Abstract
Interleukin 17 (IL-17) is an important pro-inflammatory cytokine and plays critical roles in the immune response to pathogens and in the pathogenesis of inflammatory and autoimmune diseases. Despite its important functions, the origin and evolution of IL-17 in animal phyla have not been characterized. As determined in this study, the distribution of the IL-17 family among 10 invertebrate species and 7 vertebrate species suggests that the IL-17 gene may have originated from Nematoda but is absent from Saccoglossus kowalevskii (Hemichordata) and Insecta. Moreover, the gene number, protein length and domain number of IL-17 differ widely. A comparison of IL-17-containing domains and conserved motifs indicated somewhat low amino acid sequence similarity but high conservation at the motif level, although some motifs were lost in certain species. The third disulfide bond for the cystine knot fold is formed by two cysteine residues in invertebrates, but these have been replaced by two serine residues in Chordata and vertebrates. One third of invertebrate IL-17 proteins were found to have no predicted signal peptide. Furthermore, an analysis of phylogenetic trees and exon–intron structures indicated that the IL-17 family lacks conservation and displays high divergence. These results suggest that invertebrate IL-17 proteins have undergone complex differentiation and that their members may have developed novel functions during evolution.
Introduction
Interleukin 17 (IL-17) is an important pro-inflammatory cytokine and is a critical component of the immune response to pathogens and in the pathogenesis of inflammatory and autoimmune diseases [1–3]. IL-17 was initially identified as a cytokine secreted by T helper 17 (TH17) cells as one of its signature cytokines, and recent findings have indicated that IL-17 is also produced by other cell types, particularly by the innate immune cell populations involved in the inflammatory process [4]. IL-17 was first cloned and identified as cytotoxic T-lymphocyte (CTL)-associated antigen 8 (CTLA-8), a T-cell-derived cytokine with 58% identity to predicted open reading frame 13, HSVS13, of the T-lymphotropic Herpesvirus saimiri (known as virus IL-17) [5, 6]. Six IL-17 family members, IL-17A (the original IL-17), IL-17B, IL-17C, IL-17D, IL-17E (also known as IL-25) and IL-17F, have since been identified, and these proteins range in size from 20 to 30 kDa [7]. Among these family members, IL-17A and IL-17F share the highest amino acid sequence identity (50%), whereas IL-17E is the most divergent, showing 16% identity with IL-17A. Moreover, a novel type of IL-17 family gene (IL-17N) has recently been identified in teleosts [8]. Amino acid similarity among the family members is higher in the C terminus and in five spatially conserved cysteine residues, four of which form a cystine knot fold that forms two intrachain disulfide bonds. This cystine knot fold is similar to the canonical cystine knot observed in growth factors such as transforming growth factor (TGF)-β, endocrine glycoprotein hormones (e.g. chorionic gonadotrophin), platelet-derived growth factors (PDGFs), nerve growth factor (NGF) and other neurotrophins with six cysteines rather than four [9, 10].
Among the IL-17 family members, IL-17A and IL-17F are the best characterized, followed by IL-17C and IL-17E, while IL-17B and IL-17D have remained understudied [1]. Mechanistically, the biologically active form of IL-17 is a 35-kDa homodimer or heterodimer whose activity is dependent on the single-pass transmembrane receptors, IL-17 receptors (IL-17Rs), which have several conserved structural features, including an extracellular fibronectin III-like domain and a cytoplasmic SEF (similar expression to FGF)/IL-17R (SEFIR) domain. The IL-17Rs, as well as the cognate IL-17 family, have little homology with any other known receptors or ligands and therefore are thought to represent a distinct ligand–receptor signaling system that is highly conserved across vertebrate evolution. However, the exact mechanisms of IL-17 signaling have not been fully elucidated [1, 11].
Despite an accumulation of knowledge of the functions of IL-17 and their regulatory pathways, the number of pathways involving the IL-17 family remains unclear [1, 2, 12]. Some members of the IL-17 family are highly conserved among vertebrate organisms, but evolutionary analysis of the family has mainly been limited to vertebrates and a handful of invertebrates [8, 13], and little is known about its origin and evolution in animal phyla. For example, given that homology among IL-17 family members is only 16–50%, perhaps the IL-17A-like genes in some phyla may be too dissimilar to be identified but, interestingly, IL-17D has shown some degree of homology with IL-17-like proteins in primitive phyla such as worms [4]. The identification of similarities and differences in the IL-17 family among animal phyla, particularly invertebrates, could facilitate the elucidation of the functional evolution of this family, as well as allowing further functional verification. The recent large-scale sequencing of the transcriptomes and genomes of invertebrate species [14], particularly non-model organisms [15–17], represents a global survey that can be used to investigate IL-17 family members. For instance, in the purple sea urchin, about thirty IL-17 genes and two receptor genes were identified. Many of the ligands are linked in tandem arrays [18]. In this study, we determined the distribution of the IL-17 family among invertebrates, analyzed their exon–intron structures and phylogenetic trees, and explored their origin and evolutionary history in animal phyla.
Materials and Methods
Ethics statement
No specific permits were required for the field studies described, and the field studies did not involve endangered or protected species.
Databases
The databases used in this study were obtained primarily from the National Center for Biotechnology Information (NCBI) Assembled RefSeq Genomes (http://www.ncbi.nlm.nih.gov/mapview/) and the DOE Joint Genome Institute (JGI) (http://genome.jgi.doe.gov/) websites. Nematostella vectensis (Cnidaria), Caenorhabditis briggsae (Nematoda), Capitella teleta (Annelida), Helobdella robusta (Annelida), Lottia gigantea (Mollusca), Daphnia pulex (Arthropoda), Trichoplax adhaerens (Placozoa) and Branchiostoma floridae (Chordata) from JGI and Amphimedon queenslandica (Porifera/Spongia), Hydra magnipapillata (Cnidaria), Caenorhabditis elegans (Nematoda), Saccoglossus kowalevskii (Hemichordata), Acyrthosiphon pisum (Insecta of Arthropoda, same as below), Apis mellifera (Insecta), Drosophila melanogaster (Insecta), Ciona intestinalis (Chordata) and Protozoa from NCBI Assembled RefSeq Genomes were individually analyzed by BLASTP. For Strongylocentrotus purpuratus (Echinodermata), BLASTP was performed utilizing both the NCBI Assembled RefSeq Genomes and Sea Urchin Genome Database (http://www.spbase.org/SpBase/) datasets. The protein data for Crassostrea gigas (Mollusca) were downloaded from NCBI, and a local BLAST protein database was constructed for the BLAST search. For Pinctada fucata (Mollusca), BLASTP was run on Pinctada fucata Genome Ver. 1.00 (http://marinegenomics.oist.jp/genomes/ncbiblast/search?project_id=20). BLASTP analysis was conducted on vertebrate taxa including Danio rerio, Oryzias latipes, Gallus gallus, Homo sapiens, and Xenopus (Silurana) tropicalis from NCBI Assembled RefSeq Genomes. For Takifugu rubripes, sequences reported by Hiroki Korenaga et al. [8] were used.
