNA preparation. AP, KR and AL wrote the manuscript. PD conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Anja Poehlein, Email: apoehle3@gwdg.de.
Karin Riegel, Email: karin.riegel@uni-ulm.de.
Sandra M König, Email: sandra.m.koenig@gmail.com.
Andreas Leimbach, Email: andreas.leimbach@uni-wuerzburg.de.
Rolf Daniel, Email: rdaniel@gwdg.de.
Peter Dürre, Email: peter.duerre@uni-ulm.de.
References
- 1.Metchnikoff E. Sur les microbes de la putréfacion intestinale. C R Hebd Seances Acad Sci. 1908;147:579–82. [Google Scholar]
- 2.Metchnikoff E. Etude sur la flore intestinale. IV. Le Bacillus sporogenes. Annales de l’Institut Pasteur. 1908;22:942–6. [Google Scholar]
- 3.Heller HH. Certain genera of the Clostridiaceae. Studies in pathogenic anaerobes. V J Bacteriol. 1922;7:1–38. doi: 10.1128/jb.7.1.1-36.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LD. Common mesophilic anaerobes, including Clostridium botulinum and Clostridium tetani, in 21 soil specimens. Appl Microbiol. 1975;29:590–4. doi: 10.1128/am.29.5.590-594.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finne G, Matches JR. Low-temperature-growing clostridia from marine sediments. Can J Microbiol. 1974;20:1639–45. doi: 10.1139/m74-255. [DOI] [PubMed] [Google Scholar]
- 6.Matches JR, Liston J. Mesophilic clostridia in Puget Sound. Can J Microbiol. 1974;20:1–7. doi: 10.1139/m74-001. [DOI] [PubMed] [Google Scholar]
- 7.Molongoski JJ, Klug MJ. Characterization of anaerobic heterotrophic bacteria isolated from freshwater lake sediments. Appl Environ Microbiol. 1976;31:83–90. doi: 10.1128/aem.31.1.83-90.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergey DH, Harrison FC, Breed RS, Hammer BW, Huntoon FM. Bergey’s Manual of Determinative Bacteriology. 1. Baltimore: The Williams & Wilkins Co.; 1923. [Google Scholar]
- 9.Hutson RA, Thompson DE, Lawson PA, Schocken-Itturino RP, Böttger EC, Collins MD. Genetic interrelationships of proteolytic Clostridium botulinum types A, B, and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Ant v Leeuwenhoek. 1993/1994;64:273–83. [DOI] [PubMed]
- 10.Bradbury M, Greenfield P, Midgley D, Li D, Tran-Dinh N, Vriesekoop F, et al. Draft genome sequence of Clostridium sporogenes PA 3679, the common nontoxigenic surrogate for proteolytic Clostridium botulinum. J Bacteriol. 2012;194:1631–2. [DOI] [PMC free article] [PubMed]
- 11.Quattrocchi G. Rare case of pyo-gaseous abscess of the liver caused by Clostridium sporogenes-Bacterium pyocyaneum. Riforma Med. 1963;77:288–91. [PubMed]
- 12.Corbett CE, Wall BM, Cohen M. Case report: empyema with hydropneumothorax and bacteremia caused by Clostridium sporogenes. Am J Med Sci. 1996;312:242–5. [DOI] [PubMed]
- 13.Malmborg AS, Rylander M, Selander H. Primary thoracic empyema caused by Clostridium sporogenes. Scand J Infect Dis. 1970;2:155–6. [DOI] [PubMed]
- 14.Gorbach SL, Thadepalli H. Isolation of Clostridium in human infections: evaluation of 114 cases. J Infect Dis. 1975;131(Suppl):S81–5. [DOI] [PubMed]
- 15.Hitchcock CR, Demello FJ, Haglin JJ. Gangrene infection: new approaches to an old disease. Surg Clin North Am. 1975;55:1403–10. doi: 10.1016/s0039-6109(16)40800-5. [DOI] [PubMed] [Google Scholar]
- 16.Inkster T, Cordina C, Siegmeth A. Septic arthritis following anterior cruciate ligament reconstruction secondary to Clostridium sporogenes; a rare clinical pathogen. J Clin Pathol. 2011;64:820–1. [DOI] [PubMed]
- 17.Möse JR, Möse G. Oncolysis by clostridia. I. Activity of Clostr. butyricum (M 55) and other nonpathogenic clostridia against the Ehrlich-carcinoma. Cancer Res. 1964;24:212–6. [PubMed]
- 18.Minton NP, Brown JM, Lambin P, Anné J. Clostridia in cancer therapy. In: Bahl H, Dürre P, editors. Clostridia – biotechnology and medical applications. Weinheim: Wiley-VCH; 2001. p. 251–70.