Identification of IL-17 genes
BLAST searching methods were used to identify IL-17 proteins. The amino acid sequences of the IL-17 domain previously identified in P. fucata IL-17 (JX971444) and C. gigas IL-17 (ABO93467) were used as query sequences to BLAST against the protein database of each genome for the species mentioned above [19, 20]. The threshold E-value was set to range from 3 to 10 with 50 maximum target sequences, to identify a maximal number of candidate sequences, and other parameters were left at the default values. After the corresponding hits were downloaded from the BLAST results, the sequences were examined using the NCBI CDS program (Batch CD-search, http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) with default cutoff parameters to remove sequences that did not contain the IL-17 domain. For the maximum target sequences obtained, the hit sequence with the maximum numeric E-value was used as the query sequence to BLAST against the protein database of the corresponding species. The sequences from the genome of each species were analyzed independently using Clustal Omega Multiple Sequence Alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/) to eliminate redundant sequences. To simplify the presentation and subsequent discussion, the longest isoform sequence was retained, and other isoforms were removed. For incomplete sequences containing the complete IL-17 domain, only the longest sequence was retained.
Sequence analysis and amino acid alignment
Batch CD-search was used to analyze the domain among the IL-17 protein sequences identified, and MEME 4.9.1 (Motif-based sequence analysis tools, http://meme.nbcr.net/meme/) was used to identify motifs in the IL-17 protein sequences. Signal peptides were predicted using SignalP 4.1 [21]. Comparison and phylogenetic analysis were performed using Clustal Omega multiple sequence alignment and the MEGA 6.06 software using neighbor-joining (NJ) methods and performing 10,000 bootstrap replications [22].
Exon-intron structure and location of IL-17 genes
For the IL-17 amino acid sequences, the corresponding nuclear sequences, including the EST and genomic sequences, were obtained. Spidey, an mRNA-to-genomic alignment program (http://www.ncbi.nlm.nih.gov/spidey/), was used to analyze exon-intron structures. Owing to the use of draft genomes, some IL-17 exon–intron structures were not available. Meanwhile, the genomic location of IL-17 genes were analyzed, using the NCBI mapview browsers.
Results
Genome-wide identification of IL-17 genes from invertebrates
By performing BLAST searches of databases encompassing a wide spectrum of organisms from Protozoa to B. floridae, a total of 54 putative IL-17 genes were identified from 10 invertebrate genomes: C. briggsae (1) (Nematoda), C. elegans (2) (Nematoda), C. teleta (6) (Annelida), L. gigantean (6) (Mollusca), C. gigas (7) (Mollusca), P. fucata (12) (Mollusca), D. pulex (1) (Arthropoda), S. purpuratus (6) (Echinodermata), C. intestinalis (5) (Chordata) and B. floridae (8) (Chordata). Detailed information on the IL-17 genes identified from each genome surveyed is listed in Table 1 and the protein sequences in S1 Dataset. IL-17 homologs could not be identified in Porifera (A. queenslandica), Cnidaria (H. magnipapillata and N. vectensis), Placozoa (T. adhaerens), Hemichordata (S. kowalevskii), Insecta (such as A. pisum, A. mellifera and D. melanogaster), and Protozoa. For comparison, IL-17 homologs from vertebrates including D. rerio (5), T. rubripes (6), Oryzias latipes (6), G. gallus (4), H. sapiens (6) and X. tropicalis (7) are also listed in Table 1. These results provide a concise picture of the IL-17 gene distribution, and reveal that the IL-17 gene arose not in a lower invertebrate such as Porifera and Cnidaria but in Nematoda species such as C. briggsae and C. elegans; it subsequently emerged in some mollusks. Not all arthropods exhibit IL-17 gene loss; D. pulex contains a sequence homologous to IL-17. However, no IL-17 homologous sequence has been identified in other arthropods. These results suggest that IL-17 genes may have originated from Nematoda.
Table 1. Summary of IL-17 genes.
| Classes | Species and putative genes name | Length | Location of CDS | E-Value | Signal peptide | Genomic location | Number of intron |
|---|---|---|---|---|---|---|---|
| Nematoda | C.briggsae XP_002637129.1 protein CBG09631 | 145 | 48–126 | 0.000000472 | / | chromosome V | 0 |
| C.elegans NP_505700.2 Protein F25D1.3 | 189 | 92–170 | 3.21E-08 | Yes | chromosome V | 2 | |
| C.elegans NP_510131.2 Protein T22H6.1 | 221 | 122–195 | 0.00502391 | NO | chromosome X | 6 | |
| Annelida | C.teleta 199819 | 233 | 150–226 | 2.19E-08 | NO | scaffold_169 | 5 |
| C.teleta 198235 | 366 | 286–360 | 0.000000032 | Yes | scaffold_451 | 1 | |
| C.teleta 206957 | 202 | 112–195 | 4.51E-09 | Yes | scaffold_22 | 1 | |
| C.teleta 216301 | 180 | 91–169 | 0.000216745 | Yes | scaffold_314 | 0 | |
| C.teleta 205055 | 166 | 81–157 | 0.0000268 | NO | scaffold_79 | 1 | |
| C.teleta 209749 | 348 | 260–338 | 0.000489314 | NO | scaffold_52 | 4 | |
| 237–278 | 0.00378139 a | ||||||
| Mollusca | L.gigantea 152638 | 187 | 97–173 | 0.000000148 | Yes | scaffold_2 | 0 |
| L.gigantea 169526 | 191 | 98–174 | 0.00246412 | Yes | scaffold_86 | 1 | |
| L.gigantea 164174 | 180 | 91–179 | 0.00000767 | Yes | scaffold_44 | 2 | |
| L.gigantea 228210 | 191 | 105–183 | 2.85E-11 | Yes | scaffold_2 | 0 | |
| L.gigantea 172928 | 154 | 71–144 | 0.000000264 | Yes | scaffold_144 | 0 | |
| L.gigantea 159302 | 186 | 94–169 | 0.00000077 | Yes | scaffold_20 | 1 | |
| C.gigas ABO93467.1 IL-17 | 200 | 96–179 | 3.88E-13 | Yes | /* | ||
| C.gigas EKC33705.1 protein CGI_10020734 | 190 | 100–170 | 9.85E-09 | Yes | scaffold_698 | 2 | |
| C.gigas EKC26195.1 protein CGI_10027182 | 167 | 81–161 | 2.1E-10 | Yes | scaffold_1599 | 2 | |
| C.gigas EKC33786.1 protein CGI_10014828 | 141 | 67–131 | 0.000172218 | NO | scaffold_689 | 1 | |
| C.gigas EKC38792.1 protein CGI_10026592 | 132 | 46–124 | 0.000000893 | NO | scaffold_313 | 0 | |
| C.gigas EKC33462.1 proteinGI_10015251 | 148 | 73–144 | 2.97E-11 | Yes | scaffold_723 | 1 | |
| C.gigas EKC32654.1 protein CGI_10004922 | 132 | 53–128 | 0.000525737 | Yes | scaffold_806 | 0 | |
| P.fucata 1712.1_51392.t1 | 208 | 121–196 | 1.06E-13 | NO | scaffold_1712.1 | 1 | |
| P.fucata 1712.1_51391.t1 | 158 | 62–137 | 0.0000021 | NO | scaffold_1712.1 | 2 | |
| P.fucata 1712.1_51394.t1 | 169 | 90–160 | 0.00000792 | / | scaffold_1712.1 | 0 | |
| P.fucata 20923.1_18751.t1 | 261 | 29–91 | 0.000000303 | / | scaffold_20923.1 | 1 | |
| P.fucata 24776.1_26199.t1 | 165 | 71–152 | 6E-16 | / | scaffold_24776.1 | 0 | |
| P.fucata 27731.1_19195.t1 | 189 | 96–170 | 4.92E-10 | / | scaffold_27731.1 | 1 | |
| P.fucata 27889.1_19207.t1 | 165 | 76–151 | 0.00161047 | NO | scaffold_27889.1 | 1 | |
| P.fucata 32457.1_48078.t1 | 146 | 65–138 | 9.08E-11 | NO | scaffold_32457.1 | 0 | |
| P.fucata 204780.1_72074.t1 | 145 | 64–137 | 0.000000303 | Yes | scaffold_204780.1 | 0 | |
| P.fucata 8564.1_24423.t1 | 96 | 2–75 | 7.74E-08 | NO | scaffold_0_8564.1 | 0 | |
| P.fucata 8564.1_24422.t1 | 206 | 103–174 | 0.0000643 | NO | scaffold_0_8564.1 | 0 | |
| P.fucata JX971444.1 IL-17 | 194 | 91–171 | 0.00000608 | Yes | scaffold_9999.1 | 1 | |
| Arthropoda | D.pulex 125692 | 233 | 156–225 | 0.00858264 | NO | scaffold_13069 | 0 |
| Echinodermata | S.purpuratus SPU_019350.1 | 537 | 454–532 | 5.37E-16 | Yes | scaffold_1105 | 2 |
| 279–351 | 2.25E-13 | ||||||
| 104–176 | 2.9E-12 | ||||||
| S.purpuratus SPU_022838.1 | 379 | 272–354 | 5.68E-16 | NO | scaffold_1325 | 4 | |
| 110–181 | 4.18E-11 | ||||||
| S.purpuratus SPU_030196.1 | 204 | 113–194 | 1.02E-14 | Yes | scaffold_2038 | 2 | |
| S.purpuratus SPU_030199.1 | 236 | 153–232 | 5.26E-13 | / | scaffold_1105 | 1 | |
| S.purpuratus SPU_030204.1 | 215 | 122–209 | 7.92E-10 | NO | scaffold_2240 | 2 | |
| S.purpuratus SPU_030198.1 | 344 | 256–338 | 1.46E-10 | Yes | scaffold_2038 | 2 | |
| Chordata | C.intestinalis XP_004227512.1 IL-17D-like | 179 | 96–173 | 8.57E-14 | Yes | unplaced scaffold | 4 |
| C.intestinalis NP_001123348.1 IL-17-3 | 186 | 103–180 | 1.72E-12 | NO | chromosome 1 | 2 | |
| C.intestinalis NP_001123346.1 IL-17-2 | 171 | 92–169 | 5.15E-30 | Yes | chromosome 1 | 2 | |
| C.intestinalis NP_001123347.1 IL-17-1 | 171 | 86–164 | 8.46E-31 | Yes | chromosome 1 | 2 | |
| C.intestinalis 203738 | 199 | 117–196 | 1.03E-12 | Yes | chromosome 1 | 2 | |
| B.floridae 91950 | 434 | 347–428 | 1.18E-24 | NO | scaffold_216 | 0 | |
| B.floridae 117645 | 177 | 93–174 | 0.00000461 | Yes | scaffold_6 | 1 | |
| B.floridae 230778 | 93 | 3–84 | 8.51E-27 | NO | scaffold_275 | 1 | |
| B.floridae 127768 | 199 | 114–194 | 9.77E-26 | Yes | scaffold_275 | 3 | |
| B.floridae 92872 | 177 | 93–171 | 5.24E-10 | Yes | scaffold_229 | 1 | |
| B.floridae 132638 | 470 | 326–435 | 6.28E-49 b | Yes | scaffold_746 | 9 | |
| 99–173 | 5.31E-14 c | ||||||
| 438–467 | 1.89E-09 | ||||||
| B.floridae 94821 | 254 | 169–248 | 0.00000556 | Yes | scaffold_258 | 2 | |
| B.floridae 66165 | 151 | 69–144 | 0.000114888 | Yes | scaffold_9 | 1 | |
| Vertebrate | D.rerio XM_002666436.1 protein LOC100329556 | 126 | 52–125 | 0.000000225 | Yes | chromosome 23 | 2 |
| D.rerio NP_001018634.1 IL-17a/f2 | 140 | 58–136 | 4.14E-21 | Yes | chromosome 17 | 2 | |
| D.rerio NP_001018625.1 IL-17D | 212 | 96–178 | 4.35E-40 | Yes | chromosome 9 | 1 | |
| D.rerio NP_001018623.1 IL-17a/f1 | 153 | 67–147 | 3.4E-35 | Yes | chromosome 17 | 2 | |
| D.rerio NP_001018626.1 IL-17a/f3 | 162 | 73–155 | 4.1E-33 | Yes | chromosome 20 | 2 | |
| T.rubripes BAI82582.2 IL-17C-2 | 160 | 73–154 | 6.57E-24 | Yes | chromosome 13 | 2 | |
| T.rubripes BAI82581.2 IL-17C-1 | 161 | 81–156 | 5.99E-17 | Yes | scaffold_430 | 2 | |
| T.rubripes BAI82580.1 IL-17A/F-3 | 158 | 58–151 | 3.32E-21 | Yes | chromosome 16 | 3 | |
| T.rubripes BAI82579.1 IL-17A/F-2 | 144 | 58–138 | 2.01E-19 | Yes | chromosome 13 | 2 | |
| T.rubripes BAI82578.1 IL-17A/F-1 | 160 | 72–153 | 3.17E-24 | Yes | chromosome 13 | 2 | |
| T.rubripes BAI82584.1 IL-17N | 139 | 55–133 | 8.57E-13 | Yes | scaffold_264 | 2 | |