- 19.Minton NP. Clostridia in cancer therapy. Nat Rev Microbiol. 2003;1:237–42. doi: 10.1038/nrmicro777. [DOI] [PubMed] [Google Scholar]
- 20.Liu SC, Minton NP, Giaccia AJ, Brown JM. Anticancer efficacy of systemically delivered anaerobic bacteria as gene therapy vectors targeting tumor hypoxia/necrosis. Gene Ther. 2002;9:291–6. doi: 10.1038/sj.gt.3301659. [DOI] [PubMed] [Google Scholar]
- 21.Lemmon MJ, van Zijl P, Fox ME, Mauchline ML, Giaccia AJ, Minton NP, et al. Anaerobic bacteria as a gene delivery system that is controlled by the tumor microenvironment. Gene Ther. 1997;4:791–6. doi: 10.1038/sj.gt.3300468. [DOI] [PubMed] [Google Scholar]
- 22.Lambin P, Theys J, Landuyt W, Rijken P, van der Kogel A, van der Schueren E, et al. Colonisation of Clostridium in the body is restricted to hypoxic and necrotic areas of tumours. Anaerobe. 1998;4:183–8. [DOI] [PubMed]
- 23.Theys J, Pennington O, Dubois L, Anlezark G, Vaughan T, Mengesha A, et al. Repeated cycles of Clostridium-directed enzyme prodrug therapy result in sustained antitumour effects in vivo. Br J Cancer. 2006;95:1212–9. [DOI] [PMC free article] [PubMed]
- 24.Liu SC, Ahn GO, Kioi M, Dorie MJ, Patterson AV, Brown JM. Optimized Clostridium-directed enzyme prodrug therapy improves the antitumor activity of the novel DNA cross-linking agent PR-104. Cancer Res. 2008;68:7995–8003. [DOI] [PMC free article] [PubMed]
- 25.Heap JT, Theys J, Ehsaan M, Kubiak AM, Dubois L, Paesmans K, et al. Spores of Clostridium engineered for clinical efficacy and safety cause regression and cure of tumors in vivo. Oncotarget. 2014;5:1761–9. [DOI] [PMC free article] [PubMed]
- 26.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. [DOI] [PMC free article] [PubMed]
- 27.Gibbons NE, Murray RGE. Proposals concerning the higher taxa of bacteria. Int J Syst Bacteriol. 1978;28:1–6. doi: 10.1099/00207713-28-1-1. [DOI] [Google Scholar]
- 28.Garrity GM, Holt JG. The road map to the manual. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey’s Manual of Systematic Bacteriology. 2. New York: Springer; 2001. pp. 119–69. [Google Scholar]
- 29.Murray RGE. The higher taxa, or, a place for everything…? In: Holt JG, editor. Bergey’s Manual of Systematic Bacteriology. 1. Baltimore: The Williams and Wilkins Co; 1984. pp. 31–4. [Google Scholar]
- 30.List Editor. List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol. 2010;60:469-72. [DOI] [PubMed]
- 31.Rainey FA. Class II. Clostridia class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, editors. Bergey’s Manual of Systematic Bacteriology, vol. 3. 2nd ed. New York: Springer; 2009. p. 736–1297.
- 32.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 33.Hauderoy P, Ehringer G, Guillot G, Magrou J, Prévot AR, Rosset D, et al. Dictionnaire des Bactéries Pathogènes. 2. Paris: Masson et Cie; 1953. [Google Scholar]
- 34.Pribram E. Klassifikation der Schizomyceten (Bakterien) Leipzig: Franz Deuticke; 1933. [Google Scholar]
- 35.Prazmowski A. Untersuchung über die Entwicklungsgeschichte und Fermentwirkung einiger Bakterien-Arten. PhD thesis. University of Leipzig; 1880. p. 366-71.
- 36.Smith LDS, Hobbs G. Genus III. Clostridium Prazmowski 1880, 23. In: Buchanan RE, Gibbons NE, editors. Bergey’s Manual of Determinative Bacteriology. 8th ed. Baltimore: The Williams and Wilkins Co; 1974. p. 551–72.