| O.latipes NP_001191715.1 IL-17A/F-3 | 157 | 60–150 | 8.62E-22 | Yes | chromosome 24 | 3 | |
| O.latipes NP_001191713.1 IL-17A/F-2 | 142 | 56–135 | 5.08E-22 | Yes | ultracontig 46 | 2 | |
| O.latipes NP_001191714.1 IL-17A/F-1 | 152 | 66–144 | 3.49E-24 | Yes | ultracontig 46 | 2 | |
| O.latipes NP_001191716.1 IL-17D | 211 | 97–179 | 1.29E-36 | Yes | chromosome 21 | 1 | |
| O.latipes NP_001191723.1 IL-17C | 165 | 72–162 | 1.08E-16 | Yes | chromosome 6 | 2 | |
| O.latipes NP_001191717.1 IL-17N | 139 | 55–134 | 2.65E-12 | Yes | chromosome 9 | 2 | |
| G.gallus XP_003641993.2 IL-17C | 188 | 97–183 | 3.87E-33 | Yes | chromosome 11 | 2 | |
| G.gallus XP_426223.4 IL-17F | 169 | 82–160 | 1.94E-33 | Yes | chromosome 3 | 2 | |
| G.gallus NP_989791.1 IL-17F precursor | 169 | 82–162 | 1.68E-34 | Yes | chromosome 3 | 2 | |
| G.gallus XP_004944893.1 IL-17B isoform X4 | 243 | 159–243 | 4.62E-24 | Yes | chromosome 13 | 2 | |
| X.tropicalis NP_001107719.1 IL-17D | 204 | 89–170 | 1.05E-42 | Yes | scaffold_2 | 2 | |
| X.tropicalis XP_004915038.1 IL-17A-like | 160 | 61–140 | 3.29E-32 | Yes | scaffold_5b | 2 | |
| X.tropicalis XP_002942041.2 IL-17C | 198 | 111–194 | 9E-35 | Yes | scaffold_4 | 2 | |
| X.tropicalis XP_002932904.1 IL-17D-like | 156 | 73–153 | 7.37E-23 | Yes | scaffold_10 | 2 | |
| X.tropicalis XP_004915036.1 IL-17F | 185 | 97–177 | 5.61E-33 | NO | scaffold_5b | 2 | |
| X.tropicalis NP_001006699.1 IL-17B | 203 | 115–203 | 1.8E-40 | Yes | scaffold_3 | 2 | |
| X.tropicalis XP_004915037.1 IL-17A-like | 149 | 62–140 | 3.71E-30 | Yes | scaffold_5b | 2 | |
| H.sapiens NP_612141.1 IL-17D | 202 | 89–171 | 9.32E-45 | Yes | chromosome 13 | 2 | |
| H.sapiens NP_443104.1 IL-17F | 163 | 76–156 | 6.29E-40 | Yes | chromosome 6 | 2 | |
| H.sapiens NP_055258.1 IL-17B | 180 | 95–180 | 1.16E-33 | Yes | chromosome 5 | 2 | |
| H.sapiens NP_002181.1 IL-17A | 155 | 68–148 | 9.03E-43 | Yes | chromosome 6 | 2 | |
| H.sapiens NP_037410.1 IL-17C | 197 | 103–193 | 1.4E-37 | Yes | chromosome 16 | 2 | |
| H.sapiens NP_073626.1 IL-25 isoform 1 | 177 | 84–172 | 4.18E-26 | Yes | chromosome 14 | 1 |
Syntenic loci in the species was indicated in bold
The extra domains besides IL-17 domain:
a. YccV-like domain;
b. RPA2b aaRSs OBFlike domain;
c. classII aaRS like core domain
Furthermore, this table indicates that the length of invertebrate IL-17 proteins generally ranges from 100 to 250 amino acids but fluctuates greatly when compared with vertebrate homologs. More drastic changes are found at the EST or exon sequence level, ranging from a few hundred to about two thousand base pairs, although the sequence data of some species mentioned above are insufficient or contain errors. The IL-17 domains generally contain approximately 70 amino acids and are located in the C-terminal region of the sequences. Interestingly, there are some exceptions: 1) three IL-17 superfamily domains with repetitive protein sequences in S. purpuratus SPU_019350.1; 2) two IL-17 superfamily domains with different protein sequences in S. purpuratus SPU_022838.1; 3) one IL-17 domain that partially overlaps with an incomplete YccV-like superfamily domain in C. teleta 209749; and 4) multi-domains with the N-terminal anticodon recognition domain of lysyl-tRNA synthetases (LysRS_N), the IL-17 superfamily, incomplete lysyl-tRNA synthetases, and the Class II tRNA amino-acyl synthetase-like catalytic core domain (LysRS_core) in B. floridae 132638. In addition, some IL-17 proteins, including C. elegans protein C44B12.6, isoform a (CDH93392.1), P. fucata 8548.1_09780.t1 and S. purpuratus SPU_030197.1, contain incomplete IL-17 domains and are listed in S1 Dataset but not Table 1. These results suggest that IL-17 protein sequences have undergone rapid and continual changes which may have led to a change in their function.
Conserved residues and motifs in IL-17 proteins
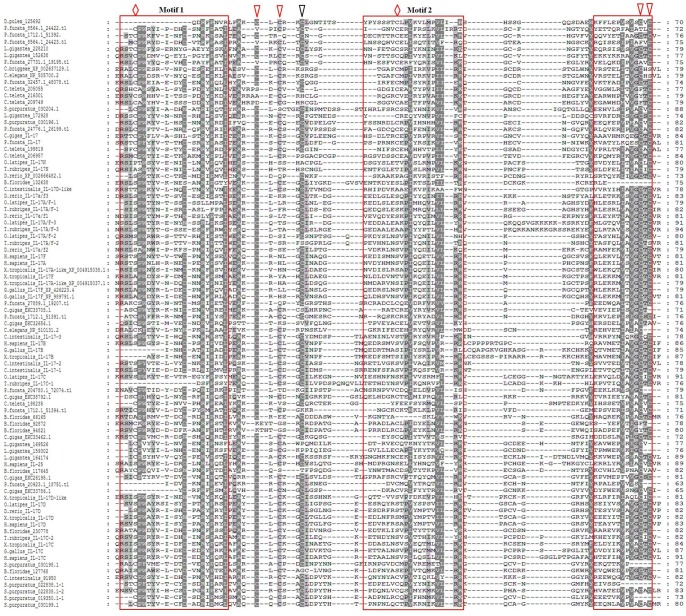
To clarify the relationships among IL-17 proteins from different species, multiple alignment analysis of the IL-17 domains was performed using Clustal Omega. The results indicated that the distribution of amino acid residues is not conserved in IL-17 domains, as illustrated in Fig 1, or in full-length invertebrate IL-17 proteins (data not shown). However, five cysteine residues (marked with arrows) were basically conserved, four (red arrows) of which are important for the cystine knot fold. Remarkably, there is a third disulfide bond for the cystine knot fold that is formed by the two cysteine residues in invertebrates, except for Chordata (B. floridae and C. intestinalis), in which the cysteine residues have been replaced by two serine residues (red rhombus).
Fig 1. Clustal Omega amino acid sequence alignment and three conserved motifs of 89 IL-17 domains.
The shading of the alignment represents different degrees of conservation among sequences. The dark shading indicates identical residues. Arrows indicate the positions of cysteine residues. A rhombus indicates the positions in which some cysteine residues have been replaced by serine residues.
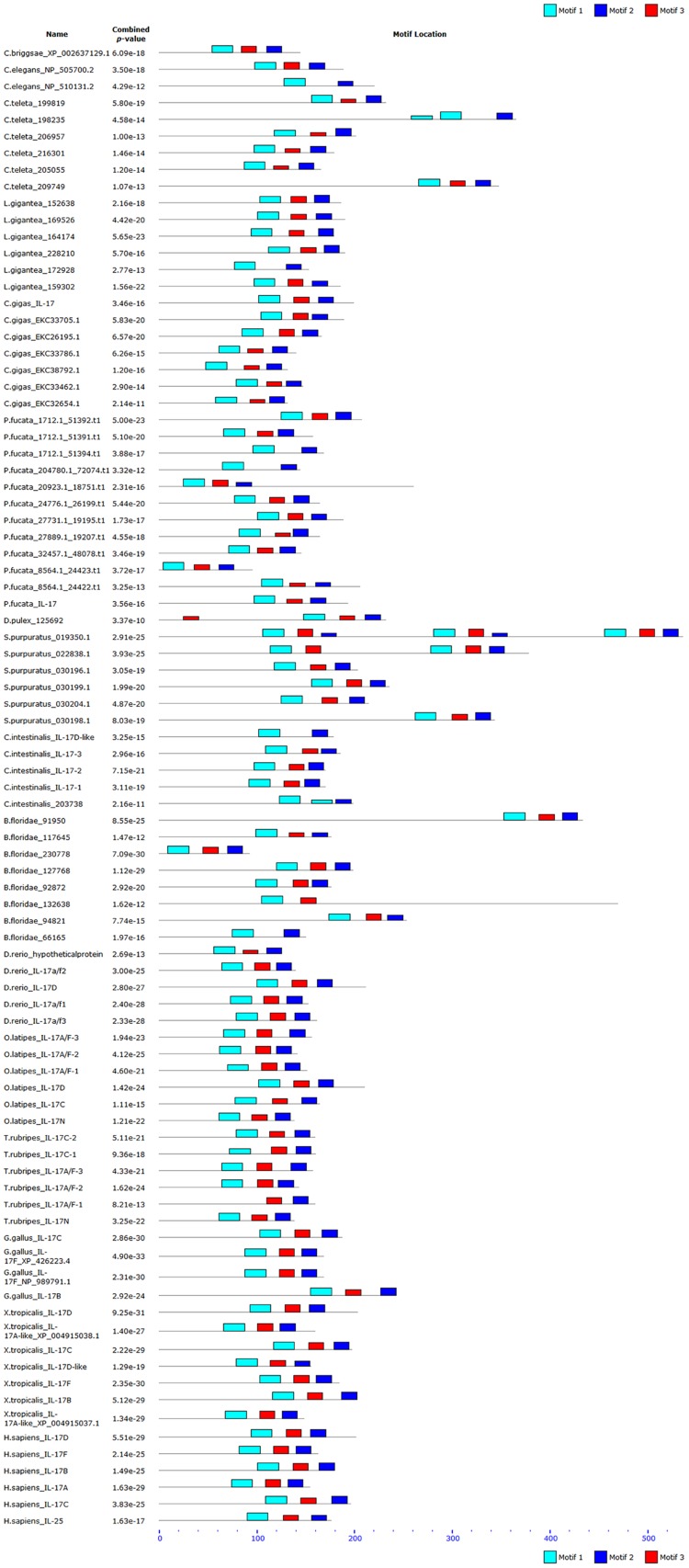
MEME was performed to discover conserved motifs within the IL-17 proteins and IL-17 domains. The sequences of the motifs in IL-17 domains are presented in Fig 2 and the combined motif block diagrams are shown in S1 Fig. From Fig 2, it can be observed that all 89 predicted IL-17 protein sequences contain the following three motifs: motif 1 (xY[VR]I[ND]xDPNR[IYF]Pxx[IL]xEA[RK]CL), motif 2 (YExxxEx[VI][APT]V[GA]CTC[VA]) and motif 3 (LN[SC]VP[IV]YQxILVLR[RK]). Similar motifs (including sequence logos) were observed in the IL-17 domains (S1B Fig); only the motif name is different, when compared with that of the full-length IL-17 proteins. Furthermore, as shown in Fig 2, motif 1 was only absent from T. rubripes IL-17A/F-1, and motif 2 was absent from the N-terminus of the IL-17 domain in S. purpuratus SPU_022838.1 and B. floridae 132638. Motif 3 was absent from C. elegans NP_510131.2, C. teleta 192928, P. fucata 1712.1_51394.t1 and 204780.1_72074.t1, C. intestinalis IL-17D-like and 203738, and B. floridae 66165. In addition, a comparison of the motifs in IL-17 proteins and IL-17 domains indicated that these motifs were primarily located in IL-17 domains, suggesting that, although the amino acid sequence identity of IL-17 proteins is rather low, they exhibit greater conservation at the motif level.
Fig 2. Combined block diagrams of three conserved motifs in IL-17 proteins.
The motifs in IL-17 protein sequences were analyzed by MEME 4.9.1. Non-overlapping sites are indicated by a p-value greater than 0.0001. The height of the motif “block” is proportional to the–log (p-value), truncated at the height of a motif with a p-value of 1e –10.
Meanwhile, SignalP was performed to predict signal peptides at the N-terminal IL-17 proteins. As shown in Table 1, in vertebrates, 33 out of 34 IL-17 proteins had a predicted signal peptide, except for X. tropicalis IL-17F. In contrast 32 out of 54 of invertebrate IL-17 proteins had the predicted signal peptide, while 1/3 (18 out of 54 IL-17 proteins) had no signal peptide, and 6 IL-17 proteins were unknown due to their incomplete protein sequences. The results indicated that many of IL-17 proteins in invertebrates have no predicted signal peptide, suggesting that they might be not be secreted proteins.