- 37.Judicial Commission of the International Committee on Systematic Bacteriology Rejection of Clostridium putrificum and conservation of Clostridium botulinum and Clostridium sporogenes-Opinion 69. Int J Syst Bacteriol. 1999;49:339. doi: 10.1099/00207713-49-1-339. [DOI] [PubMed] [Google Scholar]
- 38.Olsen I, Johnson JL, Moore LVH, Moore WEC. Rejection of Clostridium putrificum and conservation of Clostridium botulinum and Clostridium sporogenes. Request for an opinion. Int J Syst Bacteriol. 1995;45:414. [DOI] [PubMed]
- 39.Cato EP, George WL, Finegold SM. Genus Clostridium Prazmowski 1880, 23AL. 69. Clostridium sporogenes. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey’s manual of systematic bacteriology, vol. 2. 1st ed. Baltimore: The Williams and Wilkins Co; 1986. p. 1191–2.
- 40.Stickland LH. Studies in the metabolism of the strict anaerobes (genus Clostridium): the chemical reactions by which Cl. sporogenes obtains its energy. Biochem J. 1934;28:1746–59. [DOI] [PMC free article] [PubMed]
- 41.Stickland LH. Studies in the metabolism of the strict anaerobes (Genus Clostridium): the reduction of proline by Cl. sporogenes. Biochem J. 1935;29:288–90. [DOI] [PMC free article] [PubMed]
- 42.Stickland LH. Studies in the metabolism of the strict anaerobes (genus Clostridium) The oxidation of alanine by Cl. sporogenes. IV. The reduction of glycine by Cl. sporogenes. Biochem J. 1935;29:889–98. [DOI] [PMC free article] [PubMed]
- 43.Nisman B. The Stickland reaction. Bacteriol Rev. 1954;18:16–42. doi: 10.1128/br.18.1.16-42.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shreeve JE, Edwin EE. Thiaminase-producing strains of Cl. sporogenes associated with outbreaks of cerebrocortical necrosis. Vet Rec. 1974;94:330. doi: 10.1136/vr.94.15.330. [DOI] [PubMed] [Google Scholar]
- 45.Balish E, Cleven D, Brown J, Yale CE. Nose, throat, and fecal flora of beagle dogs housed in “locked” or “open” environments. Appl Environ Microbiol. 1977;34:207–21. doi: 10.1128/aem.34.2.207-221.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lovitt RW, Kell DB, Morris JG. The physiology of Clostridium sporogenes NCIB 8053 growing in defined media. J Appl Bacteriol. 1987;62:81–92. [DOI] [PubMed]
- 48.Fildes P, Richardson GM. The amino-acids necessary for the growth of Cl. sporogenes. Br J Exp Pathol. 1935;16:326–35. [Google Scholar]
- 49.Schull GM, Peterson WH. The nature of the sporogenes vitamin and other factors in the nutrition of Clostridium sporogenes. Arch Biochem. 1948;18:69–83. [PubMed]
- 50.Schull GM, Thoma RW, Peterson WH. Amino acid and unsaturated fatty acid requirements of Clostridium sporogenes. Arch Biochem. 1949;20:227–41. [PubMed]
- 51.Campbell LL, Jr, Frank HA. Nutritional requirements of some putrefactive anaerobic bacteria. J Bacteriol. 1956;71:267–9. doi: 10.1128/jb.71.3.267-269.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovitt RW, Morris JG, Kell DB. The growth and nutrition of Clostridium sporogenes NCIB 8053 in defined media. J Appl Bacteriol. 1987;62:71–80. [DOI] [PubMed]
- 53.Monticello DJ, Costilow RN. Interconversion of valine and leucine by Clostridium sporogenes. J Bacteriol. 1982;152:946–9. [DOI] [PMC free article] [PubMed]
- 54.Monticello DJ, Hadioetomo RS, Costilow RN. Isoleucine synthesis by Clostridium sporogenes from propionate or alpha-methylbutyrate. J Gen Microbiol. 1984;130:309–18. [DOI] [PubMed]
- 55.Costilow RN. Selenium requirement for the growth of Clostridium sporogenes with glycine as the oxidant in Stickland reaction systems. J Bacteriol. 1977;131:366–8. [DOI] [PMC free article] [PubMed]
- 56.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, et al. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J Bacteriol. 2007;189:818–32. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peck MW, Stringer SC, Carter AT. Clostridium botulinum in the post-genomic era. Food Microbiol. 2011;28:183–91. [DOI] [PubMed]
- 58.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez DJ, Lee SW, Hensler ME, Markley AL, Dahesh S, Mitchell DA, et al. Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J Biol Chem. 2010;285:28220–8. [DOI] [PMC free article] [PubMed]
- 61.Oultram JD, Loughlin M, Swinfeld TJ, Brehm JK, Thomson DE, Minton NP. Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol Lett. 1988;56:83–8. doi: 10.1111/j.1574-6968.1988.tb03154.x. [DOI] [Google Scholar]
- 62.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–10. [PubMed]
- 63.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc German Conf Bioinformatics. 1999;99:45–56. [Google Scholar]
- 64.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–8. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 66.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucl Acids Res. 2012;40:D115–22. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–30. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 68.Tech M, Merkl R. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 2003;3:441–51. [PubMed] [Google Scholar]
- 69.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zdobnov EM, Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 72.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 73.Fonknechten N, Chaussonnerie S, Tricot S, Lajus A, Andreesen JR, Perchat N, et al. Clostridium sticklandii, a specialist in amino acid degradation: revisiting its metabolism through its genome sequence. BMC Genomics. 2010;11:555. [DOI] [PMC free article] [PubMed]
- 74.Bednarski B, Andreesen J, Pich A. In vitro processing of the proproteins GrdE of protein B of glycine reductase and PrdA of D-proline reductase from Clostridium sticklandii: formation of a pyruvoyl group from a cysteine residue. Eur J Biochem. 2001;268:3538–44. [DOI] [PubMed]
- 75.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poehlein A, Gottschalk G, Daniel R. First insights into the genome of the Gram-negative, endospore-forming organism Sporomusa ovata strain H1 DSM 2662. Genome Announc. 2013;1:e00734–13. [DOI] [PMC free article] [PubMed]
- 77.Poehlein A, Andreesen JR, Daniel R. Complete genome sequence of amino acid-utilizing Eubacterium acidaminophilum al-2 (DSM 3953). Genome Announc. 2014;2:e00573–14. [DOI] [PMC free article] [PubMed]
- 78.Gursinsky T, Jäger J, Andreesen JR, Söhling B. A selDABC cluster for selenocysteine incorporation in Eubacterium acidaminophilum. Arch Microbiol. 2000;174:200–12. [DOI] [PubMed]
- 79.Turton LJ, Drucker DB, Ganguly LA. Effect of glucose concentration in the growth medium upon neutral and acidic fermentation end-products of Clostridium bifermentans, Clostridium sporogenes, Peptostreptococcus anaerobius. J Med Microbiol. 1983;16:61–7. [DOI] [PubMed]
- 80.Leja K, Czaczyk K, Myszka K. Biotechnological synthesis of 1,3-propanediol using Clostridium sp. Afr J Biotechnol. 2011;10:11093–101.
- 81.Chen CK, Blaschek HP. Effect of acetate on molecular and physiological aspects of Clostridium beijerinckii NCIMB 8052 solvent production and strain degeneration. Appl Environ Microbiol. 1999;65:499–505. [DOI] [PMC free article] [PubMed]
- 82.Kosaka T, Nakayama S, Nakaya K, Yoshino S, Furukawa K. Characterization of the sol operon in butanol-hyperproducing Clostridium saccharoperbutylacetonicum strain N1-4 and its degeneration mechanism. Biosci Biotechnol Biochem. 2007;71:58–68. [DOI] [PubMed]
- 83.Fischer RJ, Helms J, Dürre P. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol. 1993;175:6959–69. [DOI] [PMC free article] [PubMed]
- 84.Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–38. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poehlein A, Hartwich K, Krabben P, Ehrenreich A, Liebl W, Dürre P, et al. Complete genome sequence of the solvent producer Clostridium saccharobutylicum NCP262 (DSM 13864). Genome Announc. 2013;1:e00997–13. [DOI] [PMC free article] [PubMed]
- 86.Poehlein A, Krabben P, Dürre P, Daniel R. Complete genome sequence of the solvent producer Clostridium saccharoperbutylacetonicum strain DSM 14923. Genome Announc. 2014;2:e01056–14. [DOI] [PMC free article] [PubMed]
- 87.Collins MD, East AK. Phylogeny and taxonomy of the foodborne pathogen Clostridium botulinum and its neurotoxins. J Appl Microbiol. 1998;84:5–17. [DOI] [PubMed]
- 88.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, et al. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–70. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 89.Lindstrom M, Korkeala H. Laboratory diagnostics of botulism. Clin Microbiol Rev. 2006;19:298–314. doi: 10.1128/CMR.19.2.298-314.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leimbach A. bac-genomics-scripts. [https://github.com/aleimba/bac-genomics-scripts.]
- 91.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]