Phylogenetic analysis and classification of invertebrate IL-17 proteins
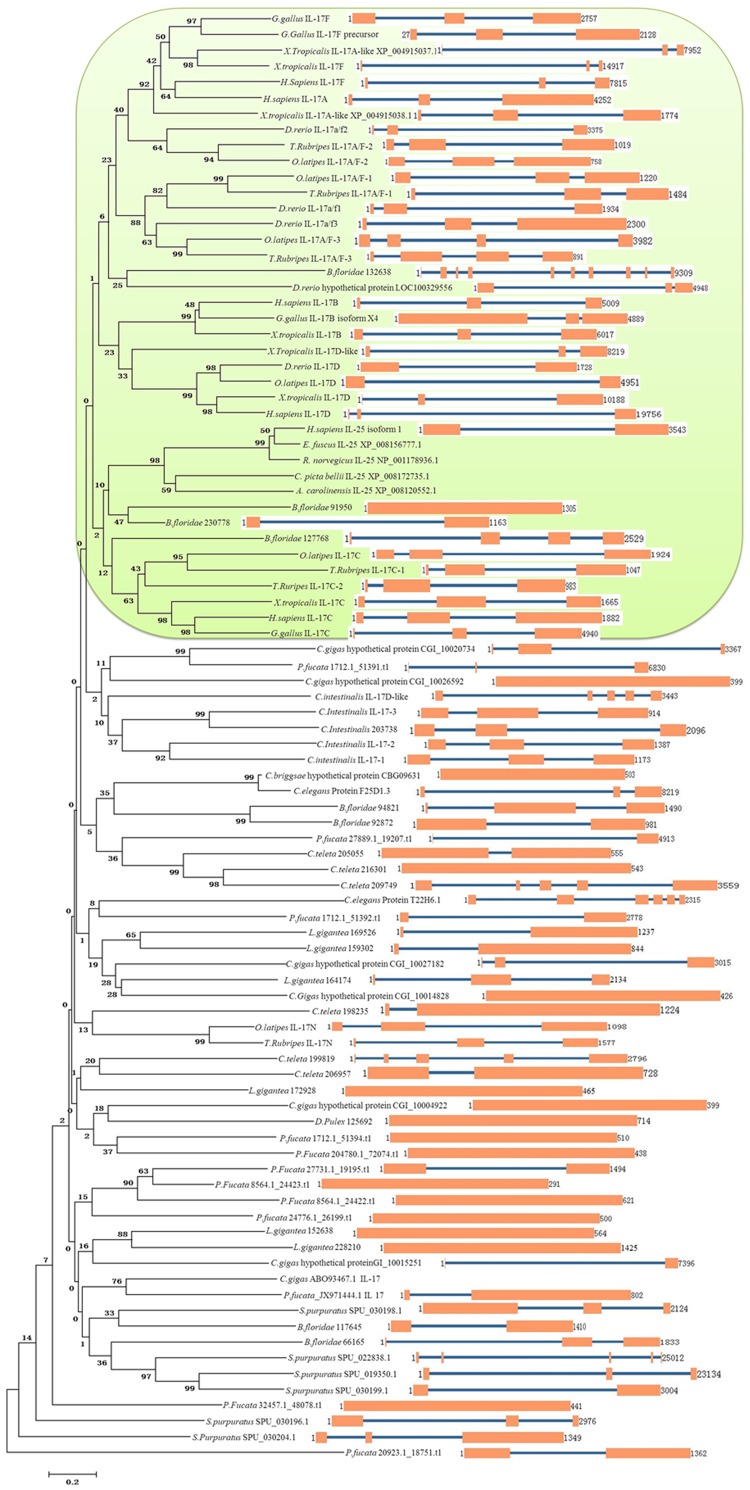
To investigate the potential evolutionary relationships of the IL-17 family, phylogenetic trees were constructed based on the amino acid sequences of the full-length proteins. The phylogenetic tree based on full-length sequences in the NJ analysis was divided into many subgroups (Fig 3). Nearly all vertebrate IL-17 proteins were located in one subgroup (the light green area), in agreement with the phylogenetic tree of vertebrate IL-17 proteins presented in S2 Fig. In addition, many of the invertebrate IL-17 proteins form a large group subsequently divided into several subgroups. In general, the IL-17 proteins from a single species were distributed over different groups. These results indicate that, during evolution, invertebrate IL-17 proteins underwent complex differentiation and include far more than the 7 members (IL-17A-F and IL-17N) found in vertebrates, suggesting that these IL-17 proteins may have developed novel functions during evolution.
Fig 3. Phylogenetic and gene structure analysis of the IL-17 gene.
The phylogenetic tree was constructed using the neighbor-joining method in the MEGA 6.06 software. Each node is represented by a number that indicates the bootstrap value for 10,000 replicates. The scale bar represents 0.2 substitutions per sequence position (left). The right side illustrates the exon–intron organization of the corresponding IL-17 genes. The exons and introns are represented by orange boxes and blue lines, respectively. The numbers indicate the length of the gene. The extra IL-17 protein sequences (Rattus norvegicus IL-25 (NP_001178936.1), Chrysemys pictabellii IL-25 (XP_008172735.1), Alligator sinensis IL-25 (XP_008120552.1)) have not been listed in S1 Dataset.
Exon-intron structure and location of IL-17genes
The exon-intron structure of IL-17 genes in invertebrates and vertebrates was examined to obtain further insight into the possible structural evolution of these genes. As shown in Table 1 and Fig 3, in vertebrates, 29 out of 34 IL-17 genes had two introns, while three members contained only one intron and two members had three introns. By contrast, in invertebrates, the intron number of IL-17 was more variable, but generally (49 of 54) ranged from 0 to 3. The exceptions were genes with 4 introns (C. teleta 209749, C. intestinalis IL-17D-like, S. purpuratus SPU_022838.1), 5 introns (C.teleta 199819), 6 introns (C. elegans NP_510131.2), 9 introns (B. floridae 132638), and C. gigas ABO93467.1, which had no corresponding genomic structure available because of the draft status of its genome. These results indicate that the number of IL-17 introns has fluctuated greatly in invertebrates but has been relatively stable in vertebrates, further indicating the complex evolution of IL-17 proteins.
Interestingly, many invertebrate IL-17 genes are found to be located in the same scaffold, including L. gigantea 152638 and 172928 in scaffold_2; P. fucata 1712.1_51392.t1, 1.0_1712.1_51391.t1 and 1712.1_51394.t1 in scaffold_1712.1, and 8564.1_24423.t1 and 8564.1_24422.t1 in scaffold_8564.1; S. purpuratus SPU_019350.1 and SPU_030199.1 in scaffold_1105, and SPU_030196.1 and SPU_030198.1 in scaffold_2038; C. intestinalis IL-17-3, IL-17-2, IL-17-1 and 203738 in chromosome 1; B. floridae 230778 and 127768 in scaffold_275. In contrast, many vertebrate IL-17 genes are on the same chromosome, such as D. rerio IL-17a/f2 and IL-17a/f1 on chromosome 17; T. rubripes IL-17C-2, IL-17A/F-1 and IL-17A/F-2 on chromosome 13; O. latipes IL-17A/F-2 and IL-17A/F-1 in ultracontig 46; G. gallus IL-17F and IL-17F precursor on chromosome 3; X. tropicalis IL-17A-like (XP_004915038.1), IL-17F, IL-17A-like (XP_004915037.1) in scaffold_5b; and H. sapiens IL-17F and IL-17A on chromosome 6. This result indicates that several IL-17 genes are present in tandem on the same chromosome and may have been derived from gene duplication.
Discussion
As an important regulatory cytokine, IL-17 is involved in and mediates cell–cell communication for many biological processes, particularly host defense responses and inflammatory diseases [1, 2]. However, the functions and characteristics of the invertebrate IL-17 family have not been well characterized [13, 14, 19, 23]. The recent release of a number of invertebrate genome databases may provide new insights into the IL-17 family. In the present study, we identified and summarized 54 IL-17-encoding genes in invertebrates and compared them with 28 vertebrate homologs, to investigate their origin and diversification. IL-17 genes were identified in invertebrates including Nematoda (C. briggsae and C. elegans), Annelida (C. teleta), Mollusca (L. gigantean, C. gigas and P. fucata), Arthropoda (D. pulex), Echinodermata (S. purpuratus) and Chordata (C. intestinalis and B. floridae) but were absent from Porifera (A. queenslandica), Cnidaria (N. vectensis and H. magnipapillata), Hemichordata (S. kowalevskii), Placozoa (T. adhaerens) and Insecta (such as A. pisum, A. mellifera, and D. melanogaster), as well as Protozoa. The number of IL-17 genes in each species was highly variable, ranging from 1 (C. briggsae) to 12 (P. fucata), which may reflect their unusually high evolutionary rate (Table 1). While the absence of the cytokine IL-17 family, which functions in cell-cell communication, in Protozoa and simple, ancient lower invertebrates such as A. queenslandica and H. magnipapillata was not unanticipated, it is puzzling that IL-17 genes were missing from Hemichordata (S. kowalevskii) and relatively high insects. This result is partially supported by a report by Simakov et al. that, although mollusks and annelids are related to flies, nematodes and flatworms within the protostomes, the genome organization, gene structure and functional content of these species are in many ways more similar to those of invertebrate deuterostomes (such as amphioxus and sea urchin) [16]. These similarities include features of bilaterian and/or metazoan genomes that have been lost or diverged in many protostome genomes.
Furthermore, immune gene families are usually under more intense evolutionary pressure, and rapid evolutionary changes are frequently observed for effector proteins such as cytokine IL-17 [24, 25]. In this study, the length and domain number of some IL-17 proteins varied greatly, suggesting broadened or reduced functions. For example, B. floridae 132638 contains not only the IL-17 domain but also the LysRS_N and incomplete LysRS core domain. LysRS_N is a beta-barrel domain (OB fold) involved in binding the tRNA anticodon stem–loop. LysRS enzymes are homodimeric class 2b aminoacyl-tRNA synthetases (aaRSs), which catalyze the specific attachment of amino acids to their cognate tRNAs during protein biosynthesis [26]. IL-17 enhances the expression of multiple pro-inflammatory cytokines, particularly members of the CXC chemokine family, through mRNA stabilization via an AUUUA/Tristetraprolin-independent sequence [27, 28]. By contrast, some IL-17 proteins contain incomplete IL-17 domains (S1 Dataset). This study also demonstrated that, although the amino acid sequence similarities of the IL-17 proteins were rather low, the motifs were highly conserved, although some motifs were lost in certain species. Given that these conserved motifs are located, to a great extent, in IL-17 domains, they provide the base for IL-17 domains and proteins. Significantly, there is a third disulfide bond for the cystine knot fold in invertebrate IL-17 proteins, suggesting that they may possess the canonical disulfides of the cystine knot, which belongs to the canonical cystine knot fold superfamily, with members such as the NGF subfamily; This is until in Chordata (B. floridae and C. intestinalis), where the two cysteine residues have been replaced by the corresponding serine residues [29, 30]. Unlike almost all vertebrate IL-17 proteins, which contain a predicted signal peptide, a significant proportion of those of invertebrates have no predicted signal peptide. The secretory signal peptide targets its passenger protein for translocation across the endoplasmic reticulum membrane in eukaryotes and the cytoplasmic membrane in prokaryotes [31]. The invertebrate IL-17 proteins without a predicted signal peptide may perform a different function from that of their vertebrate counterparts. Furthermore, some IL-17 genes were found to exhibit conserved synteny, which reveals a close evolutionary relationship between two genes or even two species and suggests that they may be derived from a common ancestor. This may also partially explain why IL-17A-like genes in some phyla may be too dissimilar to be identified. These results suggest that IL-17 proteins and their functions have been continuously undergoing dynamic change through evolution.
Previous studies of genomic organization involving phylogenetic analysis have revealed that the genomic organization of the vertebrate IL-17 family has been basically conserved through evolution [8, 13]. In mammals, the IL-17 family is generally divided into six members (IL-17A–F) or subgroups, and IL-17N is also present in fish. Furthermore, each member of the IL-17 family has different functions, with the exception of IL-17A and IL-17F. In this study, phylogenetic analysis indicated that there are many subgroups of the IL-17 family in invertebrates that likely produce numerous IL-17 family members, far more than the 7 known members in vertebrates (IL-17A-F and IL-17N), which suggests that the invertebrate proteins have undergone high divergence, including in their function. Additionally, introns may affect gene expression by increasing the time required to transcribe the gene, and intron-containing and intronless versions of otherwise identical genes can exhibit dramatically different expression profiles [32, 33]. While there is no universal intron requirement for eukaryotic gene expression, in many cases transgene expression can be dramatically increased by the addition of just one generic intron to the cDNA [34, 35]. This may give a partial explanation for the change in the number of IL-17 introns from invertebrates to vertebrates. Although intron evolution is a dynamic process in eukaryotes [36], the comparison of IL-17 family gene organization revealed that the IL-17 family gene has not been very highly conserved throughout evolution. The more drastic changes in the exons also strengthen this observation. In general, from the perspective of both phylogenetics and genomic organization, the IL-17 family lacks conservation and exhibits high divergence, suggesting that invertebrate IL-17 proteins have undergone complex differentiation and that their members may have developed novel functions during evolution.
In the progression from unicellular protozoans to multicellular animals, the capability for more advanced and complicated communication and cooperation among cells was acquired. Some cytokines, such as tumor necrosis factor (TNF)-α, appeared early in primitive invertebrates [37, 38] and, therefore, it is likely that the emerging IL-17 gene family may have fulfilled the increased demand for more complex regulation in relatively high multicellular animals. New genes must be integrated with other novel and existing genes to evolve expanded or modified biochemical pathways and/or regulatory networks [39]. Accordingly, the IL-17 family functions via its receptor IL-17R, a specific cell surface receptor, thus forming a distinct ligand-receptor signaling system to induce downstream signaling. In mollusks, IL-17 family genes participate in the immune response to stimulation [19, 23]. Therefore, IL-17 may also play a vital role in invertebrate inflammatory reactions. Inexplicably, other IL members have only arisen in lower vertebrates and not invertebrates, whereas some ILRs are found only in invertebrates [14]. However, why the IL-17 gene and not another IL member was selected during early evolution remains unclear.
So far, five members of the IL-17R family (IL-17RA–IL-17RE) have been identified, and are thought to consist of homodimers or heterodimers. Among them, the heterodimer of IL-17RA and IL-17RC is a receptor for homodimers and heterodimers of IL-17A and IL-17F, whereas the heterodimer consisting of IL-17RA and IL-17RB serves as a receptor for IL-17E. IL-17B binds to IL-17RB, and IL-17C was recently reported to bind to IL-17RE and to activate NF-κB. The receptor for IL-17RD has yet to be identified [10, 40]. Specifically, a mechanism of complex formation has been presented, such that two fibronectin-type domains of IL-17RA engage IL-17F in a groove within the IL-17F homodimer interface [41]. The IL-17R family mediates a signal pathway that serves as a bridge between innate and adaptive immune responses [40]. However, these receptors are rarely isolated from invertebrates. The signal transduction pathway mediated by IL-17 and IL-17R remains poorly defined, particularly in invertebrates, and there is still much to learn about the structures and functions of IL-17 and IL-17R and their characteristics and nature during evolution.
In conclusion, this study provided a global survey to investigate the distribution of the IL-17 family among invertebrates, revealed the features of their motifs and signal peptides. Meanwhile, phylogenetic trees and their exon-intron structures were analyzed, and their origin and evolutionary history in animal phyla were explored. The results of this study suggest that, during evolution, invertebrate IL-17 proteins have undergone complex differentiation, and that their members may have developed novel functions. The findings provide direction for future studies of the functions of the IL-17 family.
Supporting Information
(DOCX)
The motifs were analyzed by MEME 4.9.1. The full-length IL-17 proteins and IL-17 domains have similar sequence logos for the motifs, but the motifs have different names.
(TIF)
The extra IL-17 protein sequences (Rattus norvegicus IL-25 (NP_001178936.1), Alligator sinensis IL-17D-like isoform X2 (XP_006022303.1)) have not been listed in S1 Dataset.
(TIF)
Acknowledgments
We are grateful to the anonymous reviewers and the PLOS One editor for their critical reading of our paper.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Science and Technology Program of China (2012AA10A410).
References
- 1. Gu C, Wu L, Li X. IL-17 family: cytokines, receptors and signaling. Cytokine. 2013. 64:477–85. 10.1016/j.cyto.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Song X, Qian Y. IL-17 family cytokines mediated signaling in the pathogenesis of inflammatory diseases. Cell Signal. 2013. 25:2335–47. 10.1016/j.cellsig.2013.07.021 [DOI] [PubMed] [Google Scholar]
- 3. Manni ML, Robinson KR, Alcorn JF. A tale of two cytokines: IL-17 and IL-22 in asthma and infection. Expert review of respiratory medicine. 2014. 8:25–42. 10.1586/17476348.2014.854167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010. 10:479–89. 10.1038/nri2800 [DOI] [PubMed] [Google Scholar]
- 5. Rouvier E, Luciani MF, Mattei MG, Denizot F, Golstein P. Ctla-8, Cloned from an Activated T-Cell, Bearing Au-Rich Messenger-Rna Instability Sequences, and Homologous to a Herpesvirus Saimiri Gene. J Immunol. 1993. 150:5445–56. [PubMed] [Google Scholar]
- 6. Yoo ZB, Painter SL, Fanslow WC, Ulrich D, Macduff BM, Spriggs MK, et al. Human Il-17—a Novel Cytokine Derived from T-Cells. J Immunol. 1995. 155:5483–6. [PubMed] [Google Scholar]
- 7. Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004. 21:467–76. [DOI] [PubMed] [Google Scholar]
- 8. Korenaga H, Kono T, Sakai M. Isolation of seven IL-17 family genes from the Japanese pufferfish Takifugu rubripes. Fish Shellfish Immunol. 2010. 28:809–18. 10.1016/j.fsi.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 9. Zhang X, Angkasekwinai P, Dong C, Tang H. Structure and function of interleukin-17 family cytokines. Protein & cell. 2011. 2:26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional Specialization of Interleukin-17 Family Members. Immunity. 2011. 34:149–62. 10.1016/j.immuni.2011.02.012 [DOI] [PubMed] [Google Scholar]
- 11. Gaffen SL. Recent advances in the IL-17 cytokine family. Curr Opin Immunol. 2011. 23:613–9. 10.1016/j.coi.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee Y. The role of interleukin-17 in bone metabolism and inflammatory skeletal diseases. BMB reports. 2013. 46:479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kono T, Korenaga H, Sakai M. Genomics of fish IL-17 ligand and receptors: A review. Fish Shellfish Immun. 2011. 31:635–43. [DOI] [PubMed] [Google Scholar]
- 14. Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008. 18:1112–26. 10.1101/gr.069674.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, et al. Draft Genome of the Pearl Oyster Pinctada fucata: A Platform for Understanding Bivalve Biology. DNA Research. 2012. 19:117–30. 10.1093/dnares/dss005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013. 493:526–31. 10.1038/nature11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012. 490:49–54. 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- 18. Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Developmental biology. 2006. 300:349–65. [DOI] [PubMed] [Google Scholar]
- 19. Roberts S, Gueguen Y, de Lorgeril J, Goetz F. Rapid accumulation of an interleukin 17 homolog transcript in Crassostrea gigas hemocytes following bacterial exposure. Developmental and comparative immunology. 2008. 32:1099–104. 10.1016/j.dci.2008.02.006 [DOI] [PubMed] [Google Scholar]
- 20. Wu SZ, Huang XD, Li Q, He MX. Interleukin-17 in pearl oyster (Pinctada fucata): molecular cloning and functional characterization. Fish Shellfish Immunol. 2013. 34:1050–6. 10.1016/j.fsi.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 21. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods. 2011. 8:785–6. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013. 30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu SZ, Huang XD, Li Q, He MX. Interleukin-17 in pearl oyster (Pinctada fucata): Molecular cloning and functional characterization. Fish Shellfish Immunol. 2013. [DOI] [PubMed] [Google Scholar]
- 24. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature reviews Genetics. 2010. 11:47–59. 10.1038/nrg2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nehyba J, Hrdlickova R, Bose HR. Dynamic Evolution of Immune System Regulators: The History of the Interferon Regulatory Factor Family. Mol Biol Evol. 2009. 26:2539–50. 10.1093/molbev/msp167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. Embo Mol Med. 2013. 5:332–43. 10.1002/emmm.201100626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hartupee J, Liu CN, Novotny M, Li XX, Hamilton T. IL-17 enhances chemokine gene expression through mRNA stabilization. J Immunol. 2007. 179:4135–41. [DOI] [PubMed] [Google Scholar]
- 28. Chiricozzi A, Nograles KE, Johnson-Huang LM, Fuentes-Duculan J, Cardinale I, Bonifacio KM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PloS one. 2014. 9:e90284 10.1371/journal.pone.0090284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McDonald NQ, Hendrickson WA. A structural superfamily of growth factors containing a cystine knot motif. Cell. 1993. 73:421–4. [DOI] [PubMed] [Google Scholar]
- 30. Hymowitz SG, Filvaroff EH, Yin JP, Lee J, Cai L, Risser P, et al. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. Embo J. 2001. 20:5332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walter P. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Harvey lectures. 1995. 91:115–31. [PubMed] [Google Scholar]
- 32. Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003. 28:215–20. [DOI] [PubMed] [Google Scholar]
- 33. Swinburne IA, Miguez DG, Landgraf D, Silver PA. Intron length increases oscillatory periods of gene expression in animal cells. Gene Dev. 2008. 22:2342–6. 10.1101/gad.1696108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gruss P, Lai CJ, Dhar R, Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979. 76:4317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991. 88:478–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006. 22:16–22. [DOI] [PubMed] [Google Scholar]
- 37. Hayward DC, Samuel G, Pontynen PC, Catmull J, Saint R, Miller DJ, et al. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci U S A. 2002. 99:8106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005. 21:633–9. [DOI] [PubMed] [Google Scholar]
- 39. Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007. 317:86–94. [DOI] [PubMed] [Google Scholar]
- 40. Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009. 9:556–67. 10.1038/nri2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ely LK, Fischer S, Garcia KC. Structural basis of receptor sharing by interleukin 17 cytokines. Nature immunology. 2009. 10:1245–51. 10.1038/ni.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
The motifs were analyzed by MEME 4.9.1. The full-length IL-17 proteins and IL-17 domains have similar sequence logos for the motifs, but the motifs have different names.
(TIF)
The extra IL-17 protein sequences (Rattus norvegicus IL-25 (NP_001178936.1), Alligator sinensis IL-17D-like isoform X2 (XP_006022303.1)) have not been listed in S1 Dataset.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.